Abstract
PIGT-CDG, an autosomal recessive syndromic intellectual disability disorder of glycosylphosphatidylinositol (GPI) anchors, was recently described in two independent kindreds [Multiple Congenital Anomalies-Hypotonia-Seizures Syndrome 3 (OMIM, #615398)]. PIGT encodes phosphatidylinositol-glycan biosynthesis class T, a subunit of the heteropentameric transamidase complex that facilitates the transfer of GPI to proteins. GPI facilitates attachment (anchoring) of proteins to cell membranes. We describe, at ages 7 and 6 years, two children of non-consanguineous parents; they had hypotonia, severe global developmental delay, and intractable seizures along with endocrine, ophthalmologic, skeletal, hearing, and cardiac anomalies. Exome sequencing revealed that both siblings had compound heterozygous variants in PIGT (NM_015937.5), i.e., c.918dupC, a novel duplication leading to a frameshift, and c.1342C>T encoding a previously described missense variant. Flow cytometry studies showed decreased surface expression of GPI-anchored proteins on granulocytes, consistent with findings in previous cases. These siblings further delineate the clinical spectrum of PIGT-CDG, reemphasize the neuro-ophthalmologic presentation, clarify the endocrine features, and add hypermobility, low CSF albumin quotient, and hearing loss to the phenotypic spectrum. Our results emphasize that GPI anchor-related congenital disorders of glycosylation (CDGs) should be considered in subjects with early onset severe seizure disorders and dysmorphic facial features, even in the presence of a normal carbohydrate-deficient transferrin pattern and N-glycan profiling. Currently available screening for CDGs will not reliably detect this family of disorders, and our case reaffirms that the use of flow cytometry and genetic testing is essential for diagnosis in this group of disorders.
Keywords: PIGT-CDG, Congenital disorder of glycosylation, Glycosylphosphatidylinositol anchor, Phenotype, Flow cytometry, Exome
1. Introduction
Glycosylphosphatidylinositol (GPI) anchors are a group of glycolipids composed of a glycan core, a phosphoethanolamine linker, and a phospholipid tail [1,2]. GPI anchors are attached during posttranslational modification to the C-terminus of certain proteins destined to attach to the outer leaflet of the cell membrane and face the extracellular environment. This permits these proteins to participate in processes such as signal transduction and the immune response [1,3].
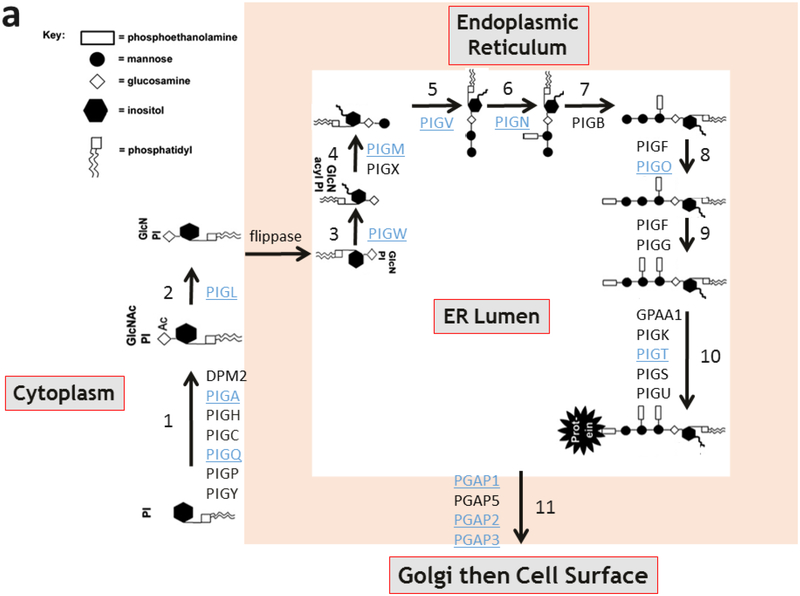
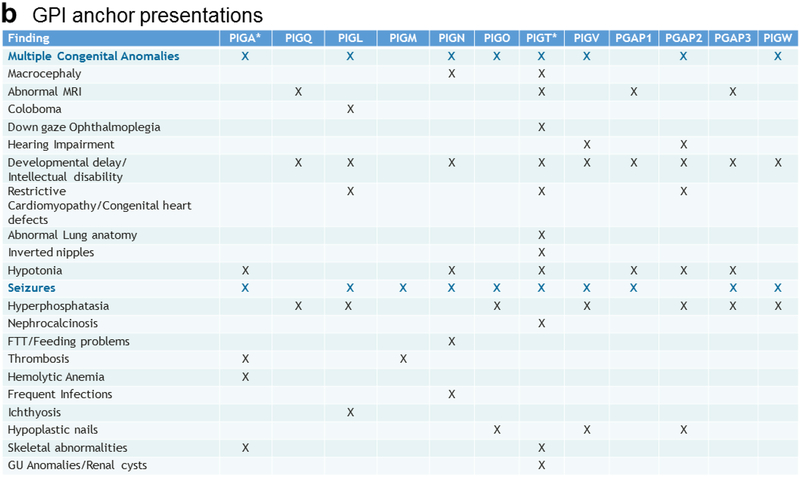
In humans, the biosynthesis and attachment of GPI anchors to proteins occurs in the endoplasmic reticulum and Golgi and involves 11 steps and protein products of at least 27 genes [4]. To date, inherited loss of function mutations in twelve of these genes have been implicated in human disease (Fig. 1a) [5–21]. Inherited congenital deficiencies in GPI anchor biosynthesis and attachment comprise a subset of congenital disorders of glycosylation (CDGs) and cause a spectrum of symptoms in humans, including seizures, intellectual disabilities, and congenital anomalies. These findings are present in most of the GPI anchor disorders (Fig. 1b). PIGT encodes phosphatidylinositol-glycan biosynthesis class T, a subunit of the heteropentameric GPI transamidase complex that facilitates the attachment of GPI anchors to proteins [22]. Recently, variants in PIGT were identified in two unrelated families with recessively inherited Multiple Congenital Anomalies-Hypotonia-Seizures Syndrome 3 (OMIM, #615398) [11,23].
Fig. 1.
GPI anchor pathway and phenotypic findings of GPI anchor disorders.
(a) The 11 steps of mammalian GPI anchor biosynthesis and protein attachment. The genes associated with each step are labeled. Underlined genes have been associated with congenital human disease. Note PIGT in step 10.
(b) Summary of the clinical features described in cases of congenital GPI anchor biosynthesis disorders. The most common features are highlighted in blue. * Note that somatic mutations in PIGA and PIGT in hematopoietic stem cells can cause paroxysmal nocturnal hemoglobinuria manifesting with hemolytic anemia, bone marrow failure, thrombosis, and smooth muscle dystonia. This is distinct from the clinical entity caused by germline mutations in these two genes. Additionally, acquired mutations in the GPI transamidase complex subunits have been implicated in human cancers.
Here we describe a third independent family with PIGT-CDG affecting two siblings who presented with seizures, intellectual disability, and congenital anomalies. The siblings were admitted to the National Institutes of Health (NIH) Clinical Center and enrolled in the NIH Undiagnosed Diseases Program protocol #76-HG-0238 [24,25]. Exome sequencing identified biallelic variants in PIGT (NM_015937.5), and molecular, protein, and flow cytometry studies verified pathogenicity of the variants. Our results expand the clinical spectrum of PIGT-CDG, verify the pathogenicity of a novel mutation, and underline the importance of using flow cytometry and molecular methods for diagnosis in GPI anchor disorders.
2. Methods
2.1. Subjects
The family was enrolled in NIH protocol #76-HG-0238 “Diagnosis and Treatment of Patients with Inborn Errors of Metabolism or Other Genetic Disorders” (http://clinicaltrials.gov/, trial NCT00369421), approved by the National Human Genome Research Institute’s Institutional Review Board. The parents gave written informed consent for their children, in accordance with the Declaration of Helsinki.
2.2. Exome and Sanger sequencing
Genomic DNA was extracted from peripheral leukocytes using the Autogen FlexStar (AutoGen, Inc. Holliston, MA) for automated genomic DNA isolation according to manufacturer’s specifications. Exome sequencing data from genomic DNA of all four family members were generated using the SureSelect Human All Exon System (Agilent Technologies Inc, Santa Clara, CA) and a GAIIx sequencer (Illumina Inc, San Diego CA). Library construction, sequence generation, sequence alignment to the reference genome, variant calling and potential pathogenic variant identification was performed as previously described [26].
