Abstract
Constructed from the consensus of short-read variant callers, existing benchmark datasets for evaluating variant calling accuracy are biased toward easy regions accessible by known algorithms. We derived a new benchmark dataset from the de novo PacBio assemblies of two human cell lines that are homozygous across the whole genome. It provides a more accurate and less biased estimate of the error rate of small variant calls in a realistic context.
Calling genomic sequence variations from resequencing data plays an important role in medical and population genetics, and has become an active research area since the advent of high-throughput sequencing. Many methods have been developed for calling single-nucleotide polymorphisms (SNPs) and short insertions/deletions (INDELs) primarily from short-read data. To measure the accuracy of these methods and ultimately to make accurate variant calls, one typically runs a variant calling pipeline on benchmark datasets where the true variant calls are known. The most widely used benchmark datasets include Genome In A Bottle1 (GIAB) and Platinum Genome2 (PlatGen) for the human sample NA12878. Both come with a set of high-quality variants and a set of confident regions where non-variant sites are deemed to be identical to the reference genome. These two datasets were constructed from the consensus of multiple short-read variant callers, with consideration of pedigree information or structural variations (SVs) found with long-fragment technologies by PacBio or 10X Genomics. A major concern with GIAB and PlatGen is that the sequencing technologies and the variant calling algorithms used to construct the data sets are the same as the ones used for testing. This strong correlation leads to biases in two subtle ways. First, variant calling with short reads is intrinsically difficult in regions with moderately diverged repeats and segmental duplications. We have to exclude such regions from the confident regions as different variant callers fail to reach consensus there. This biases GIAB and PlatGen toward “easy” genomic regions. In fact, both benchmark datasets conclude competent variant callers make a SNP error every few million bases (Figure 1a), while other work suggests we can only achieve an error rate of one per 100–200 thousand bases in wider genomic regions3, nearly an order of magnitude higher. The bias toward easy regions directs the progress in the field to focus on trivial errors while overlooking the major error modes in real applications. Second, as GIAB and PlatGen were constructed using the existing algorithms, they may penalize more advanced algorithms and hamper future method development. These caveats suggest we can only comprehensively evaluate the accuracy of short-read variant calling by constructing benchmark datasets with methods largely orthogonal to and more powerful than short-read sequencing technologies and variant calling algorithms.
Fig. 1.
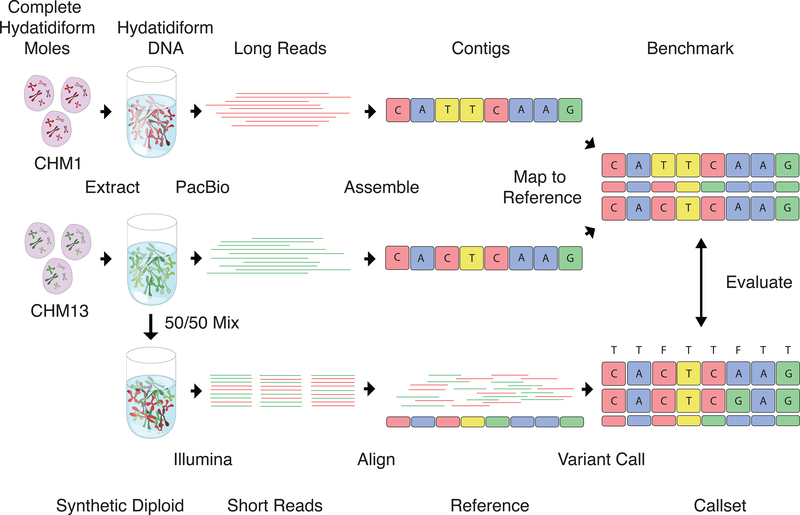
Constructing the Syndip benchmark dataset. CHM1 and CHM13 cell lines were sequenced with PacBio and de novo assembled independently. Assembly contigs were aligned to the human reference genome. Differences in the alignment were taken as ‘true’ SNPs and INDELs; regions covered by exactly one contig from each CHM assembly were identified as confident regions where true variants can be called to high accuracy. For the evaluation of diploid variant calling with short reads, equal quantities of DNA from the two cell lines were experimentally mixed. A PCR-free library was constructed from the mix and sequenced to ~45-fold coverage with 151bp paired-end reads. Variants called from the short reads were compared to the PacBio variants as truth to measure variant caller accuracy.
