Abstract
Large non-coding RNAs fold into their biologically functional structures via compact yet disordered intermediates, which couple the stable secondary structure of the RNA with the emerging tertiary folding. The specificity of the collapse transition, which coincides with the assembly of helical domains, depends on RNA sequence and counterions. It determines the specificity of the folding pathways and the magnitude of the free energy barriers to the ensuing search for the native conformation. By coupling helix assembly with nascent 3D interactions, compact folding intermediates in RNA also play a crucial role in ligand and protein recognition.
Keywords: ribozyme, energy landscape, collapse transition, SAXS, single-molecule FRET, hydroxyl radical footprinting
INTRODUCTION
Many families of non-coding RNA fold into a specific three-dimensional (3D) shapes in order to function in the cell. Despite their diverse architectures and sizes ranging from 30 to ≥ 3000 nt, common rules govern their self-assembly. Combined advances in biochemical footprinting, NMR, SAXS, single molecule spectroscopy, force denaturation and computational modeling are producing a remarkably coherent picture of how RNAs fold (for recent reviews, see (7, 19, 50, 79, 103)).
One theme is that the hierarchical division of RNA structures into secondary and tertiary interactions is more plastic than previously realized, because the 2D and 3D interactions are coupled during the formation of compact folding intermediates. A second theme is that RNA folding pathways are heterogeneous and parallel, owing to the stability of the 2D interactions and the electrostatic forces driving the formation of compact structures. The partitioning of molecules between different folding pathways is exquisitely sensitive to small changes in the sequence of the RNA. Thus, it is important to know what conditions maximize the fidelity of the folding process, and how the intermediates are remodeled. This review will discuss recent work on the folding pathways in RNA, and how compact but disordered intermediates contribute to the fidelity and cooperativity of the folding process.
STRUCTURAL HIERARCHY IN RNA
Folding Energetics
Stable Watson-Crick interactions between complementary RNA strands provide a dominant organizing force in RNA, that is followed by weaker tertiary interactions between elements of the secondary structure (8). Although nucleation of a double helix or hairpin loop is unfavorable, the propagation of base pairs is exothermic, owing to the large negative enthalpy change associated with stacking of adjacent base pairs. Typical nearest neighbor free energy terms for adding a base pair range to a helix range from −1 to −3.6 kcal/mol (1 M NaCl, 37 °C) (33).
By contrast, RNA tertiary structures are maintained by a smaller number of weak interactions that are usually not Watson-Crick (8, 49). Kissing loops are among the most stable, with ΔG37 ranging from −6 to −15 kcal/mol, depending on the number of base pairs between the loops (41, 47, 102). Docking of GAAA tetraloops with their helical receptors (ΔG of −2 to −4 kcal/mol; (26, 106) are also driven by a favorable ΔH ~ −17 kcal/mol (97). Other common tertiary interactions, such as ribose zippers and A-minor interactions, are less energetically favorable (80).
Despite these energetically favorable interactions, net folding free energies can be as small as −2 to −4 kcal/mol (16, 47, 76, 106). One reason is the enormous electrostatic force opposing the juxtaposition of phosphates, which come much closer together in the 3D fold than in a double helix. Consequently, the stability of RNA 3D structure depends strongly on the presence of multivalent ions, and ΔG for folding is diminished by the unfavorable entropy associated with localizing ions around the RNA (27).
A second reason is the contextual nature of the 3D interaction motifs (49). By their very nature, RNA functional sites lie within internal loops or at helix junction, where irregular twists of the RNA backbone create intricate pockets for ligand recognition and catalysis. These pockets are not only less stable than Watson-Crick helices, but are often formed at the expense of Watson-Crick base pairs. The net ΔG for folding is reduced by the energy required to deform or reorganize the secondary structure.
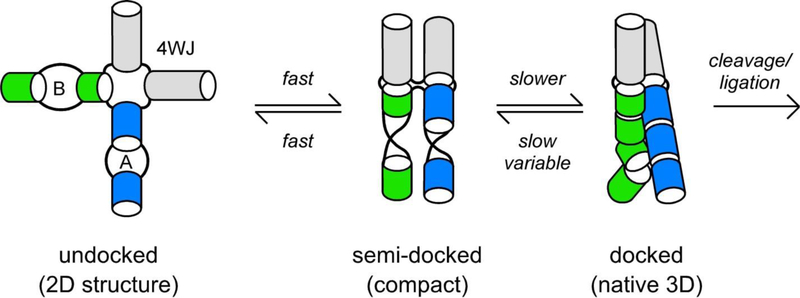
A well-studied example is the hairpin ribozyme, in which the active site is created by the association of internal loops from two different helical domains (72) (Figure 1). Both loops have different structures in isolation, and tertiary docking requires the sacrifice of Watson-Crick base pairs not present in the native ribozyme (12, 13). In other examples, tertiary folding of the P5abc RNA and the Varkud Satellite (VS) ribozyme is coupled to a shift in secondary structure (2, 105). In these cases, the tertiary structure must be stable enough to offset the ΔG gained by the change in secondary structure (90).
Figure 1. Folding of hairpin ribozyme.
Internal loops A and B dock in the native state (72). Rapid fluctuations of the four-way junction (4WJ) produces a compact intermediate; conformational changes in loops A and B lead to the native docked state. Docking is slower when the 4WJ is replaced by a two helix junction (2WJ). Adapted from (65, 88, 110).
Timescales for RNA folding
The timescales for RNA folding correlate with the stability of the RNA interactions and the number of interactions that join residues distant from one another in the RNA chain. Temperature-jump and NMR studies showed that hairpins with short loops have closing times of 10 to 100 μs, with the time-scale for adding base pairs to an existing helix on the order of 1 μs (21). This is followed by the assembly of double helices into compact intermediates, in which tertiary interactions between helices are initially established. As discussed below, collapse transitions occur in ≤ 10 ms in the presence of multivalent ions (30, 74).
The collapse to a compact state is followed by a slower, diffusive conformational search leads to the lowest ΔG state, which in most cases is the native state (91, 92). The final search for the tertiary structure, which in some cases requires significant reorganization of the RNA structure, requires anywhere from 10 ms in tRNA (22) and stable ribozymes (17, 30) to 1–1000 s in larger RNAs with complex topologies and multiple domains (78, 86, 107). The timescale for this final stage of the folding process depends on the specificity of the collapse transition, and the degree to which the compact intermediates refold.
KINETIC PARTITIONING OF FOLDING PATHWAYS
Folding of proteins and RNA can be modeled as a diffusive and stochastic search among probable conformations toward the most stable state (usually the native state). This search occurs within an “energy landscape” defined by the free energies of the conformations and the energy barriers between them (e.g., (19, 91)). The stability of RNA secondary structure and the low complexity of the primary sequence means that many RNA sequences can form stable secondary structures that are not compatible with the native 3D fold (38, 92). Because base pairing interactions in RNA are stable, the energy barriers between correct and incorrect structures are high. As a result, the free energy landscape for RNA folding is rough, with many local minima that compete with the global minimum or native state (92). Molecules that fall into these local minima (representing misfolded structures) become kinetically trapped on their way to the native structure.
A key prediction of energy landscape theories is that individual molecules follow different trajectories to the native state, due to stochastic fluctuations in the molecular interactions. When the folding landscape is “rough”, the folding trajectories lead individual RNAs through different intermediates, explaining the stretched folding times often seen in RNA. Because folding times of minutes or hours are likely incompatible with biological activity, interactions that are formed early during the folding process can determine the fate of an RNA. As we shall see below, the fraction of molecules folding directly to the native state depends on the potential for alternative secondary structures and the stability of the tertiary interactions.
