Abstract
Objective.
Most continuous cell lines with erythroid characteristics are derived from patients with myelogenous leukemia or erythroleukemia. Among them, a few cell lines have been reported to be positive for CD36. We tried to establish a continuous erythroid cell line from the primary CD36+ erythroid progenitor cells (EPCs) by the lentivirus-mediated gene transduction system.
Materials and Methods.
A lentiviral vector carrying SV40T, hTERT, or the human papillomavirus type 16 (HPV16) E6 and E7 (E6/E7) viral oncogenes, was introduced into CD36+ EPCs, singularly or combined. Transformed cells were characterized in terms of histology, phenotype, karyotype, and gene expression profile.
Results.
The lentiviral vector carrying HPV16 E6/E7 genes successfully transformed CD36+ EPCs, creating a continuous cell line, CD36E. Immunophenotype analysis revealed that the CD36E cells had characteristics of erythroid progenitors, among which about 27% of the cell population produced hemoglobin. Colony-forming cell assay demonstrated that the CD36E cells were capable of forming erythroid colonies. Using cytokines or chemical agents, attempts were made to induce differentiation of the CD36E cells but were ineffective, indicating the irreversible erythroid lineage commitment of the cells. The gene expression profile of the CD36E cells displayed a marked difference from that of the CD36+ EPCs.
Conclusions.
The continuous CD36E cell line is an erythroid progenitor cell line possessing the ability to produce hemoglobin. The CD36E cell line would be an excellent tool for applied research involving erythroid lineage cells and comparative studies with primary CD36+ EPCs.
Studies involving erythroid progenitor cells have been limited to primary cells or cell lines established from leukemia. Recent advances in differentiating and generating large numbers of specific lineages of hematopoietic cells from CD34+ hematopoietic stem cells (HSCs) have provided valuable tools and extensive uses. Differentiating cells from CD34+ HSCs toward the erythroid lineage increasingly express the CD36 transmembrane protein, a member of the scavenger receptor family, on the surface [1]. However, as erythroid progenitors evolve into more mature and differentiated cells, the CD36 expression decreases [2,3]. Therefore, the CD36 expression is defined as a determinant of erythroid progenitors [3–5]. In addition to erythroid progenitors, the CD36 expression is also detected on the surface of different hematopoietic cells, including platelets and megakaryocytes [5], and various other cell types, such as endothelial cells, mast cells, smooth muscle cells, and dendritic cells.
There are various cell lines that display erythroid characteristics, such as K562 [6], KU812Ep6 [7], JK-1 [8], KMOE [9], TF1 [10], HEL [11], B1647 [12], MB-02 [13], M-07 [14], and UT7/erythropoietin (EPO) [15]. These cell lines are derived from patients with leukemia, some of which produce varying amounts of hemoglobin innately or under induction.
We previously developed a culture system (a modification of Freyssinier et al. [16] and Giarratana et al. [17]) that generated a large and relatively pure population of erythroid progenitor cells (EPCs) from peripheral blood–derived CD34+ HSCs [3]. In this study, we sought to generate an EPC line from primary CD36+ EPCs by a lentivirus-mediated gene transduction and successfully established a CD36E cell line using the human papillomavirus type 16 (HPV16) E6 and E7 (E6/E7) genes.
Materials and methods
Cell culturing, cell proliferation monitoring, and cell sorting
CD34+ cells were purified from granulocyte colony-stimulating factor–mobilized peripheral blood stem cells using the Isolex 300i Magnetic Cell Selection System (Baxter Healthcare Corporation, Deerfield, IL, USA) [3]. CD34+ cells (104 cells/mL) were initiated in the serum-free expansion medium composed of α–minimum essential medium (Mediatech, Manassas, VA, USA) with 100 ng/mL recombinant human (rhu) stem cell factor (SCF) (Stem Cell Technologies, Vancouver, British Columbia, Canada), 5 ng/mL rhu interleukin-3 (IL-3; R&D Systems, Minneapolis, MN, USA), 3 IU/mL rhuEPO (Amgen, Thousand Oaks, CA, USA), and other factors described previously. UT7/EPO-S1 and K562 cells were cultured as described previously [18]. Cell proliferation was monitored in triplicate using the Nexcelom Cellometer Auto T4 (Isogen Life Science, De Meern, Netherlands). Single cell sorting into 96-well plates was performed using the MoFlo Cytometer (Beckman Coulter, Fort Collins, CO, USA).
Induction of CD36E cells with various cytokines and chemical agents
CD36E cells were grown in the expansion medium, but with different combinations or concentrations of cytokines (EPO, SCF, IL-3, and thrombopoietin). Further, CD36E cells were cultured with hemin, dimethyl sulfoxide (DMSO), or phorbol-12- myristate-13-acetate (PMA) (Sigma-Aldrich, St Louis, MO, USA) at the indicated concentrations.
Generation of lentiviral vector carrying SV40T, hTERT, or HPV16 E6/E7 genes
A lentiviral vector plasmid carrying SV40T alone, hTERT alone, or SV40T plus hTERT was developed by CDI Technologies (Madison, WI, USA). To generate a lentiviral vector plasmid with HPV16 E6/E7 genes, a HPV16 E6/E7-coding region was amplified by polymerase chain reaction (PCR) from a p16HH plasmid (kind gift from John Schiller) [19] using primers modified from Akimov et al. [20], followed by insertion into BamHI and EcoRI sites of a pCDF1 plasmid, creating pCDF1-E6/E7.
Pseudotyped lentiviral vectors were created using the Lentiviral Packaging System (CDI Technologies), according to manufacturer’s instruction.
CD36+ EPCs (106 cells) were infected with SV40T alone, hTERT alone, HPV16 E6/E7 alone, SV40T plus hTERT (in the same vector), SV40T plus hTERT (in separate vectors), or HPV16 E6/E7 plus hTERT (in separate vectors) on 3 consecutive days. Half of the culture was subjected to puromycin selection at a concentration of 1 μg/mL for 2 weeks, as the lentiviruses had a puromycin-resistant gene.
Histology, immunophenotyping, spectral karyotyping, and chromosome banding
Cells (2 × 104 to 5 × 104) were stained with Wright-Giemsa using the MidasIII Automatic Slide Stainer (EMD; Gibbstown, NJ, USA) or with 1% benzidine (Sigma-Aldrich), followed by microscopy.
Approximately 105 cells were subjected to flow cytometry analysis using the Beckman Coulter Cytomics FC500 System. Fluorescein isothiocyanate–conjugated antibodies were purchased through BD Biosciences (Franklin Lakes, NJ, USA): CD2, CD3, CD19, CD33, CD36, CD41a, and CD61. R-phycoerythrin–conjugated antibodies were obtained as follows: CD34, CD49e, CD44, CD10, CD71, CD110, and CD235a (glycophorin A) from BD Biosciences; CD133 from Miltenyi Biotec (Auburn, CA, USA); CD235 phycoerythrin/Cy5 from BioLegend (San Diego, CA, USA). Unconjugated anti-globoside antibody (Metreya, Pleasant Gap, PA, USA) was used with secondary antibody, fluorescein isothiocyanate goat anti-rabbit IgG antibody (Invitrogen).
Metaphase spreads were prepared from exponentially growing CD36E cells. Spectral karyotyping was carried out by fluorescence in situ hybridization with chromosome-specific painting probes labeled with different fluorochromes. Chromosome Giemsa-banding (G-banding) was performed to display a unique banding pattern of each chromosome. These analyses were conducted by the Comparative Molecular Cytogenetics Core Facility (http://web.ncifcrf.gov/research/cytogenetics/).