It may be tempting to construct a new benchmark dataset from a whole genome assembly based on PacBio data4. However, recent work shows that while PacBio assembly is accurate at the base-pair level for haploid genomes5, it is not accurate enough to confidently call heterozygotes in diploid mammalian genomes6. To derive a comprehensive truth dataset, we turned to the de novo PacBio assemblies of two complete hydatidiform mole (CHM) cell lines7,8. CHM cell lines are almost completely homozygous across the whole genome. This homozygosity enables accurate PacBio consensus sequences of each cell line in two ways. First, the PacBio consensus tool works better with homozygous sequences. Second, we can identify and mask PacBio errors by mapping Illumina short-read contigs to the PacBio contigs. Because for each CHM sample we are calling small base changes on a single haplotype, we can avoid most artifacts in diploid variant calling against a reference genome, which involves three haplotypes and complex genetic variations beyond the capability of short reads.
Using the error-masked contigs of each cell line, we combined the two homozygous calls at each locus into a synthetic diploid call, resulting in the new phased benchmark dataset: Syndip (synthetic diploid; Figure 1). This callset consists of 3.57 million SNPs and 0.58 million INDELs in 2.71 gigabases (Gbp) of confident regions, covering 95.5% of the autosomes and X chromosome of GRCh37 excluding assembly gaps.
In order to compare Syndip with existing benchmarks and re-evaluate popular short-read variant callers, we evenly mixed DNA from the two CHM cell lines and sequenced the mix with Illumina HiSeq X Ten (Figure 1). By counting supporting reads at heterozygous SNPs after variant calling, we estimated 50.7% of DNA in the mixture comes from one cell line and 49.3% from the other, concluding that the mixture is a good representative of a naturally diploid sample. We mapped the reads from the synthetic-diploid samples to the human genome with BWA-MEM-0.7.159, Bowtie2–2.2.210 and minimap2–2.511, and called variants on the synthetic-diploid samples with FreeBayes-1.0.212, Platypus- 0.8.113, Samtools-1.314 and GATK-3.515, including the HaplotypeCaller (HC) and UnifiedGenotyper (UG) algorithms. We included multiple variant callers to avoid overemphasizing caller-specific effects. We optionally filtered the initial variant calls with the following rules3: 1) variant quality ≥ 30; 2) read depth at the variant is below with d being the average read depth; 3) the fraction of reads supporting the variant allele ≥ 30% of total read depth at the variant; 4) Fisher strand p-value ≥0.001; 5) the variant allele is supported by at least one read from each strand.
We used RTG’s vcfeval16 to evaluate the variant calling accuracy. Given a truth and a test callset, a true positive (TP) is a true allele found in the test callset; a false negative (FN) is a true allele not found in the test callset; a false positive (FP) is an allele in the test callset not found in the truth set. We define %FNR=100×FN/(TP+FN) and FPPM=106×FP/L, where L is the total length of confident regions. We took FPPM as a metric instead of the more widely used metric “precision” [=TP/(TP+FN)], because FPPM does not depend on the rate of variation and is thus comparable across datasets of different populations or species.