Experimental evidence for kinetic partitioning
Experimental evidence for kinetic partitioning in RNA initially came from native gel experiments on the Tetrahymena self-splicing RNA (56). Most of the self-splicing RNA or ribozyme folds in 1–2 min through metastable intermediates in which the P3/P7 pseudoknot region is mispaired (58). However, ~8% of the population folded more rapidly, bypassing the metastable I’s. Single molecule FRET experiments showed more directly that 6% of transitions from a low FRET (unfolded) state to a high FRET (folded) state occurred directly with an average rate constant of 1 s−1, while the remaining transitions were slower and included an intermediate FRET state (109). Consistent with the presence of kinetically trapped, misfolded I’s, urea accelerated refolding of the Tetrahymena self-splicing RNA and the B. subtilis RNase P ribozyme (56, 62), while Mg2+ and tertiary interactions that stabilize the I’s lower the refolding rate (59, 69, 95).
Since then, heterogeneous folding pathways have been observed in many RNAs, and are clearly intrinsic to the dynamics of RNA structures (7). In some cases, individual molecules persist in the same type of dynamic behavior for minutes or even hours (55, 110). In the hairpin ribozyme, such “memory effects” resist denaturation of the RNA (25), and must either represent an extraordinarily high barrier to refolding or a covalent modification that has so far eluded detection.
Pathway diversity in small RNAs
Even the simplest elements of RNA structure exhibit the complex dynamics characteristic of a conformational search among competing structures. Temperature-jump experiments showed that hairpin closing times are dominated by an entropic search for the correct loop conformation (3). The search is lengthened by alternative loop conformations (3) and by interactions within the single-stranded loop (52, 82). Stochastic jumps between folding paths have also been observed by exquisitely precise force-denaturation experiments (reviewed in (93, 103)).
COLLAPSED STATES IN RNA
There is now ample evidence that the final search for the native structure occurs within compact intermediates, in which the helices interact but are not yet stably packed (104). “Collapsed” intermediates, which can form in physiological salts, provide a context in which tertiary interactions can emerge. In doing so, they form a crucial bridge between the secondary structure and the native state.
Size exclusion chromatography and native gel electrophoresis first showed that the bI5 group I ribozyme from yeast mitochondria formed compact structures in 3–7 mM MgCl2, even though 40 mM MgCl2 is needed to bury the RNA backbone from solvent and for ribozyme activity (9). Rapid photocrosslinking of the bI5 RNA showed that the intermediate could be chased to the native state and contained at least transient tertiary interactions, as well as non-native interactions (10).
SAXS experiments on the RNase P C-domain and the Tetrahymena group I ribozyme provided physical evidence for a collapse transition in RNA (28, 73). Compaction of the C-domain correlated with a large increase in the UV hypochromicity of the RNA, indicating a significant increase in base stacking (30). This step was sensitive to urea, as expected for a transition that involves a burial of RNA surface area (29). Stopped-flow spectroscopy and SAXS showed that the collapse transition occurs within a millisecond after Mg2+ is added to the RNase P C domain and begins within 10 ms for Tetrahymena ribozyme (30, 74).
Partial cleavage by ribonuclease T1 and small angle neutron scattering (SANS) on the Azoarcus group I ribozyme showed that the formation of compact intermediates (IC) in 0.3 mM MgCl2 correlates with the assembly of core helices, including a triple helix and the P3 pseudoknot (63, 68). A second transition in 2 mM MgCl2 led to protection of the RNA backbone from hydroxyl radical cleavage and the onset of catalytic activity (68). Mutations that disrupt tertiary interactions between the helical domains destabilized IC, causing it to form at higher Mg2+ concentrations (16). Thus, like the bI5 and Tetrahymena ribozymes, collapse of the Azoarcus ribozyme produces an intermediate that contains some tertiary structure, even though the RNA backbone remains solvent accessible.
Tertiary interactions direct helix assembly
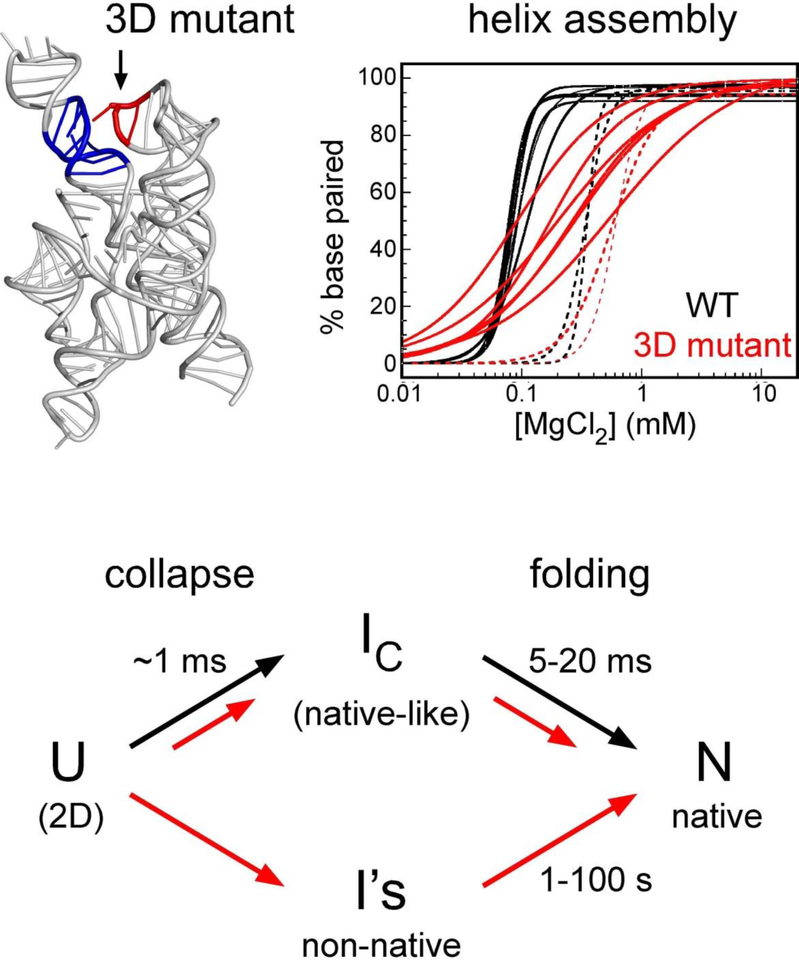
Unlike the Tetrahymena group I ribozyme, the smaller Azoarcus ribozyme can fold in 10–20 ms at 37 ˚C without becoming trapped in metastable states (15, 17). This is due in part to its stable, GC-rich secondary structure. However, tertiary interactions between helices also significantly reduce misfolding of the RNA by increasing the cooperativity of helix assembly (Figure 2) (17). Mutation of the L9 GAAA tetraloop, which docks with a receptor in P5, destabilized IC as well as N (16). More intriguing was that base pairing of G’s in the core helices spread out over a much wider range of Mg2+ concentrations in the mutant ribozyme (17).
Figure 2. Tertiary interactions increase the cooperativity of helix assembly.
A loop mutation (GAAA to GUAA) in the Azoarcus ribozyme (1) makes pairing of core helices less concerted, as revealed by partial digestion with ribonuclease T1 (WT, black lines; mutant, red lines). As helix assembly becomes less specific, the fraction of slow folding RNA folding rises. Adapted from (17).
