Abstract
Dysregulated expression of the cell surface protein, CD146, has been implicated in various types of cancer in humans, including in lung cancer. The present study aimed to clarify the mechanism underlying abnormal CD146 expression in human pulmonary large cell neuroendocrine carcinoma (LCNEC) cell lines (NCI-H460 and NCI-H810). The functions of CD146 were investigated by measuring cell migration and viability following CD146 knockdown or overexpression via small interference RNA and plasmid transfection. The findings demonstrated that decreased protein expression of CD146 could inhibit migration and viability in LCNEC cells. Furthermore, CD146 was determined to influence the expression of epithelial-mesenchymal transition markers (epithelial cadherin, vimentin and Snail) and promoted AKT phosphorylation. The present results imply CD146 may function in the migration and proliferation of pulmonary LCNEC cells.
Keywords: cluster of differentiation 146, pulmonary large cell neuroendocrine carcinoma, epithelial-mesenchymal transition, AKT
Introduction
Pulmonary large cell neuroendocrine carcinoma (LCNEC) is categorized as a large cell carcinoma. The clinical and biological characteristics of LCNEC are similar to those of small cell lung carcinomas (SCLCs), and the disease exhibits aggressive phenotypes of frequent recurrence and high metastatic potential (1,2). The optimal treatment strategies and molecular features of LCNEC remain largely unknown. Therefore, to improve the prognosis of patients with LCNEC, characterization of its molecular characteristics is required (3,4).
Cluster of differentiation (CD)146 is a cell adhesion molecule belonging to the immunoglobulin superfamily, which is located on the human adipose-derived stem cell surface (5,6). CD146 has been reported to be involved in cell adhesion by binding other cells or with the extracellular matrix (7). Moreover, abnormal CD146 expression has been identified in several types of cancer, such as breast cancer and prostate cancer, in which it was associated with cancer cell motility, the state of epithelial-mesenchymal transition (EMT), angiogenesis and prognosis (7,8). In non-small cell lung cancer, CD146 overexpression is a useful marker in predicting poor prognosis, though the reason for this remains largely unknown; likewise, in the context of pulmonary LCNEC (9,10).
In the present study, the role of CD146 in pulmonary LCNEC was investigated. CD146 expression was detected in pulmonary LCNEC cell lines (NCI-H460 and NCI-H810), and the association of CD146 overexpression with migration and proliferation of the cells was determined.
Materials and methods
Cell lines
The LCNEC cell lines, NCI-H460 and NCI-H810, were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) (11). Human umbilical vein endothelial cells (HUVECs) were obtained from Lonza (Walkersville, MD, USA; cat. no. C2517A) and maintained in endothelial basal medium-2 (Lonza). NCI-H460/H810 cells were maintained in RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C in a humidified environment with 10% CO2.
Silencing of CD146 using small interfering RNA (siRNA)
Gene silencing was performed using siRNAs (Qiagen GmbH, Hilden, Germany) directed against human CD146 (8). The siRNA sequences were as follows: siRNA-1 sense, 5′-GGGAGAGAAAUACAUCGAUTT-3′ and antisense, 5′-AUCGAUGUAUUUCUCUCCCTG-3′); siRNA-2 sense, 5′-GGAACUACUGGUGAACUAUTT-3′ and antisense, 5′-AUAGUUCACCAGUAGUUCCTG-3′. Qiagen AllStar siRNA (Qiagen GmbH) was used as a negative control. Based on western blotting results, NCI-H460 cells were selected for transfection with siRNA (20 nM) using Lipofectamine 2000 (Qiagen GmbH, Hilden, Germany), according to the manufacturer's protocol. All cells were used in subsequent experiments at 24 h following transfection. Cell morphology means to observe the change of cell-shape through a fluorescence microscope (magnification, ×200; BZ-II analyser; Keyence, Osaka, Japan) at 72 h following transfection, 20 cells were observed at a randomly selected microscopic field of view.
Plasmid transfection
A CD146 expression plasmid, CD146-HaloTag vector, was obtained from Promega Corporation (Madison, WI, USA). NCI-H460 and NCI-H810 cells were transiently transfected with this plasmid (0.015 µg/µl) or a HaloTag (HT) control vector (0.015 µg/µl; cat. no. G6591; Promega Corporation) using Fugene® HD transfection reagent (Promega Corporation), according to the manufacturer's protocol (8).
