Abstract
Osteosarcoma (OS) is the most common primary malignant bone tumor. Numerous studies have strongly implicated the ectopic expression of microRNAs (miRNAs/miRs), including miR-885-5p, which is aberrantly expressed in several cancer types, in multiple cancer-related processes. However, the role of miR-885-5p in OS remains unknown. In the present study, it was found that the expression of miR-885-5p was markedly upregulated in OS cell lines and clinical tissues. Moreover, high expression of miR-885-5p was significantly associated with the development of OS. The human OS MG-63 cell line was transfected with recombinant lentivirus to regulate miR-885-5p expression. Overexpressed miR-885-5p significantly promoted the proliferation and migration of MG-63 cells in vitro, while downregulating miR-885-5p expression reversed these effects. Furthermore, bioinformatic analysis was used to predict the potential target genes of miR-885-5p, and cell division cycle protein 73 homolog (CDC73) was identified as a novel and direct target of miR-885-5p. This interaction was further confirmed using reverse transcription-quantitative polymerase chain reaction, western blotting and luciferase activity assays. These findings suggest that miR-885-5p serves a critical role in facilitating OS proliferation and migration, and can regulate CDC73 expression in OS cells and tissues. Thus, miR-885-5p could be a promising novel therapeutic biomarker for OS.
Keywords: osteosarcoma, microRNA-885-5p, proliferation, migration, cell division cycle protein 73 homolog
Introduction
Osteosarcoma (OS), the most frequently occurring primary malignant bone tumor in children and adolescents, normally occurs in the metaphysis of the long bones and is characterized by a high level of relapse and a poor prognosis (1). The estimated worldwide annual incidence of OS is ~4 million cases, with the highest prevalence occurring in adolescents between the ages of 15 and 19 years (2). Despite the combination of adjuvant chemotherapy and advanced surgery, the cumulative 5-year survival rate of patients is only 55–68, and ~80% of patients eventually develop metastatic disease and face a poor outcome (3,4). Therefore, it is imperative to investigate the potential molecular mechanisms of OS progression and seek an effective molecular therapy for the treatment of the disease.
MicroRNAs (miRNAs/miRs) are a small class of endogenous non-coding RNAs that range in length from 18 to 25 nucleotides. miRNAs regulate gene expression at the post-transcriptional level by binding to partially complementary sites located in the 3′-untranslated regions (3′-UTR) of target messenger RNAs (mRNAs) (5–7). In cancer, numerous miRNAs locate within or close to fragile chromosomal sites that are often lost or amplified (8). Increasing evidence has suggested that miRNAs serve a crucial role in specific cellular processes, including differentiation, morphogenesis and tumorigenesis. Thus, these key effects demonstrate that miRNAs may contribute to the regulation of the pathological development of numerous diseases, particularly in human cancer (9,10). Furthermore, in OS, aberrant expression of miRNA has been demonstrated to contribute towards the development and metastasis of the cancer by promoting the expression of oncogenes or by inhibiting tumor suppressor genes.
It was recently reported that miR-885-5p is aberrantly upregulated in the liver metastases of colorectal cancer, renal cell carcinoma and Hürthle cell thyroid carcinoma (11–13). By contrast, another study reported that the expression of miR-885-5p is negatively correlated with the malignant progression of human hepatocellular carcinoma (HCC) tissue samples and cell lines via targeting of the catenin β1 gene, leading to decreased activity of the Wnt/β-catenin signaling pathway (14). Recently, we searched for GEO databases regarding miR-885-5p expression and one dataset, GSE65071, indicated that miR-885-5p was aberrantly expressed in the plasma of patients with OS compared with that in normal individuals (15). However, the biological role of miR-885-5p and its molecular mechanism in OS remains unknown.
In the present study, the expression of miR-885-5p in OS cells and tissues was investigated, and the biological behavior of miR-885-5p in OS cells was observed. In addition, target genes were predicted by bioinformatics analysis to clarify the role of miR-885-5p in the development of osteosarcoma.
Materials and methods
Human tissue
Paired human OS tissues (prior to the administration of neoadjuvant chemotherapy) and matched adjacent normal tissues were surgically acquired from patients (mean age, 22.19 years) at the Department of Orthopedics, First Affiliated Hospital of Fujian Medical University (Fuzhou, Fujian, China) between September 2015 and August 2017. The distance between the normal tissue and the tumor was ~5 cm. Following surgical removal, all the tissues were snap-frozen in liquid nitrogen and stored at −80°C until further use. All clinical samples were collected with written informed consent from the patients, and the study was approved by the Ethical Committee of the First Affiliated Hospital of Fujian Medical University.