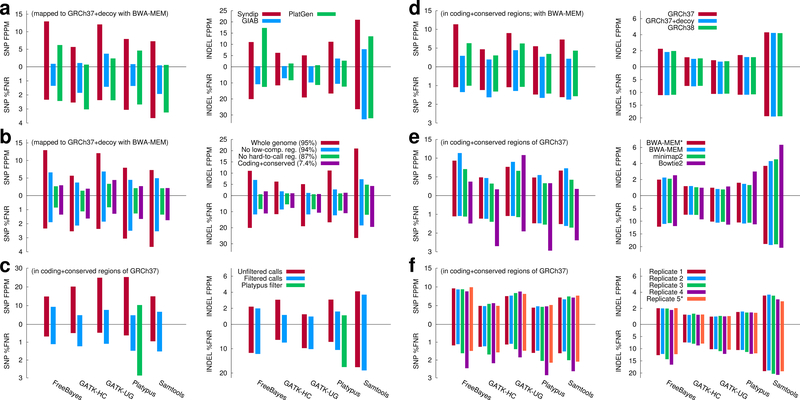
Figure 2 shows the results of evaluating variant calling pipelines with various benchmarks and conditions. Figure 2a reveals that the FPPM of SNPs estimated from Syndip is often 5–10 times higher than FPPM estimated from GIAB or PlatGen. Looking into the Syndip FP SNPs, we found most of them are located in CNVs that are evident in PacBio data in the context of long flanking regions, but look dubious in short-read data alone. GIAB-3.3.2 and PlatGen-1.0 exclude these false positives from the truth variant set based on the pedigree information or orthogonal data. However, in real applications, we often only have access to Illumina data and thus cannot achieve the accuracy suggested by the two benchmark datasets.
Fig. 2.
Evaluating variant calling accuracy with Syndip. %FNR denotes percent false negative rate, and FPPM is the number of false positives per million bases. (a) Comparison of Syndip, GIAB and PlatGen benchmark datasets on filtered calls. For GIAB and PlatGen, variants were called from the HiSeq X Ten run ‘NA12878_L7_S7’ available from the Illumina BaseSpace. (b) Effect of evaluation regions. Low-complexity regions were identified with the symmetric DUST algorithm. The ‘hard-to-call’ regions include low-complexity regions, regions unmappable with 75bp single-end reads and regions susceptible to common copy number variations. Panels (c)–(f) only show metrics in ‘coding+conserved’ regions. (c) Effect of variant filters. Green bars applied Platypus built-in filters. (d) Effect of the human genome reference build. Decoy sequences17 are real human sequences that are missing from GRCh37. (e) Effect of the mapping algorithms and post-processing. BWA-MEM* represents alignment post-processed with base quality recalibration and INDEL realignment; other alignments were not processed with these steps. (f) Effect of replicates. Replicate 1–4 were sequenced from four independent libraries, respectively, by mixing equal amount of DNA prior to library construction. Replicate 5* was generated by computationally subsampling and mixing reads sequenced from the two CHM cell lines separately. Replicate 1 is used in panels (a)–(e). Numerical data and the script to generate the figure are available as Supplementary Data.
In our evaluation, we used post-filtered variant calls instead of raw calls. For GATK-HC, filtering reduces sensitivity by only 0.5%, but reduces the number of FPs by four folds (Figure 2c), reduces the number of coding SNPs absent from the 1000 Genomes Project17 by 58%, and reduces the number of loss-of-function (LoF) calls by 30%. We manually inspected 20 filtered LoF calls in IGV18 and confirmed that all of them are either false positives or fall outside confident regions; those outside confident regions look spurious as well. False positives are enriched among LoF calls because real LoF mutations are subjected to strong selection but errors are not. For functional analyses, such as in the study of Mendelian diseases, we recommend applying stringent filtering to avoid variant calling artifacts. We note that the popular metric F1-score, which is the average of sensitivity [=TP/(TP+FP)] and precision, is usually higher for unfiltered calls. For example, on GIAB, the F1-score of unfiltered GATK-HC SNP calls is 0.997, higher than that of filtered calls 0.990. The F1 metric may not reflect the accuracy in many applications.
Consistent with our previous finding3, most FP INDELs come from low-complexity regions19,20 (LCRs), 2.3% of human genome (Figure 2b). While this finding helps to guide our future development, it over-emphasizes a class of INDELs that often have unknown functional implications. To put the evaluation in a more practical context, we compiled a list of potentially functional regions, which consist of coding regions with 20bp flanking regions, regions conserved in vertebrate or mammalian evolution and variants in the ClinVar or GWAScatalog databases with 100bp flanking regions. Only 0.5% of these regions intersect with LCRs. As a result, the FPPM of INDELs in these regions is much lower.
