Abstract
Head and neck squamous cell carcinoma (HNSCC) is an aggressive cancer. Early detection and management of HNSCC may prevent progression of the disease. Long non-coding RNAs (lncRNAs) may serve as prognostic biomarkers for various cancer types. The current study downloaded an RNA-Seq dataset containing 43 tumor-normal pairs. An independent t-test identified that the expression level of lncRNA LOC541471 was significantly increased in tumor tissues compared with healthy tissues. Additionally, the current study demonstrated that high lncRNA LOC541471 expression was significantly associated with increasing lymph node metastasis classification and perineural invasion. A multivariate Cox regression analysis revealed that high lncRNA LOC541471 expression levels were an independent predictor for reduced overall survival (n=487) and relapse-free survival (n=355). According to the anatomic neoplasm subdivision, HNSCC samples were classified as oropharyngeal carcinoma (n=297), oral carcinoma (n=80), laryngeal carcinoma and hypopharyngeal carcinoma (n=123). A negative association was revealed between lncRNA LOC541471 expression and overall survival in all subtypes of HNSCC. Therefore, lncRNA LOC541471 is significantly negatively associated with overall survival and relapse-free survival of patients with HNSCC and may be considered a potential prognostic factor for HNSCC.
Keywords: head and neck squamous cell carcinoma, long non-coding RNA, prognostic biomarker, invasion, overall survival
Introduction
Long non-coding RNAs (lncRNAs) are a class of RNA molecules, longer than 200 nucleotides, with no protein-coding capacity (1,2) Previously, studies have identified that lncRNAs are aberrantly expressed in different cancer types and are closely associated with different disease processes (3,4). LncRNAs have been explored as a prognostic tumor biomarker as they can easily be detected in tissues and serum (5,6). Many lncRNAs have been identified to serve a role as prognostic biomarkers for various cancer types. Downregulation of lncRNA SDPR-AS is associated with poor prognosis in renal cell carcinoma (7). lncRNA CCAT1 promotes metastasis and is associated with poor prognosis in epithelial ovarian cancer (8). In addition, several molecular tools, including reverse transcription-quantitative polymerase chain reaction and RNA-fluorescent in situ hybridization, are used for examining the lncRNA expression levels in the saliva, plasma, urine, serum and tissues (5,9). Several lncRNAs, including PCA3, are routinely used in the clinic as a grade reclassification predictor in prostate cancer (10). Interest in therapeutic strategies against cancer based on lncRNAs continues to increase.
Head and neck squamous cell carcinoma (HNSCC), the sixth most common cancer worldwide, is an aggressive cancer type that has a high morbidity (11,12). Early detection and management of HNSCC may prevent progression of the disease. A previous study has demonstrated that a panel of three lncRNAs (KTN1-AS1, LINC00460 and RP5-894A10.6) may be a novel biomarker for the accurate prediction of the prognosis of patients with HNSCC (13).
In the present study, next-generation sequencing and clinical data of patients with HNSCC from The Cancer Genome Atlas (TCGA) were utilized. RNA-Sequencing (RNA-Seq) datasets for 43 paired tumor and adjacent normal tissues were downloaded from TCGA and significant differences in lncRNA expression between normal and cancer tissues were investigated. A novel lncRNA LOC541471 was revealed to be significantly upregulated in HNSCC. The lncRNA LOC541471 mRNA expression levels in various cases of HNSCC were also analyzed to evaluate its prognostic value. Overall, the potential of lncRNA LOC541471 as a biomarker and a therapeutic target in high-risk HNSCC was investigated in the current study.