Migration assays
The migration capacity of cancer cells was assessed by counting the number of cells migrating through Transwell chambers (8 µm pore size; Corning Incorporated, Corning, NY, USA) as described previously (12). Cells were maintained in 10% FBS/Dulbecco's modified Eagle's medium (Invitrogen; Thermo Fisher Scientific, Inc.) during these assays. Cells were transfected with siRNAs or plasmids 48 h prior to experimentation, and migration was determined at 24 h following transfection.
Cell viability assay
A cell viability assay was performed as described previously (8). Briefly, cancer cells (1.5×103 cells/well) were seeded in 96-well plates 24 h after transfection in the aforementioned culture conditions. Cell viability was examined using a CellTiter-Glo Luminescent Cell Viability assay kit (cat. no. G7570; Promega Corporation) with a luminometer (Infinite 200, Tecan, Switzerland) at 24, 47, 72 and 96 h following transfection. Background was subtracted using the values of wells containing only culture medium.
Western blot analysis
Cancer cells were lysed in PRO-PREP™ Protein Extraction Solution (iNtRON Biotechnology, Seongnam, Korea), and proteins were separated on 12% SDS-polyacrylamide gels and transferred onto mini polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 1X TBST with 5% non-fat dry milk for 1 h at room temperature. Membranes were incubated overnight at 4°C with the following primary antibodies: Anti-CD146 (1:10,000; cat. no. ab75769; Abcam, Cambridge, UK), anti-epithelial (E)-cadherin (1:1,000; cat. no. 3195), anti-vimentin (1:1,000; cat. no. 5741), anti-Snail (1:1,000; cat. no. 4719), anti-AKT (1:1,000; cat. no. 4691), anti-phosphorylated AKT (1:2,000; cat. no. 4060; all from Cell Signalling Technology, Inc., Danvers, MA, USA) and anti-β-actin (1:5,000; cat. no. ab8227; Abcam). Membranes were then incubated with Anti-rabbit/mouse IgG, HRP-linked Antibody (1:2,000; cat. no. 7074/7076; from Cell Signalling Technology, Inc.) for 1 h at room temperature. An electrochemiluminescence western blotting analysis system (Amersham Biosciences, Little Chalfont, UK) was used to visualize the proteins, according to the manufacturer's protocol. Densitometric analysis was performed with ImageJ 1.48v software (National Institutes of Health, Bethesda, MD, USA). The protein level of CD146 was also assessed in HUVECs as a positive control. β-actin was used as the loading control.
Statistical analysis
Data are expressed as the mean ± standard deviation. Comparisons between multiple groups were conducted by one-way analysis of variance (ANOVA) with Dunnet's post-hoc test. P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed using JMP 11.0.0 software (SAS Institute, Inc., Cary, NC, USA).
Results
Analysis of CD146 expression in LCNEC cells
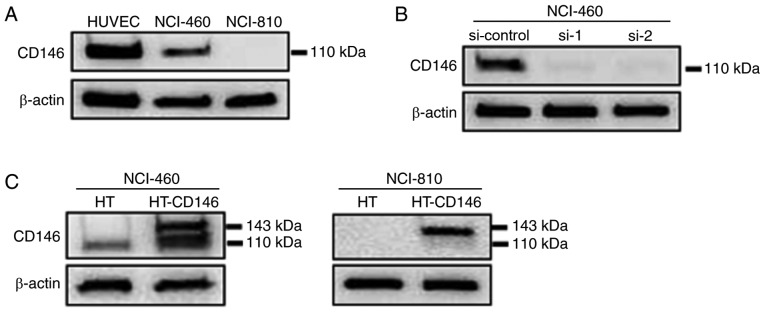
The protein expression level of CD146 in two LCNEC cell lines (NCI-H460 and NCI-H810) was analyzed by western blotting. HUVECs were used as a positive control as they express high levels of CD146 (8). High protein levels of CD146 were detected in NCI-460 cells, but CD146 expression was not detected in NCI-810 cells (Fig. 1A). To investigate the function of CD146, it was knocked down in NCI-460 cells and upregulated in NCI-H460 and NCI-H810 cells using siRNA and plasmids. The efficiencies of knockdown (Fig. 1B) and overexpression (Fig. 1C) of CD146 were then confirmed. Since overexpression of CD146 was induced by transfection with CD146-HaloTag vector, endogenous CD146 (110 kDa) and exogenous CD146 (plus 33-kDa HaloTag) expressions occurred simultaneously in the NCI-H460 cells.