This reduced cooperativity of helix assembly correlated with increased misfolding of the RNA population in time-resolved hydroxyl radical footprinting experiments (17). While 80–90% of the wild type Azoarcus ribozyme folds in 10–30 ms at 37 ˚C, only half the mutant RNA did so (Figure 2). The remaining population reached the native state very slowly (100 s), through I’s in which the P3 pseudoknot as well as other interactions within the ribozyme core were malformed (15). Mutations that disrupt other tertiary interactions yield similar results (R. Behrouzi and S.W, unpublished data).
Therefore, tertiary interactions can influence the cooperativity of base pairing, via the process of helix assembly in compact I states. One explanation is that there exists an ensemble of base paired I states, and this ensemble is biased toward native-like structures by tertiary interactions between helices. When the collapse transition (and helix assembly) favors native-like I’s, the folding process becomes more specific.
Specificity of collapse
If the accuracy of helix assembly controls the homogeneity of the folding paths, what factors make this step more specific? Polyelectrolyte effects from the counterions and RNA tertiary interactions both contribute to the formation of compact intermediates.
Time-resolved SAXS experiments on the Tetrahymena ribozyme revealed a series of transitions to more compact populations (23, 74). The earliest step (~10 ms) was unaffected by mutations that disrupted five long-range tertiary contacts, suggesting that this stage depends only on neutralization of the phosphate charge by counterions (Figure 3) (23). However, these mutations prohibited subsequent steps (100 ms and ≥1 s), which clearly depend on sequence-specific folding (23, 45). Thus, full compaction to a native-like state requires the capacity to form tertiary structure. A similar picture was obtained from time-resolved experiments on the P4–P6 domain of the Tetrahymena ribozyme (77).
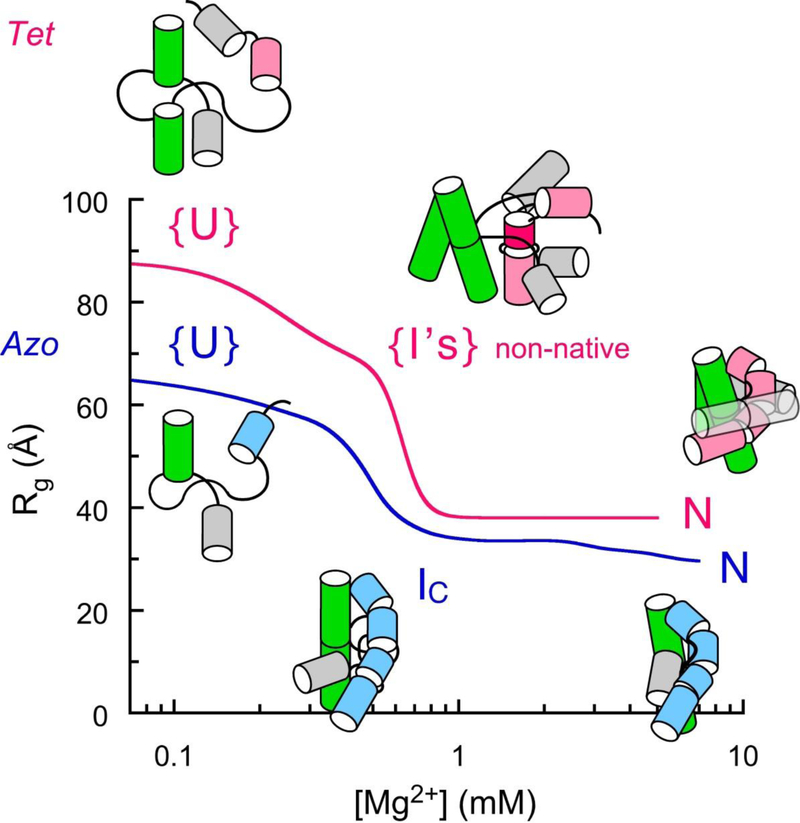
Figure 3. Specificity of collapse from equilibrium SAXS.
The Azoarcus ribozyme collapses to native-like intermediates (IC) while the Tetrahymena ribozyme collapses to an ensemble of non-native intermediates (I’s) (16, 23, 74). In both RNAs, specific transitions to a native-like fold are sensitive to counterion charge density. Redrawn from (53).
The equilibrium collapse transitions in the Azoarcus and Tetrahymena group I ribozymes showed that both RNAs experience specific and non-specific contraction of the RNA in the presence of counterions (53). In both RNAs, the neutralization of the phosphate charge upon condensation of counterions around the RNA creates a driving force for the collapse transition. However, the different stabilities of the RNA tertiary interactions lead to very different outcomes (Figure 3). In the stable Azoarcus ribozyme, specific collapse occurs in the same window of Mg2+ concentration as charge neutralization, resulting in a sharp transition to the native-like IC state. In the Tetrahymena ribozyme, charge neutralization produces non-native I’s (60–64 Å) (23, 53) that must undergo a second specific collapse transition at higher Mg2+ to form the native ribozyme (38 Å; (53)).
The specificity of the collapse transition correlates strongly with the time required to reach the native state (91). The Azoarcus and RNase P C domains both form compact intermediates that are similar in size to the native state (28, 63), and both RNAs fold within 5–50 ms under native conditions (30, 68). By contrast, non-specific collapse of the Tetrahymena ribozyme means that more than 90% of the RNA folds through misfolded I’s that require extensive refolding to become fully native (56, 109).
Counterion charge density and compact states of RNA
As the counterions also contribute to the stability of RNA tertiary structure, how might they affect the specificity of collapse? In general, multivalent cations stabilize compact forms of the RNA more efficiently than monovalent ions (reviewed in (19, 27)). However, compaction of the RNA also depends on ion size, with larger ions stabilizing the folded state less effectively than small ions (11, 35). This is because small, multivalent ions interact closely with the RNA, and produce stronger ion-ion correlations that enhance helix association (70, 89).
Titrations of the Azoarcus and Tetrahymena ribozymes with tri-, di- and monovalent ions showed that the initial contraction of the unfolded RNA depends on the valence of the counterion, but not on its size (53). However, the transition to native-like structures was sensitive to ion size, requiring higher concentrations of larger ions such as Sr2+ and Ba2+ than smaller ions such as Mg2+. This outcome was consistent with native PAGE experiments showing that the folding free energy of the Tetrahymena ribozyme in divalent metal ions diminished linearly with the size of the ion (42). This is not simply due to changes in ion hydration or direct coordination, because similar trends are observed in polyamines and can be simulated by coarse grained models that ignore the details of ion hydration and chelation (42, 43). A similar trend in the stability of the RNase P C domain in divalent metal ions was observed in ensemble fluorescence experiments (31).
Specific collapse transitions are more sensitive to ion size (or charge density) because the ion-ion interactions become more pronounced as the RNA structure becomes more compact (42). The folded RNA must be stabilized by tightly localized ions, yet the ions are confined to a smaller volume. A recent comparison showed that RNAs with complex 3D folds that bury parts of the RNA backbone are significantly more stable in Li+ and Na+ than in Cs+ (47). Molecular dynamics simulations on the TAR-TAR* kissing complex explained that small ions are able to enter deep, negatively charged pockets on the surface of the RNA and have a greater probability of exchanging their water ligands for ligands within the RNA (18).