Sanger sequencing was performed on DNA of all four family members to confirm the presence and inheritance of the identified PIGT variants. PCR amplification of genomic regions around the variants was performed using Qiagen HotStarTaq master mix (Qiagen, Valencia, CA) using the primer pairs 5’-GTCCCTCTTCCGGATGTTCT-3’ and 5’-GGAATAGCTACAGTCAGCAGCA-3’ for c.918dupC, and 5’-CTGGCTTCTATGCTCCCAAG-3’ and 5’-AAGAGGGGACTCTCTTCCCA-3’ for c.1342C>T. PCR amplification conditions were: 1 cycle of 95°C for 5 min, followed by 39 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 30 s, and a final extension at 72°C for 5 min. Unincorporated primers and nucleotides were removed using ExoSAP-IT reagent (USB, Cleveland, OH, USA). Sanger dideoxy sequencing of the PCR products was performed by Macrogen (Rockville, MD). The sequences were aligned and analyzed using Sequencher v.5.0.1 (Gene Codes, Ann Arbor, MI). Genetic variant interpretation analysis was conducted using Alamut 2.0 (Interactive Biosoftware, San Diego, CA).
2.3. mRNA expression analysis
Primary fibroblasts were cultured from forearm skin biopsies. Total RNA was isolated from control and patients’ fibroblasts, cultured in Dulbecco’s Modified Eagle Medium (DMEM), High Glucose (Invitrogen, Carlsbad, CA) to confluence in a 10-cm dish, using the RNeasy Mini-Kit (Qiagen, Valencia, CA) including on-column treatment with DNA-free DNase (Qiagen). 2μg of RNA were reverse transcribed with the Omniscript RT Kit (Qiagen) using random nonamer primers. qPCR was performed utilizing the PIGT Taqman Assay-On-Demand, Hs00988032_g1, located at the PIGT NM_015937.5 exon 4–5 boundary (Applied Biosystems, Foster City, CA), and the human GAPD (GAPDH) Endogenous Control (VIC®/TAMRA™ probe, primer limited; 4310884E, Life Technologies, Grand Island, NY). PCR amplifications were performed on 100 ng of cDNA using TaqMan Gene Expression Master Mix reagent (Applied Biosystems) and were carried out on an ABI 7500 FAST Real-Time PCR System (Applied Biosystems). Results were analyzed with the comparative CT method using the manufacturer’s software (v.2.0.1) accompanying the ABI 7500 FAST Real-Time PCR System (Applied Biosystems). All assays were performed in triplicate.
2.4. Immunoblotting
Fibroblasts, were grown to confluence in a 15-cm dish, washed twice with phosphate buffered saline, and collected in 600μL RIPA buffer containing Protease inhibitor cocktail (Roche, Manheim, Germany). Samples were sonicated for 10 min in an ultrasonic bath. 25 μg of total protein, as determined by the DC Protein assay (BioRad, Hercules, VA), was loaded on a 4% stacking – 12–15% resolving polyacrylamide gel for immunoblotting with PIGT, PIGS, PIGK, PIGU and GPAA1antibodies. Proteins were transferred to 0.45-μm Immobilon-FL PVDF transfer membranes (Millipore, Billerica, MA) and probed with anti-PIGT (ab80888), anti-PIGU [EPR16424]-C-terminal (ab192255) produced in rabbit (Abcam Inc., Cambridge, MA), anti-PIGS (sc-373701), anti-PIGK (sc-398611), or anti-GPAA1 (sc-373710) produced in mouse (Santa Cruz Biotechnology, Santa Cruz, CA) and monoclonal anti-GAPDH antibody produced in mouse, (G8795, Sigma-Aldrich, St. Louis, MO) or anti-GAPDH produced in rabbit (NB300–327, Novus Biologicals, Littleton, CO). Li-cor Donkey anti-Rabbit IRDye 680RD (Li-cor Inc., Lincoln, NE) and Li-cor Donkey anti-Mouse IRDye 800CW (Li-cor Inc.) were used as secondary antibodies and fluorescence was measured on a 9140 Odyssey CLx Infrared Imaging system (Li-cor Inc.). Band density was analyzed using Image studio Software (Li-cor Inc.).
2.5. Flow cytometry analysis
Granulocytes isolated from whole blood were analyzed for surface expression of GPIanchored proteins by flow cytometric analysis. Granulocyte surface expression of total GPIanchored proteins was quantified with FITC-conjugated inactivated aerolysin (FLAER; Pinewood Scientific, Victoria, BC, Canada). Cell surface expression of specific GPI-anchored proteins was examined using antibodies to CD16 (BD Biosciences, San Jose, CA), CD55 and CD59 (BD Pharmingen, San Diego, CA), and CD66b (Beckman Coulter, Indianapolis, IN). Multiparameter flow cytometric analysis was performed on a FACSCanto II flow cytometer (Becton Dickinson, San Diego, CA). Data were analyzed using DIVA and FSC Express software. The results were expressed as geometric mean fluorescence intensity (MFI). The expression of granulocyte surface markers was evaluated by comparison of geometric means of fluorescence intensity between normal controls and patients.
2.6. Magnetic Resonance Imaging (MRI)
MRI exams (under sedation) were performed on either 3 T (NIH) or 1.5 T (other institutions) Philips scanners and included standard clinical images. There were 7 imaging studies that included either 3D TFE or 3D MPRAGE images with 1 × 1 × 1 mm (or better) resolution; these studies were processed with the NeuroQuant software (CorTechs Labs, San Diego, CA) to segment the brain images and calculate the volumes of each anatomical region. Total volumes of the cerebrum and of the cerebellum were analyzed in comparison to normative curves previously published in the literature [27].
2.7. Magnetic Resonance Spectroscopy (MRS)
Single voxel spectroscopy was performed on voxels graphically prescribed from either MPRAGE or 3D-TFE images (PRESS localization; CHESS water suppression; TE=38 ms; TR=2000 ms; 128 NEX). An unsuppressed water spectrum (TR=5000 ms, TE=38 ms, 16 NEX) with identical gain and shim settings was also acquired for each voxel. Four voxel locations were acquired during each exam: left cerebellar white matter (LCWM), left centrum semiovale (LCSO), midline parietal gray matter (PGM), and either the pons or left thalamus. Cerebrospinal fluid (CSF) included within the voxels was corrected for, as previously described [28, 29]. An identical MRS acquisition and CSF correction protocol was performed on a group of normal adult volunteers and on a group of near-normal children acquired from a natural history of Niemann-Pick disease type C (NPC) [30].
Raw MRS data was exported to LCModel [31]. The LCModel software produces a first estimate of the metabolite concentration in the tissue. These estimates were corrected for estimated concentration of water and for T1 of metabolites within the tissues, using published T1 values [32]. Correction for T2 decay was not applied, as neither the PIGT-deficient patients nor the reference group demonstrated abnormal signal on the T2 weighted images. The analyzed spectroscopic peaks included NAA+NAAG at 2.0 ppm (referred to as NAA), creatine at 3.0 ppm, choline-containing compounds at 3.2 ppm (referred to as choline), myo-inositol at 3.5 ppm, and glutamine+glutamate+GABA at 2.2–2.4 ppm (referred to as Glx). A power curve (of the form y=AxB) was fit to the reference measurements for each metabolite at each location, resulting in an expected concentration as a function of age; the variance of the reference measurements from the fitted curve was calculated and used to generate a Z score for each of the PIGT-deficient patient measurements.
3. Results
3.1. Clinical characteristics
The proband and her affected brother initially presented to the Undiagnosed Diseases program at the NIH with hypotonia, severe global developmental delay, and intractable seizures.
Patient 1
Patient 1 (proband), a 7-year old girl, was born at 31 5/7 weeks’ gestation via induced vaginal delivery to a Caucasian mother and an African American father. The pregnancy was complicated by pre-eclampsia, preterm labor, and a second trimester screen strongly positive for risk of open spinal bifida (AFP 7.79 multiples of the mean); a level 2 ultrasound and amniocyte karyotype were normal. Her birth weight was 1519 grams (10%); birth length was 41 cm (1025%), and birth head circumference was 28 cm (<10%). APGAR scores were 9 at both 1 and 5 minutes of life. She was admitted to the neonatal intensive care unit for 44 days; cardiorespiratory and feeding support was required. There was no seizure activity, and the newborn hearing screen, head ultrasound, brain MRI, and ophthalmologic examination were all normal. Her echocardiogram showed a patent foramen ovale and small muscular ventricular septal defect. Her initial newborn screen showed low T4 but a repeat screen was normal.
At 3 months of life, she developed strabismus. At 5 months, she manifested epilepsy, especially during febrile illnesses, and required repeated hospitalizations. These seizures were largely myoclonic and tonic multifocal frontal or occipital seizures that generalized; they were poorly controlled despite combinations of levetiracetam, topiramate, phenobarbital, clonazepam, midazolam, diazepam, clonidine and a ketogenic diet. Development was normal until the onset of seizures at 5 months; there were no significant developmental advances subsequently. At 7 years of age a formal developmental assessment identified her developmental age equivalent to a 3–4 month old in all categories.