Materials and methods
Patients and methods
The clinical and RNA-Seq expression data for HNSCC were obtained from TCGA (https://tcga-data.nci.nih.gov) and the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). To investigate differential lncRNA expression between normal and tumor tissues, the expression profile analysis was based on 43 paired tumor and adjacent normal tissues from TCGA (Table I). The RNA-Seq data were matched with clinical data and used to study expression profiles and search for lncRNAs associated with HNSCC. Prior to analysis, the HNSCC RNA expression data downloaded from TCGA were processed and normalized using the TCGA RNA-seq system (version 2,TCGA) and the Illumina HiSeq (v7) and Illumina GA microRNA (v7) sequencing platforms (Illumina, Inc., Hayward, CA, USA). A total of 511 HNSCC samples were included in the analysis of lncRNA LOC541471 and its association with clinicopathological features and patient survival. Clinical characteristics of the total HNSCC cohort used are provided in Table II. Patients were classified into two groups (high and low expression) according to the median lncRNA LOC541471 expression value. Known genes located close to LOC541741 were analyzed using the University of California, Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu).
Table I.
The ID of 43 paired tumor and adjacent normal tissues from TCGA.
| Tumor tissue ID | Normal tissue ID |
|---|---|
| TCGA-CV-6933-01 | TCGA-CV-6933-11 |
| TCGA-CV-6934-01 | TCGA-CV-6934-11 |
| TCGA-CV-6935-01 | TCGA-CV-6935-11 |
| TCGA-CV-6936-01 | TCGA-CV-6936-11 |
| TCGA-CV-6938-01 | TCGA-CV-6938-11 |
| TCGA-CV-6939-01 | TCGA-CV-6939-11 |
| TCGA-CV-6943-01 | TCGA-CV-6943-11 |
| TCGA-CV-6955-01 | TCGA-CV-6955-11 |
| TCGA-CV-6956-01 | TCGA-CV-6956-11 |
| TCGA-CV-6959-01 | TCGA-CV-6959-11 |
| TCGA-CV-6960-01 | TCGA-CV-6960-11 |
| TCGA-CV-6961-01 | TCGA-CV-6961-11 |
| TCGA-CV-6962-01 | TCGA-CV-6962-11 |
| TCGA-CV-7091-01 | TCGA-CV-7091-11 |
| TCGA-CV-7097-01 | TCGA-CV-7097-11 |
| TCGA-CV-7101-01 | TCGA-CV-7101-11 |
| TCGA-CV-7103-01 | TCGA-CV-7103-11 |
| TCGA-CV-7177-01 | TCGA-CV-7177-11 |
| TCGA-CV-7178-01 | TCGA-CV-7178-11 |
| TCGA-CV-7183-01 | TCGA-CV-7183-11 |
| TCGA-CV-7235-01 | TCGA-CV-7235-11 |
| TCGA-CV-7238-01 | TCGA-CV-7238-11 |
| TCGA-CV-7242-01 | TCGA-CV-7242-11 |
| TCGA-CV-7245-01 | TCGA-CV-7245-11 |
| TCGA-CV-7250-01 | TCGA-CV-7250-11 |
| TCGA-CV-7252-01 | TCGA-CV-7252-11 |
| TCGA-CV-7255-01 | TCGA-CV-7255-11 |
| TCGA-CV-7261-01 | TCGA-CV-7261-11 |
| TCGA-CV-7406-01 | TCGA-CV-7406-11 |
| TCGA-CV-7416-01 | TCGA-CV-7416-11 |
| TCGA-CV-7423-01 | TCGA-CV-7423-11 |
| TCGA-CV-7424-01 | TCGA-CV-7424-11 |
| TCGA-CV-7425-01 | TCGA-CV-7425-11 |
| TCGA-CV-7432-01 | TCGA-CV-7432-11 |
| TCGA-CV-7434-01 | TCGA-CV-7434-11 |
| TCGA-CV-7437-01 | TCGA-CV-7437-11 |
| TCGA-CV-7438-01 | TCGA-CV-7438-11 |
| TCGA-CV-7440-01 | TCGA-CV-7440-11 |
| TCGA-H7-A6C4-01 | TCGA-H7-A6C4-11 |
| TCGA-HD-8635-01 | TCGA-HD-8635-11 |
| TCGA-HD-A6HZ-01 | TCGA-HD-A6HZ-11 |
| TCGA-HD-A6I0-01 | TCGA-HD-A6I0-11 |
| TCGA-WA-A7GZ-01 | TCGA-WA-A7GZ-11 |
TCGA, The Cancer Genome Atlas.