Cell culture
The normal human osteoblastic hFOB1.19 cell line, and the human OS MG-63 and U2OS cell lines (Shanghai Cell Bank, Chinese Academy of Sciences, Shanghai, China) were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc. Waltham, MA, USA) with 10% fetal bovine serum (FBS; Hyclone; GE Healthcare Life Sciences, Logan, UT, USA), 100 U/ml penicillin and 100 mg/ml streptomycin in a humidified incubator with 5% CO2 at 37°C.
Transfection and generation of stable cell lines
Recombinant lentivirus containing miR-885-5p mimic (miR-885-5p-up; 5′-UCCAUUACACUACCCUGCCUCUAGGCAGGGUAGUGUAAUGGAUU-3′), miR-885-5p inhibitor (miR-885-5p-down; 5-AGAGGCAGGGUAGUGUAAUGGA-3′) and negative control (NC; 5′-CAGUACUUUGUGUAGUACAA-3′) were synthesized by GenePharma (Shanghai, China) to generate either upregulated or downregulated expression of miR-885-5p. The MG-63 cell line was selected for following experiments as it has previously been used to confirm a correlation between OS and miRNA, and MG-63 cells are easily transfected for stable expression of miRNA (16). For transfection, 20 nM miRNA were transfected into MG-63 cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
To generate stable cell lines with miR-885-5p-up/down, a promoter-driven green fluorescent protein (GFP) reporter was used to detect the transfection efficiency. The cells were selected with 0.5 µg/ml puromycin for 2 weeks. When the GFP expression was observed up to 80%, MG-63 cells stably expressing miR-885-5p-up, miR-885-5p-down and NC were used in the following cell experiments.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from frozen tissues and cultured cell lines using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The concentration and purity of total RNA were identified by ultraviolet spectrophotometry and gel-electrophoresis, and reverse transcribed into cDNA using the PrimeScript reverse transcription kit (Takara Bio, Inc., Otsu, Japan). RT-qPCR was performed using SYBR Green Supermix (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and analyzed on an ABI 7500 thermocycler (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: 95°C for 5 min, 45 cycles of 95°C for 15 sec and 60°C for 45 sec. The relative expression levels of miR-885-5p (forward, 5′-GTCCATTACACTACCCTGCCTC-3′ and reverse, 5′-CGCGAGCACAGAATTAATACG-3′) were evaluated and normalized to that of U6 (forward, 5′-CTCGTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′) using the 2−ΔΔCq method (17).
3-(4,5-dimethyl-2-thiazol)-2,5-diphenyl-2-H-tetrazolium bro-mide (MTT) and colony formation assays
The MTT assay was used to evaluate cell proliferation, according to the manufacturer's protocols. Transfected MG-63 cells (five replicates) were plated at a density of 5×103 cells per well in 96-well culture plates for 24 h. Subsequent to 24, 48 or 72 h of incubation, 20 µl MTT (5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added to each well for an additional incubation period of 4 h. Subsequently, the MTT solution was discarded and 150 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) was added into each well and the plate was agitated gently for 10 min. The absorbance was measured using a microplate reader SpectraMax i3× (Molecular Devices, LLC, Sunnyvale, CA, USA) at a wavelength of 490 nm.
For the colony formation assay, cells were seeded in a 12-well plate at 500 cells per well (three replicates). Subsequent to 2 weeks of incubation at 37°C the colonies were fixed with methanol for 30 min at room temperature and stained with 0.5% crystal violet for 30 min at room temperature. Each experiment was repeated five times.
Transwell migration assay
Migration assays were performed in triplicate using a 24-well Transwell chamber (Costar 3422; Corning Inc., Corning, NY, USA). MG-63 cells transfected with miR-885-5p-up, miR-885-5p-down and NC, respectively, were seeded in the upper chamber at 2×104 cells per well in serum-free DMEM. DMEM supplemented with 10% FBS was added to the bottom well at a volume of 0.6 ml. Following incubation for 24 h, the cells on the top filter were gently removed with cotton swabs and cells that had migrated to the lower surface were fixed with methanol for 30 min at room temperature and stained with 0.5% crystal violet for 30 min at room temperature. The values for migration were obtained by counting three fields under a light microscope. The experiment was repeated three times over multiple days.
Luciferase activity assay
MG-63 cells were seeded in 24-well plates 1 day prior to transfection, followed by co-transfection with constructed pGL3 vectors (GenePharma) containing the wild-type 3′-UTR of CDC73 or its 3′-UTR mutant, and miR-885-5p mimic or the miRNA NC using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were harvested for 36 h after transfection and luciferase activity was measured by the Dual-Luciferase assay (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocol. The firefly luciferase activity was then normalized to the Renilla luciferase activity. Each experiment was performed in duplicate three times.