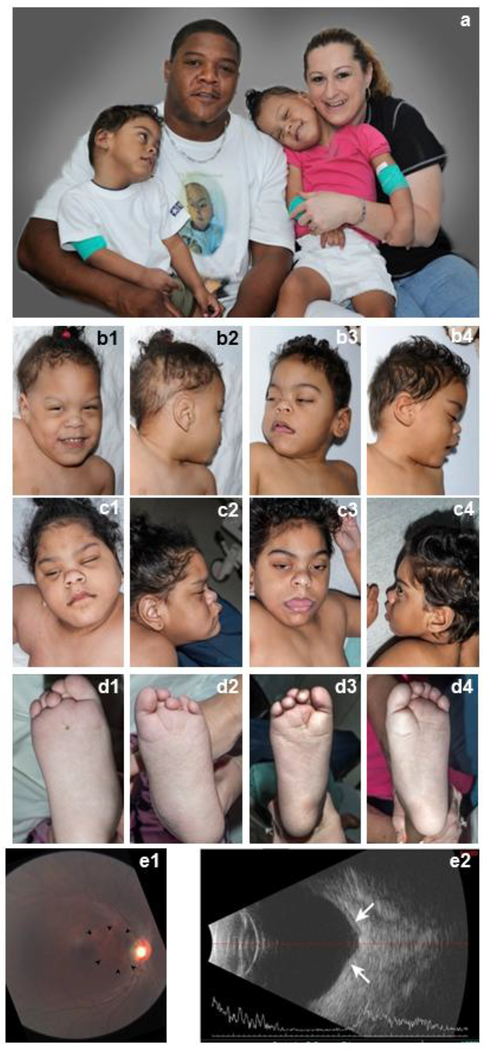
She also exhibited joint hypermobility, dysmorphic features (Fig. 2), progressive cerebellar and cerebral atrophy (Fig. 3), cortical visual dysfunction with hyperopic astigmatism and nystagmus. Other problems included hearing dysfunction, obesity, precocious puberty at 6 years of age, skeletal abnormalities (Fig. 4), osteopenia, gastroesophageal reflux, constipation, poor oral-motor skills with high aspiration risk (requiring a Nissen fundoplication and complete G-tube dependence from age 20 months), and hypertriglyceridemia (Table 1).
Fig. 2.
Photographs of the family displaying dysmorphic features in the affected siblings.
For familial comparison, parents are depicted with both siblings in (a). Additional photographs are shown of each of the siblings when Patient 1 was 36 months old (b1, b2) and Patient 2 was 24 months old (b3, b4), and when Patient1 was 91 months old (c1, c2; d1, d2) and Patient 2 was 79 months old (c3, c4; d3, d4). Among the dysmorphic features, note the depressed nasal bridge, high forehead, bitemporal narrowing, rounded nasal tip that was correlated to very soft cartilage, large ears also with soft cartilage, overlapping toes, and deep plantar creases in both affected siblings.
RetCAM 3 fundus image of the right eye of Patient 2 is shown (e1). The staphyloma noted on exam is located between the optic nerve head and the nasal edge of the macula (arrow-heads). This finding was confirmed in a B-scan ultrasound documenting the protrusion of the staphylomatous area (e2).
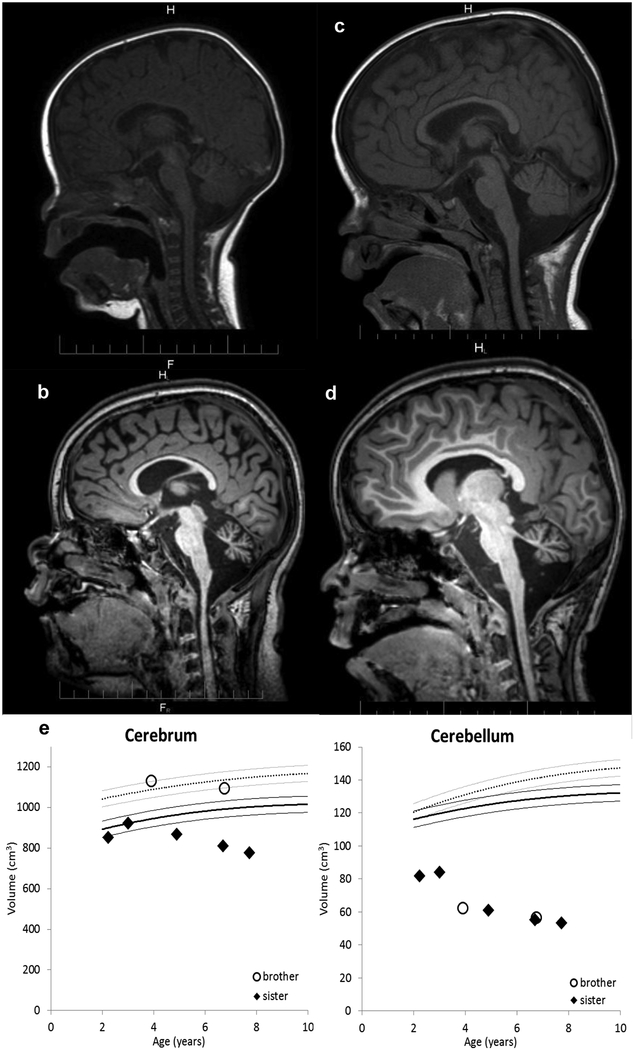
Fig. 3.
T1 sagittal brain magnetic resonance imaging (MRI) of the two affected patients.
MRI scans of Patient 1 were taken at 6 months (a), and 91 months (b). MRI scans of Patient 2 were taken at 12 months (c), and 79 months (d). Brain volume measurements on the Patient 1 (diamonds) and Patient 2 (open circles) are displayed along with published normal population means [27] (dotted curve = boys, solid curve = girls, lighter lines = 95% CI) (e). Progressing volume loss is evident in both the cerebrum and cerebellum for both patients. Atrophy is more profound in the cerebellum than in the cerebrum. Atrophy in the cerebellum proceeded at about the same rate for both patients, whereas in the cerebrum, atrophy seemed to progress more rapidly in the sister.
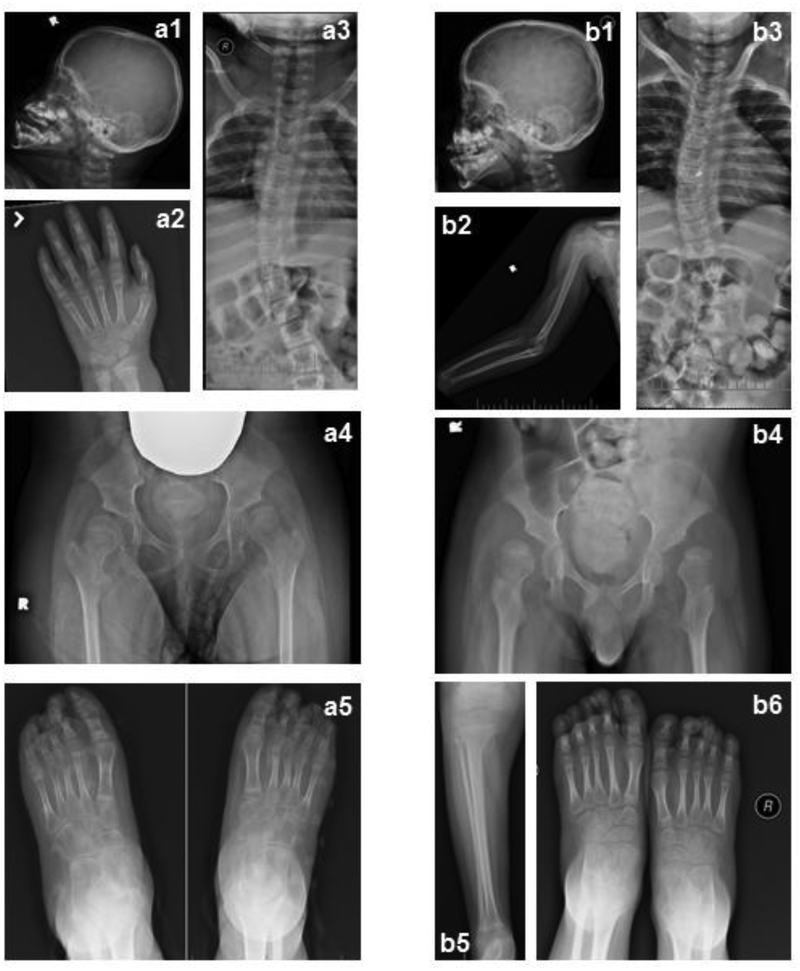
Fig. 4.
Skeletal radiographs.
Skeletal survey of Pt1(a), and Pt2 (b) performed at 91 months and 79 months of age, respectively. a1 and b1 illustrate brachycephaly, a2 shows advanced bone age (bone age 10 years +/− 20 months) of Patient 1, b2 and b5 illustrates the slender osteopenic long bones that both patients have, b2 also shows dislocation of elbow and subluxation of shoulder, a3 and b3 illustrate S-shaped scoliosis, a4 and b4 show bilateral dislocated hips with severe coxa valga-and shallow acetabulum, and a5 is unusual for pointy distal phalanxes bilaterally especially on the first digit that is not seen in b6.
Table 1.
