Abstract
Aberrant Hedgehog (HH) signaling leads to various types of cancer and birth defects. N-terminally palmitoylated HH initiates signaling by binding its receptor Patched-1 (PTCH1). A recent 1:1 PTCH1–HH complex structure visualized a palmitate-mediated binding site on HH, which was inconsistent with previous studies that implied a distinct, calcium-mediated binding site for PTCH1 and HH co-receptors. Here, our 3.5-Å resolution cryo-EM structure of native Sonic Hedgehog (SHH-N) in complex with PTCH1 at a physiological calcium concentration reconciles these disparate findings and demonstrates that one SHH-N molecule engages both epitopes to bind two PTCH1 receptors in an asymmetric manner. Functional assays using PTCH1 or SHH-N mutants that disrupt the individual interfaces illustrate that simultaneous engagement of both interfaces is required for efficient signaling in cells.
Introduction
Appropriate Hedgehog (HH) signaling is important for human health: excessive HH signaling leads to various cancers, while insufficient HH signaling results in birth defects (1–6). Nascent HH proteins enter the secretory pathway. After signal peptide cleavage, HH undergoes autocatalytic processing between its N and C-terminal domain (HH-N and HH-C) in the endoplasmic reticulum (7). During this reaction, cholesterol is covalently linked to the carboxyl terminus of HH-N, which is necessary for its trafficking and distribution (8). Hedgehog acyltransferase (Hhat) subsequently adds palmitate to the α-amino group of the N-terminal cysteine of HH-N (9, 10). This modification is indispensable in activating the HH pathway (9–14). Activation occurs when the lipid-modified HH-N binds to its receptor, Patched-1 (PTCH1) protein (15). Notably, a palmitoylated SHH-N-terminal peptide bound to PTCH1 partially stimulated HH signaling while the globular portion of HH could increase the binding to PTCH1 and facilitated PTCH1 export from cilia (16). In the absence of HH-N binding, PTCH1 represses the HH pathway presumably by regulating the transport of a ligand of its downstream protein, Smoothened (SMO) (2, 17, 18).
We recently determined the cryo-EM structure of a functional human PTCH1 protein (PTCH1*) alone and in complex with Sonic hedgehog (SHH-N) (19). PTCH1* lacks the internal loop between TM6 and TM7 and the C-terminal cytoplasmic domain but can still repress the HH pathway and be regulated by SHH-N, comparable to wild-type full-length PTCH1 in Ptch1−/−mouse embryo fibroblasts (MEFs) (19, 20). The structure shows that PTCH1* comprises two structurally homologous extracellular domains (ECDs) and 12 transmembrane helices (TMs). Notably, an endogenous ligand observed in the Sterol-Sensing domain (SSD) consisting of TMs 2–5, suggests that the SSD may play a role in the transport and/or regulation of the SMO ligand. The structure of the complex shows a palmitate-dominated interface between SHH-N and PTCH1 (19); this interface is opposite the interface of SHH-N with its co-receptors (21), which had been implicated in PTCH1 binding in previous studies (22–25) and can be regulated by calcium (21). Consistent with this, we reported biochemical assays showing that SHH-N lacking palmitate interacts with PTCH1 through the Ca2+-mediated interface. However, unpalmitoylated SHH-N fails to stimulate HH signaling in the cell (16, 19).
Assembly of 2:1 PTCH1*–SHH-N complex
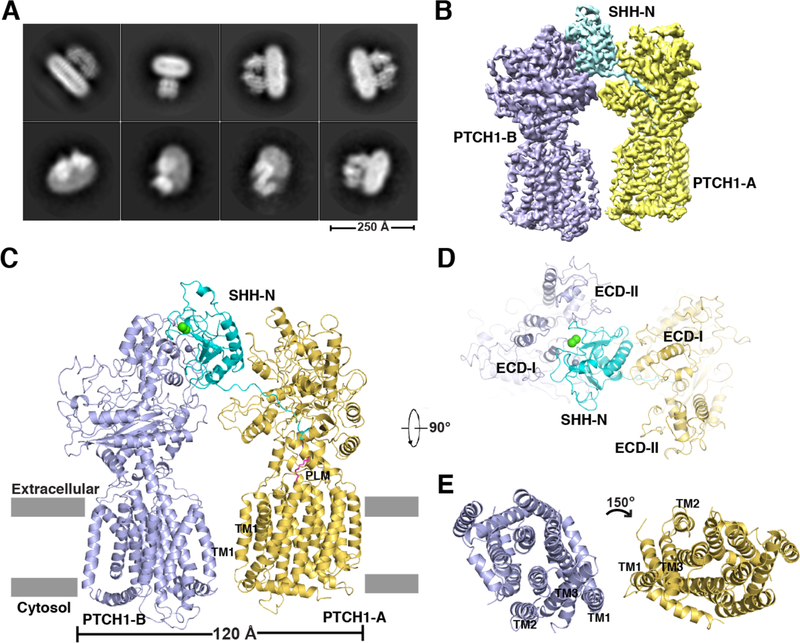
The extracellular Ca2+ concentration is typically in the range of 1–2 mM (26) and mutations on either the palmitate-dominated interface (19) or the Ca2+-mediated interface (27) attenuated HH signaling, indicating that both SHH-N interfaces are necessary for maximal HH signaling in the cell. Previously, Ca2+ was not included in our purification buffer. To enable the formation of either or both interfaces, we mixed PTCH1* with native SHH-N containing its N-terminal palmitate and C-terminal cholesterol modifications in the presence of 1 mM CaCl2. A 2:1 PTCH1*–SHH-N complex was eluted ahead of the 1:1 complex by gel filtration and subjected to cryo-EM studies (Figs. 1A and fig. S1A).
Fig. 1. Overall structure of 2:1 PTCH1*–SHH-N complex.
(A) The cryo-EM 2D classification from RELION. (B) The cryo-EM map after Frealign refinement sharpened using BFACTOR.EXE (author: Nikolaus Grigorieff) with a resolution limit of 3.5 Å and a B-factor value of −100 Å2. (C) Ribbon representation of the structure viewed from the side of the membrane. Calcium and zinc are shown as spheres in green and gray, respectively. Palmitate is shown in magenta and labeled “PLM”. (D) The ECDs from the top view. (E) The transmembrane domains from the top view.