Figure 1.
CD146 expression in large cell neuroendocrine carcinoma cells and the transfection efficiency of siRNAs and overexpression plasmid. (A) Endogenous CD146 protein levels were determined by western blotting. (B) Western blotting was used to determine the protein level of CD146 in NCI-460 cells following siRNA transfection. (C) Western blot showing the protein levels of CD146 in NCI-460 and NCI-810 cells following transfection with CD146 expression plasmid. Each experiment was performed three times. CD146, cluster of differentiation 146; si/siRNA, small interfering RNA; HUVEC, human umbilical vein endothelial cell.
CD146 expression enhances the migration ability of LCNEC cells
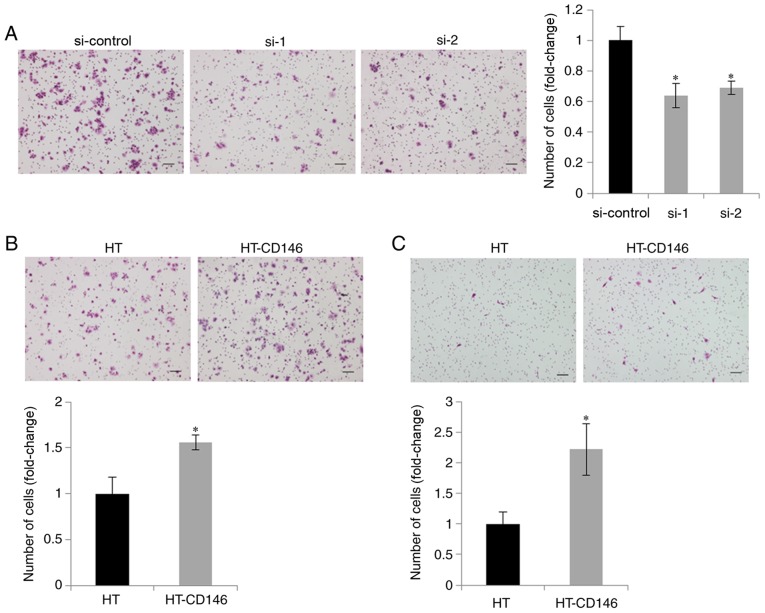
As CD146 has been reported to be involved in the migration of cancer cells (13), migration assays were performed following knockdown of CD146 in NCI-460 cells expressing high endogenous levels of CD146. It was demonstrated that cell migration ability was decreased upon CD146 knockdown, when compared with cells transfected with negative control siRNA (P<0.05; Fig. 2A). Conversely, migration ability was increased upon overexpression of CD146 in the two LCNEC cell lines, when compared with those cells transfected with the HT control vector (P<0.05; Fig. 2B and C). These results suggest that CD146 was involved in the migration of LCNEC cells.
Figure 2.
CD146 expression increases the migratory ability of large cell neuroendocrine carcinoma cells. (A) Migration of NCI-460 cells following transfection with control or CD146-specific siRNAs. (B) Migration of NCI-460 cells following plasmid transfection. (C) Migration of NCI-810 cells following plasmid transfection. Representative photomicrographs are presented for each group (scale bar, 100 µm; magnification, ×100). The bar charts show quantification of the cell migration. The bars represent the mean cell counts ± standard deviation, and are normalized to the control group. The experiment was performed three times. *P<0.05, vs. si-control or HT alone group). CD146, cluster of differentiation 146; si/siRNA, small interfering RNA; HT, HaloTag vector.