Colony-forming cell assay
CD36E cells (5 × 104) were subjected to colony-forming cell assay using two different methylcellulose-based media (Stem Cell Technologies) according to manufacturer’s instructions: MethoCult GF+ (Catalog #H4435) complete methylcellulose medium supports hematopoietic colony formation of erythroid, granulocyte, macrophage, and multipotential progenitors; MethoCult (catalog #H4230) methylcellulose medium with a cocktail used for the expansion medium (supplemented MethoCult) supports erythroid colony formation. On day 12, colony cultures were stained with Wright-Giemsa or benzidine.
PCR amplification and Southern blotting
PCR amplification and Southern blotting were performed to examine whether the E6/E7 genes were integrated in CD36E genomic DNA. PCR was carried out using 100 ng DNA and the same PCR primers used for the E6/E7 gene amplification (described here), according to manufacturer’s instruction, followed by detection on a 1% agarose gel.
After digestion with BamHI or EcoRI, genomic DNA (20 μg) was subjected to Southern blot analysis using a 32P-labeled probe and then analyzed by autoradiography. The probe was prepared by excising a BamHI–EcoRI fragment from the pCDF1-E6/E7 plasmid and labeled with 32P using the Prime-It Random Primer Labeling kit (Stratagene/Agilent Technologies, Santa Clara, CA, USA).
Real-time reverse transcriptase (RT)-PCR
A custom PCR array targeting hematopoietic genes and a modified-cataloged cytokine array (to include EPO, EPOR, and CSF genes) were created by SABiosciences (Frederick, MD, USA). Total RNA extracted using the Qiagen microRNA Extraction kit was converted to complementary DNA and subjected to real-time RT-PCR following manufacturer’s instructions. Data were analyzed by the ΔΔCt method using the SABiosciences Data Analysis Web Portal (http://www.SABiosciences.com/pcrarraydataanalysis.php), and genes with fivefold or greater differences were extracted. Fold changes represent ratios of gene expression levels between CD36E cells and parental CD36+ EPCs.
Immunoblotting
Whole cell lysates were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (4–12%) and immunoblot analysis was performed using primary and then secondary antibodies. Signals were detected by chemiluminescent detection method. The following antibodies were used: hemoglobin-α (H-80, sc-21005), hemoglobin-β (37-8, sc-21757), hemoglobin-γ (51-7, sc-21756), and glyceraldehyde phosphate dehydrogenase (sc-47724) from Santa Cruz Biotechnology (Santa Cruz, CA, USA); HRP-rabbit anti-mouse IgG antibody (61-6520) from Invitrogen.
Results
Immortalization of CD36+ EPCs using the HPV16 E6/E7 lentiviral vector
CD34+-selected cells cultured for 8 days (typically >95% CD36+ cells) were transduced by infection on 3 consecutive days with the pseudotyped lentiviral vectors. At 2 weeks post transduction, half of the culture was subjected to puromycin selection. As the puromycin-treated cells exhibited significant growth retardation, possibly due to its cytotoxic effect on CD36+ EPCs, these cell cultures were terminated.
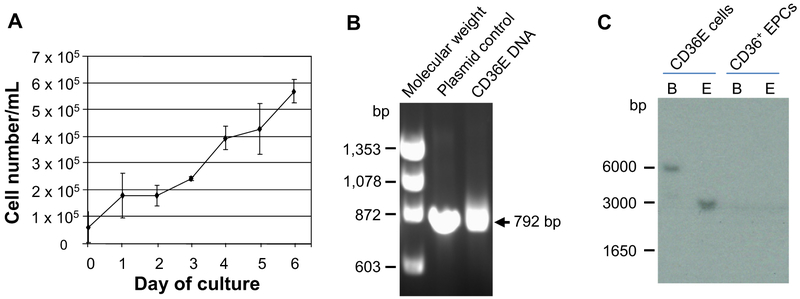
Nontransduced CD36+ EPCs generally proliferated for ~1 month, and survived up to 1.5 months. Compared with nontransduced cells, the lifespan of CD36+ EPCs was not significantly extended by infection with the lentiviral vector carrying SV40T alone, hTERT alone, or SV40T plus hTERT (both in the same lentiviral vector), or by coinfection with lentiviral vectors containing SV40T and hTERT (as a separate vector) (data not shown). In contrast, cells infected with the HPV16 E6/E7 lentiviral vector or co-infected with the HPV16 E6/E7 and the hTERT lentiviral vectors continued to proliferate. Using these two kinds of cells, attempts were made to generate single cell clones by single cell deposition into individual wells of 96-well plates by cell sorting with or without CD36+ EPC feeder cells, but failed to produce viable clones. As a next trial, 10 or 20 cells were seeded into individual wells of ten 96-well plates and allowed to incubate until colonies appeared. After 2 to 4 weeks, 53 of these cultures were expanded into 24-well plates and evaluated for their proliferation capabilities and appearance of hemoglobin. Clone 35 derived from the HPV16 E6/E7 alone transduced cells displayed the greatest proliferative capability with the strongest appearance of hemoglobin. Therefore, the clone 35 termed CD36E was selected to use in further experiments. As shown in Figure 1A, a doubling time of the CD36E cells in the expansion medium ranged from 3 to 4 days, depending on the cell density. HPV16 E6/E7 gene integration into CD36E genomes was confirmed by PCR amplification with specific primers (Fig. 1B) and by Southern blotting (Fig. 1C). Attempts were again made to isolate a single cell clone from CD36E cells, but were unsuccessful.
Figure 1.
Growth curve of CD36E cells and detection of HPV16 E6/E7-gene integration in the genomes. (A) CD36E cells were cultured in the serum-free expansion medium. Cell number and viability were assessed by the Trypan blue-dye exclusion. Each value is the mean ± SD of three experiments. (B) Genomic DNA isolated from CD36E cells or CD36+ EPCs was PCR-amplified to examine the integration of HPV16 E6/E7 genes into the genomes using the adequate primer set. An arrow represents a position of the amplified fragment from the HPV16 E6/E7 genes. DNA size markers (bp) are denoted on the left. (C) Genomic DNA isolated from CD36E cells or CD36+ EPCs was digested with BamHI or EcoRI, and subjected to Southern blot analysis. Numbers on the left indicate molecular weight marker positions.
CD36E cells display characteristic features of EPCs
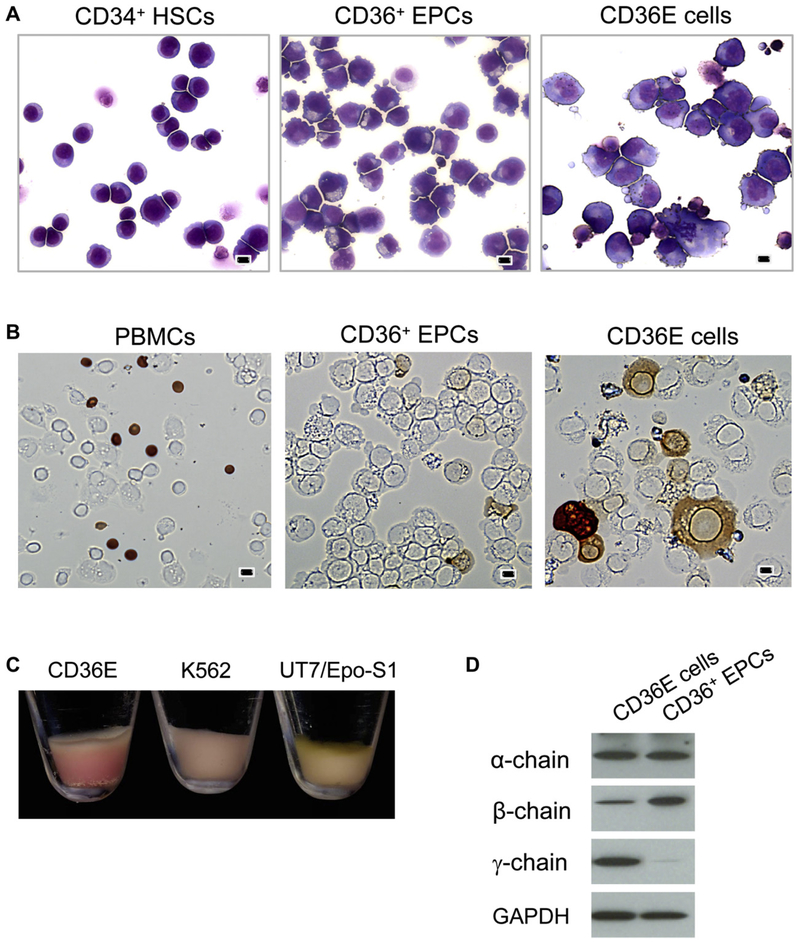
CD36E cells were stained with Wright-Giemsa and compared with CD34+ HSCs and parental CD36+ EPCs (Fig. 2A). CD34+ HSCs were round and uniform in size and shape. By incubation in the expansion medium, CD34+ HSCs were forced to differentiate toward CD36+ EPCs, resulting in a gradual increase of their sizes. On day 8, more than half of CD36+ EPCs developed microvilli on the surface. Compared with parental CD36+ EPCs, the CD36E cells were larger with round nuclei, were less homogenous in size and shape, and more than half had microvilli on the surface. The CD36E cells grew in clusters and some were semi-adherent in liquid culture, but easily disassociated.