Western blotting
Total protein was isolated from cells using RIPA lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) and the protein concentrations were measured using the bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology). A total of 20 µg of protein were resolved with 10% SDS-PAGE gel, transferred to polyvinylidene difluoride membranes and blocked in 5% skimmed dry milk in Tris-buffered saline (pH 7.4), containing 0.05% Tween-20. Subsequently, the membrane was incubated with anti-CDC73 antibody (catalog no. PB0587; 1:1,000; Boster Biological Technology, Wuhan, China) overnight at 4°C and then with anti-GAPDH (catalog no. BM3876; 1:3,000; Boster Biological Technology) for 2 h at room temperature as a loading control. Signals were detected by secondary antibodies labeled with horseradish peroxidase (HRP) and were visualized using an enhanced chemiluminescent kit (Beyotime Institute of Biotechnology).
Statistical analysis
Data are presented as the mean ± standard deviation for at least three independent experiments. One-way analysis of variance or an unpaired Student's t-test was used for the statistical analysis. The association between clinicopathological features and miR-885-5p was estimated by Fisher's exact test. The association between CDC73 and miR-885p expression was explored by Pearson's correlation coefficient. P<0.05 was considered to be statistically significant. All analyses were performed using SPSS version 23.0 (IBM, Corp., Armonk, NY, USA).
Results
miR-885-5p is significantly upregulated in human OS tissues and cell lines
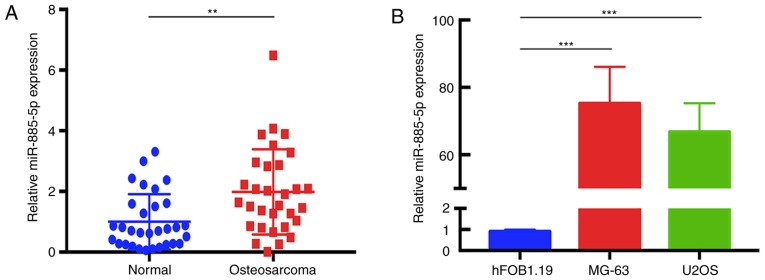
To determine the expression levels of miR-885-5p in human OS tissues, RT-qPCR was performed in 31 pairs of OS tissues compared with adjacent normal tissues, and in the human OS MG-63 and U2OS cell lines compared with the normal osteoblast hFOB1.19 cell line. The data revealed that miR-885-5p was significantly upregulated in human OS tissues compared with that in adjacent normal tissues (Fig. 1A). In addition, similar results were observed between MG-63 and hFOB1.19, and between U2OS and hFOB1.19 (Fig. 1B).
Figure 1.
Expression of miR-885-5p in OS tissues and cell lines. (A) Reverse transcription-quantitative polymerase chain reaction analysis of relative miR-885-5p expression between 31 pairs of OS tissues and adjacent normal tissues. (B) Differences in the expression levels of miR-885-5p between OS MG-63 and U2OS cell lines compared with the human normal osteoblastic hFOB1.19 cell line. U6 was used as an internal control. **P<0.01 and ***P<0.001. OS, osteosarcoma; miR, microRNA.
Upregulation of miR-885-5p is associated with advanced clinicopathological features of OS
To evaluate the clinical relevance of miR-885-5p in OS samples, the high and low expression of miR-885-5p was defined based on the median expression levels. Measuring tumor size on computed tomography, 5 cm was defined as the cut-off line (18). OS stage was determined by Enneking staging system (19). A fusion of stage IIB and stage III was to indicate the higher malignance than stage IIA (20). As shown in Table I, high miR-885-5p expression was significantly associated with tumor size (P=0.017) and clinical stage (P=0.003). No significant difference was found between the expression of miR-885-5p and patient age (P=0.458), gender (P=0.677), anatomic location (P=0.174) and metastasis (P=0.999). The Enneking stage of each sample was judged by two experienced surgeons and further confirmed by pathological results.
Table I.
Association of miR-885-5p expression with the clinicopathological features of osteosarcoma cases (n=31).
| miR-885-5p expression | ||||
|---|---|---|---|---|
| Characteristics | Number of cases | High, n | Low, n | P-value |
| Age, years | 0.458 | |||
| <18 | 15 | 9 | 6 | |
| ≥18 | 16 | 12 | 4 | |
| Gender | 0.677 | |||
| Male | 21 | 14 | 7 | |
| Female | 10 | 8 | 2 | |
| Tumor size, cm | 0.017a | |||
| <5 | 5 | 2 | 4 | |
| ≥5 | 26 | 21 | 5 | |
| Anatomical location | 0.174 | |||
| Femur | 19 | 11 | 8 | |
| Tibia | 7 | 6 | 1 | |
| Humerus | 5 | 4 | 0 | |
| Clinical stage (Enneking) | 0.003a | |||
| IIA | 8 | 2 | 6 | |
| IIB/III | 23 | 20 | 3 | |
| Metastasis | 0.999 | |||
| Yes | 11 | 8 | 3 | |
| No | 20 | 14 | 6 | |
Statistically significant P-value. miR, microRNA.