CD146 promotes EMT in LCNEC cells
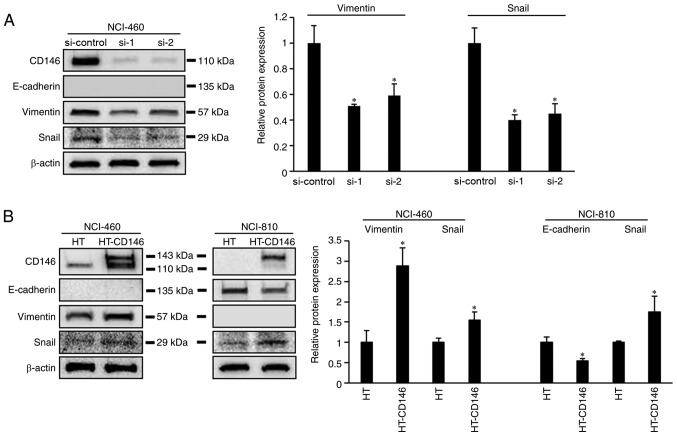
The process of cancer cell migration requires epithelial cancer cells to undergo EMT (14); therefore, the association between the expression of CD146 and EMT markers in LCNEC cells was investigated. It was demonstrated that expression of vimentin and Snail was decreased in NCI-460 cells following knockdown of CD146 (P<0.05). Meanwhile, vimentin and Snail expression was increased following overexpression of CD146 in NCI-460 cells (P<0.05; Fig. 3B). In NCI-810 cells, overexpression of CD146 resulted in downregulated E-cadherin expression and upregulated Snail expression (P<0.05; Fig. 3B). However, changes in cell morphology were not observed following knockdown or overexpression of CD146 (data not shown).
Figure 3.
CD146 expression is correlated with EMT marker levels. (A) Western blot showing the levels of EMT marker proteins in NCI-460 cells following siRNA transfection. (B) Western blot showing the levels of EMT maker proteins in NCI-460 and NCI-810 cells following transfection with CD146 expression plasmid. Each experiment was performed three times. *P<0.05, vs. si-control or HT alone group. CD146, cluster of differentiation 146; EMT, epithelial-mesenchymal transition; si/siRNA, small interfering RNA; HT, HaloTag vector.
CD146 increases the proliferative ability of LCNEC cells
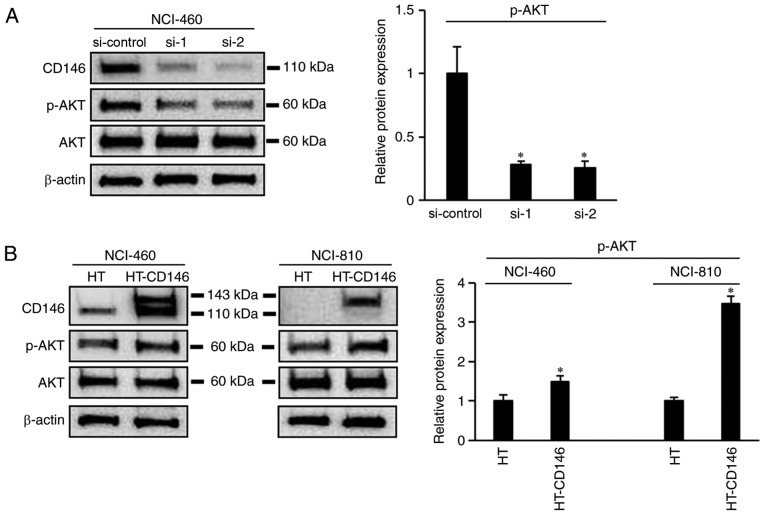
The effect of CD146 on the proliferation of LCNEC cells was also examined via a cell viability assay. The results revealed that cell viability was significantly decreased following knockdown of CD146 in NCI-460 cells by day 4 post-transfection (P<0.05; Fig. 4A) and that cell viability was increased upon overexpression of CD146 in NCI-460 and NCI-810 cells by ≥2 days post-transfection (P<0.05; Fig. 4B). Moreover, the level of phosphorylation of AKT decreased following knockdown of CD146 in NCI-460 cells (P<0.05; Fig. 5A). The opposite result was apparent following overexpression of CD146 in NCI-460 and NCI-810 cells (Fig. 5B).
Figure 4.
CD146 expression increases proliferation in LCNEC cells. The viability of LCNEC cells was determined using a CellTiter-Glo luminescent cell viability assay following (A) CD146 knockdown and (B) CD146 overexpression. *P<0.05, **P<0.01, vs. si-control or HT alone group. CD146, cluster of differentiation 146; LCNEC, large cell neuroendocrine carcinoma; si, small interfering RNA; HT, HaloTag vector.
Figure 5.