Clinical features of Pt1 and Pt2 compared to two other published PIGT-CDG cases.
| Physiologic System | Clinical Feature | 4 cases [11] Kvarnung et al. 2014 | 1 case [16] Nakashima et al.2014 | Patient 1 | Patient 2 |
|---|---|---|---|---|---|
| Neonatal | Gestational Age | 37–40 weeks | 40 weeks | 31 5/7 weeks | 31 3/7 weeks |
| Birth Weight | 69–99% | 50–90% | 10% | 90% | |
| Birth Length | 93–99% | 50% | 10–25% | 90% | |
| Birth Head Circumference | 84–99% | 50–90% | <10% | 50–75% | |
| Other | n/a | Polyhydramnios | Elevated AFP on 2nd trimester screen | n/a | |
| Neurological | Intellectual Disability | 4/4 | Profound | Profound | Profound |
| Seizures | 4/4 (2/4 myoclonic, ¼ generalized tonic clonic, 1/4 absence + head jerks & blinking) | Myoclonic, tonic with apnea that can generalize | Myoclonic, tonic, and tonic clonic that occasionally generalize | Myoclonic, tonic, and tonic clonic that occasionally generalize | |
| Hypotonia | 4/4 | Yes | Yes | Yes | |
| Brain Imaging – Global cerebral and cerebellaratrophy with predominant vermis and cerebellar atrophy | 3/4 | Yes | Yes | Yes | |
| Brain Imaging – Possible neuronal migration defect | 2/4 | Not reported | No | No | |
| EEG - No seizure activity initially | 4/4 | Yes | Yes | Yes | |
| EEG - Theta waves | 1/4 | Not reported | Yes | Yes | |
| EEG - Multifocal epileptiform abnormalities | 3/4 | Yes | Yes | Yes | |
| CSF (glucose, protein, cell count, oligoclonal bands) | Not reported | Not reported | Normal | Normal | |
| CSF Albumin & Albumin Quotient | Not reported | Not reported | Low | Low | |
| Peripheral Nerve Conduction Studies | Not reported | Not reported | Normal | Normal | |
| Ophthalmologic | Cortical Visual Impairment | 4/4 | Yes | Yes | Yes |
| Strabismus, Nystagmus | 4/4 | Yes | Yes | Yes | |
| Hyperopia | 4/4 | Not reported | Yes | No | |
| Myopia | 0/4 | Not reported | No | Yes | |
| Other | 1/4 down gaze palsy | Not reported | astigmatism | posterior staphyloma | |
| Audiologic | Hearing loss | 0/4 | Not Reported | Yes | Yes |
| Cardiologic | Patent Ductus Arteriosus | 1/4 | Yes | No | No |
| Restrictive Cardiomyopathy | 1/4 | No | No | No | |
| Increased Atrial Load | 1/4 | No | No | Yes | |
| Atrial Septal Defect | 0/4 | No | No | Yes | |
| Borderline Long QT | Not Reported | Yes | No | ||
| Respiratory | 1/4 atypical lung lobulation | Not Reported | No abnormalities | Mixed central and obstructive sleep apnea | |
| Gastro-intestinal | Obesity | 0/4 | Not Reported | Yes | Yes |
| Gastroesophageal Reflux | Not Reported | Not Reported | Yes | Yes | |
| Aspiration Risk + Secondary G-tube Dependence | Not Reported | Not Reported | Yes (at 20 months) | Yes (at 20 months) | |
| Hypertriglyceridemia | Not Reported | Not Reported | Yes | No | |
| Urologic/Renal | Nephrocalcinosis | 4/4 | No (but + urolithiasis) | No | No |
| Urine Calcium | High | Normal | Normal | Normal | |
| Ureteral Dilation | 3/4 | Yes | No | No | |
| Renal Cysts and Dysplasia | 1/4 | No | No | No | |
| Musculoskeletal | Premature Loss of Teeth | 2/4 | No | No | No |
| Slender Long Bones | 4/4 | Not Reported | Yes | Yes | |
| Scoliosis | 2/4 | Yes | Yes | Yes | |
| Craniosynostosis | 2/4 | No | Unknown | Unknown | |
| Brachycephaly | Yes | Not Reported | Yes | Yes | |
| Short Arms | 4/4 | Not Reported | No | No | |
| Pectus Excavatum | 1/4 | Not Reported | No | Yes | |
| Joint Hypermobility | Not Reported | Not Reported | Yes | Yes | |
| Electromyogram | Not Reported | Not Reported | Reduced muscle bulk, but normal motor units | Reduced muscle bulk, but normal motor units | |
| Endocrine | Bone Age | Delayed – 4/4 | Not Reported | Advanced | Normal |
| Precocious Puberty | 0/4 | Not Reported | Yes | No | |
| Osteopenia | 4/4 | Yes | Yes | Yes | |
| Plasma Alkaline Phosphatase | Low | Low | Normal | Normal | |
| Plasma Calcium | High/high normal | Normal | Normal | Normal | |
| Plasma Phosphate | Normal | Not Reported | Normal | Normal | |
| Parathyroid Hormone Values | Low | Not Reported | Normal | Normal | |
| Thyroid Function Tests | Normal | Not Reported | Normal | Normal | |
| Immunologic | IgA and IgM Deficiency | Not Reported | Not Reported | No | Yes |
| Other Immune Deficiencies | Not Reported | Not Reported | No | No | |
| Hematologic | Hyper/hypo Coagulability | Not Reported | Not Reported | No | No |
| Factor excess or deficiency | Not Reported | Not Reported | Slightly decreased Factors V & XI | Mildly increased Factor VIII | |
| Dysmorphia | Inverted Mamillae | 2/4 | Not Reported | No | No |
| Other | 4/4 High forehead, bitemporal narrowing, broad nasal root, antevered nose, depressed nasal bridge, long philtrum with a deep groove, cupid bow lips | Low set ears, micrognathia, malar flattening, upslanting palpebral fissures, depressed nasal bridge, anteverted nose, downturned corners of mouth, tented lip, high arched palate | High forehead, bitemporal narrowing, anteverted nose, depressed nasal bridge, long philtrum downturned corners of the mouth, high-arched palate | High forehead, bitemporal narrowing, anteverted nose, depressed nasal bridge, long philtrum downturned corners of the mouth, high-arched palate | |
| Age at most recent reported | 12 years | 7 years 8 months | 6 years 8 months |
Patient 2
Patient 2 (affected brother), a 6-year-old boy, was born at 31 3/7 weeks’ gestation via vaginal delivery. The pregnancy was complicated by gestational diabetes and preterm labor. His birth weight was 2038 grams (90%); birth length was 45 cm (90%), and birth head circumference was 30 cm (50–75%). His APGAR scores were 4, 5 and 7 at 1, 5 and 10 minutes of life. He was admitted to the neonatal intensive care unit for 43 days and required phototherapy, cardiorespiratory support, and feeding support. He had a small umbilical and inguinal hernia. He had tremors and jitteriness but no seizures. His newborn hearing and bloodspot screens, head ultrasound, and ophthalmologic examinations were normal. His echocardiogram showed a dilated right atrium and an atrial septal defect with a left to right shunt; the latter was treated with an Amplatzer device at 25 months. At age 5 months, he had his first tonic-clonic seizure. His seizures progressed to intractable, poorly controlled epilepsy requiring hospitalizations, especially during febrile illnesses.
His development was normal until the onset of seizures at age 5 months. After this, there were no significant gains. In addition to seizures and developmental delay, he also had joint hypermobility, dysmorphic features (Fig. 2), progressive cerebellar and cerebral atrophy (Fig. 3), cortical visual dysfunction with bilateral staphyloma and nystagmus (Fig. 2e), and skeletal abnormalities (Fig. 4) (Table 1).
3.2. Clinical biochemical laboratory findings
Patient 1
Biochemical testing of patient 1 showed mild abnormalities including variably elevated blood lactate, up to 4.2 mmol/L (normal 0.3–1.3 mmol/L) with a mildly elevated lactate to pyruvate ratio at 32.3 (normal 8–20) at age 6 months. At 16 months, her creatine phosphokinase was 95 IU/L (0–91), and her acylcarnitine profile revealed elevated C0, C2, C6, C10, C10:1; urine amino acids demonstrated multiple elevations in a non-specific pattern. A urine mucopolysaccharide screen at 27 months was slightly elevated at 98.5 μg/mg creatinine (normal 16–90 μg/mg creatinine). At 32 months, alanine and tyrosine in plasma were slightly decreased. Urine organic acid profile, sialic acid, sulfite, purines and pyrimidines were normal. Plasma testing with normal results included amino acid profile, uric acid, very long chain fatty acid profile, ammonia, and 7-dehydrocholesterol screen by gas chromatography-mass spectrometry. Normal CSF testing included lactate, pyruvate, amino acid profile, and neurotransmitters. Also, white blood cell palmitoyl protein thioesterase enzyme analysis was normal.
A muscle biopsy at 16 months showed no significant abnormalities with PAS, modified Gomori trichrome, NADH, and ATPase histochemical staining. At 91 months, her CSF neurotransmitters, amino acids, pterins, lactate, MTHF, pyridoxal 5-phosphate, alphaaminoadipic semialdehyde, protein, oligoclonal bands, and IgG index and level were normal. The CSF albumin quotient was 2.88 (normal 3.2–9.0) with a normal plasma albumin.