Transfection efficiency of the generated stable cell lines
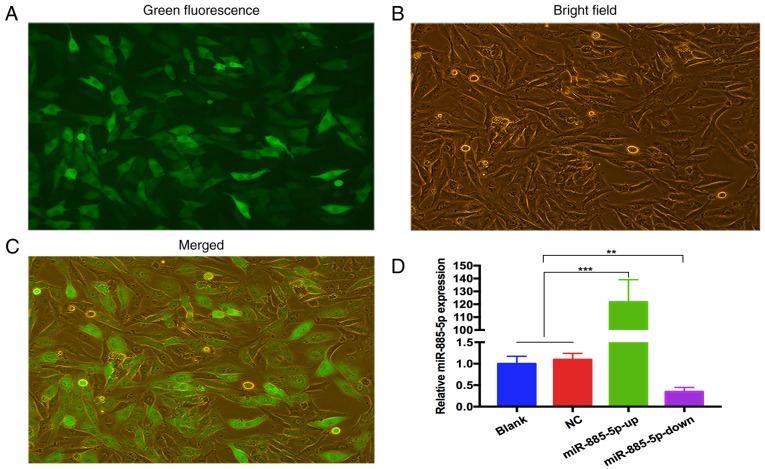
Following transfection and selection of cells, the expression of GFP-labeled oligonucleotides was detected using fluorescence microscopy (Fig. 2A-C). RT-qPCR was used to validate the transfection efficiency. The results revealed that miR-885-5p expression levels were significantly higher in the miR-885-5p-up group and lower in the miR-885-5p-down group compared with the blank and NC groups. There was no difference between the blank group and the NC groups. The data indicated that lentivirus containing miR-885-5p-up/down was successfully transfected in MG-63 cells (Fig. 2D).
Figure 2.
miR-885-5p lentivirus was transfected into MG-63 cells and the relative expression of miR-885-5p was quantified by RT-qPCR. (A) Expression of green fluorescent protein-labeled MG-63 cells was evaluated under a fluorescence inverted microscope. Magnification, ×400. (B) Observations of MG-63 cells were made using an optical microscope. Magnification, ×400. (C) The merging of (A) and (B) Magnification, ×400. (D) The histogram revealed the relative expression of miR-885-5p in the different groups as analyzed by RT-qPCR. U6 was used as an internal control. **P<0.01 and ***P<0.001. miR, microRNA; NC, negative control; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; up, mimic; down, inhibitor.
miR-885-5p promotes proliferation of OS cells
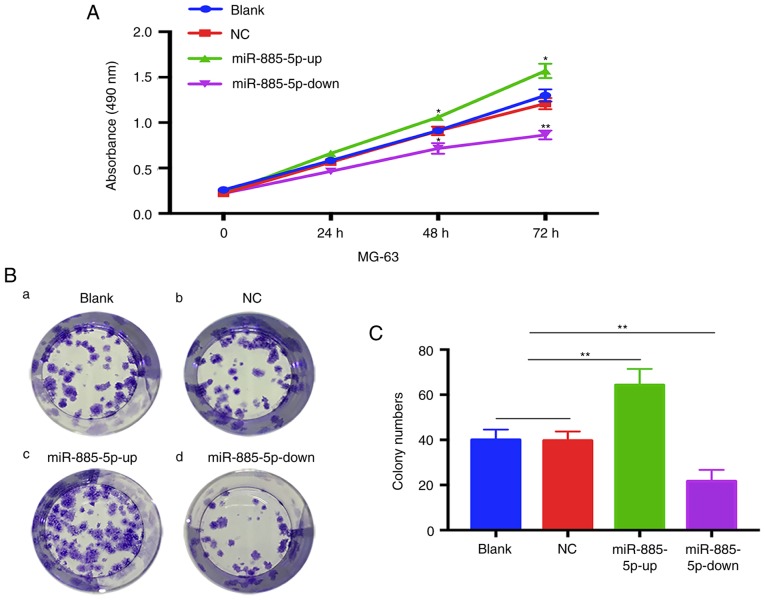
The MTT assay showed that MG-63 cells in the miR-885-5p-up group exhibited a significant increase in proliferation compared with the other three groups, while the cells in the miR-885-5p-down group exhibited a decrease in proliferation. No statistical differences were found between the blank and NC groups (Fig. 3A). The results of the colony formation assay also presented a similar trend in which the colony formation was significantly decreased in the miR-885-5p-down group of MG-63 cells compared with the blank and NC groups (Fig. 3B and C). By contrast, cells transfected with miR-885-5p-up exhibited significantly increased colony formation compared with the other groups.