We determined this structure at 3.5-Å resolution (Figs. 1B, fig. S1B-D, and table S1). One SHH-N forms a complex with two PTCH1* molecules (PTCH1-A and PTCH1-B) using two distinct interfaces (Fig. 1C). The SHH-N palmitate inserts into the ECD cavity of PTCH1-A, and its Ca2+-mediated interface directly binds the ECDs of PTCH1-B (Fig. 1C). PTCH1-A employs its ECD-I to bind the globular portion of SHH-N, while PTCH1-B engages both of its ECDs to bind the Ca2+-mediated epitope (Fig. 1D). The interface between PTCH1-A and SHH-N is ~2300 Å2 (including 350 Å2 to accommodate the palmitate), while the interface between PTCH1-B and SHH-N is ~1100 Å2. The transmembrane domains of the two PTCH1* molecules are arranged asymmetrically, with the TMs of PTCH1-B rotated about 150° relative to the TMs of PTCH1-A (Fig. 1E).
Interfaces between SHH-N and two PTCH1* molecules
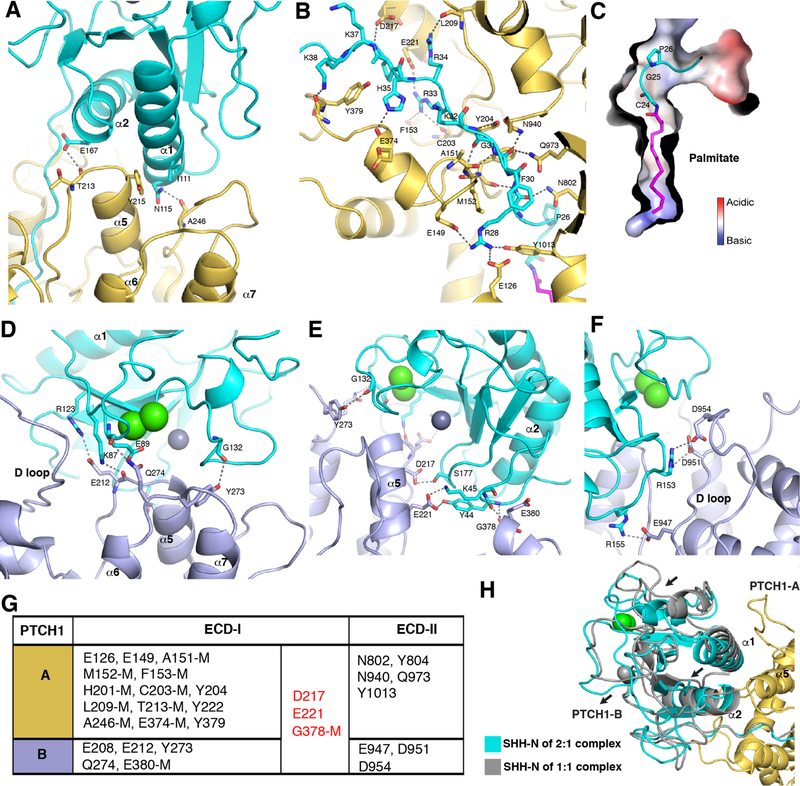
The current map allows us to visualize the interactions between SHH-N and the two PTCH1* molecules (figs. S2 and S3). There are several hydrogen bonds and salt bridges between the N-terminal residues of SHH-N and the PTCH1-A ECDs, which form the palmitate-dominated interface. Only five residues of ECD-II (N802, Y804, N940, Q973, and Y1013) interact with the N-terminal peptide (Np) and the palmitate of SHH-N; however, over ten residues from ECD-I contribute to this interaction (Fig. 2A and B). A previous study showed that the mutation P26A on SHH-N impairs signaling (16). Our structure reveals the molecular basis of this effect: the proline facilitates palmitate insertion into the hydrophobic cavity by keeping the N-terminus of SHH-N in a bent position (Fig. 2C). In addition to the palmitate insertion, the palmitate-dominated interface includes α5 of PTCH1-A recognizing α1 and the N-terminus of α2 of SHH-N (Fig. 2A). We showed previously that mutations on α5 of PTCH1 abolished PTCH1*–SHH-N binding and HH signaling (19).
Fig. 2. The two interfaces of SHH-N bound to PTCH1*.
(A) Details of α1 and α2 of SHH-N binding to PTCH1-A. (B) Details of the interaction between the N-terminus of SHH-N and PTCH1-A (PLM: stick representation in magenta). (C) The bend in the N-terminus of SHH-N. (D-F), Interaction detail of the Ca2+ -mediated interface of SHH-N to PTCH1-B residues (Zn, Ca: sphere representation in gray, green). (G) Summary of the residues of PTCH1* bound to the distinct interfaces of SHH-N, M: main chain involved interaction. (H) Structural comparison of SHH-N in 2:1 and 1:1 PTCH1*–SHH-N complexes.
The PTCH1-B molecule also employs its α5 helix and several loops of ECD-I to form the Ca2+-mediated interface of SHH-N (Fig. 2D and E). In addition, three residues (E947, D951 and D954) in an ECD-II loop (D loop) form salt bridges with R153 and R155 of SHH-N. The D-loop is not used in the PTCH1-A palmitate-mediated interaction (Fig. 2F). In the context of SHH-N without palmitate, mutation of R153 causes loss of binding to PTCH1 (20). In the context of SHH-N with palmitate, the mutant also fails to stimulate HH signaling (27), consistent with the requirement for both interfaces. A structure of PTCH1 with unpalmitoylated SHH-N was recently published (28), which is consistent with our observation of the interface between PTCH1-B and native SHH-N
Overall across both interfaces, residues in ECD-I contribute to over 80% of the interactions with SHH-N. Residues D217 and E221 in helix α5 and G378 in the loop connecting α3 and α3’ function in both interfaces; however, no residues in ECD-II are used in both interfaces (Fig. 2G). Residues in ECD-II bind to the globular portion of SHH-N on the Ca2+-mediated interface and different residues in ECD-II are involved in the palmitate insertion. This distinction introduces an asymmetric arrangement of two PTCH1* molecules in the complex. Superimposing PTCH1-A and SHH-N from the 2:1 complex with the PTCH1*– SHH-N 1:1 complex (PDB code: 6D4J) reveals a slight change in the palmitate-dominated interface. SHH-N in the 2:1 complex has moved away from α5 of PTCH1-A due to the interaction of PTCH1-B with the Ca2+-mediated interface (Fig. 2H).