Figure 2.
Comparison of morphological features and hemoglobin production. CD36E cells were cytocentrifuged on glass slides for staining. (A) After Wright-Giemsa staining, morphological features of CD36E cells were observed by microscopy at 200× magnification, and compared with those of CD34+ HSCs and CD36+ EPCs. Scale bars: 10 μm. (B) Hemoglobin production in CD36E cells, peripheral blood mononuclear cells (PBMCs), and CD36+ EPCs was examined by benzidine staining followed by microscopy. Scale bars: 10 μm. (C) Shown are pictures of cell pellets prepared from CD36E, K562, and UT7/EPO-S1 (a subclone of UT7/EPO). (D) Whole cell lysates obtained from CD36E cells and CD36+ EPCs were analyzed by immunoblot using antibodies against α-, β-, and γ-globin chains, and glyceraldehyde phosphate dehydrogenase (an internal control).
Hemoglobin production of CD36E cells was examined by benzidine staining and compared with that of parental CD36+ EPCs using peripheral blood cells as a positive control (Fig. 2B). A higher ratio of hemoglobin-positive cells was observed in the CD36E cells (~27%) compared with the parental CD36+ EPCs (~2%). CD36E cell pellet was redder in color than those of K562 and UT7/EPO-S1, indicating the highest hemoglobin production among the three cell lines (Fig. 2C). Immunoblot analysis was performed to identify α-, β-, and γ-globin chains. When compared with the parental CD36+ EPCs, the CD36E cells produced a higher level of γ-chain, whereas an α- or β-chain level was similar or lower, respectively (Fig. 2D).
Surface phenotype of CD36E cells was characterized by flow cytometry using various monoclonal antibodies against the surface antigens of human hematopoietic cells (Table 1). The CD36E cells and parental CD36+ EPCs were clearly not HSCs, as indicated by the loss of CD34 on their cell surface. The number of cells positive for CD33 (a myeloid cell marker) was much lower in the CD36E cells than the CD36+ EPCs. None of the following lymphoid surface antigens were detected in the CD36E cells or CD36+ EPCs: CD2 (T cells), CD3 (T cells), and CD19 (B cells). The CD36, CD44, and CD71 surface antigens marking erythroid lineage cells remained positive in the majority of the CD36E cells, which was similar to the parental CD36+ EPCs. As for other erythroid lineage markers, the CD36E cells positive for CD235a (glycophorin A) were considerably increased (96%) compared with the CD36+ EPCs (64%), whereas globoside-positive CD36E cells were decreased to 81%, compared with the CD36+ EPCs (99%). When the CD36E cells were evaluated for surface antigens for megakaryocytes, in this case CD41a, CD61, and CD110, cell populations with these antigens were lower than those of the CD36+ EPCs: CD41a and CD61, <1%; CD110, ~20%.
Table 1.
Flow cytometry analysis of cellular surface antigens
| Antigen | Description | CD34+ (day 0) | CD36+ (day 8) | CD36E | UT7/ EPO-S1 |
|---|---|---|---|---|---|
| CD2 | T-cell surface antigen T11, E-rosette receptor | <1 | 0 | 0 | 0 |
| CD3 | T-cell antigen receptor complex | <1 | 0 | 0 | 0 |
| CD10 | Common acute lymphocytic leukemia antigen | 0 | 0 | 6.1 | 0 |
| CD19 | B-lymphocyte surface antigen B4 | <1 | 0 | 0 | 0 |
| CD33 | Sialic acid-binding Ig-like lectin 3 | 46.2 | 58.6 | 30.0 | 91.3 |
| CD34 | Hematopoietic progenitor cell antigen | 96.6 | 1.0 | 0 | 0 |
| CD36 | Collagen type I recepto, thrombospondin receptor | 11.1 | 97.9 | 99.5 | 99.0 |
| CD41a | Alpha IIb integrin chain | 2.9 | <1 | ||
| CD44 | Extracellular matrix receptor III | 98.9 | 98.0 | 98.5 | 99.0 |
| CD61 | Integrin β3-subunit | 2.4 | <1 | ||
| CD71 | Transferrin receptor | 61.3 | 97.0 | 99.0 | 96.5 |
| CD110 | Thrombopoietin receptor | 58.0 | 19.3 | ||
| CD235a | Glycophorin A | <1 | 63.9 | 96.0 | 26.6 |
| Globoside | Glycosphingolipid with N-acetylgalactosaminea | 4.7 | 99.1 | 81.2 | 57.6 |
Values represent percentages of cells.
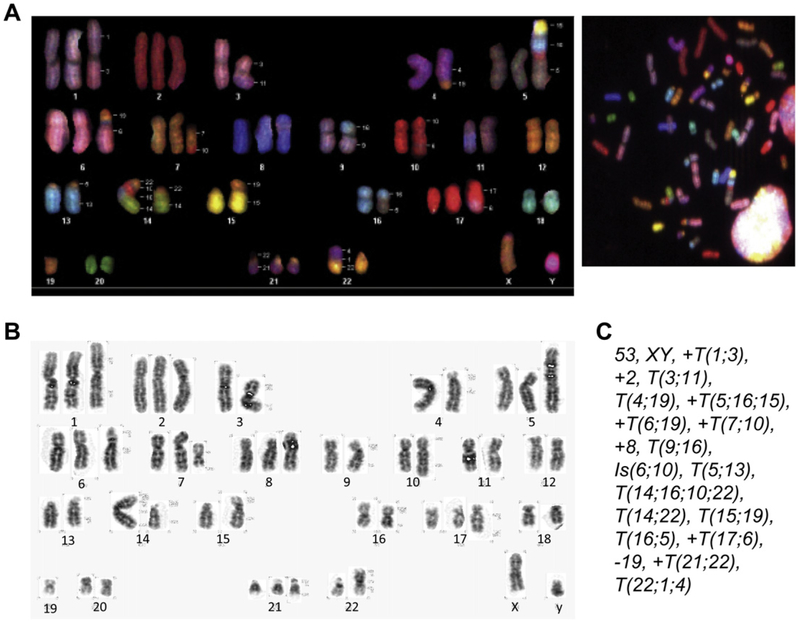
Karyotype analysis of CD36E cells
CD36E cells were evaluated for chromosomal abnormalities by spectral karyotyping and G-banding at 52 weeks (Fig. 3). Analysis of 10 cells revealed that the CD36E cells showed some chromosomal instability. The majority of abnormalities seemed to result from translocations with losses and gains of chromosomal materials, such as T(1;3), translocation involving chromosomes 1 and 3 and Is(6;10), insertion of a fragment from chromosomes 6 into 10. Chromosomes 1, 2, 5, 6, 7, 8, 17, and 21 were trisomy, while chromosome 19 was monosomy. The chromosome number of the CD36E cells ranged between 43 and 53, with a modal number of 53. Although the CD36E cells were originated from the 20-cell sorted culture, the cells were highly clonal as determined by karyotyping, implicating that a progressively expanded clone eventually took over the culture or that the initial culture was comprised of clonal cells when expanded.