Figure 3.
miR-885-5p promotes the growth of MG-63 in vitro. (A) The proliferation rate of MG-63 cells was determined by MTT assay from 1 to 3 days (*P<0.05 vs. blank). (B) Representative images of colony formation from each group. (C) Histogram showing the number of colonies formed in the different groups. n=5 independent experiments. **P<0.01. miR, microRNA; NC, negative control; up, mimic; down, inhibitor.
miR-885-5p promotes migration of OS cells
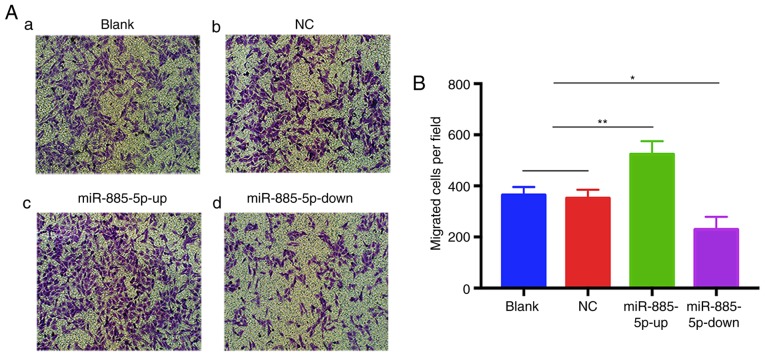
To further corroborate the effect of miR-885-5p on MG-63 cell migration, a Transwell migration assay was performed. The results revealed that the numbers of migrated cells in the blank and NC groups were not significantly different, while the miR-885-5p-up group exhibited significantly increased migration compared with the other groups. Conversely, the number of migrated cells transfected with miR-885-5p-down was significantly suppressed (Fig. 4A and B). These results indicated that miR-885-5p promotes the migration of OS cells in vitro.
Figure 4.
miR-885-5p promotes the migration of MG-63 cells in vitro. (A) A representative image of a Transwell assay was obtained for each group. The migrated cells were stained with crystal violet and observed under a light microscope. Original magnification, ×100. (B) Histogram showing the number of cells that passed through the Transwell chambers. n=3 independent experiments. *P<0.05 and **P<0.01. mi, microRNA; NC, negative control; up, mimic; down, inhibitor.
miR-885-5p directly targets and inhibits CDC73 protein expression
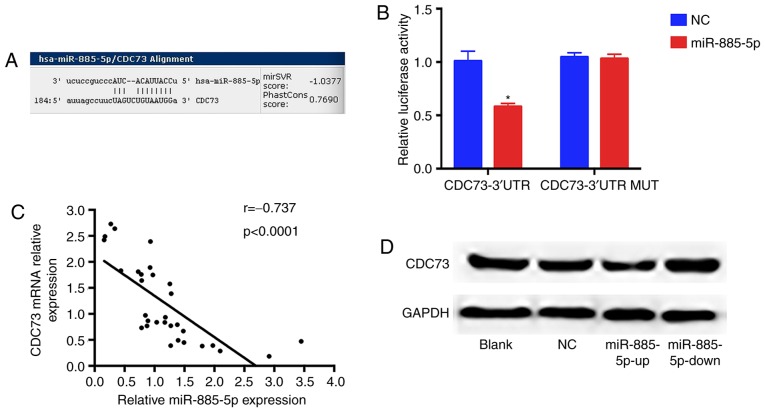
To elucidate the underlying miR-885-5p-mediated mechanisms, TargetScan (targetscan.org), miRanda (microrna.org) and miRBaseTargets (mirbase.org) were used to find potential target genes of miR-885-5p. Based on bioinformatics algorithms, a highly-conserved miR-885-5p targeting sequence was observed in the CDC73 3′-UTR, which suggested that CDC73 could be a candidate target of miR-885-5p. Dual luciferase reporter assay results showed that luciferase activity was markedly reduced in MG-63 cells when CDC73 3′-UTR wild-type construct was co-transfected with miR-885-5p-up compared with the mutant group. Furthermore, miR-885-5p and CDC73 expression levels were examined in 31 pairs of OS tissues and the association between miR-885-5p and CDC73 was investigated. As demonstrated in Fig. 5, mRNA and protein levels of CDC73 were significantly decreased and increased in response to miR-885-5p-up and miR-885-5p-down groups, respectively. These data support the hypothesis that miR-885-5p directly targets the 3′-UTR of CDC73 mRNA, leading to mRNA degradation and inhibiting its expression.