Patient 2
Patient 2 underwent a skin biopsy and lumbar puncture at 11 months. Fibroblast electron transport chain complexes II, III, and IV activity and pyruvate dehydrogenase complex activities were normal. His CSF neurotransmitter metabolites, urine organic acids, mucopolysaccharide screen, and urine sialic acid were normal. At 79 months, CSF neurotransmitters, amino acids, pterins, lactate, MTHF, pyridoxal 5-phosphate, alpha-aminoadipic semialdehyde were normal. He also had a low CSF albumin quotient (1.96) despite a normal plasma albumin. CSF protein, oligoclonal bands and IgG levels were normal with an IgG index slightly elevated at 0.64 (normal 0.26–0.62).
Various screening tests for congenital disorders of glycosylation were performed on both siblings (Table 2).
Table 2.
Biochemical test results for congenital disorders of glycosylation in Patient 1 and Patient 2.
| Test | Age in months (Pt1, Pt2) | Patient 1 | Patient 2 |
|---|---|---|---|
| MALDI-TOF1 serum carbohydrate deficient transferrin | 47, 35 | Normal | Normal |
| MALDI-TOF/TOF urine oligosaccharide and glycan screen | 47, 35 | Normal | Normal |
| 73, 61 | Mild increase of several fucosylated and phosphate oligosaccharides2 | Same finding | |
| 92, 80 | Previous findings present but less prominent. | Same finding | |
| MALDI-TOF/TOF blood N-glycan and transferrin panel | 73, 61 | Normal | Normal |
| 92, 80 | Normal | Normal | |
| MALDI-TOF blood O-glycan profile and quantification | 73, 61 | Normal | Normal |
| 92, 80 | Normal | Normal | |
| Affinity Chromatography-Mass Spectrometry serum carbohydrate deficient transferrin | 36, 24 | Underglycosylated transferrin slightly elevated3 | Normal |
| 73, 61 | Normal | Normal | |
| 92, 80 | Normal | Normal |
MALDI-TOF = matrix assisted laser desorption/ionization-time of flight.
Fucosylated oligosaccharides at m/z 692, 1029, and 1070 were mildly increased with a phosphated oligosaccharide at m/z 1111. Clinical significance of these findings is unknown.
mono/di oligo 0.101 (<0.074), a/di-oligo 0.034 (<=0).
3.3. Cytogenetic and molecular findings
Karyotype, FMR-1 analysis, oligo chromosomal microarray analysis, MitoMet oligo array comparative genomic hybridization analysis, and maternal cystic fibrosis 32 mutation analysis were all normal for Pt1.
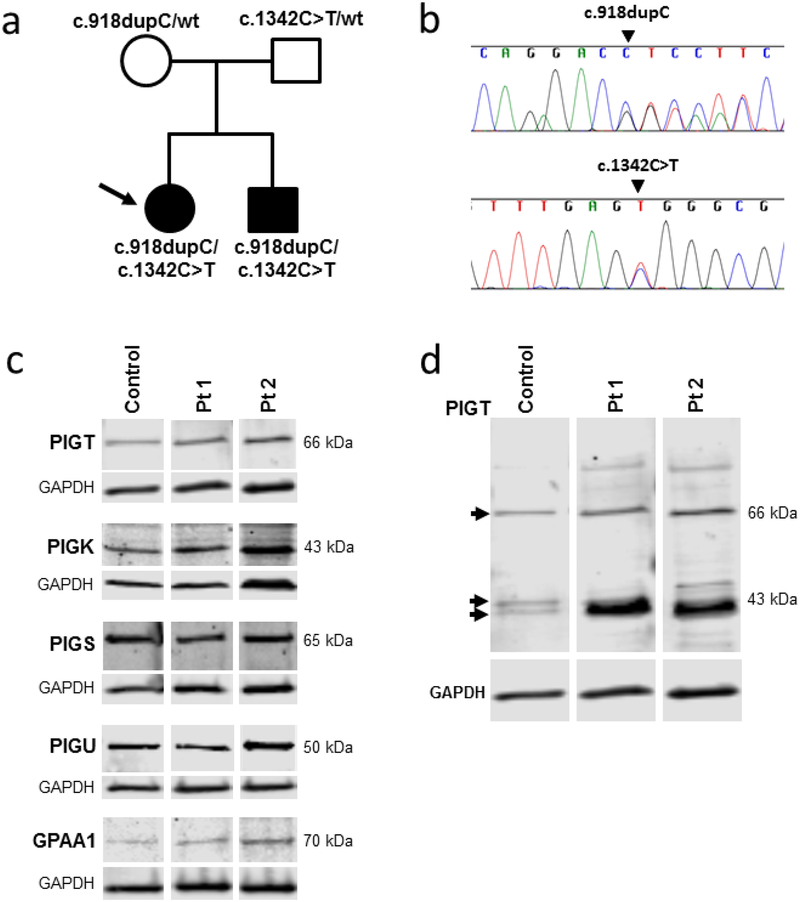
Exome sequencing identified two compound heterozygous variants in PIGT, NM_015937.5:c.[918dupC];[1342C>T], and were validated by Sanger sequencing. The patients carried both variants, whereas the parents each carried one variant and a wild type allele (Fig. 5a,b). Neither variant was found in 6500 ESP (Exome Sequencing Project) or 1000 genomes [33, 34].
Fig. 5.
Molecular data and protein expression results.
(a) Pedigree of the family with identified PIGT variants in each individual indicated.
(b) Sanger sequencing validation of the PIGT (NM_015937.5) variants c.918dupC and c.1342C>T.
(c) Immunoblots of fibroblast lysates from control, Pt1 and Pt2 were probed with antibodies to the GPI transamidase complex subunits and GAPDH (loading control). The bands corresponding to the major isoform of each GPI transamidase subunit are shown. The predicted molecular weight of each band, calculated by gel imaging software (Image studio Software, Li-cor Inc.) is indicated in the Figure. The calculated molecular weight of each subunit based on amino acid sequence are listed in Supplemental Table 1, and full images of the immunoblots are in Supplemental Fig. 2.
(d) Image of the entire PIGT antibody-probed immunoblot from (c). The indicated molecular weights are calculated by gelimaging software. The major protein bands are indicated by arrowheads. Note the protein band with increased intensity in both patients, compared to control around ~40 kDa, which corresponds to the predicted molecular weight of the truncated protein resulting from NM_015937.5:c.918dupC.
NM_015937.5:c.1342C>T, recently reported in a Japanese patient [23], is predictive of p.Arg448Trp. NM_015937.5:c.1342C>T, involves a weakly conserved nucleotide (c.1342C) and a moderately conserved amino acid (p.Arg488); p.Arg448Trp creates a moderate physicochemical difference (Grantham distance = 101), considered probably disease-causing by Polyphen-2, deleterious by SIFT (score 0.01, median 2.92) and disease causing by MutationTaster (p-value = 1).
NM_015937.5:c.918dupC is a novel variant predictive of a frameshift leading to a premature stop-codon (p.Val307Argfs*13). Although the truncated mRNA, resulting from the NM_015937.5:c.918dupC allele, might be a target for nonsense-mediated decay, quantitative real-time PCR analysis (targeting the PIGT mRNA NM_015937.5 exon 4–5 boundary) showed no significant changes in PIGT mRNA expression in fibroblasts from the patients compared to a control (Supplemental Fig. 1).
3.4. Protein findings
On immunoblotting of fibroblasts with antibodies against PIGT (Fig. 5c), no significant deficiency of the major PIGT protein isoform (66 kDa, NP_057021.2) expression was detected in the patients’ extracts compared to the control.
Since defects in one subunit can destabilize an entire multiprotein complex, we assessed protein expression of the other four subunits (PIGK, PIGS, PIGU and GPAA1) of the GPI transamidase complex in the patients’ fibroblast extracts. Immunoblotting demonstrated normal expression of all four subunits in both patients (Fig. 5c); none of the extraneous bands on the blots showed a different size or concentration in the patients compared to control cells (Supplemental Fig. 2, Supplemental Table 1).
Human PIGT has 11 predicted mRNA splice variants, six encode protein of unknown biological functions (Supplemental Table 1). On PIGT immunoblots of fibroblast extracts, three protein bands were visible (Fig. 5d, arrows). The 66 kDa band represents the major PIGT isoform variant 1 (NP_057021.2, Supplemental Table 1), and the two lower molecular weight bands in control fibroblast lysate (each ~43 kDa) likely represent two other PIGT protein isoforms, PIGT variant X4 (XP_005260487.1; 46.4 kDa predicted weight) and PIGT variant X6 (XP_005260489.1; 36.1 kDa). The discrepancies in molecular weight could be due to posttranslational modifications or to electrophoretic conditions. In both patients, a protein migrating similarly to the 36.1 kDa PIGT isoform showed considerably higher expression compared to the control. As suggested by the qPCR data (Supplemental Fig. 1) this band likely represents the truncated protein, p.Val307Argfs*13, which has a predicted molecular weight of 36.2 kDa.