Figure 3.
Chromosome analyses of CD36E cells by spectral karyotyping and G-banding. Metaphase spreads were prepared from exponentially growing CD36E cells. Chromosomal analyses were conducted using 10 metaphase spreads and one representative is shown. (A) Spectral karyotyping was performed by fluorescence in situ hybridization with chromosome-specific painting probes. Structural chromosomal abnormalities are designated on the left side of each chromosome. (B) G-banding was carried out to generate dark and light banding patterns unique to individual chromosomes. (C) Detailed numerical and structural descriptions of the CD36E karyotype.
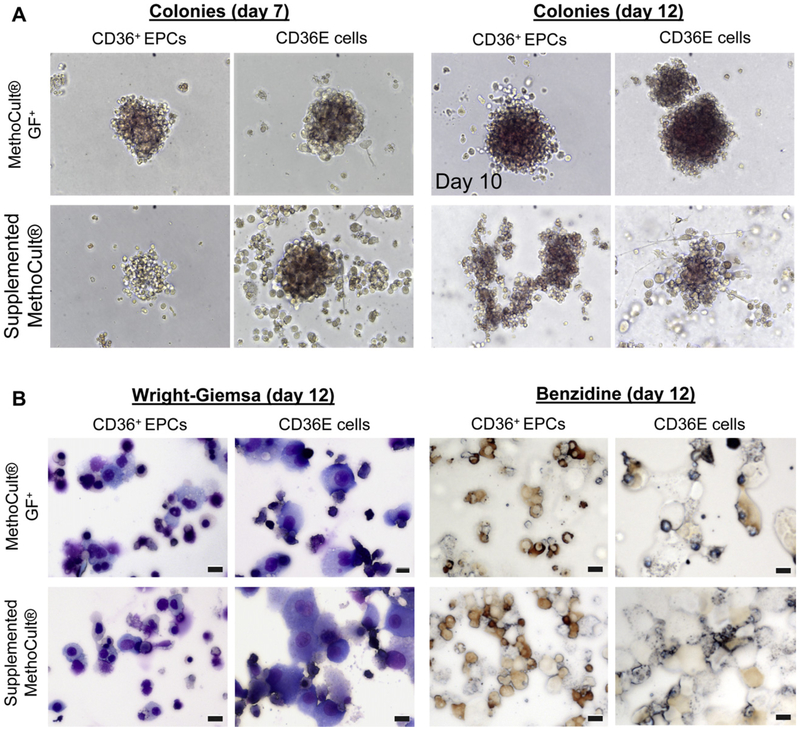
Erythroid colony-forming ability of CD36E cells
The colony-forming cell assay was performed to examine a functional potentiality of CD36E cells to proliferate and differentiate into colonies in semi-solid media in response to cytokine stimulation. CD36E cells and CD36+ EPCs were cultured in two kinds of methylcellulose-based media and evaluated on days 7, 10, and 12: the MethoCult GF+ complete methylcellulose medium for hematopoietic colony formation of erythroid, granulocyte, macrophage, and multipotential progenitors; the supplemented MethoCult methylcellulose medium for erythroid colony formation. On day 7, erythroid colonies were observed in cultures of the CD36E cells and the CD36+ EPCs in either the MethoCult GF+ or the supplemented MethoCult, although colonies were generally too small to count (Fig. 4A). On day 10, colonies were increased in size and allowed to continue to day 12. By day 12, all cultures of the CD36E cells and CD36+ EPCs in the MethoCult GF+ or the supplemented MethoCult had <20 erythroid colonies/well, with no significant difference in the number of colonies in each culture (Fig. 4A). However, the CD36+ EPCs in the MethoCult GF+ appeared more granular and were decreased in size. Also not observed in cultures of the CD36E cells or CD36+ EPCs were colonies of granulocytes, macrophages, or eosinophils that can be cultured in the MethoCult GF+.
Figure 4.
Colony-forming cell assay of CD36E cells. CD36E cells were cultured in two different methylcellulose-based media: the MethoCult GF+ for various kinds of hematopoietic colony formation; the supplemented MethoCult for erythroid colony formation. (A) Using a light microscope at 200× magnification, colonies were evaluated on days 7 and 12, but colonies from CD36+ EPCs cultured in the MethoCult GF+ were evaluated on day 10, instead of day 12, due to overgrowth. (B) On day 12, individual colony cultures were harvested, cytocentrifuged onto glass slides, and stained with Wright-Giemsa or benzidine followed by microscopy. Scale bars: 10 μm.
To examine the morphology and hemoglobin production of colony-forming cells, all 12-day colonies in individual wells cultured in the MethoCult GF+ or the supplemented MethoCult were stained with Wright-Giemsa or benzidine (Fig. 4B). Wright-Giemsa staining showed that colonies derived from CD36+ EPCs in either culture medium contained differentiated erythroid cells in addition to undifferentiated cells. Small erythroid cells were also observed in specimens from the CD36E-derived colonies in either culture medium, while a relatively high number of undifferentiated CD36E cells (large in size) were also seen. To check hemoglobin production of the cells, benzidine staining was performed, confirming that colonies from the CD36E cells and CD36+ EPCs contained hemoglobinpositive cells. Collectively, the CD36E cells were capable of erythroid colony formation in semi-solid culture with EPO, but not other types of hematopoietic colonies. This was highly conceivable because the parental CD36+ EPCs were erythroid progenitors.
CD36E cells are growth factor–dependent
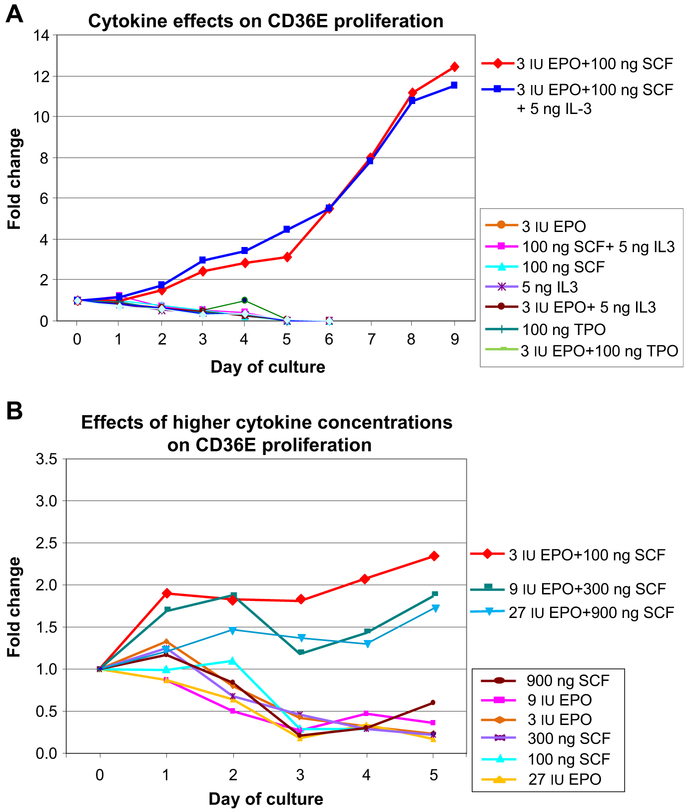
Effects of cytokines on the survival and proliferation of CD36E cells were assessed by culture in various conditions where cytokines, alone or in combination, were excluded from the original expansion medium (3 IU/mL EPO + 100 ng/mL SCF + 5 ng/mL IL-3). In addition, an effect of thrombopoietin (100 ng/mL) was also examined. CD36E cells proliferated continuously in the presence of EPO and SCF with or without IL-3, indicating that IL-3 was not required for growth (Fig. 5A and Supplementary Table E1; online only, available at www.exphem.org). CD36E cells cultured in any other conditions showed decreased proliferation and fewer viable cells, indicating both EPO and SCF were indispensable for CD36E growth. Thrombopoietin (100 ng/mL) appeared to have no enhancing effect on the cells. We next examined whether increased concentrations of EPO and/or SCF improved CD36E cell proliferation (Fig. 5B and Supplementary Table E2; online only, available at www.exphem.org). Higher concentrations (three- and nine-fold) of EPO plus SCF inhibited cell proliferation. Removal of a specific cytokine or higher cytokine concentrations appeared to have no stimulating effects on cell differentiation.