CD146 expression is associated with AKT activity. (A) Western blot and quantified results showing AKT phosphorylation level in NCI-460 cells following siRNA transfection. (B) Western blot and quantified results showing AKT phosphorylation level in NCI-460 and NCI-810 cells following transfection with CD146 expression plasmid. Each experiment was performed three times. *P<0.05, vs. si-control or HT alone group. CD146, cluster of differentiation 146; si/siRNA, small interfering RNA; HT, HaloTag vector; p-, phosphorylated.
Discussion
CD146, also known as melanoma cell adhesion molecule, is a transmembrane glycoprotein belonging to the immunoglobulin superfamily (15). The expression of CD146 has been detected in multiple types of human carcinoma (7), and its overexpression has been associated with poor overall survival in non-small cell lung cancer (9). In the present study, the role of CD146 in LCNEC cell lines (NCI-460 and NCI-810) was evaluated. Endogenous CD146 expression was detected in NCI-460 cells, and exogenous overexpression was demonstrated to enhance migratory ability in NCI-460 and NCI-810 cells. Previous studies have reported that CD146 promotes breast cancer progression via induction of EMT due to upregulated expression of the EMT transcription factor, Slug (13,16). In this study, CD146 was also demonstrated to regulate the expression of EMT markers, namely vimentin, E-cadherin and Snail. These findings suggest that CD146 expression is associated with cell migration via regulation of EMT in LCNEC cells.
The effect of CD146 on cell proliferation was also investigated, which revealed that CD146 increased the viability of LCNEC cells and increased AKT phosphorylation. The AKT kinases are key members of various signaling pathways that regulate cellular processes, involved in control of cell growth, proliferation and survival (17). A previous study reported that CD146 promotes tumor proliferation and survival through the phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)/AKT pathway, and that the expression level of CD146 is reciprocally regulated by PI3K/AKT signaling in melanoma (18). Taken together, these data suggest that CD146 promotes LCNEC cell proliferation and may be involved in modulation of the AKT pathway.
Although the exact mechanism underlying the regulation of AKT activity by CD146 remains unclear, the association between CD146 and AKT may indicate how CD146 increases the viability of LCNEC cells. Improving the existing understanding of CD146 function in signal transduction will require further study of its crosstalk with members of other signaling pathways (7), including those in EMT induction. The clinical significance of CD146 expression in LCNEC was not investigated in the present study, and should be a focus of future study.
In conclusion, the present study determined that CD146 served a critical role in controlling the migration and proliferation of pulmonary LCNEC cells. Further exploration of the molecular mechanisms underlying the interaction between CD146 and AKT signaling, and EMT, in LCNEC cells may aid the development of novel therapies for LCNEC. Further investigation is required to elucidate the association between CD146 expression and the clinicopathological characteristics of pulmonary LCNEC, as well as prognosis.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Heilongjiang Postdoctoral Science Foundation (grant no. LBH-Z16157) and the Medical Scientific Research Foundation of Guangdong Province, China (grant no. A2018015).
Availability of data and materials
The datasets generated and/or analysed during this study are available from the corresponding author on reasonable request.
Authors' contributions
BZ designed the research. YP, HG and YG performed the research. HG and ZQ contributed to data collection and statistical analysis. YP, YG and BZ wrote the manuscript. All the authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Iyoda A, Hiroshima K, Nakatani Y, Fujisawa T. Pulmonary large cell neuroendocrine carcinoma: Its place in the spectrum of pulmonary carcinoma. Ann Thorac Surg. 2007;84:702–707. doi: 10.1016/j.athoracsur.2007.03.093. [DOI] [PubMed] [Google Scholar]