An interesting question is how much the counterions affect the local and global dynamics of the RNA. The Tetrahymena ribozyme, for example, refolds faster in monovalent salts or large multivalent cations (34, 46). This has been attributed to less stable I’s and a broader transition state ensemble in low charge density ions, such as K+, Ba2+ or putrescine (44). As discussed below, ions can also participate directly in the last stages of refolding (31). At the same time, the initial TSE for the collapse transition likely also depends on how well the counterions promote helix assembly. Time-resolved footprinting on the Tetrahymena ribozyme in Na+ and Mg2+ suggests that RNA sequence is more important for partitioning among different folding pathways than ions (46), but more work is needed to resolve this question.
SEARCH FOR THE NATIVE STRUCTURE
Ions provide a strong driving force for helix assembly and RNA compaction. As discussed above, however, a further search among 3D contacts is needed to achieve the native structure. For molecules that collapse to native-like I’s, local structural changes should be all that are needed to reach the native state (91, 92). For molecules that collapse non-specifically, the resulting I’s are more likely to contain non-native interactions that require extensive remodeling of the RNA structure.
Helix docking and induced fit
What kinds of structural rearrangements typically limit RNA folding rates? Single-molecule FRET experiments on the hairpin ribozyme found that docking of the two helical domains was faster when the concentration of Mg2+ or Na+ was raised, while the rate of undocking changed very little (6). Therefore, the transition state for docking is compact and likely involves a slow reorganization of base stacking interactions in the docked state (Figure 1). This is consistent with a significant and unfavorable activation entropy for folding (64) and sensitivity of the folding and cleavage kinetics to mutations throughout the ribozyme (71). Rapid orientation of the four-way helical junction which is present in natural forms of the hairpin ribozyme speeds up docking 500 fold, yet formation of the catalytically active state still requires internal reorganization of docked intermediates (65, 88).
Like the hairpin ribozyme, folding of the P4–P6 domain of the Tetrahymena ribozyme requires docking of two helical domains, but in this case, docking is achieved by a 150° bend in the J5/5a internal loop (14). When P4–P6 RNA is refolded from low ionic strength, interactions within the J5/5a “hinge” limit docking (87), and the folding time is ~1 s (24, 77, 81). Moreover, most docking events are non-native, in that they inhibit refolding of the P5abc subdomain (24). If the RNA is first incubated in Na+ before Mg2+ is added, however, pre-structuring of the J5/5a hinge lowers the barrier to docking and shortens the folding time (20–50 ms; (24, 77, 81)). Thus, helix assembly is a necessary for 3D folding, but is typically followed by a further search for more stable 3D interactions.
Slow refolding of the group I ribozyme core
For the complete Tetrahymena ribozyme, the slow steps of compaction coincide with the formation of specific tertiary interactions (45). Early studies showed that tertiary interactions in the P3–P9 domain emerge much more slowly than those in the stable P4–P6 domain (78, 108), due to mispairing of the P3/P7 pseudoknot and nucleotides near the 5’ splice site (58). Pseudoknots form slowly in many other RNAs as well, in part because the pseudoknot base pairs are easily replaced by less topologically constrained secondary structures (36, 39, 60).
In the Tetrahymena ribozyme, footprinting, single-molecule FRET and SAXS experiments show that the ribozyme core often refolds after peripheral tertiary interactions have formed (75, 78). Mutations that destabilize these peripheral tertiary interactions increase the refolding rate, indicating that they stabilize the misfolded I’s and must open during reorganization of the ribozyme core (59, 95, 96). Thus, rescue of these I’s not only requires many base pairs to exchange, but also relaxation of the surrounding tertiary interactions. This explains why the folding rate is slow and decreases in higher Mg2+ (57, 69).
As the rate of helix assembly and the stability of core tertiary interactions contribute to RNA folding pathways, it should not be surprising that the topology of the RNA also influences the folding energy landscape. One way that circular permutation of the RNA sequence can change the preferred folding pathway is by changing the connections between domains (37, 62). One permutant of the Tetrahymena ribozyme nicked a linker between the P4–P6 and P3–P9 domains. Their folding was totally decoupled, so that the stable P4–P6 domain folded even more rapidly than before, but the P3 pseudoknot and the ribozyme core folded much more slowly, diminishing the yield of active RNA (48). A similar decoupling between domains was obtained by permuting the sequence of RNase P ribozyme (60).
Local conformational change in RNase P C-domain
For RNAs that avoid misfolding, what local structural changes might determine their folding rates? The RNase P C-domain folds through at least three intermediates, with an overall folding time < 200 ms at 37°C (30). The slowest step of folding occurs after collapse and is insensitive to urea, consistent with a local rearrangement of the structure (30). However, the rate-determining step is very sensitive to the metal ions. Although the C-domain is stabilized best by the smaller Mg2+ and Ca2+, folding was faster in the larger Sr2+ and Ba2+ (31). The ΔH‡ for folding was directly proportional to the ion charge density and ΔH‡ for metal dehydration, suggested that the rate determining step for folding of the C domain involves consolidation of the RNA around one or more metal ion binding sites (31). This event could involve direct chelation of a metal ion, or indirect RNA-metal ion interactions as suggested by us for the Tetrahymena ribozyme (42).
Recent single molecule FRET experiments show how such energy barriers can trap molecules in different regions of the free energy landscape under non-equilibrium conditions (66). As the Mg2+ concentration is raised, fluctuations between low FRET and high FRET conformations slow down and folding becomes multistage, consistent with an increasingly rough free energy landscape (66). As the Mg2+ concentration is cycled between denaturing and native conditions, sub-populations with different dynamics (but the same FRET value) emerge. These subpopulations are separated by a hidden energy barrier and cannot equilibrate within the 10 s Mg2+ pulse. These experiments illustrate how non-equilibrium measurements, along with pulse-chase labeling schemes (51), can tease out folding energy landscapes in more detail.
Folding barriers in group II ribozymes
The aI5γ group II ribozyme from yeast mitochondria represents a different example in which a small region controls the overall folding rate. Group II ribozymes are composed of 6 helical domains that associate via an extensive network of tertiary interactions that includes tetraloop-receptor and kissing loop motifs (20, 94).
As the ribozymes discussed so far, domains 1, 3 and 5 (D135) of aI5γ go through a collapse transition in the presence of counterions. Although the Stoke’s radius (RH) shortens dramatically (~75% of the total) in 0.5 M KCl, Mg2+ is needed to form a native-like intermediate (32, 84). Hydroxyl radical footprinting and hydrodynamic measurements showed that Mg2+-dependent folding steps are very slow (min to hr) (32, 84, 86), indicating a high energy barrier to forming the native structure.
What is unusual about aI5γ, however, is that the entire tertiary structure forms at the same rate (84). The folding rate increases with MgCl2 concentration and is unaffected by sub-denaturing urea (32, 84, 86). These results are seemingly inconsistent with a kinetically trapped intermediate. They suggest firstly that refolding does not involve opening buried surface area, and secondly, that Mg2+ stabilizes the transition state or helps populate a high energy intermediate. Single-molecule FRET trajectories also revealed at least one obligatory folding intermediate, although there may be other transitions (83).
Footprinting and nucleotide analog interference (NAIM) studies showed that once the domain 1 folds, the other domains dock quickly (85, 99). The rate-limiting step of folding correlates with a Mg2+-dependent rearrangement of contacts in the κ-ζ region of domain 1 (100). This region positions the catalytically essential domain 5 within the folded ribozyme, explaining why it is required to dock the other domains.