We found that mapping reads to GRCh38 leads to slightly better results than mapping to GRCh37 (Figure 2d), potentially due to the higher quality of the latest build. Although mapping to GRCh37 with decoy sequences further helps to reduce FP calls, this often comes at a minor loss in sensitivity. The choice of read mapping pipelines affects variant calling accuracy more (Figure 2e). Bowtie2 alignment often yields lower FPPM because Bowtie2 intentionally lowers mapping quality of reads with excessive mismatches, which helps to avoid FPs caused by divergent CNVs, but may lead to a bias against regions under balancing selection or reduce sensitivity for species with high heterozygosity. It would be preferable to implement a post-alignment or post-variant filter instead of building the limitation into the mapper. We observed comparable FPPM but varying sensitivity across four independent experiments (Figure 2f). Replicate 4 has the lowest coverage and base quality as well as the lowest variant calling sensitivity. Importantly, replicate 5* in Figure 2f suggests that computationally subsampling and mixing reads sequenced from each CHM cell line separately, which is an easier technical exercise than experimentally mixing DNA to a precise fraction, is adequate for the evaluation of short variant calling.
We have manually inspected FPs and FNs called by each variant caller. GATK-HC performs local re-assembly and consistently achieves the highest INDEL sensitivity (Figure 2). However, it may assemble a spurious haplotype around a long INDEL in a long LCR and call a false allele. We believe this can be improved with a better assembly algorithm3. FreeBayes is efficient and accurate for SNP calling. However, it does not penalize reads with intermediate mapping quality as much as other variant callers, which may lead to high FPPM in regions affected by CNVs. Platypus and SAMtools also demonstrate good SNP accuracy. Nonetheless, they both suffer from an error mode in which they may call a weakly supported false INDEL that is similar but not identical to a correct INDEL call nearby. This affects their FPPM. It is not obvious how to filter such false INDELs without looking at the underlying alignments.
Syndip is a special benchmark dataset that has been constructed from high-quality PacBio assemblies of two independent, homozygous cell lines. It leverages the power of long-read sequencing technologies while avoiding the difficulties in calling complex heterozygous variants from relatively noisy data. Syndip is the first benchmark dataset that does not heavily depend on short-read data and short-read variant callers, and thus more honestly reflects the true accuracy of such variant callers. On the other hand, Syndip also has weakness: the PacBio consensus of homozygous genomes is still associated with a small error rate. We have to use Illumina short reads to avoid PacBio errors being identified as wrong FNs in evaluation. Better PacBio assembly would eliminate this step and produce a benchmark dataset fully orthogonal to Illumina data.
Methods
Identifying erroneous regions on PacBio contigs
We acquired CHM1 and CHM13 PacBio assemblies8 (accession GCA_001297185 and GCA_000983455, respectively) from NCBI and downloaded Illumina short reads from SRA (accession SRR2842672 and SRR3099549 for CHM1; SRR2088062 and SRR2088063 for CHM13). We assembled the Illumina CHM1 reads with FermiKit21 and mapped the short-read unitigs to the corresponding PacBio assembly to call the sequence differences between the Illumina and the PacBio assemblies. These differences may come from three sources: incorrect PacBio consensus, somatic cell line mutations and collapse of segmental duplications.
An incorrect PacBio consensus base leads to a homozygous difference between the Illumina and PacBio assemblies. We found ~62k such homozygous differences from each sample, most of which are 1bp INDELs. A somatic mutation appears to be an isolated heterozygote with read depth close to the genome average. We identified such erroneous heterozygotes by requiring 1) each allele is supported by at least 9 Illumina read bases; 2) read depth is no greater than 50 and 3) the distance between adjacent somatic calls is above 10,000bp. We found 10,498 potential somatic calls in CHM1 and 4,701 in CHM13.
Collapse of segmental duplications in the PacBio assemblies lead to elevated read depth and clustered heterozygotes. To pinpoint such regions, we hierarchically clustered heterozygous events as follows: we merged two clusters adjacent on the PacBio assembly if 1) the minimal distance between them is within 10kb and 2) the density of heterozygotes in the merged cluster is at least 1 per 1kb. This resulted in about 3,000 clusters containing three or more heterozygotes from each PacBio assembly.