- 2.Bodey B, Bodey B, Jr, Groger AM, Siegel SE, Kaiser HE. Invasion and metastasis: The expression and significance of matrix metalloproteinases in carcinomas of the lung. In Vivo. 2001;15:175–180. [PubMed] [Google Scholar]
- 3.Makino T, Mikami T, Hata Y, Otsuka H, Koezuka S, Isobe K, Tochigi N, Shibuya K, Homma S, Iyoda A. Comprehensive biomarkers for personalized treatment in pulmonary large cell neuroendocrine carcinoma: A comparative analysis with adenocarcinoma. Ann Thorac Surg. 2016;102:1694–1701. doi: 10.1016/j.athoracsur.2016.04.100. [DOI] [PubMed] [Google Scholar]
- 4.Miyoshi T, Umemura S, Matsumura Y, Mimaki S, Tada S, Makinoshima H, Ishii G, Udagawa H, Matsumoto S, Yoh K, et al. Genomic profiling of large-cell neuroendocrine carcinoma of the lung. Clin Cancer Res. 2017;23:757–765. doi: 10.1158/1078-0432.CCR-16-0355. [DOI] [PubMed] [Google Scholar]
- 5.Okumura S, Kohama K, Kim S, Iwao H, Miki N, Taira E. Induction of gicerin/CD146 in the rat carotid artery after balloon injury. Biochem Biophys Res Commun. 2004;313:902–906. doi: 10.1016/j.bbrc.2003.12.028. [DOI] [PubMed] [Google Scholar]
- 6.Mohsen-Kanson T, Hafner AL, Wdziekonski B, Villageois P, Chignon-Sicard B, Dani C. Expression of cell surface markers during self-renewal and differentiation of human adipose-derived stem cells. Biochem Biophys Res Commun. 2013;430:871–875. doi: 10.1016/j.bbrc.2012.12.079. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Yan X. CD146, a multi-functional molecule beyond adhesion. Cancer Lett. 2013;330:150–162. doi: 10.1016/j.canlet.2012.11.049. [DOI] [PubMed] [Google Scholar]
- 8.Zheng B, Ohuchida K, Chijiiwa Y, Zhao M, Mizuuchi Y, Cui L, Horioka K, Ohtsuka T, Mizumoto K, Oda Y, et al. CD146 attenuation in cancer-associated fibroblasts promotes pancreatic cancer progression. Mol Carcinog. 2016;55:1560–1572. doi: 10.1002/mc.22409. [DOI] [PubMed] [Google Scholar]
- 9.Kristiansen G, Yu Y, Schluns K, Sers C, Dietel M, Petersen I. Expression of the cell adhesion molecule CD146/MCAM in non-small cell lung cancer. Anal Cell Pathol. 2003;25:77–81. doi: 10.1155/2003/574829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oka S, Uramoto H, Chikaishi Y, Tanaka F. The expression of CD146 predicts a poor overall survival in patients with adenocarcinoma of the lung. Anticancer Res. 2012;32:861–864. [PubMed] [Google Scholar]
- 11.Odate S, Nakamura K, Onishi H, Kojima M, Uchiyama A, Nakano K, Kato M, Tanaka M, Katano M. TrkB/BDNF signaling pathway is a potential therapeutic target for pulmonary large cell neuroendocrine carcinoma. Lung Cancer. 2013;79:205–214. doi: 10.1016/j.lungcan.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Zheng B, Ohuchida K, Cui L, Zhao M, Shindo K, Fujiwara K, Manabe T, Torata N, Moriyama T, Miyasaka Y, et al. TM4SF1 as a prognostic marker of pancreatic ductal adenocarcinoma is involved in migration and invasion of cancer cells. Int J Oncol. 2015;47:490–498. doi: 10.3892/ijo.2015.3022. [DOI] [PubMed] [Google Scholar]
- 13.Zeng Q, Li W, Lu D, Wu Z, Duan H, Luo Y, Feng J, Yang D, Fu L, Yan X. CD146, an epithelial-mesenchymal transition inducer, is associated with triple-negative breast cancer. Proc Natl Acad Sci USA. 2012;109:1127–1132. doi: 10.1073/pnas.1111053108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 15.Lehmann JM, Riethmuller G, Johnson JP. MUC18, a marker of tumor progression in human melanoma, shows sequence similarity to the neural cell adhesion molecules of the immunoglobulin superfamily. Proc Natl Acad Sci USA. 1989;86:9891–9895. doi: 10.1073/pnas.86.24.9891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imbert AM, Garulli C, Choquet E, Koubi M, Aurrand-Lions M, Chabannon C. CD146 expression in human breast cancer cell lines induces phenotypic and functional changes observed in epithelial to mesenchymal transition. PLoS One. 2012;7:e43752. doi: 10.1371/journal.pone.0043752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–7464. doi: 10.1038/sj.onc.1209085. [DOI] [PubMed] [Google Scholar]
- 18.Li G, Kalabis J, Xu X, Meier F, Oka M, Bogenrieder T, Herlyn M. Reciprocal regulation of MelCAM and AKT in human melanoma. Oncogene. 2003;22:6891–6899. doi: 10.1038/sj.onc.1206819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analysed during this study are available from the corresponding author on reasonable request.