Intriguingly, there is a rod-like density of unknown identity among the α5-α7 helices of ECD-I in both PTCH1* molecules (fig. S4). This ligand may stabilize α5 of ECD-I in the conformation that binds SHH-N or could be a ligand that is regulated by PTCH1 in the HH pathway; however, this molecule and its precise function require identification and further analysis.
Structural comparison of PTCH1-A and PTCH1-B after SHH-N binding
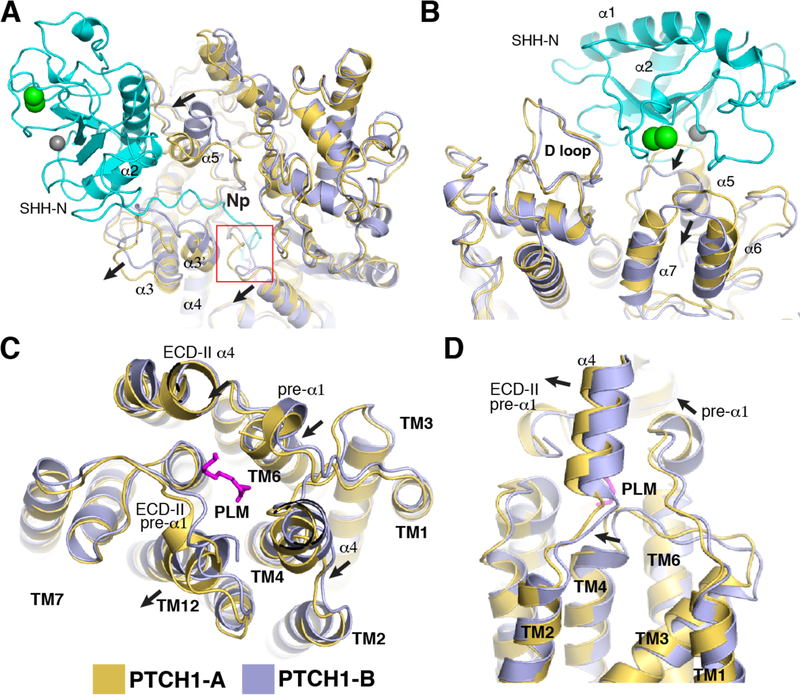
PTCH1-A employs its ECD-I for binding the globular domain of SHH-N and PTCH1-B uses a similar area of its ECD-I for SHH-N recruitment (Fig. 1C). To compare the conformations of the two PTCH1* molecules in the complex, we superimposed PTCH1-A and PTCH1-B: the α3 and α3’ helices of ECD-I in PTCH1-A are moved outward relative to PTCH1-B to accommodate the N-terminus of SHH-N that inserts into the cavity of PTCH1-A (Fig. 3A). The Np and the palmitate of SHH-N trigger a loop in ECD-I to move, allowing insertion (Fig. 3A, red frame). The α5 in ECD-I is in distinct conformations in PTCH1-A and PTCH1-B because it is involved in different interfaces with SHH-N (Figs. 2A, 2E and 3A). The loop between α6 and α7 in ECD-I of PTCH1-B is shifted due to its interaction with E89 in SHH-N (Figs. 2D and 3B). In the transmembrane domain, TM4 and TM12 have moved slightly in PTCH1-A relative to PTCH1-B (Fig. 3C and D). There are some conformational changes between the two small α-helices following TM1 and TM7 (pre-α1), and α4 of ECD-I, which form the cavity that accommodates the palmitate (Fig. 3C and fig. S3).
Fig. 3. Structural comparison of the two PTCH1* molecules.
(A) Movement of structural elements of ECD-I in PTCH1-A after SHH-N insertion. (B) Superimposition of the ECDs of PTCH1* molecules from top view. (C) Superimposition of transmembrane domains of PTCH1* molecules from the top view. (D) Superimposition of transmembrane domains of PTCH1* molecules from the side of the membrane.
As seen in the 1:1 complex (19), two endogenous sterol-like ligand densities can be detected in the transmembrane domain of each PTCH1* molecule, one in the SSD and the other close to TM12 (figs. S2 and S5). The palmitate moiety in the cavity between the ECDs together with TM4 and TM6 provides a hydrophobic environment for the endogenous ligand (Fig. 3D). The palmitate may affect the dynamics of the SSD by inserting into the center of the protein; a similar mechanism was postulated to regulate cholesterol egress from the lysosome in the homologous NPC1 protein (29–31). The cavity between the ECDs might also serve as part of a tunnel for the transport of endogenous ligands from ECD-I or the TM domain (figs. S4 and S5). Blocking the tunnel by palmitate could then inactivate PTCH1.