Table II.
Demographics and clinical characteristics of the cohort of patients with head and neck squamous cell carcinoma downloaded from The Cancer Genome Atlas (n=487).
| Characteristics | Sample no, n |
|---|---|
| Sex | |
| Male | 356 |
| Female | 131 |
| Age | |
| ≤60 | 258 |
| >61 | 229 |
| T classification | |
| T1-2 | 180 |
| T3-4 | 307 |
| N classification | |
| N1-2 | 180 |
| N3-4 | 307 |
| Clinical stage | |
| I–II | 115 |
| III–IV | 372 |
T, primary tumor stage; N, lymph node metastasis.
Statistical analysis
A paired Student's t-test was used to identify any significant differences in lncRNA LOC541471 expression between tumor and tumor-adjacent normal tissues according to data from TCGA and GEO, including 42 paired samples from TCGA and 24 paired samples from GEO (one sample lacked the corresponding clinical data). An independent sample t-test was used to identify significant differences in lncRNA LOC541471 expression according to data from patients with HNSCC with different lymph node metastasis (N) classification and perineural invasion. Significant differences in lncRNA LOC541471 expression between the groups were presented by box plots (mean ± standard deviation). Univariate Cox regression analysis was used to evaluate the overall survival (OS) and relapse-free survival of patients with clinical characteristics as categorical dependent variables. Furthermore, multivariate Cox analysis was employed to assess the association of lncRNA LOC541471 expression with the OS and relapse-free survival of patients together with other clinical factors, including age, sex, primary tumor stage (T), N and clinical TNM staging. Survival curves were constructed using Kaplan-Meier analysis and a log-rank test was performed to assess differences between the groups. All data were analyzed with SPSS software (version 20.0; IBM Corp., Armonk, NY, USA) P<0.05 was considered to indicate a statistically significant difference.
Results
LncRNA LOC541471 upregulation in HNSCC and association with perineural invasion
RNA-Seq datasets for 43 tumor-adjacent normal pairs were downloaded and significant differences in lncRNA expression were identified in HNSCC samples compared with healthy samples (Table III). It was revealed that lncRNA LOC541471 expression was significantly higher in the HNSCC samples compared with the adjacent healthy samples, as presented in Fig. 1A. In addition, lncRNA LOC541471 was identified to be significantly upregulated in patients with HNSCC and perineural invasion compared with patients without perineural invasion from TCGA (Fig. 1B). Furthermore, high lncRNA LOC541471 expression was significantly associated with increased N classification (Fig. 1C). Previously, studies have revealed that perineural invasion and N classification are prognostic factors for HNSCC (14,15). Therefore, the current study analyzed whether lncRNA LOC541471 may be a biomarker and potential therapeutic target for high-risk HNSCC. Following clarification, HNSCC gene expression data and corresponding clinical data were obtained from the GEO database. The results revealed that lncRNA LOC541471 was significantly upregulated in tongue squamous cell carcinoma (GSE51700, n=18), oral squamous cell carcinoma (GSE84805, n=12) and laryngeal squamous cell carcinoma (GSE84957, n=18), as presented in Fig. 1D-F, respectively.
Table III.
Dysregulated long non-coding RNAs identified in head and neck squamous cell carcinoma.