Figure 5.
miR-885-5p directly targets CDC73 mRNA and inhibits its expression. (A) miR-885-5p and its putative binding sequence in the 3′-UTR of CDC73 mRNA. (B) Co-transfection of MG-63 cells with miR-885-5p and wild-type CDC73 3′-UTR led to a significant decrease in luciferase activity, whereas co-transfection with CDC73 3′-UTR MUT and miR-885-5p-up showed no difference with the control group. The normalized luciferase activity in the control group was set as relative luciferase activity. (C) Spearman's correlation analysis between miR-144 expression and CDC73 mRNA levels. (D) Upregulated miR-885-5p decreased the expression of the CDC73 protein and downregulated miR-885-5p increased the level of the CDC73 protein in the MG-63 cells, while GAPDH was used as an internal control. *P<0.05. miR, microRNA; CDC73, cell division cycle protein 73 homolog; NC, negative control; up, mimic; down, inhibitor; UTR, untranslated region; MUT, mutant-type.
Discussion
OS is the most common primary solid tumor of the bone. The malignancy is characterized by malignant mesenchymal cells, which generate osteoid and/or immature bone (21). One previous study demonstrated that the dysregulation of miRNAs contributes to proliferation and migration of numerous types of human cancer (22). Moreover, the expression of several miRNAs, including miR-9, miR-21, miR-93 and miR-603, was shown to be increased in OS tissues and cells (23–26). Several studies have confirmed that miR-885-5p is aberrantly expressed in various types of cancer. Furthermore, miR-885-5p functions as a negative regulator of osteogenic differentiation of bone marrow-derived mesenchymal stem cells, leading to terminal differentiation failure. OS development may also be associated with defects in osteogenic differentiation (27,28). Thus, for a better understanding of the underlying gene network orchestrated by miRNAs involved in OS tumor initiation, development and progression, further studies are required to find effective biomarkers for the diagnosis and treatment of OS.
The present study first investigated the effect of miR-885-5p on OS tumor growth and migration. Consistent with previous studies that demonstrated a role for miR-885-5p in certain cancer types, including colorectal cancer, renal cell carcinoma and Hurthle cell carcinoma (11–13) the present study observed that the expression of miR-885-5p was significantly higher in OS tissues compared with that in corresponding adjacent normal tissues. In addition, OS cells lines (MG-63 and U2OS) were compared with a normal human osteoblastic cell line, hFOB1.19, and it was found that the expression of miR-885-5p was significantly upregulated. The upregulated miR-885-5p expression was strongly associated with malignant clinicopathological features of OS patients, including tumor size and clinical stage. It was observed that the overexpression of miR-885-5p promoted the proliferation and migration of MG-63 cells, while the downregulation of miR-885-5p expression showed the opposite results.
CDC73, also known as hyperparathyroidism 2, encodes a tumor suppressor gene that is involved in regulating transcriptional and post-transcriptional control pathways. Mutations in this gene are associated with hyperparathyroidism-jaw tumor syndrome and parathyroid carcinomas (29,30). A previous study reported that loss of heterozygosity of the wild-type CDC73 allele can be observed in sporadic human renal tumors, including, papillary, clear cell and chromophobe renal cell carcinomas, Wilms' tumors and oncocytomas (31). Overexpression of CDC73 inhibits the formation of colonies and the proliferation of cells, and induces arrest of the cell cycle in the G1 phase, suggesting that it has a key role in cell growth and proliferation (32). Moreover, it was reported that Bruton's tyrosine kinase increased the abundance of CDC73 when stimulation was not present and that CDC73 functioned as a repressor of β-catenin-mediated transcription in human colorectal cancer cells and B cells (33). These findings indicate that CDC73 serves a critical role in the development and progression of certain types of malignant tumors. However, the potential function of CDC73 and associated miRNAs in OS remain to be established. In the present study, bioinformatics analysis was used to identify CDC73 as a putative target of miR-885-5p. miR-885-5p negatively modulates CDC73 mRNA and protein expression in MG-63 cells. In the luciferase activity assay, it was observed that overexpression of miR-885-5p markedly reduced the luciferase activity of wild-type CDC73 3′-UTR. These data demonstrated that miR-885-5p directly targets CDC73 by interacting with its 3′-UTR. Although this does not fully explain how miR-885-5p induces OS cell proliferation and migration, the TargetScan program predicted hundreds of potential targets of miR-885-5p. Indeed, a single miRNA can target multiple mRNAs to regulate gene expression (34). It is possible that other potential targets may be involved in the proliferation and migration of OS cells. Therefore, further research should be conducted to comprehensively understand the role of miR-885-5p in OS tumorigenesis.
Taken together, the results of the present study demonstrated that miR-885-5p is highly upregulated in OS tissues and cell lines, and that its increased expression is strongly associated with malignant clinicopathological features. Moreover, we speculate that the overexpression of miR-885-5p acts as an oncogene that promotes cell proliferation and migration. It was also confirmed that miR-885-5p directly targets CDC73 and inversely regulates its expression. Collectively, it may be concluded that the ectopic expression of miR-885-5p may serve a critical role in the progression of OS, and may act as a novel diagnostic biomarker and potential therapeutic target for OS.