Physiological importance of the Ca2+-mediated interface of SHH-N
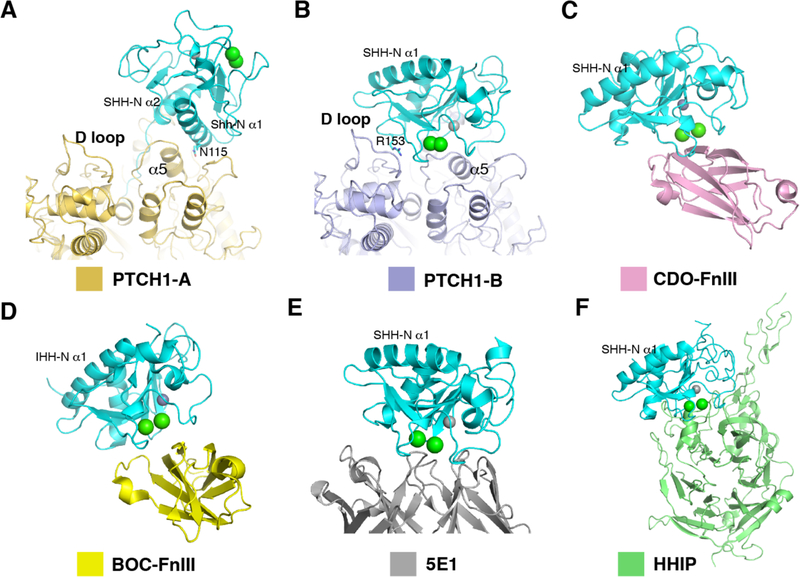
The palmitate of SHH-N facilitates an interaction with PTCH1 that is distinct from interactions with other HH-N partners (Fig. 4A). In contrast, the Ca2+-mediated interface of SHH-N, which is required for PTCH1-B binding, has been reported to recruit other HH-N binders such as the co-receptors CDO and BOC (Fig. 4B-F) (21). Consistent with this, previous experiments showed that the fibronectin type (Fn) III domain of CDO competes with PTCH1 for HH-N binding in vitro (24). HH co-receptors (e.g. CDO and BOC) are essential for HH-mediated cell proliferation (27, 32): by binding the Ca2+-mediated interface of HH-N, they may increase the local concentration of HH-N, which could promote the PTCH1–HH-N interaction (Fig. 4C and D).
Fig. 4. Physiological importance of the Ca2+-mediated interface of SHH-N.
(A) The interaction details of PTCH1-A and the globular portion of SHH-N. (B) The interaction details of PTCH1-B and SHH-N. (C) The structure of SHH-N with FNIII domain of CDO (PDB code: 3D1M). (D) The structure of IHH-N with FNIII domain of BOC (PDB code: 3N1M). (E) The structure of SHH-N with 5E1 Fab (PDB code: 3MXW). (F) The structure of SHH-N with Hedgehog-interacting protein (HHIP) (PDB code: 3HO5).
5E1 is a commonly used monoclonal antibody that binds to the Ca2+-mediated interface of SHH-N, blocking HH signaling (33, 34). Structural analysis indicates that the interface between 5E1 and SHH-N overlaps with that of PTCH1-B and SHH-N (Fig. 4B and E). However, 5E1 does not compete with the palmitate-dominated interface—we previously detected a PTCH1*–SHH-N–5E1 complex in pull-down assays (19). Hedgehog-interacting protein (HHIP) serves as a negative regulator of the HH pathway and blocks HH signaling (35) by directly binding the Ca2+-mediated interface of SHH-N (25, 36) (Fig. 4F). Therefore, we speculate that the Ca2+-mediated interface rather than the palmitate-dominated interface may be primarily regulated by the different HH binders in cells to prevent or promote the formation of the 2:1 complex, affecting signaling.
2:1 PTCH1–SHH-N complex in HH signaling
The structural analysis shows that most of the SHH-N surface is occupied by two PTCH1 molecules and the N-terminus of SHH-N is buried in the cavity formed by the ECDs of PTCH1-A (Fig. 1C). Interestingly, PTCH1-A and PTCH1-B employ a very similar area to recognize the globular portion of SHH-N, making it impossible for SHH-N to mediate the formation of a PTCH1 oligomer of higher order than a dimer. Our finding suggests that 2:1 PTCH1–HH complex could be the functional organization in HH signaling.
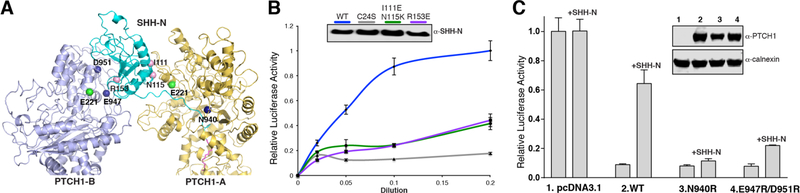
To investigate this further, we performed cell-based assays. SHH-light II cells are transfected with GLI-responsive firefly luciferase reporter and a constitutive renilla-luciferase expression vector for the quantitative measurement of biologically active HH proteins. The palmitoylated SHH-N and its variants were collected from the medium and detected by anti-SHH antibody. Using the SHH-N with either I111E/N115K or R153E mutations—located in the two interfaces in our 2:1 complex—substantially reduces signaling in cells (Fig. 5A and B). The C24S palmitoylation site SHH-N mutant completely loses its activity for HH signaling (Fig. 5B). Together, these studies show that both interfaces of SHH-N and its palmitate moiety are necessary for maximal HH signaling in cells and the palmitate moiety is absolutely required.
Fig. 5. Functional characterization of HH signal biogenesis.
(A) Location of the key residues for HH signal biogenesis. (B) Mutation on either interface or palmitoylation site of SHH-N abolishes HH signaling. HH activity was measured by luciferase assay. Data are mean ± s.d. (n = 3). SHH-N in conditioned medium was detected by western blotting. (C) HH signaling in Ptch1−/−MEFs transfected with PTCH1 and treated with SHH-N ligand. Luciferase activity measured as described in Methods section. Data are mean ± s.d. (n = 3).
Using Ptch1−/−MEFs with the same luciferase reporting system we expressed full-length human PTCH1 with either N940R or E947R/D951R mutations on ECD-II to specifically block the interaction on either side of SHH-N (Figs. 2G and 5A).
Expression of either of the mutants alone repressed the HH pathway, and in neither case could the repression be released by introducing SHH-N (Fig. 5C). Additionally, our previous work showed that PTCH1 with an E221 mutation, a residue that is necessary for the PTCH1–SHH-N interaction in both interfaces (Fig. 5A), can repress the HH pathway but does not respond to HH-N (19), supporting our current observations. Therefore, these assays validated our structure-based proposal that the 2:1 PTCH1–SHH-N complex is essential for maximal HH signaling.