| GeneID | Symbol | Fold-change | Style | P-value | FDR |
|---|---|---|---|---|---|
| 112597 | LINC00152 | 2.07 | up | 0.0000036 | 0.000182 |
| 606 | NBEAP1 | 2.06 | up | 0.0002079 | 0.00649 |
| 440101 | FLJ12825 | 2.06 | up | 0.0000421 | 0.00165 |
| 29774 | POM121L9P | 2.05 | up | 0.0000261 | 0.00108 |
| 400508 | CRYM-AS1 | 2.05 | up | 0.0000048 | 0.000236 |
| 100128191 | TMPO-AS1 | 2.05 | up | 0.0000005 | 0.0000307 |
| 440081 | DDX12P | 2.04 | up | 0.0000001 | 0.00000747 |
| 3136 | HLA-H | 2.03 | up | 0.0000001 | 0.00000747 |
| 344967 | LOC344967 | 2.02 | up | 0.0000053 | 0.000258 |
| 541471 | LOC541471 | 2.02 | up | 0.0000001 | 0.00000747 |
| 2679 | GGT3P | 2.00 | up | 0.0009594 | 0.0227 |
| 10984 | KCNQ1OT1 | 1.99 | up | 0.0000067 | 0.000319 |
| 114043 | TSPEAR-AS2 | 1.96 | up | 0.0001926 | 0.00608 |
| 3139 | HLA-L | 1.95 | up | 0.0000018 | 0.0000971 |
| 54718 | BTN2A3P | 1.94 | up | 0.0000070 | 0.000332 |
| 145978 | LINC00052 | 1.94 | up | 0.0005988 | 0.0155 |
| 645166 | LOC645166 | 1.94 | up | 0.0000367 | 0.00146 |
| 692099 | FAM86DP | 0.66 | Down | 0.0000353 | 0.00113 |
| 729614 | FLJ37453 | 0.66 | Down | 0.0000694 | 0.00204 |
| 84981 | MIR22HG | 0.65 | down | 0.0011390 | 0.0251 |
| 284593 | FAM41C | 0.65 | down | 0.0004447 | 0.0107 |
| 84852 | ATP1A1-AS1 | 0.64 | down | 0.0000839 | 0.00241 |
| 197187 | SNAI3-AS1 | 0.64 | down | 0.0005752 | 0.0135 |
| 400752 | LINC01144 | 0.64 | down | 0.0015342 | 0.0329 |
| 100093630 | SNHG8 | 0.64 | down | 0.0000012 | 0.000065 |
| 57061 | HYMAI | 0.63 | down | 0.0000991 | 0.00279 |
| 282566 | LINC00515 | 0.63 | down | 0.0000965 | 0.00273 |
| 389741 | GLIDR | 0.63 | down | 0.0000010 | 0.000056 |
| 653162 | RPSAP9 | 0.63 | down | 0.0000372 | 0.00118 |
| 114814 | GNRHR2 | 0.62 | down | 0.0000001 | 0.00000149 |
| 645683 | RPL13AP3 | 0.62 | down | 0.0006937 | 0.016 |
| 84837 | ARHGAP5-AS1 | 0.61 | down | 0.0003332 | 0.00826 |
| 284835 | LINC00323 | 0.61 | down | 0.0011373 | 0.0251 |
| 574036 | SERTAD4-AS1 | 0.61 | down | 0.0014364 | 0.031 |
| 100128782 | LINC00476 | 0.61 | down | 0.0000001 | 0.00000889 |
| 84983 | FAM222A-AS1 | 0.6 | down | 0.0002945 | 0.00739 |
| 387066 | SNHG5 | 0.6 | down | 0.0000029 | 0.000135 |
FDR, false discovery rate.
Figure 1.
LncRNA LOC541471 expression in HNSCC and association with perineural invasion. (A) LncRNA LOC541471 expression was significantly higher in HNSCC tumor compared with normal tissue. (B) LncRNA LOC541471 expression was significantly higher in patients with HNSCC with perineural invasion compared with patients with no perineural invasion. (C) LncRNA LOC541471 expression was significantly higher in patients with a greater N classification. LncRNA LOC541471 was significantly higher in (D) tongue squamous cell carcinoma, (E) oral squamous cell carcinoma and (F) laryngeal squamous cell carcinoma compared with normal tissue. LncRNA, long non-coding RNA; HNSCC, head and neck squamous cell carcinoma; N, lymph node metastasis. *P<0.05, **P<0.01, ***P<0.001.