A crystal structure of a group II ribozyme from Oceanobacillus iheyensis shows that domain 1 forms an A-frame structure that folds over the domain 5 stem-loop, contacting it on both sides (94). This unique architecture raises the question of how the structure of domain 1 opens to allow rapid docking of domain 5. The κ-ζ region forms two sharp bends, shaping the helices on one side of the A-frame into a jelly roll (94). Two nucleotides adjacent to the ζ motif form another interaction (ω-ω’) that helps hold the “jelly roll” in place. Interestingly, the analogous residues in aI5γ appear to change conformation in the native-like compact state (32). An intriguing possibility is that Mg2+-dependent assembly of the jelly roll limits folding of domain 1.
PROSPECTS FOR RNA BIOLOGY
Much remains to be learned on how RNAs fold in the cell. Yet, indirect evidence suggests that RNAs do misfold in ways that impair their cellular function. Point mutations that increase misfolding of the Tetrahymena self-splicing RNA in vitro reduced the extent of splicing in yeast and E. coli (40, 54), and the half-life of the unspliced RNA is consistent with the turnover of misfolded RNAs within a limited window after transcription (40).
Compact folding intermediates are not only important for guiding the self-assembly of RNA structures. They are also necessary for protein binding, ligand recognition, and conformational switching. Many large RNAs, including bI5 and aI5γ, exist only as the collapsed state in physiological levels of Mg2+ (~1–2 mM) in the absence of proteins. These intermediate states of RNA can be engaged and driven towards the native state by specific RNA-binding proteins (5, 101). For example, CBP2 protein binds bI5 RNA in yeast mitochondria. When the protein is added to native-like intermediates of bI5, the yield of active complex is much higher than when the protein is added to unfolded RNA (101). Non-specific interactions with CBP2 increase the RNA dynamics and may facilitate folding (5). Similarly, ribosomal proteins perturb the average structure of early 16S rRNA folding intermediates, even under conditions in which specific protein-RNA complexes cannot yet form (67).
We are only beginning to understand how osmolytes, RNA binding proteins, chaperones, and the process of transcription may increase the specificity of helix assembly and accelerate recovery of misfolded transcripts (4, 61, 98). The work reviewed here demonstrates how compact, disordered states in RNA ensure the productive coupling of secondary and tertiary interactions during the folding process.
Summary.
Counterions trigger the collapse of RNA chains into compact folding intermediates, in which the double helices are fully assembled and engage in partial or dynamic tertiary interactions.
Secondary and tertiary folding steps are coupled via helix assembly during the collapse transition, blurring the concept of folding hierarchy.
Specific collapse produces native-like compact states and increases the fraction of RNA that folds directly (and rapidly) to the native state. Non-specific collapse increases the population of misfolded intermediates.
Collapse is rapid (1–10 ms or less) and followed bya rate-determining search for stable tertiary interactions and the native structure.
Specific folding transitions that produce compact structures are most sensitive to the valence and size of the counterions.
ACKNOWLEDGEMENTS
I apologize in advance to those colleagues whose work was not cited here due to space limitations. I thank D. Thirumalai and R. Briber and the members of my laboratory for their collaboration and helpful discussions, and acknowledge the NIH and NIST for research support.
Glossary
- Compact intermediate.
An intermediate stage of RNA folding containing secondary structure and transient or dynamic tertiary structure. Also called “collapsed state”
- Energy landscape.
A statistical thermodynamical model for protein and RNA folding, in which molecules diffuse along a hyper-surface constructed from energies of plausible conformational states and the barriers between them. For illustrations, chain conformation is commonly depicted by a horizontal plane and free energy by the vertical dimension.
- Helix docking.
Tertiary interactions between two or more double-stranded RNA helices that align the helices with each other in a fixed orientation.
- Hydroxyl radical footprinting.
Chemical method for probing tertiary interactions in RNA. The solvent exposure of each ribose sugar is evaluated based on the frequency of strand nicking in the presence of hydroxyl radical.
- Kinetic partitioning.
The separation of folding molecules into different folding pathways defined by the energy landscape for the reaction.
- Kissing loops.
An RNA tertiary structure motif formed by base pairing between complementary sequences in two terminal hairpin loops.
- Pseudoknot.
An RNA tertiary structure motif, in which a single-stranded segment of the RNA folds back and base pairs with bases in a loop.
- Tetraloop.
A four nucleotide loop at the end of a hairpin, often belonging to one of several sequence motifs such as 5’ GAAA (GNRA) or 5’ UNCG.
Acronyms
- 2D
secondary structure (of RNA)
- 3D
tertiary structure (of RNA)
- 2WJ
Two-way (helical) junction
- 4WJ
Four-way (helical) junction
- FRET
fluorescence resonance energy transfer
- SANS
small angle neutron scattering
- SAXS
small angle X-ray scattering
- UV
ultraviolet light
REFERENCES
- 1.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. 2004. Crystal structure of a self-splicing group I intron with both exons. Nature 430: 45–50 [DOI] [PubMed] [Google Scholar]
- 2.Andersen AA, Collins RA. 2001. Intramolecular secondary structure rearrangement by the kissing interaction of the Neurospora VS ribozyme. Proc. Natl. Acad. Sci. U.S.A 98: 7730–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansari A, Kuznetsov SV, Shen Y. 2001. Configurational diffusion down a folding funnel describes the dynamics of DNA hairpins. Proc. Natl. Acad. Sci. U.S.A 98: 7771–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaskaran H, Russell R. 2007. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature 449: 1014–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bokinsky G, Nivon LG, Liu S, Chai G, Hong M, et al. 2006. Two distinct binding modes of a protein cofactor with its target RNA. J Mol Biol 361: 771–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, et al. 2003. Single-molecule transition-state analysis of RNA folding. Proc. Natl. Acad. Sci. U.S.A [DOI] [PMC free article] [PubMed] [Google Scholar]; Analysis of transition states for individual folding pathways for the hairpin ribozyme showed they are stabilized by Mg2+ and are similar to the native state.
- 7.Bokinsky G, Zhuang X. 2005. Single-molecule RNA folding. Acc Chem Res 38: 566–73 [DOI] [PubMed] [Google Scholar]
- 8.Brion P, Westhof E. 1997. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct 26: 113–37 [DOI] [PubMed] [Google Scholar]
- 9.Buchmueller KL, Webb AE, Richardson DA, Weeks KM. 2000. A collapsed, non-native RNA folding state. Nat. Struct. Biol 7: 362–66 [DOI] [PubMed] [Google Scholar]
- 10.Buchmueller KL, Weeks KM. 2003. Near native structure in an RNA collapsed state. Biochemistry 42: 13869–78 [DOI] [PubMed] [Google Scholar]; Together with Ref. 9, this paper provided early evidence for compact, native-like intermediates in RNA using photo-crosslinking and other biochemical methods.