We marked the three types of errors plus 10bp flanking regions on the PacBio contigs. At a later step, we lift these regions over to the human reference coordinate and exclude them from the final list of confident regions. We note that the procedure above is aggressive in that regions we identified may not be associated with PacBio errors. These false regions will not lead to wrong FP/FN classification in evaluation.
Constructing the truth call set and confident regions
We aligned each CHM PacBio assembly to GRCh37 with minimap2. To call pseudo-diploid variants from PacBio assemblies, we merged the assembly-to-reference alignments of CHM1 and CHM13. We discarded alignments with mapping quality below 5, dropped aligned segments shorter than 50kb and made an unfiltered call set by calling the alignment differences between each PacBio contig and GRCh37.
We constructed the initial set of confident regions from the same alignment. For each PacBio assembly, we say a region on GRCh37 is orthologous to the assembly if 1) the region is covered by one PacBio alignment longer than 50kb with mapping quality at least 5; 2) the region is not covered by another PacBio alignment longer than 10kb, with mapping quality at least 5. In downstream evaluation, we later noticed that if a small region harbors excessive variant calls, the region tends to be enriched with errors potentially due to misalignments or structural variations. We thus applied another hierarchical clustering to spot clusters of variations. More precisely, we hierarchically merged two clusters if 1) the minimal distance between two variants is within 250bp and 2) the density of variants in the merged cluster is at least 1 per 50bp. We collected clusters consisting of 10 or more variants and excluded the related regions together with erroneous PacBio regions from the orthologous regions. This gives us the list of confident regions for each sample.
The final list of confident regions of Syndip is the intersection of confident regions from each sample. It covers 96.0% of GRCh37, or 95.5% when we also excluded poly-A runs ≥10bp. We applied a similar procedure to both GRCh37 with decoy contigs and GRCh38.
To confirm the quality of the Syndip data set, we manually inspected a few hundred discordant calls in IGV18. We observed that a few percent of false positive and false negative INDEL calls made by HaplotypeCaller appear to have strong support from Illumina reads. Most of them are associated with 1bp deletions in poly-C. They may be remaining PacBio consensus errors we failed to identify. Regardless, a few percent of discrepancy between Illumina and PacBio evidence would not change our general conclusions or the relative performance between calling methods as PacBio contig errors and somatic mutations are not biased toward a particular calling method.
Quantification, normalization and mixing of the CHM samples
Initial sample quantification was performed using the Invitrogen Quant-It broad range dsDNA quantification assay kit (Thermo Scientific Catalog: Q33130) with a 1:200 PicoGreen dilution. Following quantification, each sample was normalized to a concentration of 10 ng/μL using a 1X Low TE pH 7.0 solution, then sample concentration was confirmed via PicoGreen. Sample mixing was then performed by combining an equal mass (ng) of each of the two samples (CHM1 & CHM13) needed to obtain enough material for the Whole Genome library preparation (500ng). The samples for creating the 4 libraries were normalized and mixed independently (Life Science Reporting Summary).
Preparation of libraries & sequencing
For PCR-free whole genomes, library construction was performed using Kapa Biosystems reagents with the following modifications: (1) initial genomic DNA input was reduced from 3μg to 500ng, and (2) custom full-length dual-indexed library adapters at a concentration of 15 uM were utilized. Following sample preparation, libraries were quantified using quantitative PCR (kit purchased from Kapa biosystems) with probes specific to adapter ends in an automated fashion on Agilent’s Bravo liquid handling platform. Based on qPCR quantification, libraries were normalized and pooled on the Hamilton MiniStar liquid handling platform. For HiSeq X Ten, pooled samples were normalized to 2nM and denatured with 0.1N NaOH for a loading concentration of 200 pM. Cluster amplification of denatured templates and paired-end sequencing was then performed according to the manufacturer’s protocol (Illumina) for the HiSeq X Ten, with the following modification: we enabled dual indexing outside of the standard HiSeq control software by altering the sequencing recipe files.