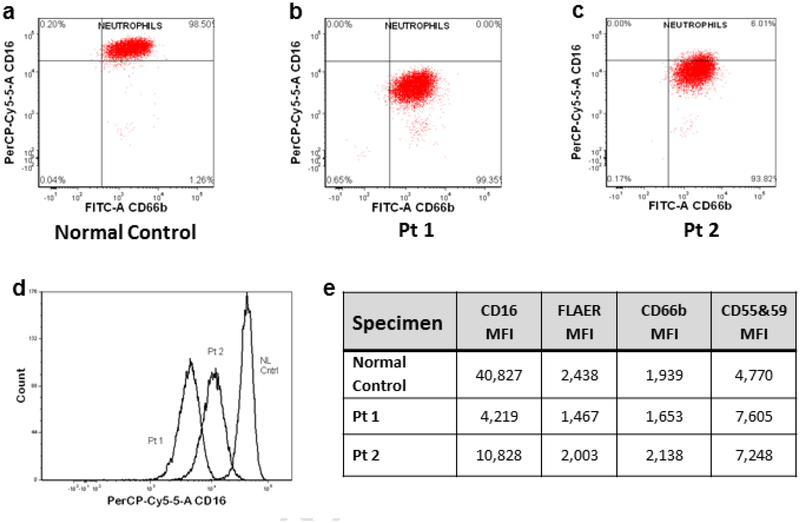
3.5. Flow cytometry findings
Flow cytometric comparison of cell surface GPI-anchored proteins between Patient 1 and Patient 2 and an unaffected control showed decreased granulocytic expression of CD16, which contains a GPI-linked Fc receptor on CD16b (Fig. 6). Binding of fluorochrome-conjugated aerolysin (FLAER), recognizing total GPI-anchored cell surface proteins was also decreased; antibody binding to other GPI-anchored proteins tested (CD55, CD59 and CD66b) was not significantly different (Fig. 6e).
Fig. 6.
Flow cytometry results of GPI-anchored proteins in patients’ granulocytes.
Scatter plots on logarithmic scale of fluorescent intensity of granulocytes tagged by antibody markers for CD66b on x axis and CD16 on y axis for normal control (a), Patient1 (b), and Patient 2 (c). Histogram overlays of normal control, Patient 1 and Patient 2 (d). The mean fluorescent intensity (MFI) determined by flow cytometry on granulocytes that were tagged with antibodies to fluorochrome conjugated aerolysin (FLAER) or antibodies to CD16, CD66b, and CD55 & 59 (e).
3.6. MRI findings
Inspection of the clinical images did not reveal any discrete lesions or abnormal signal in the brain. However, cerebellar atrophy was apparent on some of the images. The measured volumes of the cerebrum and cerebellum are presented in Figure 3. Cerebral volumes in both patients were not clearly abnormal until about 5 years of age; volume loss seems to have started earlier in Patient 1 than in Patient 2, and also seems to be progressing faster in Patient 1 compared to Patient 2 (-31 cm3/year versus −13 cm3/year). Although the cerebellum was qualitatively normal on the MRI exam performed at 6.5 months of age, the images were not suitable for volume calculation; the earliest available quantitative measurements on both patients demonstrated profound cerebellar atrophy. Cerebellar atrophy followed an exponential pattern, with rapid loss initially and more gradual loss on the later studies.
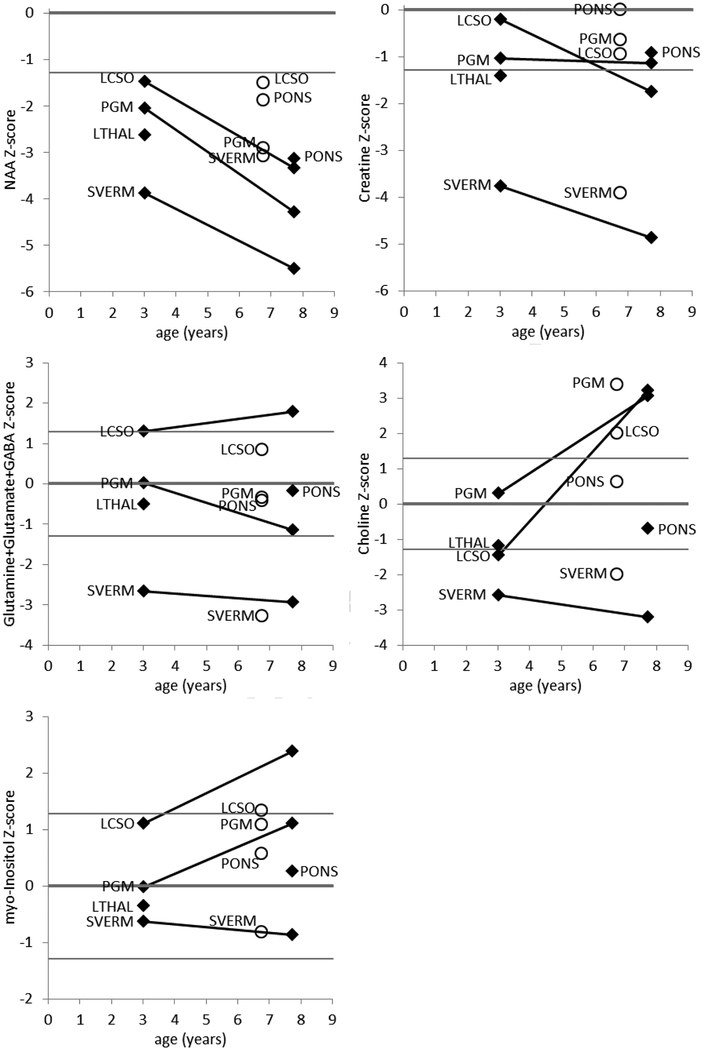
3.7. MRS findings
The measured metabolite concentrations are presented in Supplemental Table 2 and plotted in Figure 7. The most striking finding is that all measurements of NAA were low (<10th percentile), regardless of the location or age, and all measurements declined dramatically on follow-up. Another striking finding is that all of the metabolites were low in the superior cerebellar vermis, and declined on follow-up. All measurements of creatine were lower than average (50th percentile or lower), and declined on follow-up. Except in the left centrum semiovale, glutamine+glutamate+GABA (Glx) was low and declined on follow-up; in the left centrum semiovale, glutamine+glutamate+GABA was higher than average and rose upon followup. Except in the superior cerebellar vermis, early measurements of choline were average or low and late measurements were average or high. Similarly, early measurements of myo-inositol were average or high and rose later.
Fig. 7.
MRS results. Measurements on the Patient1 (diamonds) and on Patient 2 (open circles) are displayed in separate plots for each metabolite. The metabolite levels are plotted as a Z-score relative to the distribution of the reference group; the horizontal lines on the plots denote the 10th, 50th, and 90th percentiles. The metabolites that are produced by healthy neurons (NAA, creatine, glutamine, glutamate, and GABA) are generally lower than normal and decline on follow-up. The metabolites that are elevated by inflammation (choline and myo-inositol) are generally high and rising in the parietal gray matter and centrum semiovale, relatively normal in the pons and thalamus, and low and falling in the superior cerebellar vermis. The spatial and temporal patterns suggest that the cerebellum is affected earlier in the disease than is the cerebrum, and that there is a transient inflammatory phase that precedes the loss of neurons.
4. Discussion
We describe the third PIGT-CDG family; their PIGT variants were recognized as pathogenic when the first case of PIGT-CDG was published [11,23]. The mixed African American and Caucasian ethnic background of our family is distinct from the initial PIGT-CDG kindreds, who were consanguineous Aramaic Turkish and Japanese [11,23].
Similar to the other PIGT-CDG families, the predominant presentation in our family was neuro-ophthalmologic with profound intellectual disability and progressive neurological features: hypotonia, intractable seizures, cortical visual impairment, and nystagmus and/or strabismus. Although the prenatal course was benign and initial EEGs showed no seizure activity, our patients’ EEG worsened in parallel with clinical seizure activity. Also, as measured by brain MRI, the cerebral and cerebellar atrophy initially progressed rapidly and then was followed by a period of stability. The temporal patterns followed by NAA, creatine, and glutamine+glutamate+GABA suggest progressive neuronal loss, as these compounds are predominantly found in neurons or their attendant astroglia. The more dramatic early deficit in the superior cerebellar vermis is consistent with the MRI observation that in the PIGT-deficient patients, atrophy in the cerebellum starts earlier and proceeds more rapidly than atrophy elsewhere in the brain. It also suggests the disease preferentially affects the cerebellum. The reason for the high and slightly rising Glx in the LCSO is unclear. The dramatic rise in choline and myo-inositol everywhere except the superior cerebellar vermis, the area of most advanced disease, suggests that the disease process initially causes inflammation or gliosis and is followed by a late burn-out phase. Indicative of a blood-CSF barrier dysfunction, the CSF albumin concentration and quotient were low in our siblings [35].