- 11.Bukhman YV, Draper DE. 1997. Affinities and selectivities of divalent cation binding sites within an RNA tertiary structure. J Mol Biol 273: 1020–31 [DOI] [PubMed] [Google Scholar]
- 12.Butcher SE, Allain FH, Feigon J. 1999. Solution structure of the loop B domain from the hairpin ribozyme. Nat. Struct. Biol 6: 212–6 [DOI] [PubMed] [Google Scholar]
- 13.Cai Z, Tinoco I Jr. 1996. Solution structure of loop A from the hairpin ribozyme from tobacco ringspot virus satellite. Biochemistry 35: 6026–36 [DOI] [PubMed] [Google Scholar]
- 14.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, et al. 1996. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science 273: 1678–85 [DOI] [PubMed] [Google Scholar]
- 15.Chauhan S, Behrouzi R, Rangan P, Woodson SA. 2009. Structural rearrangements linked to global folding pathways of the Azoarcus group I ribozyme. J. Mol. Biol 386: 1167–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chauhan S, Caliskan G, Briber RM, Perez-Salas U, Rangan P, et al. 2005. RNA tertiary interactions mediate native collapse of a bacterial group I ribozyme. J. Mol. Biol 353: 1199–209 [DOI] [PubMed] [Google Scholar]
- 17.Chauhan S, Woodson SA. 2008. Tertiary interactions determine the accuracy of RNA folding. J. Am. Chem. Soc 130: 1296–303 [DOI] [PMC free article] [PubMed] [Google Scholar]; Rapid time-resolved footprinting experiments show that peripheral tertiary interactions improve the cooperativity of base pairing in the Azoarcus ribozyme core and favor partitioning of the RNA into folding pathways that are fast and direct.
- 18.Chen AA, Draper DE, Pappu RV. 2009. Molecular simulation studies of monovalent counterion-mediated interactions in a model RNA kissing loop. J Mol Biol 390: 805–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen SJ. 2008. RNA folding: conformational statistics, folding kinetics, and ion electrostatics. Annu Rev Biophys 37: 197–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa M, Deme E, Jacquier A, Michel F. 1997. Multiple tertiary interactions involving domain II of group II self- splicing introns. J. Mol. Biol 267: 520–36 [DOI] [PubMed] [Google Scholar]
- 21.Crothers DM. 2001. RNA conformational dynamics In RNA, ed. Söll D, Nishimura S, Moore P, pp. 61–70. Oxford, UK: Elsevier [Google Scholar]
- 22.Crothers DM, Cole PE, Hilbers CW, Shulman RG. 1974. The molecular mechanism of thermal unfolding of Escherichia coli formylmethionine transfer RNA. J. Mol. Biol 87: 63–88 [DOI] [PubMed] [Google Scholar]
- 23.Das R, Kwok LW, Millett IS, Bai Y, Mills TT, et al. 2003. The fastest global events in RNA folding: electrostatic relaxation and tertiary collapse of the Tetrahymena ribozyme. J. Mol. Biol 332: 311–9 [DOI] [PubMed] [Google Scholar]
- 24.Deras ML, Brenowitz M, Ralston CY, Chance MR, Woodson SA. 2000. Folding mechanism of the Tetrahymena ribozyme P4–P6 domain. Biochemistry 39: 10975–85 [DOI] [PubMed] [Google Scholar]
- 25.Ditzler MA, Rueda D, Mo J, Hakansson K, Walter NG. 2008. A rugged free energy landscape separates multiple functional RNA folds throughout denaturation. Nucleic Acids Res 36: 7088–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Downey CD, Fiore JL, Stoddard CD, Hodak JH, Nesbitt DJ, Pardi A. 2006. Metal ion dependence, thermodynamics, and kinetics for intramolecular docking of a GAAA tetraloop and receptor connected by a flexible linker. Biochemistry 45: 3664–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Draper DE, Grilley D, Soto AM. 2005. Ions and RNA folding. Annu Rev Biophys Biomol Struct 34: 221–43 [DOI] [PubMed] [Google Scholar]
- 28.Fang X, Littrell K, Yang XJ, Henderson SJ, Siefert S, et al. 2000. Mg2+-dependent compaction and folding of yeast tRNAPhe and the catalytic domain of the B. subtilis RNase P RNA determined by small-angle X-ray scattering. Biochemistry 39: 11107–13 [DOI] [PubMed] [Google Scholar]
- 29.Fang X, Pan T, Sosnick TR. 1999. A thermodynamic framework and cooperativity in the tertiary folding of a Mg(2+)-dependent ribozyme. Biochemistry 38: 16840–46 [DOI] [PubMed] [Google Scholar]
- 30.Fang XW, Pan T, Sosnick TR. 1999. Mg2+-dependent folding of a large ribozyme without kinetic traps. Nat. Struct. Biol 6: 1091–95 [DOI] [PubMed] [Google Scholar]
- 31.Fang XW, Thiyagarajan P, Sosnick TR, Pan T. 2002. The rate-limiting step in the folding of a large ribozyme without kinetic traps. Proc. Natl. Acad. Sci. U.S.A 99: 8518–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fedorova O, Waldsich C, Pyle AM. 2007. Group II intron folding under near-physiological conditions: collapsing to the near-native state. J. Mol. Biol 366: 1099–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, et al. 1986. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl. Acad. Sci. U.S.A 83: 9373–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heilman-Miller SL, Pan J, Thirumalai D, Woodson SA. 2001. Counterion condensation in folding of the Tetrahymena ribozyme. II. Counterion dependence of folding kinetics. J. Mol. Biol 309: 57–68 [DOI] [PubMed] [Google Scholar]
- 35.Heilman-Miller SL, Thirumalai D, Woodson SA. 2001. Role of counterion condensation in folding of the Tetrahymena ribozyme. I. Equilibrium stabilization by cations. J. Mol. Biol 306: 1157–66 [DOI] [PubMed] [Google Scholar]
- 36.Heilman-Miller SL, Woodson SA. 2003. Effect of transcription on folding of the Tetrahymena ribozyme. RNA 9: 722–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heilman-Miller SL, Woodson SA. 2003. Perturbed folding kinetics of circularly permuted RNAs with altered topology. J Mol Biol 328: 385–94 [DOI] [PubMed] [Google Scholar]
- 38.Herschlag D 1995. RNA chaperones and the RNA folding problem. J. Biol. Chem 270: 20871–74 [DOI] [PubMed] [Google Scholar]
- 39.Holmes KL, Culver GM. 2004. Mapping structural differences between 30S ribosomal subunit assembly intermediates. Nat Struct Mol Biol 11: 179–86 [DOI] [PubMed] [Google Scholar]
- 40.Jackson SA, Koduvayur S, Woodson SA. 2006. Self-splicing of a group I intron reveals partitioning of native and misfolded RNA populations in yeast. RNA 12: 2149–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim CH, Tinoco I Jr., 2000. A retroviral RNA kissing complex containing only two G.C base pairs. Proc. Natl. Acad. Sci. U.S.A 97: 9396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koculi E, Hyeon C, Thirumalai D, Woodson SA. 2007. Charge density of divalent metal cations determines RNA stability. J. Am. Chem. Soc 129: 2676–82 [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Ref. 53, first showed that the stability of the Tetrahymena ribozyme increases with the charge density of divalent cations and that non-sequence-specific polyelectrolyte effects are sufficient to explain this trend.
- 43.Koculi E, Lee NK, Thirumalai D, Woodson SA. 2004. Folding of the Tetrahymena ribozyme by polyamines: importance of counterion valence and size. J. Mol. Biol 341: 27–36 [DOI] [PubMed] [Google Scholar]
- 44.Koculi E, Thirumalai D, Woodson SA. 2006. Counterion charge density determines the position and plasticity of RNA folding transition states. J. Mol. Biol 359: 446–54 [DOI] [PubMed] [Google Scholar]
- 45.Kwok LW, Shcherbakova I, Lamb JS, Park HY, Andresen K, et al. 2006. Concordant Exploration of the Kinetics of RNA Folding from Global and Local Perspectives. J. Mol. Biol 355: 282–93 [DOI] [PubMed] [Google Scholar]
- 46.Laederach A, Shcherbakova I, Jonikas MA, Altman RB, Brenowitz M. 2007. Distinct contribution of electrostatics, initial conformational ensemble, and macromolecular stability in RNA folding. Proc. Natl. Acad. Sci. U.S.A 104: 7045–50 [DOI] [PMC free article] [PubMed] [Google Scholar]; Rapid hydroxyl radical footprinting and semi-automated model fitting show how the folding pathways of the Tetrahymena ribozyme and kinetic partitioning among intermediates are influenced by ions and by folding conditions.