The putative mechanism of palmitoylated HH mediated signaling
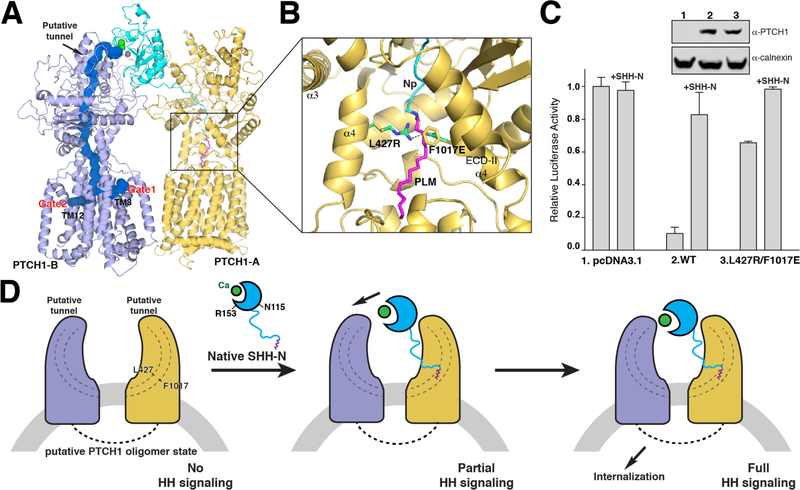
The palmitate of SHH-N inserts into the cavity created by several hydrophobic residues of the ECDs, preventing any access of a putative ligand through the protein. Interestingly, MOLE calculations (37) show that there is a ~150 Å length tunnel with a minimum radius of 4 Å that stretches throughout the PTCH1-B protein but not PTCH1-A, which interacts with the palmitate of SHH-N (Fig. 6A). The path of this tunnel includes the area where the unidentified ligand is located in ECD-I (fig. S4), plus the cavity of the ECDs that accommodates the palmitate and the transmembrane region. There are two gates of this tunnel in the transmembrane domain: one is near TM3 of the SSD and the other is near TM12. Notably, two unidentified sterol-like densities were detected in these two areas in both the 2:1 PTCH1–SHH-N complex (fig. S5) and in the 1:1 PTCH1–SHH-N complex (19), implying that these gates are involved in the transport and/or regulation of the sterol-like molecules.
Fig. 6. Model of palmitoylated HH mediated signaling.
(A) Structure of the 2:1 PTCH1*–SHH-N complex with the putative tunnel in blue, and the two gates in the transmembrane domain are indicated. (B) Location of the residues for mimicking the palmitate insertion to block the ECDs’ cavity. (C) HH signaling in Ptch1−/−MEFs transfected with PTCH1 and treated with SHH-N ligand. Luciferase activity measured as described in Methods section. Data are mean ± s.d. (n = 3). (D) Model for HH binding and signal generation. Native SHH-N initially binds to one PTCH1 molecule to form a transitional state presenting partial HH signaling. Then, the second PTCH1 molecule recognizes the globular portion of SHH-N to trigger the internalization of PTCH1, promoting maximal HH signaling.
To test the function of the palmitate insertion, we introduced two point mutations (L427R and F1017E) on full-length human PTCH1 to block this tunnel by a ~3Å salt bridge instead of a palmitate insertion (Fig. 6B). This variant was transfected to Ptch1−/−MEFs, and unlike wild-type PTCH1, it was not able to repress the HH pathway (Fig. 6C). This finding supports our hypothesis that the palmitate could abolish PTCH1 repression of HH signaling by blocking the cavity of the ECDs.
Since SHH-N has two interfaces that interact with PTCH1 molecules, it is interesting to analyze which interface PTCH1 will bind to first. Three pieces of evidence support the palmitate-dominated interface being the preferred interface of PTCH1 to native SHH-N: 1) in the 1:1 complex, only the palmitate-dominated interface can be detected in the presence of the native SHH-N (19); 2) PISA (Proteins, Interfaces, Structures and Assemblies) server analysis (38) suggests that the ΔiG and ΔiG P-value between the palmitate and its dominated interface to PTCH1-A are −12.1 kcal/mol and 0.232 respectively, more stable than the calcium-mediated interface to PTCH1-B (−1.1 kcal/mol and 0.717); 3) palmitoylated SHH-N mutants with a defective globular portion still present a little biological activity to stimulate HH signaling more than unpalmitoylated SHH-N (Fig. 5B). This evidence implies that the palmitate of SHH-N should be recognized before its globular portion; therefore, the palmitate-dominated interface should be the preferred contact to PTCH1 along with the palmitate insertion.
Notably, before the HH-N recognizes PTCH1, co-receptors might capture the Ca2+-mediated interface of HH-N by their flexible extracellular domains (21). This would increase the likelihood of forming a PTCH1–HH-N complex, a function reminiscent of the interleukin-2 receptor α-subunit (39). It is also possible for a PTCH1 molecule to engage the Ca2+-mediated interface if the palmitoylated N-terminus of HH-N is bound to other binders, such as various Heparan Sulfate Proteoglycans (40, 41). This would allow PTCH1 to serve as a possible co-receptor to promote extracellular transport of HH-N. Therefore, the Ca2+-mediated interface of HH-N could be recognized by different binders prior to signaling generation.
Here, we propose a mechanism for palmitoylated HH mediated signaling (Fig. 6D). PTCH1 serves as a functional suppressor to maintain the HH pathway in the inactive state, presumably as an oligomer through the interaction between the cytosolic domains of PTCH1 (20, 42). It might transport or regulate a small molecule by its internal tunnel (Fig. 6A). The palmitate insertion blocks the tunnel of one subunit of the PTCH1 oligomer, while the subunit without palmitate inserted could still be active. This transitional state with partial HH signal can be emulated by adding the palmitoylated SHH-N with a defective interface to PTCH1 (Fig. 5B) or without the globular portion of SHH-N (16). After the palmitate interface binds to PTCH1, the second PTCH1 molecule recognizes the Ca2+-mediated interface on the globular portion of SHH-N at a physiological calcium concentration, which has been shown to be important for PTCH1–HH interaction (20, 22–25). Binding to the globular portion of SHH-N could increase the binding affinity between PTCH1 and SHH-N; moreover, it triggers the internalization of PTCH1 (16), creating a PTCH1-free environment that causes an accumulation of the putative ligand of SMO (18). This ligand might then be transported and/or regulated by PTCH1 to stimulate SMO, promoting HH signaling.