Acknowledgements
The authors would like to thank Professor Lurong Zhang, Dr Ruiqing Chen, Dr Fei Huang, Dr Ruilong Lan, Dr Junying Chen and Dr Jinrong Chen (Central Laboratory, The First Affiliated Hospital of Fujian Medical University, Fuzhou, China) for providing excellent technical assistance and useful discussions.
Funding
This study was supported by grants from the National Natural Science Foundation of China (no. 31571292) and the Science Foundation for Youths of Province (no. 2016J05182).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
FY and ZW conducted the experiment and drafted the manuscript. XW and SW contributed to statistical analysis and manuscript writing. XL and QH participated in cell experiments and clinical data collection. JL conceived the present study and helped revise the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Institutional Review Board of The First Affiliated Hospital of Fujian Medical University (Fuzhou, China) and the protocols conformed to the ethical guidelines of the Declaration of Helsinki. All participants involved in this study provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kobayashi E, Hornicek FJ, Duan Z. MicroRNA involvement in osteosarcoma. Sarcoma. 2012;2012:359739. doi: 10.1155/2012/359739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the surveillance, epidemiology, and end results program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 4.Marina N, Gebhardt M, Teot L, Gorlick R. Biology and therapeutic advances for pediatric osteosarcoma. Oncologist. 2004;9:422–441. doi: 10.1634/theoncologist.9-4-422. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Zamore PD, Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 7.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: Functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin S, Ai F, Ji WF, Rao W, Zhang HC, Yao WJ. miR-19a promotes cell growth and tumorigenesis through targeting SOCS1 in gastric cancer. Asian Pac J Cancer Prev. 2013;14:835–840. doi: 10.7314/APJCP.2013.14.2.835. [DOI] [PubMed] [Google Scholar]
- 10.Jones KB, Salah Z, Del Mare S, Galasso M, Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL, et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;72:1865–1877. doi: 10.1158/0008-5472.CAN-11-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam CS, Ng L, Chow AK, Wan TM, Yau S, Cheng NS, Wong SK, Man JH, Lo OS, Foo DC, et al. Identification of microRNA 885-5p as a novel regulator of tumor metastasis by targeting CPEB2 in colorectal cancer. Oncotarget. 2017;8:26858–26870. doi: 10.18632/oncotarget.15844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshino H, Yonemori M, Miyamoto K, Tatarano S, Kofuji S, Nohata N, Nakagawa M, Enokida H. microRNA-210-3p depletion by CRISPR/Cas9 promoted tumorigenesis through revival of TWIST1 in renal cell carcinoma. Oncotarget. 2017;8:20881–20894. doi: 10.18632/oncotarget.14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petric R, Gazic B, Goricar K, Dolzan V, Dzodic R, Besic N. Expression of miRNA and occurrence of distant metastases in patients with Hürthle cell carcinoma. Int J Endocrinol. 2016;2016:8945247. doi: 10.1155/2016/8945247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z, Yin J, Yang J, Shen W, Zhang C, Mou W, Luo J, Yan H, Sun P, Luo Y, et al. miR-885-5p suppresses hepatocellular carcinoma metastasis and inhibits Wnt/β-catenin signaling pathway. Oncotarget. 2016;7:75038–75051. doi: 10.18632/oncotarget.12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen-Rhoades W, Kurenbekova L, Satterfield L, Parikh N, Fuja D, Shuck RL, Rainusso N, Trucco M, Barkauskas DA, Jo E, et al. Cross-species identification of a plasma microRNA signature for detection, therapeutic monitoring, and prognosis in osteosarcoma. Cancer Med. 2015;4:977–988. doi: 10.1002/cam4.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao G, Cai C, Yang T, Qiu X, Liao B, Li W, Ji Z, Zhao J, Zhao H, Guo M, et al. MicroRNA-221 induces cell survival and cisplatin resistance through PI3K/Akt pathway in human osteosarcoma. PLoS One. 2013;8:e53906. doi: 10.1371/journal.pone.0053906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Shi K, Lan RL, Tao X, Wu CY, Hong HF, Lin JH. Vitronectin significantly influences prognosis in osteosarcoma. Int J Clin Exp Pathol. 2015;8:11364–11371. [PMC free article] [PubMed] [Google Scholar]