Figure 5.
Effects of cytokines on CD36E proliferation. (A) CD36E cells were cultured under various conditions in which cytokines alone or in combination were excluded from the original serum-free expansion medium or replaced by 100 ng/mL thrombopoietin. These cultures were compared with cells grown in the standard cytokine cocktail (3 IU/mL EPO + 100 ng/mL SCF + 5 ng/mL IL-3). Cell proliferation was measured at indicated time points in triplicate by Trypan blue staining. Indicated cytokine amounts, 3 IU, 100 ng, and 5 ng, are per mL. (B) CD36E cells were grown in serum-free expansion medium supplemented with higher concentrations (three- or nine-fold) of EPO and/or SCF alone or in combination. Because IL-3 had no effect on CD36E proliferation, the cytokine was kept at a constant 5 ng/mL in all conditions. The cultures were compared with the standard cytokine concentrations of 3 IU/mL EPO and 100 ng/mL SCF. Cell proliferation was monitored on various days in triplicate culture conditions.
To examine whether hemin, DMSO, or PMA induced CD36E differentiation, cells were cultured with each of these reagents at two different concentrations as described in previous studies [21–26]. The treated cells were subjected to measurement of cell proliferation, microscopic analysis, and immunoblotting (Supplementary Figure E1 and Supplementary Table E3; online only, available at www.exphem.org). From these results, it appears that the three reagents had no inducible effects on CD36E differentiation.
Gene expression profiling of CD36E cells using real-time RT-PCR arrays
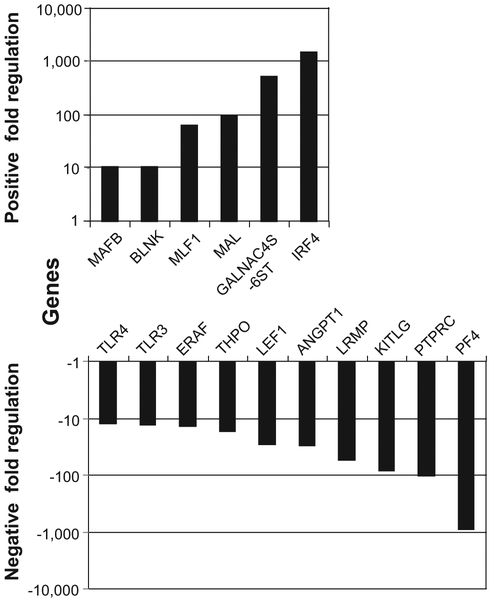
We carried out real-time RT-PCR array analysis to investigate a gene expression profile of CD36E cells compared with that of parental CD36+ EPCs. The CD36E cells expressed a number of genes with greater differences from those in the CD36+ EPCs (Fig. 6 and Table 2). The following genes were expressed with > 50-fold differences: IRF4 (1,450-fold), GALNAC4S-6ST (520-fold), FAM3B (138-fold), MAL (100-fold), and MLF1 (63-fold) were upregulated; PF4 (869-fold), PTPRC (100-fold), KITLG (82-fold), and LRMP (54-fold) were downregulated. Among these, IRF4, GALNAC4S-6ST, MAL, PTPRC, and LRMP are lymphoid-relating factors. As for lymphocyte-relating factors with < 50-fold differences, BLNK and CD86 molecules were considerably upregulated, while LEF1, TLR3, TLR4, and IRF8 were highly downregulated. Among erythroid-relating factors, ERAF expression was fairly decreased (13-fold) in the CD36E cells compared with the CD36+ EPCs. Within the interleukin group, expression levels of several factors, including IL-1A, IL-4, IL-6, IL-10, IL-11, IL-12A, IL-13, and IL-15, were highly elevated.
Figure 6.
Gene expression profiling of CD36E cells using the real-time RT-PCR arrays. Total RNA was isolated from CD36E cells using the Qiagen microRNA Extraction kit and converted to complementary DNA, followed by real-time RT-PCR assay using the Custom PCR array targeting hematopoietic genes and the modified-cataloged cytokine array. Fold changes represent ratios of gene expression levels with fivefold or greater differences between the CD36E cells and the parental CD36+ EPCs.
Table 2.
Up- and downregulation of selected genes analyzed by realtime RT-PCR
| Gene symbol | Fold changea | Description |
|---|---|---|
| IRF4 | 1,449.5 | Interferon regulatory factor 4 |
| GALNAC4S-6ST | 520.2 | B-cell RAG associated protein |
| FAM3B | 137.5 | Family with sequence similarity 3, member B |
| MAL | 99.6 | Mal, T-cell differentiation protein |
| MLF1 | 63.1 | Myeloid leukemia factor 1 |
| GDF2 | 38.2 | Growth differentiation factor 2 |
| TGFB2 | 28.8 | Transforming growth factor, β2 |
| BLNK | 10.7 | B-cell linker |
| MAFB | 10.6 | V-maf musculoaponeurotic fibrosarcoma oncogene homolog B |
| IL12A | 10.6 | Interleukin 12A |
| IL15 | 9.2 | Interleukin 15 |
| IL4 | 9.1 | Interleukin 4 |
| HDAC9 | 9.0 | Histone deacetylase 9 |
| IL6 | 8.1 | Interleukin 6 (interferon, β2) |
| CD86 | 8.1 | CD86 molecule |
| IL13 | 7.4 | Interleukin 13 |
| IL1A | 7.0 | Interleukin 1, α |
| IL10 | 6.9 | Interleukin 10 |
| TNF | 6.4 | Tumor necrosis factor (TNF superfamily, member 2) |
| GDF9 | 5.7 | Growth differentiation factor 9 |
| IL11 | 5.7 | Interleukin 11 |
| PDGFA | 5.6 | Platelet-derived growth factor α polypeptide |
| BMP1 | 5.5 | Bone morphogenetic protein 1 |
| CD34 | −5.7 | CD34 molecule |
| LMO2 | −6.0 | LIM domain only 2 (rhombotin-like 1) |
| ETV6 | −7.9 | Ets variant gene 6 (TEL oncogene) |
| PECAM1 | −8.2 | Platelet/endothelial cell adhesion molecule (CD31 antigen) |
| IRF8 | −8.6 | Interferon regulatory factor 8 |
| MAP4K1 | −9.8 | Mitogen-activated protein kinase kinase kinase kinase 1 |
| TLR4 | −12.3 | Toll-like receptor 4 |
| TLR3 | −12.9 | Toll-like receptor 3 |
| ERAF | −13.2 | Erythroid associated factor |
| THPO | −16.7 | Thrombopoietin |
| LEF1 | −28.1 | Lymphoid enhancer-binding factor 1 |
| ANGPT1 | −29.8 | Angiopoietin 1 |
| LRMP | −53.7 | Lymphoid-restricted membrane protein |
| KITLG | −81.8 | KIT ligand |
| PTPRC | −99.5 | Protein tyrosine phosphatase, receptor type, C |
| PF4 | −868.8 | Platelet factor 4 (chemokine (C-X-C motif) ligand 4) |
More than fivefold expression changes.