Conclusions
In this manuscript, we report a structure of a signaling-competent complex consisting of SHH-N and an asymmetric arrangement of two PTCH1* molecules reconciling the disparate findings of how HH-N interacts with Patched. Our cell biological assays revealed that both PTCH1–SHH-N interactions are required for efficient signaling. The CTD and cytosolic loop between TM6 and TM7 have been removed from PTCH1*; therefore, further investigation is required to determine whether the complex results in any coupling between the cytosolic regions of full-length PTCH1.
The structural analysis reveals a tunnel in the PTCH1 molecule. Interestingly, the palmitate of SHH-N directly disrupts this tunnel when bound to PTCH1 (Fig. 6A). Mutations in the ECDs that mimic the palmitate insertion by blocking the ECD cavity, abolish the repression of the HH pathway by PTCH1 (Fig. 6B). The specific identity of the endogenous sterol-like molecules in ECD-I and the transmembrane region of PTCH1 would substantially facilitate our understanding of PTCH1 function and SMO activation.
Previous studies also show that HH-N co-receptors are necessary for HH signaling by binding to HH-N and forming a complex with PTCH1 (27, 32, 43–45). These HH-N co-receptors might play a role in increasing the local concentration of HH-N; therefore, promoting PTCH1–HH-N interaction and HH signaling. The structure of PTCH1–HH-N in complex with its co-receptors will be informative in furthering our understanding of their function in the HH pathway.
Materials and Methods
Protein expression and purification
Human PTCH1* and SHH-N proteins were purified according to our previously published protocol (19). To assemble the PTCH1* and SHH-N complex, native SHH-N (purchased from R&D Systems, Cat # 8908-SH/CF) was mixed with purified PTCH1* at a 1:2 molar ratio and purified by size-exclusion chromatography using a Superdex-200 column (GE Healthcare) in buffer A (20 mM Hepes pH 7.5, 150 mM NaCl) with 0.06% Digitonin and 1mM CaCl2. The peak fractions were collected and concentrated to 5 mg/ml for grid preparation. The mutated and truncated DNA constructs were generated using Gibson Assembly (NEB) or QuikChange Mutagenesis kit (Agilent).
EM Sample Preparation and Imaging
A freshly purified protein sample was added to Quantifoil R1.2/1.3 400 mesh Au holey carbon grids (Quantifoil), blotted using a Vitrobot Mark IV (FEI), and frozen in liquid ethane. The grids were imaged in a 300 keV Titan Krios (FEI) with a Gatan K2 Summit direct electron detector (Gatan). Data were collected at 1 Å/pixel with a dose rate of 8 electrons per physical pixel per second. Images were recorded for 10 s exposures in 50 subframes to give a total dose of 80 electrons per Å2.
Imaging Processing and 3D reconstruction
Dark subtracted images were first normalized by gain reference that resulted in a pixel size of 1 Å/pixel. Drift correction was performed using the program MotionCorr (46). The contrast transfer function (CTF) was estimated using CTFFIND4 (47). To generate 2:1 PTCH1*–SHH-N templates for automatic picking, around 3000 particles were manually picked and classified by 2D classification in RELION (48). After auto-picking in RELION, the low-quality images and false-positive particles were removed manually. About 275k particles were extracted for subsequent 2D and 3D classification. We used the cryo-EM structure of human PTCH1* (EMD-7795) low-pass filtered to 60 Å as the initial model for 3D classification in RELION. The PTCH1* model of best class after 3D classification was used as the initial model for the final 3D classification in RELION. The best class, containing ~77k particles, provided a 5 Å map after 3D auto-refinement in RELION. Refinement was performed in FREALIGN (49) using this best class as the initial model. The global search was performed once without mask followed by another global search using the mask, which was generated using “relion_mask_create” with 6 Å extensions excluding the micelle. The final map is estimated to be 3.5 Å using the 0.143 cutoff criteria.
Model Construction
To obtain better side-chain densities for model building, we sharpened the map using BFACTOR.EXE (author: Nikolaus Grigorieff) with a resolution limit of 3.5 Å and a B-factor value of −100 Å2. The structure of human PTCH1*–SHH-N complex was docked to the map. The density of residues 1–75 (N-terminal domain), 608–618 and 721–730 (TM6-TM7 linker), and 1177–1188 (C-terminus) of PTCH1* is not resolved nor built. Residues 890–915 of PTCH1* were built with poly-alanine due to limited local resolution.
Model Refinement and Validation
The model was refined in real space using PHENIX (50) and also in reciprocal space using Refmac with secondary-structure restraints and stereochemical restraints (51, 52). Structure factors were calculated from a half-map (working) using the program SFall (53). Fourier shell correlations (FSCs) were calculated between the two half maps, the model against the working map, the other (free) half map, and full (sum) map (54). Local resolutions were estimated using Blocres (55). MolProbity (56) was used to validate the geometries of the model. Structure Figurers were generated using PyMOL (http://www.pymol.org) and Chimera (57).
HH Reporter Assays
Human SHH-N (24–197) was constructed into pcDNA3.1 vector with the signal sequence of human calreticulin at the N-terminus as described before (16). Secreted SHH-N was produced in HEK293 cells by transient transfection for 72 hours and were collected in DMEM, with 0.5% Fetal Bovine Serum (FBS). SHH Light II cells, a stable cell line expressing firefly luciferase with an 8X-Gli promoter and Renilla luciferase with a constitutive promoter (from Drs. B. Chen and J. Kim), were used to measure HH pathway activity. SHH Light II cells were treated with the conditioned medium diluted in fresh DMEM with 0.5% Newborn Calf Serum for 30 hours. To measure the activity of PTCH1 variants in HH signaling, the 8X-Gli-Firefly luciferase reporter transgene, a constitutive Renilla luciferase transgene, and a pcDNA3.1 vector encoding PTCH1 variants were transfected to Ptch1−/−MEFs (from Drs. B. Chen and J. Kim) using TransIT reagent (Mirus Bio LLC). After 24 hours, cells were serum-starved in DMEM with 0.5% FBS. 24 hours later, cells were treated with SHH-N conditioned medium for another 24 hours. Firefly and Renilla luciferase activity were measured using the Dual-Luciferase® Reporter Assay System (Promega). All reporter assays were performed at least three times. The conditioned medium added was normalized based on western blotting with anti-SHH antibody. The expression of PTCH1 variants and internal calnexin in MEFs cells were detected by western blotting with anti-PTCH1 antibody (GeneTex, 83771) and anti-calnexin antibody (Novus, NB100–1965), respectively. Each assay was reproduced at least three times.