- 47.Lambert D, Leipply D, Shiman R, Draper DE. 2009. The influence of monovalent cation size on the stability of RNA tertiary structures. J. Mol. Biol 390: 791–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lease RA, Adilakshmi T, Heilman-Miller S, Woodson SA. 2007. Communication between RNA folding domains revealed by folding of circularly permuted ribozymes. J Mol Biol 373: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lescoute A, Westhof E. 2006. The interaction networks of structured RNAs. Nucleic Acids Res 34: 6587–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li PT, Vieregg J, Tinoco I Jr. 2008. How RNA unfolds and refolds. Annu. Rev. Biochem 77: 77–100 [DOI] [PubMed] [Google Scholar]
- 51.Liu S, Bokinsky G, Walter NG, Zhuang X. 2007. Dissecting the multistep reaction pathway of an RNA enzyme by single-molecule kinetic “fingerprinting”. Proc. Natl. Acad. Sci. U.S.A 104: 12634–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma H, Proctor DJ, Kierzek E, Kierzek R, Bevilacqua PC, Gruebele M. 2006. Exploring the energy landscape of a small RNA hairpin. J. Am. Chem. Soc 128: 1523–30 [DOI] [PubMed] [Google Scholar]; Temperature-jump spectroscopy, mutagenesis and lattice simulations were combined to obtain a detailed energy landscape for a tetraloop hairpin. The results show that even small RNA structures have rugged folding landscapes.
- 53.Moghaddam S, Caliskan G, Chauhan S, Hyeon C, Briber RM, et al. 2009. Metal Ion Dependence of Cooperative Collapse Transitions in RNA. J. Mol. Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nikolcheva T, Woodson SA. 1999. Facilitation of group I splicing in vivo: misfolding of the Tetrahymena IVS and the role of ribosomal RNA exons. J Mol Biol 292: 557–67 [DOI] [PubMed] [Google Scholar]
- 55.Okumus B, Wilson TJ, Lilley DM, Ha T. 2004. Vesicle encapsulation studies reveal that single molecule ribozyme heterogeneities are intrinsic. Biophys. J 87: 2798–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan J, Thirumalai D, Woodson SA. 1997. Folding of RNA involves parallel pathways. J. Mol. Biol 273: 7–13 [DOI] [PubMed] [Google Scholar]
- 57.Pan J, Thirumalai D, Woodson SA. 1999. Magnesium-dependent folding of self-splicing RNA: exploring the link between cooperativity, thermodynamics, and kinetics. Proc. Natl. Acad. Sci. U.S.A 96: 6149–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pan J, Woodson SA. 1998. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J. Mol. Biol 280: 597–609 [DOI] [PubMed] [Google Scholar]
- 59.Pan J, Woodson SA. 1999. The effect of long-range loop-loop interactions on folding of the Tetrahymena self-splicing RNA. J. Mol. Biol 294: 955–65 [DOI] [PubMed] [Google Scholar]
- 60.Pan T, Fang X, Sosnick T. 1999. Pathway modulation, circular permutation and rapid RNA folding under kinetic control. J. Mol. Biol 286: 721–31 [DOI] [PubMed] [Google Scholar]
- 61.Pan T, Sosnick T. 2006. RNA Folding During Transcription. Annu. Rev. Biophys. Biomol. Struct [DOI] [PubMed] [Google Scholar]
- 62.Pan T, Sosnick TR. 1997. Intermediates and kinetic traps in the folding of a large ribozyme revealed by circular dichroism and UV absorbance spectroscopies and catalytic activity. Nat. Struct. Biol 4: 931–38 [DOI] [PubMed] [Google Scholar]
- 63.Perez-Salas UA, Rangan P, Krueger S, Briber RM, Thirumalai D, Woodson SA. 2004. Compaction of a bacterial group I ribozyme coincides with the assembly of core helices. Biochemistry 43: 1746–53 [DOI] [PubMed] [Google Scholar]
- 64.Pljevaljcic G, Klostermeier D, Millar DP. 2005. The tertiary structure of the hairpin ribozyme is formed through a slow conformational search. Biochemistry 44: 4870–6 [DOI] [PubMed] [Google Scholar]
- 65.Pljevaljcic G, Millar DP, Deniz AA. 2004. Freely diffusing single hairpin ribozymes provide insights into the role of secondary structure and partially folded states in RNA folding. Biophys. J 87: 457–67 [DOI] [PMC free article] [PubMed] [Google Scholar]; Evidence that rapid equilibrium between undocked and quasi-docked states of the hairpin ribozyme precede stable docking of helical domains.
- 66.Qu X, Smith GJ, Lee KT, Sosnick TR, Pan T, Scherer NF. 2008. Single-molecule nonequilibrium periodic Mg2+-concentration jump experiments reveal details of the early folding pathways of a large RNA. Proc. Natl. Acad. Sci. U.S.A 105: 6602–7 [DOI] [PMC free article] [PubMed] [Google Scholar]; Rapid cycling between native and non-native conditions revealed hidden barriers to refolding of the catalytic domain from RNase P.
- 67.Ramaswamy P, Woodson SA. 2009. Global Stabilization of rRNA Structure by Ribosomal Proteins S4, S17, and S20. J Mol Biol: 666–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rangan P, Masquida B, Westhof E, Woodson SA. 2003. Assembly of core helices and rapid tertiary folding of a small bacterial group I ribozyme. Proc Natl Acad Sci U S A 100: 1574–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rook MS, Treiber DK, Williamson JR. 1999. An optimal Mg(2+) concentration for kinetic folding of the tetrahymena ribozyme. Proc. Natl. Acad. Sci. U.S.A 96: 12471–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rouzina I, Bloomfield VA. 1996. Influence of ligand spatial organization on competitive electrostatic binding to DNA. Journal of Physical Chemistry 100: 4305–13 [Google Scholar]
- 71.Rueda D, Bokinsky G, Rhodes MM, Rust MJ, Zhuang X, Walter NG. 2004. Single-molecule enzymology of RNA: essential functional groups impact catalysis from a distance. Proc. Natl. Acad. Sci. U.S.A 101: 10066–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rupert PB, Ferre-D’Amare AR. 2001. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature 410: 780–6. [DOI] [PubMed] [Google Scholar]
- 73.Russell R, Millett IS, Doniach S, Herschlag D. 2000. Small angle X-ray scattering reveals a compact intermediate in RNA folding. Nat. Struct. Biol 7: 367–70 [DOI] [PubMed] [Google Scholar]
- 74.Russell R, Millett IS, Tate MW, Kwok LW, Nakatani B, et al. 2002. Rapid compaction during RNA folding. Proc. Natl. Acad. Sci. U.S.A 99: 4266–71 [DOI] [PMC free article] [PubMed] [Google Scholar]; First application of stopped-flow and continuous-flow SAXS to monitor early phases of collapse in real time in the Tetrahymena ribozyme. Together with Ref. 53, showed that the earliest steps depend on polyelectrolyte effects that than specific folding.