Discussion
Some reports have demonstrated that stable expression of hTERT, a catalytic subunit of telomerase, is sufficient to extend the lifespans of various human cells [27,28]. However, other reports have described that several types of cells, including corneal keratinocytes [29] and mammary epithelial cells [30], require additional genetic hits to achieve immortality, such as viral oncogenes or dominant negative-acting transgenes either alone or in combination with hTERT expression. Three viral oncogenes, SV40T, HPV16 E6, and HPV16 E7, are well-documented for cellular immortalization and transformation properties and widely used to establish cell lines [31,32]. SV40T binds to and inactivates Rb and p53 tumor suppressor proteins. Similarly, HPV16 E6 is involved in targeted degradation of p53 and thereby facilitates cell proliferation. HPV16 E7 binds to Rb by displacing the E2F transcription factor, resulting in the activation of E2F and subsequently cell-cycle progression.
In this work, we successfully generated immortalized cells from the CD36+ EPCs by introducing HPV16 E6/E7 alone or HPV16 E6/E7 plus hTERT, but failed to immortalize the cells by transducing with SV40T alone, hTERT alone, or SV40T plus hTERT. Similarly, a previous work that used CD34+ stem/progenitor cells from human cord blood (CB) has reported the immortalization and transformation of the cells by transduction with lentiviral vectors containing HPV16 E6/E7 alone or in concert with hTERT, but not with hTERT alone [20]. Considering this work in addition to ours, hTERT transduction alone appears to be insufficient for the immortalization of hematopoietic stem/progenitor cells, and additional elements, such as the viral HPV16 E6/E7 oncoproteins, are required for successful transformation.
Zhang et al. [33] serially examined cytogenetic events of HPV16 E6/E7-immortalized esophageal epithelial cells; there was a progressive increase of chromosomal aberrations during the early stage of crisis; in the later stage, the karyotypes were more convergent with an increased karyotypic complexity. It is generally suggested that most of the chromosomal rearrangements are randomly generated during the early stage of the crisis (a state characterized by genomic instability, cell death, and the eventual emergence of rare replicatively immortal variants) [34–37], and some chromosomal aberrations may accelerate cellular proliferation, whereby cells that acquired these aberrations can overcome the crisis. The serial steps may contribute to transforming the CD36+ EPCs cells to establish the CD36E cells. In fact, the CD36E cells possessed various chromosomal abnormalities and aneuploidy, although they were still dependent on EPO and retained other features of erythroid progenitors. As for other hematopoietic cell lines, various cell lines derived from leukemic patients also display chromosomal aberrations with aneuploidy and a number of translocations; for example, the modal chromosome numbers are 54 in the TF-1 erythroleukemic cells [10]; 45 in the MB-02 megakaryoblastic cells [13]; and 92 in the UT7 megakaryocytic cells, a parental cell to the UT7/EPO cells [15]. In the HPV16 E6/E7- or HPV16 E6/E7 plus hTERT-transduced cell lines derived from human CB CD34+ cells, numerous chromosomal alterations are also observed [20], but with much less than those seen in the CD36E cells. Because the adult CD36+ EPCs are more mature and differentiated compared with the CB CD34+ cells, it is highly possible that the CB CD34+ cells may have great plasticity intracellularly, allowing immortalization without radical changes of chromosomal structure and number. In addition to the hematopoietic cell lines, chromosomal abnormalities are consistently observed in other cell types transformed with the HPV16 E6/E7 genes [38], which display progressively increased chromosomal aberrations during the early stage of cellular crisis. Among the E6/E7-transduced cells, the nonrandom occurrence of chromosome-20 gain or chromosome-20q amplification has been frequently reported in immortalized epithelial lineage cells [33,39]. Similarly, trisomy of chromosome 20 has been described in one of the cell lines derived from the HPV16 E6/E plus hTERT-transduced CB CD34+ cells [20]. However, this phenomenon was not seen in the CD36E cells.
One of the remarkable characteristics of the CD36E cells was the ability to produce hemoglobin with ~27% of benzidine-positive cells, a higher percentage than the parental CD36+ EPCs. Compared with the CD36E cells, other cell lines had a lower ratio of benzidine-positive cells in their normal culture conditions. The JK-1 cell line derived from a chronic myelogenous leukemia patient is shown to have the highest number of benzidine-positive cells (~20%) [8]. The UT7/EPO cells, which are committed more to erythroid lineage than the UT-7 cells, have 4% benzidine-positive cells [15]. K562 [40], KMOE [9], and HEL [41] have <1% to 5% of benzidine-positive cells in normal culture condition.
There are various hematopoietic cell lines with erythroid features derived from leukemic patients, most of which have immunoprofiles typical of immature erythroid cells. Most cell lines, including K562, KMOE, and HEL, grow autonomously. Other cell lines are constitutively cytokine-dependent; mainly on granulocyte macrophage colony-stimulating factor or IL-3 like UT7, TF-1, and MB-02; strictly on EPO like UT7/EPO. Compared with these cell lines, the CD36E cells appear distinctive due to the strict dependency on both EPO and SCF for viability and proliferation. However, higher concentrations (three- and nine-fold) of EPO plus SCF inhibited cell growth, probably due to selection or adaptation of the cells during culture at the original cytokine concentrations.
EPO is uniquely associated with erythroid lineage differentiation. As reported previously, EPO plays a role as a differentiation inducer to promote erythropoiesis for some hematopoietic cell lines, such as UT7, TF-1 [42], M-TAT [43], MB-02 [13], and NS-Meg [44]. In the case of the CD36E cells, there were no apparent signs of differentiation induction with EPO, even at a ninefold higher concentration, alone or in combination with SCF. As the CD36E cells evolved from the culture supplemented with EPO, SCF, and IL-3, it is highly plausible that the main actions of EPO for the cells are to promote proliferation and to maintain viability, but not to induce differentiation.
Hemin, DMSO, and PMA are chemicals known to induce differentiation of various hematopoietic cell lines. CD36E cells treated with hemin, PMA, or DMSO did not exhibit the desired result of differentiation as reported with some erythroleukemic cell lines, such as K562 [22,25], HEL [25] UT7 [15], U937 [24], and HL-60 [24]. DMSO rather negatively affected cell viability.
In cell lines derived from patients with hematological malignancies, acquired elevation of hemoglobin (Hb) F has been reported with or without induction by measuring HbF or γ-globin chain expression: for example, HbF is a major hemoglobin subtype in UT7/EPO [15] and JK-1 [8]; HbF is induced in K562 [45]. In the CD36E cells, enhanced γ-chain expression concomitant with reduced β-chain expression was observed, whereas α- and β-chains were predominant in the CD36+ EPCs with only a trace amount of γ-chain. The results indicate that switching of HbA to HbF occurred during the establishment of the CD36E cells from the CD36+ EPCs. Previous studies have reported that an increased HbF production was observed in hemin-treated cells, such as primary erythroid progenitors [45] and K562 cells [45,46]. However, treatment of the CD36E cells with hemin or PMA had no clear effect on HbF production when evaluated by γ-chain expression.