Supplementary Material
Acknowledgements
The authors would like to dedicate this work to Dr. Günter Blobel. We thank M. Ebrahim and J. Sotiris at The Rockefeller University Evelyn Gruss Lipper Cryo-Electron Microscopy Resource Center for data collection and D. Stoddard at the UT Southwestern Cryo-EM Facility (funded in part by the CPRIT Core Facility Support Award RP170644) for grid screening; L. Beatty and A. Hassan for technical help and M. Brown, E. Debler, J. Goldstein, J. Jiang for discussion during manuscript preparation.
Funding: This work was supported by the Endowed Scholars Program in Medical Science of UT Southwestern Medical Center, O’Donnell Junior Faculty Funds, Welch Foundation (I-1957) (to X.L.) and NIH grant P01 HL020948 (to Tissue Culture Core), by The Rockefeller University (to E.C.). X.L. is the Rita C. and William P. Clements, Jr. Scholar in Biomedical Research at UT Southwestern Medical Center.
Footnotes
Competing Interests: The authors declare no competing financial interests.
Data and Materials Availability: The 3D cryo-EM density maps have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-8955. Atomic coordinates for the atomic model have been deposited in the Protein Data Bank under the accession numbers 6E1H.
References
- 1.Taipale J, Beachy PA, The Hedgehog and Wnt signalling pathways in cancer. Nature 411, 349–354 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Ingham PW, McMahon AP, Hedgehog signaling in animal development: paradigms and principles. Genes & development 15, 3059–3087 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Rubin LL, de Sauvage FJ, Targeting the Hedgehog pathway in cancer. Nature reviews. Drug discovery 5, 1026–1033 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Jiang J, Hui CC, Hedgehog signaling in development and cancer. Developmental cell 15, 801–812 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briscoe J, Therond PP, The mechanisms of Hedgehog signalling and its roles in development and disease. Nature reviews. Molecular cell biology 14, 416–429 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Petrov K, Wierbowski BM, Salic A, Sending and Receiving Hedgehog Signals. Annual review of cell and developmental biology 33, 145–168 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Lee JJ, Ekker SC, von Kessler DP, Porter JA, Sun BI, Beachy PA, Autoproteolysis in hedgehog protein biogenesis. Science 266, 1528–1537 (1994). [DOI] [PubMed] [Google Scholar]
- 8.Porter JA, Young KE, Beachy PA, Cholesterol modification of hedgehog signaling proteins in animal development. Science 274, 255–259 (1996). [DOI] [PubMed] [Google Scholar]
- 9.Williams KP, Rayhorn P, Chi-Rosso G, Garber EA, Strauch KL, Horan GS, Reilly JO, Baker DP, Taylor FR, Koteliansky V, Pepinsky RB, Functional antagonists of sonic hedgehog reveal the importance of the N terminus for activity. Journal of cell science 112, 4405–4414 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Chamoun Z, Mann RK, Nellen D, von Kessler DP, Bellotto M, Beachy PA, Basler K, Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science 293, 2080–2084 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Pepinsky RB, Zeng C, Wen D, Rayhorn P, Baker DP, Williams KP, Bixler SA, Ambrose CM, Garber EA, Miatkowski K, Taylor FR, Wang EA, Galdes A, Identification of a palmitic acid-modified form of human Sonic hedgehog. The Journal of biological chemistry 273, 14037–14045 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Kohtz JD, Lee HY, Gaiano N, Segal J, Ng E, Larson T, Baker DP, Garber EA, Williams KP, Fishell G, N-terminal fatty-acylation of sonic hedgehog enhances the induction of rodent ventral forebrain neurons. Development 128, 2351–2363 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Dawber RJ, Hebbes S, Herpers B, Docquier F, van den Heuvel M, Differential range and activity of various forms of the Hedgehog protein. BMC developmental biology 5, 21 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goetz JA, Singh S, Suber LM, Kull FJ, Robbins DJ, A highly conserved amino-terminal region of sonic hedgehog is required for the formation of its freely diffusible multimeric form. The Journal of biological chemistry 281, 4087–4093 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Marigo V, Davey RA, Zuo Y, Cunningham JM, Tabin CJ, Biochemical evidence that patched is the Hedgehog receptor. Nature 384, 176–179 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Tukachinsky H, Petrov K, Watanabe M, Salic A, Mechanism of inhibition of the tumor suppressor Patched by Sonic Hedgehog. Proceedings of the National Academy of Sciences of the United States of America 113, E5866–E5875 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denef N, Neubuser D, Perez L, Cohen SM, Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell 102, 521–531 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Taipale J, Cooper MK, Maiti T, Beachy PA, Patched acts catalytically to suppress the activity of Smoothened. Nature 418, 892–897 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Qi X, Schmiege P, Coutavas E, Wang J, Li X, Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature, (2018); published online EpubJul 11 ( 10.1038/s41586-018-0308-7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleet A, Lee JP, Tamachi A, Javeed I, Hamel PA, Activities of the Cytoplasmic Domains of Patched-1 Modulate but Are Not Essential for the Regulation of Canonical Hedgehog Signaling. The Journal of biological chemistry 291, 17557–17568 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beachy PA, Hymowitz SG, Lazarus RA, Leahy DJ, Siebold C, Interactions between Hedgehog proteins and their binding partners come into view. Genes & development 24, 2001–2012 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuse N, Maiti T, Wang B, Porter JA, Hall TM, Leahy DJ, Beachy PA, Sonic hedgehog protein signals not as a hydrolytic enzyme but as an apparent ligand for patched. Proceedings of the National Academy of Sciences of the United States of America 96, 10992–10999 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pepinsky RB, Rayhorn P, Day ES, Dergay A, Williams KP, Galdes A, Taylor FR, Boriack-Sjodin PA, Garber EA, Mapping sonic hedgehog-receptor interactions by steric interference. The Journal of biological chemistry 275, 10995–11001 (2000). [DOI] [PubMed] [Google Scholar]