- 75.Russell R, Zhuang X, Babcock HP, Millett IS, Doniach S, et al. 2002. Exploring the folding landscape of a structured RNA. Proc. Natl. Acad. Sci. U.S.A 99: 155–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sattin BD, Zhao W, Travers K, Chu S, Herschlag D. 2008. Direct measurement of tertiary contact cooperativity in RNA folding. J. Am. Chem. Soc 130: 6085–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schlatterer JC, Kwok LW, Lamb JS, Park HY, Andresen K, et al. 2008. Hinge stiffness is a barrier to RNA folding. J. Mol. Biol 379: 859–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. 1998. RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science 279: 1940–43 [DOI] [PubMed] [Google Scholar]
- 79.Shcherbakova I, Mitra S, Laederach A, Brenowitz M. 2008. Energy barriers, pathways, and dynamics during folding of large, multidomain RNAs. Curr. Opin. Chem. Biol 12: 655–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Silverman SK, Cech TR. 1999. Energetics and cooperativity of tertiary hydrogen bonds in RNA structure. Biochemistry 38: 8691–702 [DOI] [PubMed] [Google Scholar]
- 81.Silverman SK, Deras ML, Woodson SA, Scaringe SA, Cech TR. 2000. Multiple Folding Pathways for the P4–P6 RNA Domain. Biochemistry 39: 12465–75 [DOI] [PubMed] [Google Scholar]
- 82.Stancik AL, Brauns EB. 2008. Rearrangement of partially ordered stacked conformations contributes to the rugged energy landscape of a small RNA hairpin. Biochemistry 47: 10834–40 [DOI] [PubMed] [Google Scholar]
- 83.Steiner M, Karunatilaka KS, Sigel RK, Rueda D. 2008. Single-molecule studies of group II intron ribozymes. Proc. Natl. Acad. Sci. U.S.A 105: 13853–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Su LJ, Brenowitz M, Pyle AM. 2003. An alternative route for the folding of large RNAs: apparent two-state folding by a group II intron ribozyme. J. Mol. Biol 334: 639–52 [DOI] [PubMed] [Google Scholar]
- 85.Su LJ, Waldsich C, Pyle AM. 2005. An obligate intermediate along the slow folding pathway of a group II intron ribozyme. Nucleic Acids Res 33: 6674–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Swisher JF, Su LJ, Brenowitz M, Anderson VE, Pyle AM. 2002. Productive folding to the native state by a group II intron ribozyme. J. Mol. Biol 315: 297–310 [DOI] [PubMed] [Google Scholar]
- 87.Szewczak AA, Cech TR. 1997. An RNA internal loop acts as a hinge to facilitate ribozyme folding and catalysis. RNA 3: 838–49 [PMC free article] [PubMed] [Google Scholar]
- 88.Tan E, Wilson TJ, Nahas MK, Clegg RM, Lilley DM, Ha T. 2003. A four-way junction accelerates hairpin ribozyme folding via a discrete intermediate. Proc. Natl. Acad. Sci. U.S.A 100: 9308–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tan ZJ, Chen SJ. 2006. Ion-mediated nucleic acid helix-helix interactions. Biophys J 91: 518–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Thirumalai D 1998. Native secondary structure formation in RNA may be a slave to tertiary folding. Proc. Natl. Acad. Sci. U.S.A 95: 11506–08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thirumalai D, Lee N, Woodson SA, Klimov D. 2001. Early events in RNA folding. Annu. Rev. Phys. Chem 52: 751–62 [DOI] [PubMed] [Google Scholar]
- 92.Thirumalai D, Woodson SA. 1996. Kinetics of folding of protein and RNA. Acc. Chem. Res 29: 433–39 [Google Scholar]
- 93.Tinoco I Jr., Li PT, Bustamante C. 2006. Determination of thermodynamics and kinetics of RNA reactions by force. Q Rev Biophys 39: 325–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Toor N, Keating KS, Taylor SD, Pyle AM. 2008. Crystal structure of a self-spliced group II intron. Science 320: 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Treiber DK, Rook MS, Zarrinkar PP, Williamson JR. 1998. Kinetic intermediates trapped by native interactions in RNA folding. Science 279: 1943–46 [DOI] [PubMed] [Google Scholar]
- 96.Treiber DK, Williamson JR. 2001. Concerted kinetic folding of a multidomain ribozyme with a disrupted loop-receptor interaction. J. Mol. Biol 305: 11–21 [DOI] [PubMed] [Google Scholar]
- 97.Vander Meulen KA, Davis JH, Foster TR, Record MT Jr., Butcher SE. 2008. Thermodynamics and folding pathway of tetraloop receptor-mediated RNA helical packing. J. Mol. Biol 384: 702–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Waldsich C, Grossberger R, Schroeder R. 2002. RNA chaperone StpA loosens interactions of the tertiary structure in the td group I intron in vivo. Genes Dev 16: 2300–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Waldsich C, Pyle AM. 2007. A folding control element for tertiary collapse of a group II intron ribozyme. Nat Struct Mol Biol 14: 37–44 [DOI] [PubMed] [Google Scholar]; Nucleotide analog interference was used to identify a small region of the aI5γ ribozyme critical to folding. A slow conformational change in this region determines the overall folding rate of this large RNA.
- 100.Waldsich C, Pyle AM. 2008. A kinetic intermediate that regulates proper folding of a group II intron RNA. J. Mol. Biol 375: 572–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Webb AE, Weeks KM. 2001. A collapsed state functions to self-chaperone RNA folding into a native ribonucleoprotein complex. Nat. Struct. Biol 8: 135–40. [DOI] [PubMed] [Google Scholar]
- 102.Weixlbaumer A, Werner A, Flamm C, Westhof E, Schroeder R. 2004. Determination of thermodynamic parameters for HIV DIS type loop-loop kissing complexes. Nucleic Acids Res 32: 5126–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Woodside MT, Garcia-Garcia C, Block SM. 2008. Folding and unfolding single RNA molecules under tension. Curr. Opin. Chem. Biol 12: 640–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Woodson SA. 2000. Compact but disordered states of RNA. Nat. Struct. Biol 7: 349–52 [DOI] [PubMed] [Google Scholar]
- 105.Wu M, Tinoco I Jr. 1998. RNA folding causes secondary structure rearrangement. Proc. Natl. Acad. Sci. U.S.A 95: 11555–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Young BT, Silverman SK. 2002. The GAAA tetraloop-receptor interaction contributes differentially to folding thermodynamics and kinetics for the P4–P6 RNA domain. Biochemistry 41: 12271–6 [DOI] [PubMed] [Google Scholar]
- 107.Zarrinkar PP, Williamson JR. 1994. Kinetic intermediates in RNA folding. Science 265: 918–24 [DOI] [PubMed] [Google Scholar]
- 108.Zarrinkar PP, Williamson JR. 1996. The kinetic folding pathway of the Tetrahymena ribozyme reveals possible similarities between RNA and protein folding. Nat. Struct. Biol 3: 432–38 [DOI] [PubMed] [Google Scholar]
- 109.Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, et al. 2000. A single-molecule study of RNA catalysis and folding. Science 288: 2048–51 [DOI] [PubMed] [Google Scholar]
- 110.Zhuang X, Kim H, Pereira MJ, Babcock HP, Walter NG, Chu S. 2002. Correlating structural dynamics and function in single ribozyme molecules. Science 296: 1473–6 [DOI] [PubMed] [Google Scholar]