Consistent with the parental CD36+ EPCs, lymphoid-specific markers, CD2, CD3, CD10, and CD19, were undetectable on the surface of the CD36E cell. Nevertheless, when measured by RT-PCR array analysis, mRNA expression levels of some lymphoid-relating factors, such as IRF4, GALNAC4S-6ST, and MAL, were extremely elevated compared with those of the parental CD36+ EPCs, although mRNA levels of other lymphoid factors such as PTPRC, LRMP, and LEF1 were markedly decreased. Of note, IRF4 expression was dramatically activated in the CD36E cells (> 1,000-fold). IRF4, which is primarily expressed in B cells and activated T cells, is an important regulator of B-cell development and inducible by antigen stimulation. Overexpression of IRF4 may facilitate the CD36E proliferation because IRF4 was reported as a master regulator of the malignancy-specific gene expressions in multiple myelomas in which IRF4 constitutes a positive autoregulatory loop with MYC [47]. The MAL (CD88 adaptor-like) protein expression level was also significantly elevated (100-fold) in the CD36E cells, similar to T- and B-lineage malignant cells [48,49] in which overexpression of MAL have been described. MAL is expressed highly in thymocytes (the CD4+ cells > CD8+ T cells) and acts to recruit MyD88 to TRL4, resulting in activation of the signaling pathway. As the TLR4 expression was fairly downregulated (12-fold) in the CD36E cells, MAL may act in a different manner in the CD36E cells. It is unclear whether the skewed expressions of lymphoid lineage markers in the CD36E cells were due to the HPV16 E6/E7 expression or they were randomly acquired by the CD36E cells over the course of chromosomal rearrangements. Previously, Gubin et al. has reported the gene expression profile of normal human erythroid cells sorted with anti-CD71 antibody using a method involving suppression subtractive hybridization [50]. Further, two groups have demonstrated gene expression profiling of a growth factor-dependent erythroblast cell line derived from fetal liver of the p53-deficient mouse using the Atlas complementary DNA arrays or custom-made hematopoietic microarrays [51,52]. Here we examined a gene expression profile of the CD36E cells using the custom real-time PCR array targeting 92 hematopoietic genes and compared it with that of the parental CD36+ EPCs. Thus, due to the differences of cell origins and experimental conditions between their experiments and ours, we were unable to directly compare gene expression profiles of the CD36E cells with those of others. However, as shown in Table 1, surface proteins, CD36, CD71, and glycophorin A (CD235a) were detected in CD36E cells, consistent with data described by Gubin et al.
To the best of our knowledge, the CD36E cells are the first cell line established from primary CD36+ EPCs. The cell line would create promising opportunities for various studies using EPCs. We would like to freely distribute the cell line upon request, or better yet, deposit the cells into a public repository, such as ATCC.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH) Intramural Research Program (Bethesda, MD, USA). We thank Andre Larochelle, Hematology Branch, National Heart, Lung and Blood Institute (NHLBI), NIH for his helpful discussion. We also thank the following people and facilities for their technical support: Leigh Samsel and Phil McCoy at the NHLBI Flow Cytometry Core Facility, NIH; Sandra Burkett at National Cancer Institute Frederick SKY facility, NIH (Frederick, MD, USA); Bey-Dih Chang and Thomas Primiano at CDI Technologies, Madison, WI, USA; and Rick Dreyfuss, Medical Arts Photography Branch, NIH (Bethesda, MD, USA).
Footnotes
Conflict of Interest Disclosure
No authors of this article have any commercial, proprietary, or financial interest in the products or companies described in this work.
References
- 1.Febbraio M, Hajjar DP, Silverstein RL. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J Clin Invest. 2001;108:785–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okumura N, Tsuji K, Nakahata T. Changes in cell surface antigen expressions during proliferation and differentiation of human erythroid progenitors. Blood. 1992;80:642–650. [PubMed] [Google Scholar]
- 3.Wong S, Zhi N, Filippone C, et al. Ex vivo-generated CD36+ erythroid progenitors are highly permissive to human parvovirus B19 replication. J Virol. 2008;82:2470–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Wolf JT, Muller EW, Hendriks DH, Halie RM, Vellenga E. Mast cell growth factor modulates CD36 antigen expression on erythroid progenitors from human bone marrow and peripheral blood associated with ongoing differentiation. Blood. 1994;84:59–64. [PubMed] [Google Scholar]
- 5.Edelman P, Vinci G, Villeval JL, et al. A monoclonal antibody against an erythrocyte ontogenic antigen identifies fetal and adult erythroid progenitors. Blood. 1986;67:56–63. [PubMed] [Google Scholar]
- 6.Lozzio CB, Lozzio BB. Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome. Blood. 1975;45:321–334. [PubMed] [Google Scholar]
- 7.Kishi K A new leukemia cell line with Philadelphia chromosome characterized as basophil precursors. Leuk Res. 1985;9:381–390. [DOI] [PubMed] [Google Scholar]
- 8.Okuno Y, Suzuki A, Ichiba S, et al. Establishment of an erythroid cell line (JK-1) that spontaneously differentiates to red cells. Cancer. 1990;66: 1544–1551. [DOI] [PubMed] [Google Scholar]
- 9.Okano H, Okamura J, Yagawa K, Tasaka H, Motomura S. Human erythroid cell lines derived from a patient with acute erythremia. J Cancer Res Clin Oncol. 1981;102:49–55. [DOI] [PubMed] [Google Scholar]
- 10.Kitamura T, Tange T, Terasawa T, et al. Establishment and characterization of a unique human cell line that proliferates dependently on GM-CSF, IL-3, or erythropoietin. J Cell Physiol. 1989;140:323–334. [DOI] [PubMed] [Google Scholar]
- 11.Martin P, Papayannopoulou T. HEL cells: a new human erythroleukemia cell line with spontaneous and induced globin expression. Science. 1982;216:1233–1235. [DOI] [PubMed] [Google Scholar]
- 12.Bonsi L, Grossi A, Strippoli P, et al. An erythroid and megakaryocytic common precursor cell line (B1647) expressing both c-mpl and erythropoietin receptor (Epo-R) proliferates and modifies globin chain synthesis in response to megakaryocyte growth and development factor (MGDF) but not to erythropoietin (Epo). Br J Haematol. 1997;98:549–559. [DOI] [PubMed] [Google Scholar]
- 13.Morgan DA, Gumucio DL, Brodsky I. Granulocyte-macrophage colony-stimulating factor-dependent growth and erythropoietin-induced differentiation of a human cell line MB-02. Blood. 1991;78:2860–2871. [PubMed] [Google Scholar]
- 14.Avanzi GC, Lista P, Giovinazzo B, et al. Selective growth response to IL-3 of a human leukaemic cell line with megakaryoblastic features. Br J Haematol. 1988;69:359–366. [DOI] [PubMed] [Google Scholar]
- 15.Komatsu N, Yamamoto M, Fujita H, et al. Establishment and characterization of an erythropoietin-dependent subline, UT-7/Epo, derived from human leukemia cell line, UT-7. Blood. 1993;82:456–464. [PubMed] [Google Scholar]
- 16.Freyssinier JM, Lecoq-Lafon C, Amsellem S, et al. Purification, amplification and characterization of a population of human erythroid progenitors. Br J Haematol. 1999;106:912–922. [DOI] [PubMed] [Google Scholar]