- 19.Enneking WF, Spanier SS, Goodman MA. A system for the surgical staging of musculoskeletal sarcoma. Clin Orthop Relat Res. 1980:106–120. [PubMed] [Google Scholar]
- 20.Han K, Zhao T, Chen X, Bian N, Yang T, Ma Q, Cai C, Fan Q, Zhou Y, Ma B. microRNA-194 suppresses osteosarcoma cell proliferation and metastasis in vitro and in vivo by targeting CDH2 and IGF1R. Int J Oncol. 2014;45:1437–1449. doi: 10.3892/ijo.2014.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arndt CA, Rose PS, Folpe AL, Laack NN. Common musculoskeletal tumors of childhood and adolescence. Mayo Clin Proc. 2012;87:475–487. doi: 10.1016/j.mayocp.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson KM, Weiss GJ. MicroRNAs and cancer: Past, present, and potential future. Mol Cancer Ther. 2008;7:3655–3660. doi: 10.1158/1535-7163.MCT-08-0586. [DOI] [PubMed] [Google Scholar]
- 23.Zhu SW, Li JP, Ma XL, Ma JX, Yang Y, Chen Y, Liu W. miR-9 modulates osteosarcoma cell growth by targeting the gcip tumor suppressor. Asian Pac J Cancer Prev. 2015;16:4509–4513. doi: 10.7314/APJCP.2015.16.11.4509. [DOI] [PubMed] [Google Scholar]
- 24.Ziyan W, Shuhua Y, Xiufang W, Xiaoyun L. MicroRNA-21 is involved in osteosarcoma cell invasion and migration. Med Oncol. 2011;28:1469–1474. doi: 10.1007/s12032-010-9563-7. [DOI] [PubMed] [Google Scholar]
- 25.Kawano M, Tanaka K, Itonaga I, Ikeda S, Iwasaki T, Tsumura H. microRNA-93 promotes cell proliferation via targeting of PTEN in Osteosarcoma cells. J Exp Clin Cancer Res. 2015;34:76. doi: 10.1186/s13046-015-0192-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma C, Zhan C, Yuan H, Cui Y, Zhang Z. MicroRNA-603 functions as an oncogene by suppressing BRCC2 protein translation in osteosarcoma. Oncol Rep. 2016;35:3257–3264. doi: 10.3892/or.2016.4718. [DOI] [PubMed] [Google Scholar]
- 27.Xu JF, Yang GH, Pan XH, Zhang SJ, Zhao C, Qiu BS, Gu HF, Hong JF, Cao L, Chen Y, et al. Altered microRNA expression profile in exosomes during osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. PLoS One. 2014;9:e114627. doi: 10.1371/journal.pone.0114627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner ER, Luther G, Zhu G, Luo Q, Shi Q, Kim SH, Gao JL, Huang E, Gao Y, Yang K, et al. Defective osteogenic differentiation in the development of osteosarcoma. Sarcoma. 2011;2011:325238. doi: 10.1155/2011/325238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carpten JD, Robbins CM, Villablanca A, Forsberg L, Presciuttini S, Bailey-Wilson J, Simonds WF, Gillanders EM, Kennedy AM, Chen JD, et al. HRPT2, encoding parafibromin, is mutated in hyperparathyroidism-jaw tumor syndrome. Nat Genet. 2002;32:676–680. doi: 10.1038/ng1048. [DOI] [PubMed] [Google Scholar]
- 30.Shattuck TM, Välimäki S, Obara T, Gaz RD, Clark OH, Shoback D, Wierman ME, Tojo K, Robbins CM, Carpten JD, et al. Somatic and germ-line mutations of the HRPT2 gene in sporadic parathyroid carcinoma. N Engl J Med. 2003;349:1722–1729. doi: 10.1056/NEJMoa031237. [DOI] [PubMed] [Google Scholar]
- 31.Zhao J, Yart A, Frigerio S, Perren A, Schraml P, Weisstanner C, Stallmach T, Krek W, Moch H. Sporadic human renal tumors display frequent allelic imbalances and novel mutations of the HRPT2 gene. Oncogene. 2007;26:3440–3449. doi: 10.1038/sj.onc.1210131. [DOI] [PubMed] [Google Scholar]
- 32.Zhang C, Kong D, Tan MH, Pappas DL, Jr, Wang PF, Chen J, Farber L, Zhang N, Koo HM, Weinreich M, et al. Parafibromin inhibits cancer cell growth and causes G1 phase arrest. Biochem Biophys Res Commun. 2006;350:17–24. doi: 10.1016/j.bbrc.2006.08.169. [DOI] [PubMed] [Google Scholar]
- 33.James RG, Biechele TL, Conrad WH, Camp ND, Fass DM, Major MB, Sommer K, Yi X, Roberts BS, Cleary MA, et al. Bruton's tyrosine kinase revealed as a negative regulator of Wnt-beta-catenin signaling. Sci Signal. 2009;2:ra25. doi: 10.1126/scisignal.2000230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.