- 24.McLellan JS, Zheng X, Hauk G, Ghirlando R, Beachy PA, Leahy DJ, The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature 455, 979–983 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosanac I, Maun HR, Scales SJ, Wen X, Lingel A, Bazan JF, de Sauvage FJ, Hymowitz SG, Lazarus RA, The structure of SHH in complex with HHIP reveals a recognition role for the Shh pseudo active site in signaling. Nature structural & molecular biology 16, 691–697 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Hurwitz S, Homeostatic control of plasma calcium concentration. Critical reviews in biochemistry and molecular biology 31, 41–100 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Izzi L, Levesque M, Morin S, Laniel D, Wilkes BC, Mille F, Krauss RS, McMahon AP, Allen BL, Charron F, Boc and Gas1 each form distinct Shh receptor complexes with Ptch1 and are required for Shh-mediated cell proliferation. Developmental cell 20, 788–801 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong X, Qian H, Cao P, Zhao X, Zhou Q, Lei J, Yan N, Structural basis for the recognition of Sonic Hedgehog by human Patched1. Science, (2018); published online EpubJun 28 ( 10.1126/science.aas8935). [DOI] [PubMed] [Google Scholar]
- 29.Lu F, Liang Q, Abi-Mosleh L, Das A, De Brabander JK, Goldstein JL, Brown MS, Identification of NPC1 as the target of U18666A, an inhibitor of lysosomal cholesterol export and Ebola infection. eLife 4, (2015) ( 10.7554/eLife.12177). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X, Wang J, Coutavas E, Shi H, Hao Q, Blobel G, Structure of human Niemann-Pick C1 protein. Proceedings of the National Academy of Sciences of the United States of America 113, 8212–8217 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Saha P, Li J, Blobel G, Pfeffer SR, Clues to the mechanism of cholesterol transfer from the structure of NPC1 middle lumenal domain bound to NPC2. Proceedings of the National Academy of Sciences of the United States of America 113, 10079–10084 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen BL, Song JY, Izzi L, Althaus IW, Kang JS, Charron F, Krauss RS, McMahon AP, Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Developmental cell 20, 775–787 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ericson J, Morton S, Kawakami A, Roelink H, Jessell TM, Two critical periods of Sonic Hedgehog signaling required for the specification of motor neuron identity. Cell 87, 661–673 (1996). [DOI] [PubMed] [Google Scholar]
- 34.Maun HR, Wen X, Lingel A, de Sauvage FJ, Lazarus RA, Scales SJ, Hymowitz SG, Hedgehog pathway antagonist 5E1 binds hedgehog at the pseudo-active site. The Journal of biological chemistry 285, 26570–26580 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chuang PT, Kawcak T, McMahon AP, Feedback control of mammalian Hedgehog signaling by the Hedgehog-binding protein, Hip1, modulates Fgf signaling during branching morphogenesis of the lung. Genes & development 17, 342–347 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bishop B, Aricescu AR, Harlos K, O’Callaghan CA, Jones EY, Siebold C, Structural insights into hedgehog ligand sequestration by the human hedgehog-interacting protein HHIP. Nature structural & molecular biology 16, 698–703 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berka K, Hanak O, Sehnal D, Banas P, Navratilova V, Jaiswal D, Ionescu CM, Svobodova Varekova R, Koca J, Otyepka M, MOLEonline 2.0: interactive web-based analysis of biomacromolecular channels. Nucleic acids research 40, W222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krissinel E, Henrick K, Inference of macromolecular assemblies from crystalline state. Journal of molecular biology 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Stauber DJ, Debler EW, Horton PA, Smith KA, Wilson IA, Crystal structure of the IL-2 signaling complex: paradigm for a heterotrimeric cytokine receptor. Proceedings of the National Academy of Sciences of the United States of America 103, 2788–2793 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bellaiche Y, The I, Perrimon N, Tout-velu is a Drosophila homologue of the putative tumour suppressor EXT-1 and is needed for Hh diffusion. Nature 394, 85–88 (1998). [DOI] [PubMed] [Google Scholar]
- 41.The I, Bellaiche Y, Perrimon N, Hedgehog movement is regulated through tout velu-dependent synthesis of a heparan sulfate proteoglycan. Molecular cell 4, 633–639 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Lu X, Liu S, Kornberg TB, The C-terminal tail of the Hedgehog receptor Patched regulates both localization and turnover. Genes & development 20, 2539–2551 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen BL, Tenzen T, McMahon AP, The Hedgehog-binding proteins Gas1 and Cdo cooperate to positively regulate Shh signaling during mouse development. Genes & development 21, 1244–1257 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao S, Lum L, Beachy P, The ihog cell-surface proteins bind Hedgehog and mediate pathway activation. Cell 125, 343–357 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Song JY, Holtz AM, Pinskey JM, Allen BL, Distinct structural requirements for CDON and BOC in the promotion of Hedgehog signaling. Developmental biology 402, 239–252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y, Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature methods 10, 584–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohou A, Grigorieff N, CTFFIND4: Fast and accurate defocus estimation from electron micrographs. Journal of structural biology 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheres SH, RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grigorieff N, Frealign: An Exploratory Tool for Single-Particle Cryo-EM. Methods in enzymology 579, 191–226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murshudov GN, Vagin AA, Dodson EJ, Refinement of macromolecular structures by the maximum-likelihood method. Acta crystallographica. Section D, Biological crystallography 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Brown A, Long F, Nicholls RA, Toots J, Emsley P, Murshudov G, Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta crystallographica. Section D, Biological crystallography 71, 136–153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eyck LFT, Efficient structure-factor calculation for large molecules by the fast fourier transform. Acta Crystallogr. A 33, 486–492 (1977). [Google Scholar]
- 54.Wang Z, Hryc CF, Bammes B, Afonine PV, Jakana J, Chen DH, Liu X, Baker ML, Kao C, Ludtke SJ, Schmid MF, Adams PD, Chiu W, An atomic model of brome mosaic virus using direct electron detection and real-space optimization. Nature communications 5, 4808 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heymann JB, Belnap DM, Bsoft: image processing and molecular modeling for electron microscopy. Journal of structural biology 157, 3–18 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC, MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.