- 17.Giarratana MC, Kobari L, Lapillonne H, et al. Ex vivo generation of fully mature human red blood cells from hematopoietic stem cells. Nat Biotechnol. 2005;23:69–74. [DOI] [PubMed] [Google Scholar]
- 18.Wong S, Brown KE. Development of an improved method of detection of infectious parvovirus B19. J Clin Virol. 2006;35:407–413. [DOI] [PubMed] [Google Scholar]
- 19.Vousden KH, Doniger J, DiPaolo JA, Lowy DR. The E7 open reading frame of human papillomavirus type 16 encodes a transforming gene. Oncogene Res. 1988;3:167–175. [PubMed] [Google Scholar]
- 20.Akimov SS, Ramezani A, Hawley TS, Hawley RG. Bypass of senescence, immortalization, and transformation of human hematopoietic progenitor cells. Stem Cells. 2005;23:1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honma Y, Onozuka Y, Okabe-Kado J, Kasukabe T, Hozumi M. Hemin enhances the sensitivity of erythroleukemia cells to 1-beta-D-arabinofuranosylcytosine by both activation of deoxycytidine kinase and reduction of cytidine deaminase activity. Cancer Res. 1991;51:4535–4538. [PubMed] [Google Scholar]
- 22.Baliga BS, Mankad M, Shah AK, Mankad VN. Mechanism of differentiation of human erythroleukaemic cell line K562 by hemin. Cell Prolif. 1993;26:519–529. [DOI] [PubMed] [Google Scholar]
- 23.Craston R, Koh M, McDermott A, Ray N, Prentice HG, Lowdell MW. Temporal dynamics of CD69 expression on lymphoid cells. J Immunol Methods. 1997;209:37–45. [DOI] [PubMed] [Google Scholar]
- 24.Depraetere S, Vanhaesebroeck B, Fiers W, Willems J, Joniau M. Polar agents with differentiation inducing capacity potentiate tumor necrosis factor-mediated cytotoxicity in human myeloid cell lines. J Leukoc Biol. 1995;57:141–151. [DOI] [PubMed] [Google Scholar]
- 25.Drexler HG, Gaedicke G, Minowada J. Erythroleukemia cell lines HEL and K-562: changes in isoenzyme profiles and morphology during induction of differentiation. Hematol Oncol. 1986;4:163–174. [DOI] [PubMed] [Google Scholar]
- 26.Komatsu N, Fujita H. Induced megakaryocytic maturation of the human leukemia cell line UT-7 results in down-modulation of erythropoietin receptor gene expression. Cancer Res. 1993;53:1156–1161. [PubMed] [Google Scholar]
- 27.Bodnar AG, Ouellette M, Frolkis M, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998; 279:349–352. [DOI] [PubMed] [Google Scholar]
- 28.Wieser M, Stadler G, Jennings P, et al. hTERT alone immortalizes epithelial cells of renal proximal tubules without changing their functional characteristics. Am J Physiol Renal Physiol. 2008;295:F1365–F1375. [DOI] [PubMed] [Google Scholar]
- 29.Pellegrini G, Dellambra E, Paterna P, et al. Telomerase activity is sufficient to bypass replicative senescence in human limbal and conjunctival but not corneal keratinocytes. Eur J Cell Biol. 2004;83: 691–700. [DOI] [PubMed] [Google Scholar]
- 30.Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. [DOI] [PubMed] [Google Scholar]
- 31.Wise-Draper TM, Wells SI. Papillomavirus E6 and E7 proteins and their cellular targets. Front Biosci. 2008;13:1003–1017. [DOI] [PubMed] [Google Scholar]
- 32.Jha KK, Banga S, Palejwala V, Ozer HL. SV40-Mediated immortalization. Exp Cell Res. 1998;245:1–7. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Jin Y, Chen X, et al. Cytogenetic aberrations in immortalization of esophageal epithelial cells. Cancer Genet Cytogenet. 2006; 165:25–35. [DOI] [PubMed] [Google Scholar]
- 34.Beausejour CM, Krtolica A, Galimi F, et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J. 2003;22:4212–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei W, Sedivy JM. Differentiation between senescence (M1) and crisis (M2) in human fibroblast cultures. Exp Cell Res. 1999;253: 519–522. [DOI] [PubMed] [Google Scholar]
- 36.Campisi J Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. [DOI] [PubMed] [Google Scholar]
- 37.Shay JW, Van Der Haegen BA, Ying Y, Wright WE. The frequency of immortalization of human fibroblasts and mammary epithelial cells transfected with SV40 large T-antigen. Exp Cell Res. 1993;209:45–52. [DOI] [PubMed] [Google Scholar]
- 38.White AE, Livanos EM, Tlsty TD. Differential disruption of genomic integrity and cell cycle regulation in normal human fibroblasts by the HPV oncoproteins. Genes Dev. 1994;8:666–677. [DOI] [PubMed] [Google Scholar]
- 39.Klingelhutz AJ, Qian Q, Phillips SL, Gourronc FA, Darbro BW, Patil SR. Amplification of the chromosome 20q region is associated with expression of HPV-16 E7 in human airway and anogenital epithelial cells. Virology. 2005;340:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang LP, Jiang JK, Tam JW, et al. Effects of Matrine on proliferation and differentiation in K-562 cells. Leuk Res. 2001;25:793–800. [DOI] [PubMed] [Google Scholar]
- 41.Hooper WC, Pruckler J, Jackson D, Evatt BL. The synergistic effect of hemin and transforming growth factor-beta on hemoglobin accumulation in HEL erythroleukemia cells. Leuk Res. 1991;15:753–758. [DOI] [PubMed] [Google Scholar]
- 42.Chretien S, Varlet P, Verdier F, et al. Erythropoietin-induced erythroid differentiation of the human erythroleukemia cell line TF-1 correlates with impaired STAT5 activation. EMBO J. 1996;15:4174–4181. [PMC free article] [PubMed] [Google Scholar]
- 43.Minegishi N, Minegishi M, Tsuchiya S, et al. Erythropoietin-dependent induction of hemoglobin synthesis in a cytokine-dependent cell line M-TAT. J Biol Chem. 1994;269:27700–27704. [PubMed] [Google Scholar]
- 44.Tsuyuoka R, Takahashi T, Suzuki A, et al. A newly established megakaryoblastic/erythroid cell line that differentiates to red cells in the presence of erythropoietin and produces platelet-like particles. Stem Cells. 1995;13:54–64. [DOI] [PubMed] [Google Scholar]
- 45.Fibach E, Kollia P, Schechter AN, Noguchi CT, Rodgers GP. Hemin-induced acceleration of hemoglobin production in immature cultured erythroid cells: preferential enhancement of fetal hemoglobin. Blood. 1995;85:2967–2974. [PubMed] [Google Scholar]
- 46.Rutherford T, Clegg JB, Higgs DR, Jones RW, Thompson J, Weatherall DJ. Embryonic erythroid differentiation in the human leukemic cell line K562. Proc Natl Acad Sci U S A. 1981;78:348–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaffer AL, Emre NC, Lamy L, et al. IRF4 addiction in multiple myeloma. Nature. 2008;454:226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsi ED, Sup SJ, Alemany C, et al. MAL is expressed in a subset of Hodgkin lymphoma and identifies a population of patients with poor prognosis. Am J Clin Pathol. 2006;125:776–782. [DOI] [PubMed] [Google Scholar]
- 49.Copie-Bergman C, Plonquet A, Alonso MA, et al. MAL expression in lymphoid cells: further evidence for MAL as a distinct molecular marker of primary mediastinal large B-cell lymphomas. Mod Pathol. 2002;15:1172–1180. [DOI] [PubMed] [Google Scholar]
- 50.Gubin AN, Njoroge JM, Bouffard GG, Miller JL. Gene expression in proliferating human erythroid cells. Genomics. 1999;59:168–177. [DOI] [PubMed] [Google Scholar]
- 51.Dolznig H, Boulme F, Stangl K, et al. Establishment of normal, terminally differentiating mouse erythroid progenitors: molecular characterization by cDNA arrays. FASEB J. 2001;15:1442–1444. [DOI] [PubMed] [Google Scholar]
- 52.Kolbus A, Blazquez-Domingo M, Carotta S, et al. Cooperative signaling between cytokine receptors and the glucocorticoid receptor in the expansion of erythroid progenitors: molecular analysis by expression profiling. Blood. 2003;102:3136–3146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.