Calling SNPs and short INDELs from Illumina data
We mapped the Illumina reads to the human genome GRCh37 with the GATK best-practice pipeline, which uses BWA-MEM for mapping and post-processes alignments with BQSR and INDEL realignment. We additionally mapped the reads from one sample with BWA-MEM to various human genome versions without post processing steps. We have also run minimap2 and Bowtie2 for the same sample. We used the default settings of various mappers, except for tuning the maximal insert size.
We called variants on the mixed synthetic-diploid samples with FreeBayes, Platypus, Samtools and GATK, including the HaplotypeCaller (HC) and UnifiedGenotyper (UG) algorithms and filtered the raw variant calls with the set of rules described in the main text. We have tried GATK’s VQSR model for filtering. However, as the VQSR training set is biased towards variants in regions with unambiguous mapping, VQSR misses many truth variants without perfect averaged mapping quality. Both GATK and Platypus come with a set of hard filters. However, by not filtering on read depth, one of the most effective filters on single-sample WGS calling, these filters lead to a low precision.
Supplementary Material
Acknowledgements.
This study was supported by NIH grant 5U54DK105566–04, 5U01HG009088–03 and 1R01HG010040–01. We are grateful to Evan Eichler for providing the DNA of CHM cell lines. We thank Andrew Carrol for testing PacBio’s new consensus caller Arrow, and thank Mark DePristo, Justin Zook and Brad Chapman for their helpful suggestions.
Footnotes
Competing Interests. The authors declare no competing interests.
Data availability. Illumina reads from this study were deposited to ENA under accession PRJEB13208. Syndip variant calls and confident regions can be acquired from https://github.com/lh3/CHM-eval.
Code availability. Evaluation scripts are available from https://github.com/lh3/CHM-eval. The variant calling pipeline and hard filters are implemented in https://github.com/lh3/unicall. Source code is also available as Supplementary Software.
References
- 1.Zook JM et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol 32, 246–251, doi: 10.1038/nbt.2835 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Eberle MA et al. A reference data set of 5.4 million phased human variants validated by genetic inheritance from sequencing a three-generation 17-member pedigree. Genome Res 27, 157–164, doi: 10.1101/gr.210500.116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics 30, 2843–2851, doi: 10.1093/bioinformatics/btu356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin CS et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods 13, 1050–1054, doi: 10.1038/nmeth.4035 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chin CS et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10, 563–569, doi: 10.1038/nmeth.2474 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Seo JS et al. De novo assembly and phasing of a Korean human genome. Nature 538, 243–247, doi: 10.1038/nature20098 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Huddleston J et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res 27, 677–685, doi: 10.1101/gr.214007.116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneider VA et al. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res 27, 849–864, doi: 10.1101/gr.213611.116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 (2013).
- 10.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359, doi: 10.1038/nmeth.1923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H Minimap2: versatile pairwise alignment for nucleotide sequences. arXiv:1708.01492 (2017). [DOI] [PMC free article] [PubMed]
- 12.Garrison E & Marth G Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 (2012).
- 13.Rimmer A et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet 46, 912–918, doi: 10.1038/ng.3036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993, doi: 10.1093/bioinformatics/btr509 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DePristo MA et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43, 491–498, doi: 10.1038/ng.806 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cleary JG et al. Comparing Variant Call Files for Performance Benchmarking of Next-Generation Sequencing Variant Calling Pipelines. bioRxiv, doi: 10.1101/023754 (2015). [DOI]
- 17.Consortium GP et al. A global reference for human genetic variation. Nature 526, 68–74, doi: 10.1038/nature15393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson JT et al. Integrative genomics viewer. Nat Biotechnol 29, 24–26, doi: 10.1038/nbt.1754 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgulis A, Gertz EM, Schaffer AA & Agarwala R A fast and symmetric DUST implementation to mask low-complexity DNA sequences. J Comput Biol 13, 1028–1040, doi: 10.1089/cmb.2006.13.1028 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Mallick S et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538, 201–206, doi: 10.1038/nature18964 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H FermiKit: assembly-based variant calling for Illumina resequencing data. Bioinformatics 31, 3694–3696, doi: 10.1093/bioinformatics/btv440 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.