The bone and endocrine features in our patients differ from reported cases. Our patients exhibited normal to advanced bone age, and, normal alkaline phosphatase, plasma calcium, plasma phosphate, urine calcium, and parathyroid hormone values. In contrast, the previously reported cases had reduced bone mineralization and scoliosis, features that could arise secondary to their neurologic complications. Our patients had normal dentition, like the Japanese subject [23] but unlike the Turkish kindred [11]. The dental findings in the Turkish PIGT-CDG kindred could be unrelated to the PIGT variants, could reflect a genotype-phenotype effect, or could arise from differences in genetic background. Unlike previously reported cases, our patients also exhibited significant joint hypermobility and normal motor unit function on electromyogram (Table 1).
Quantitative PCR analysis of our patients’ mRNA showed normal PIGT mRNA expression (Supplemental Fig. 1), strongly suggesting that their frameshift allele (NM_015937.5: c.918dupC predictive of p.Val307Argfs*13) was not targeted for nonsense-mediated RNA decay. Indeed, expression of the frameshift allele was confirmed at the protein level. Immunoblotting showed that both patients expressed an aberrant protein around the predicted size of the truncated protein p.Val307Argfs*13 (Fig. 5d), which lacks the C-terminal PIGT transmembrane domain necessary for the function of the GPI-anchor protein [22]. In fibroblasts, our patients’ second mutant PIGT allele (NM_015937.5: c.918dupC predictive of p.Arg448Trp) produced the major 66 kDa PIGT isoform as well as 2 minor isoforms (46.1 and 36.1 kDa), similar to control cells. The patients’ 36.1 kDa PIGT isoform was, however, ‘overshadowed’ by the 36.2 kDa truncated protein band (Fig. 5d). Both minor PIGT isoforms contain the p.Arg448 amino acid, but their biologic functions remain unknown. Although PIGT plays a critical role in stabilizing the GPI transamidase complex [22], we found normal protein expression of all 5 subunits in both patients (Fig. 5c). This indicated that neither the PIGT p.Arg448Trp missense variant nor the truncated p.Val307Argfs*13 PIGT protein rendered other GPI transamidase complex subunits unstable. The GPI transamidase complex activity, however, was diminished based on the decreased surface expression of the GPI anchored protein CD16 (containing a GPI linked Fc receptor of CD 16b [36]) on our patients’ granulocytes (Fig. 6a); however, the normal expression of other GPI-anchored proteins, detected by FLEAR and antibody staining (Fig. 6), indicated residual GPI transamidase activity in the patients’ cells. Similar residual GPI anchored protein expression was found in the recently reported Japanese patient who also had the p.Arg448Trp missense variant and a nonsense variant [23]. In that report, expression of the PIGT p.Arg448Trp protein in PIGT-deficient cells partially restored expression of GPI anchored proteins.
All 3 reported cases with PIGT-CDG have at least one missense variant, and all had reduced but not absent GPI-anchored proteins on their cell surfaces. We hypothesize that residual GPI transamidase activity is necessary for life. In fact, all reported mutation pairs for the GPI biosynthesis pathway have at least one ‘mild’ allele that leads to only partial loss of protein function, i.e., reduced but measurable amounts of GPI-anchored cell surface proteins [11, 23]. Furthermore, complete disruption of the GPI synthesis pathway by targeted deletion of PIGA is embryonic lethal in mice [37].
There are no current FDA approved therapies for GPI anchor disorders, but the progressive nature of PIGT-CDG is attractive for therapies that slow or halt the neurologic deterioration [21]. Identification of additional PIGT-CDG patients should further define the clinical spectrum and assist in developing diagnostic criteria. Current testing methods for CDGs may need to be expanded to diagnose PIGT-CDG because it is only detectable by flow cytometry and genetic testing.
Supplementary Material
Highlights:
We describe the third kindred of PIGT-CDG caused by biallelic variants in PIGT.
We clarify and expand clinical phenotype of PIGT-CDG.
We identify a novel mutation, c.918dupC predictive of p.Val307Argfs*13, in PIGT.
We emphasize current screening methods for CDGs cannot detect PIGT-CDG.
We explore flow cytometry as a possible screening test in GPI anchor disorders.
Acknowledgments
We would like to thank the family for participating in this study. We appreciate the expert laboratory assistance of Carla Ciccone, the Magnetic Resonance technologists who scanned the patients (Mastaneh Owhadi and Bonita Damaska), and Dr. F.D. Porter for the use of his patients as a reference group. This study was supported by the by the Common Fund, Office of the Director and the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors declare no conflict of interest.
References
- [1].Paulick MG, Bertozzi CR, The glycosylphosphatidylinositol anchor: a complex membrane-anchoring structure for proteins, Biochemistry 47 (2008) 6991–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nosjean O, Briolay A, Roux B, Mammalian GPI proteins: sorting, membrane residence and functions, Biochim. Biophys. Acta 1331 (1997) 153–186. [DOI] [PubMed] [Google Scholar]
- [3].Ferguson MAJ, Kinoshita T, Hart GW, Glycosylphosphatidylinositol Anchors, in: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME (Eds), Essentials of Glycobiology, 2nd edition, Cold Spring Harbor (NY), Cold Spring Harbor Laboratory Press, 2009, Chapter 11. [PubMed] [Google Scholar]
- [4].Kinoshita T, Fujita M, Maeda Y, Biosynthesis, remodelling and functions of mammalian GPI-anchored proteins: recent progress, J. Biochem 144 (2008) 287–294. [DOI] [PubMed] [Google Scholar]
- [5].Johnston JJ, Gropman AL, Sapp JC, Teer JK, Martin JM, Liu CF, Yuan X, Ye Z, Cheng L, Brodsky RA, Biesecker LG, The phenotype of a germline mutation in PIGA: the gene somatically mutated in paroxysmal nocturnal hemoglobinuria, Am. J. Hum. Genet 90 (2012) 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Swoboda KJ, Margraf RL, Carey JC, Zhou H, Newcomb TM, Coonrod E, Durtschi J, Mallempati K, Kumanovics A, Katz BE, Voelkerding KV, Opitz JM, A novel germline PIGA mutation in Ferro-Cerebro-Cutaneous syndrome: a neurodegenerative X-linked epileptic encephalopathy with systemic iron-overload, Am. J. Med. Genet. A 164A (2014) 17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Almeida AM, Murakami Y, Layton DM, Hillmen P, Sellick GS, Maeda Y, Richards S, Patterson S, Kotsianidis I, Mollica L, Crawford DH, Baker A, Ferguson M, Roberts I, Houlston R, Kinoshita T, Karadimitris A, Hypomorphic promoter mutation in PIGM causes inherited glycosylphosphatidylinositol deficiency, Nat. Med 12 (2006) 846851. [DOI] [PubMed] [Google Scholar]
- [8].Krawitz PM, Schweiger MR, Rodelsperger C, Marcelis C, Kolsch U, Meisel C, Stephani F, Kinoshita T, Murakami Y, Bauer S, Isau M, Fischer A, Dahl A, Kerick M, Hecht J, Kohler S, Jager M, Grunhagen J, de Condor BJ, Doelken S, Brunner HG, Meinecke P, Passarge E, Thompson MD, Cole DE, Horn D, Roscioli T, Mundlos S, Robinson PN, Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome, Nat. Genet 42 (2010) 827829. [DOI] [PubMed] [Google Scholar]
- [9].Maydan G, Noyman I, Har-Zahav A, Neriah ZB, Pasmanik-Chor M, Yeheskel A, Albin-Kaplanski A, Maya I, Magal N, Birk E, Simon AJ, Halevy A, Rechavi G, Shohat M, Straussberg R, Basel-Vanagaite L, Multiple congenital anomalies-hypotoniaseizures syndrome is caused by a mutation in PIGN, J. Med. Genet 48 (2011) 383–389. [DOI] [PubMed] [Google Scholar]
- [10].Krawitz PM, Murakami Y, Hecht J, Kruger U, Holder SE, Mortier GR, Delle Chiaie B, De Baere E, Thompson MD, Roscioli T, Kielbasa S, Kinoshita T, Mundlos S, Robinson PN, Horn D, Mutations in PIGO, a member of the GPI-anchor-synthesis pathway, cause hyperphosphatasia with mental retardation, Am. J. Hum. Genet 91 (2012) 146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kvarnung M, Nilsson D, Lindstrand A, Korenke GC, Chiang SC, Blennow E, Bergmann M, Stodberg T, Makitie O, Anderlid BM, Bryceson YT, Nordenskjold M, Nordgren A, A novel intellectual disability syndrome caused by GPI anchor deficiency due to homozygous mutations in PIGT, J. Med. Genet 50 (2013) 521–528. [DOI] [PubMed] [Google Scholar]
- [12].Hansen L, Tawamie H, Murakami Y, Mang Y, ur Rehman S, Buchert R, Schaffer S, Muhammad S, Bak M, Nothen MM, Bennett EP, Maeda Y, Aigner M, Reis A, Kinoshita T, Tommerup N, Baig SM, Abou Jamra R, Hypomorphic mutations in PGAP2, encoding a GPI-anchor-remodeling protein, cause autosomal-recessive intellectual disability, Am. J. Hum. Genet 92 (2013) 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chiyonobu T, Inoue N, Morimoto M, Kinoshita T, Murakami Y, Glycosylphosphatidylinositol (GPI) anchor deficiency caused by mutations in PIGW is associated with West syndrome and hyperphosphatasia with mental retardation syndrome, J. Med. Genet 51 (2014) 203–207. [DOI] [PubMed] [Google Scholar]
- [14].Ng BG, Freeze HH, Human genetic disorders involving glycosylphosphatidylinositol (GPI) anchors and glycosphingolipids (GSL), J. Inherit. Metab. Dis 38 (2014) 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Krawitz PM, Hochsmann B, Murakami Y, Teubner B, Kruger U, Klopocki E, Neitzel H, Hoellein A, Schneider C, Parkhomchuk D, Hecht J, Robinson PN, Mundlos S, Kinoshita T, Schrezenmeier H, A case of paroxysmal nocturnal hemoglobinuria caused by a germline mutation and a somatic mutation in PIGT, Blood 122 (2013) 1312–1315. [DOI] [PubMed] [Google Scholar]
- [16].Alazami AM, Patel N, Shamseldin HE, Anazi S, Al-Dosari MS, Alzahrani F, Hijazi H, Alshammari M, Aldahmesh MA, Salih MA, Faqeih E, Alhashem A, Bashiri FA, Al-Owain M, Kentab AY, Sogaty S, Al Tala S, Temsah MH, Tulbah M, Aljelaify RF, Alshahwan SA, Seidahmed MZ, Alhadid AA, Aldhalaan H, AlQallaf F, Kurdi W, Alfadhel M, Babay Z, Alsogheer M, Kaya N, Al-Hassnan ZN, AbdelSalam GM, Al-Sannaa N, Al Mutairi F, El Khashab HY, Bohlega S, Jia X, Nguyen HC, Hammami R, Adly N, Mohamed JY, Abdulwahab F, Ibrahim N, Naim EA, AlYounes B, Meyer BF, Hashem M, Shaheen R, Xiong Y, Abouelhoda M, Aldeeri AA, Monies DM, Alkuraya FS, Accelerating novel candidate gene discovery in neurogenetic disorders via whole-exome sequencing of prescreened multiplex consanguineous families, Cell Rep 10 (2015) 148–161. [DOI] [PubMed] [Google Scholar]
- [17].Martin HC, Kim GE, Pagnamenta AT, Murakami Y, Carvill GL, Meyer E, Copley RR, Rimmer A, Barcia G, Fleming MR, Kronengold J, Brown MR, Hudspith KA, Broxholme J, Kanapin A, Cazier JB, Kinoshita T, Nabbout R, Bentley D, McVean G, Heavin S, Zaiwalla Z, McShane T, Mefford HC, Shears D, Stewart H, Kurian MA, Scheffer IE, Blair E, Donnelly P, Kaczmarek LK, Taylor JC, Clinical whole-genome sequencing in severe early-onset epilepsy reveals new genes and improves molecular diagnosis, Hum. Mol. Genet 23 (2014) 3200–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Murakami Y, Tawamie H, Maeda Y, Buttner C, Buchert R, Radwan F, Schaffer S, Sticht H, Aigner M, Reis A, Kinoshita T, Jamra RA, Null mutation in PGAP1 impairing Gpi-anchor maturation in patients with intellectual disability and encephalopathy, PLoS Genet 10 (2014) e1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Novarino G, Fenstermaker AG, Zaki MS, Hofree M, Silhavy JL, Heiberg AD, Abdellateef M, Rosti B, Scott E, Mansour L, Masri A, Kayserili H, Al-Aama JY, Abdel-Salam GM, Karminejad A, Kara M, Kara B, Bozorgmehri B, Ben-Omran T, Mojahedi F, Mahmoud IG, Bouslam N, Bouhouche A, Benomar A, Hanein S, Raymond L, Forlani S, Mascaro M, Selim L, Shehata N, Al-Allawi N, Bindu PS, Azam M, Gunel M, Caglayan A, Bilguvar K, Tolun A, Issa MY, Schroth J, Spencer EG, Rosti RO, Akizu N, Vaux KK, Johansen A, Koh AA, Megahed H, Durr A, Brice A, Stevanin G, Gabriel SB, Ideker T, Gleeson JG, Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders, Science 343 (2014) 506511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Howard MF, Murakami Y, Pagnamenta AT, Daumer-Haas C, Fischer B, Hecht J, Keays DA, Knight SJ, Kolsch U, Kruger U, Leiz S, Maeda Y, Mitchell D, Mundlos S, Phillips JA 3rd, Robinson PN, Kini U, Taylor JC, Horn D, Kinoshita T, Krawitz PM, Mutations in PGAP3 impair GPI-anchor maturation, causing a subtype of hyperphosphatasia with mental retardation, Am. J. Hum. Genet 94 (2014) 278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ng BG, Freeze HH, Human genetic disorders involving glycosylphosphatidylinositol (GPI) anchors and glycosphingolipids (GSL), J. Inherit. Metab. Dis 38 (2015) 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ohishi K, Inoue N, Kinoshita T, PIG-S and PIG-T, essential for GPI anchor attachment to proteins, form a complex with GAA1 and GPI8, EMBO J 20 (2001) 4088–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nakashima M, Kashii H, Murakami Y, Kato M, Tsurusaki Y, Miyake N, Kubota M, Kinoshita T, Saitsu H, Matsumoto N, Novel compound heterozygous PIGT mutations caused multiple congenital anomalies-hypotonia-seizures syndrome 3, Neurogenetics 15 (2014) 193–200. [DOI] [PubMed] [Google Scholar]
- [24].Gahl WA, Markello TC, Toro C, Fajardo KF, Sincan M, Gill F, Carlson-Donohoe H, Gropman A, Pierson TM, Golas G, Wolfe L, Groden C, Godfrey R, Nehrebecky M, Wahl C, Landis DM, Yang S, Madeo A, Mullikin JC, Boerkoel CF, Tifft CJ, Adams D, The National Institutes of Health Undiagnosed Diseases Program: insights into rare diseases, Genet. Med 14 (2012) 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gahl WA, Tifft CJ, The NIH Undiagnosed Diseases Program: lessons learned, Jama 305 (2011) 1904–1905. [DOI] [PubMed] [Google Scholar]
- [26].Adams DR, Sincan M, Fuentes Fajardo K, Mullikin JC, Pierson TM, Toro C, Boerkoel CF, Tifft CJ, Gahl WA, Markello TC, Analysis of DNA sequence variants detected by high-throughput sequencing, Hum. Mutat 33 (2012) 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tiemeier H, Lenroot RK, Greenstein DK, Tran L, Pierson R, Giedd JN, Cerebellum development during childhood and adolescence: a longitudinal morphometric MRI study, Neuroimage 49 (2010) 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Baker EH, Basso G, Barker PB, Smith MA, Bonekamp D, Horska A, Regional apparent metabolite concentrations in young adult brain measured by (1)H MR spectroscopy at 3 Tesla, J. Magn. Reson. Imaging 27 (2008) 489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Horska A, Calhoun VD, Bradshaw DH, Barker PB, Rapid method for correction of CSF partial volume in quantitative proton MR spectroscopic imaging, Magn. Reson. Med 48 (2002) 555–558. [DOI] [PubMed] [Google Scholar]
- [30].Yanjanin NM, Velez JI, Gropman A, King K, Bianconi SE, Conley SK, Brewer CC, Solomon B, Pavan WJ, Arcos-Burgos M, Patterson MC, Porter FD, Linear clinical progression, independent of age of onset, in Niemann-Pick disease, type C, Am. J. Med. Genet. B. Neuropsychiatr. Genet 153B (2010) 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Provencher SW, Automatic quantitation of localized in vivo 1H spectra with LCModel, NMR Biomed 14 (2001) 260–264. [DOI] [PubMed] [Google Scholar]
- [32].Srinivasan R, Sailasuta N, Hurd R, Nelson S, Pelletier D, Evidence of elevated glutamate in multiple sclerosis using magnetic resonance spectroscopy at 3 T, Brain 128 (2005) 1016–1025. [DOI] [PubMed] [Google Scholar]
- [33].1000 Genomes Project Consortium, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA, A map of human genome variation from population-scale sequencing, Nature 467 (2010) 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA, An integrated map of genetic variation from 1,092 human genomes, Nature 491 (2012) 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Deisenhammer F, Bartos A, Egg R, Gilhus NE, Giovannoni G, Rauer S, Sellebjerg F, Guidelines on routine cerebrospinal fluid analysis. Report from an EFNS task force, Eur. J. Neurol 13 (2006) 913–922. [DOI] [PubMed] [Google Scholar]
- [36].Selvaraj P, Rosse WF, Silber R, Springer TA, The major Fc receptor in blood has a phosphatidylinositol anchor and is deficient in paroxysmal noctural hemoglobinuria, Nature 333 (1988) 565–567. [DOI] [PubMed] [Google Scholar]
- [37].Kawagoe K, Kitamura D, Glycosylphosphatidylinositol-anchor-deficient mice: implications for clonal dominance of mutant cells in paroxysmal nocturnal hemoglobinuria. Blood 87 (1996) 3600–3606. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.