LncRNA LOC541471 as an independent predictor of OS and relapse-free survival of patients
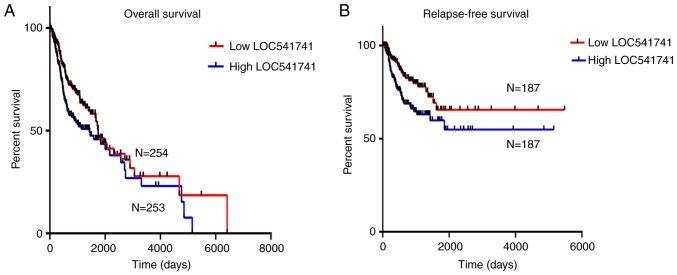
The lncRNA LOC541471 mRNA expression level was analyzed in cases of HNSCC from TCGA and evaluated for its prognostic value and potential as a prognostic biomarker for HNSCC. Kaplan-Meier survival analysis was performed according to the expression levels of lncRNA LOC541471. The results revealed that high expression of lncRNA LOC541471 was negatively associated with poor OS of patients compared with low expression of lncRNA LOC541471 (Fig. 2A). Univariate Cox regression analysis identified that lncRNA LOC541471 expression and age significantly affected OS of patients. Meanwhile, multivariate Cox regression analysis revealed that high lncRNA LOC541471 expression may be an independent predictor for reduced OS of patients with HNSCC (Table IV). Similarly, the results indicated that high lncRNA LOC541471 expression was negatively associated with relapse-free survival of patients compared with high lncRNA LOC541471 expression (Fig. 2B). A univariate analysis and multivariate Cox regression analysis also identified that high lncRNA LOC541471 expression may be an independent predictor for reduced relapse-free survival (Table V). These results indicate that lncRNA LOC541471 may be an independent predictor for OS and relapse-free survival of patients with HNSCC.
Figure 2.
High lncRNA LOC541471 expression is negatively associated with the (A) overall and (B) relapse-free survival of patients with head and neck squamous cell carcinoma (P<0.05). LncRNA, long non-coding RNA; N, number of samples.
Table IV.
Univariate and multivariate analysis of prognostic factors for overall survival using Cox proportional hazards regression model (n=487).
| Overall survival | ||||
|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | |||
| Variable | HR | 95% CI | HR | 95% CI |
| Age | ||||
| ≤60 | 0.722a | 0.541–0.963 | 0.772 | 0.571–1.043 |
| >60 | 1 | 1 | ||
| Sex | ||||
| Male | 0.753 | 0.555–1.023 | 0.801 | 0.583–1.102 |
| Female | 1 | 1 | ||
| Primary tumor stage | ||||
| T1-2 | 0.798 | 0.586–1.088 | 0.761 | 0.452–1.283 |
| T3-4 | 1 | 1 | ||
| Lymph node metastasis | ||||
| N0-1 | 0.807 | 0.591–1.101 | 0.753 | 0.533–1.064 |
| N2-3 | 1 | 1 | ||
| Clinical TNM staging | ||||
| I–II | 0.835 | 0.590–1.180 | 1.159 | 0.624–2.152 |
| III–IV | 1 | 1 | ||
| LOC541471 expression level | ||||
| Low | 0.677a | 0.507–0.904 | 0.722a | 0.539–0.968 |
| High | 1 | 1 | ||
P<0.05. HR, hazard ratio; CI, confidence interval.
Table V.
Univariate and multivariate analysis of prognostic factors for relapse-free survival using Cox proportional hazards regression model (n=355).
| Overall survival | ||||
|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | |||
| Variable | HR | 95% CI | HR | 95% CI |
| Age | ||||
| ≤60 | 0.937 | 0.604–1.454 | 0.974 | 0.617–1.536 |
| >60 | 1 | 1 | ||
| Sex | ||||
| Male | 1.026 | 0.619–1.703 | 1.075 | 0.637–1.814 |
| Female | 1 | 1 | ||
| Primary tumor stage | ||||
| T1-2 | 0.956 | 0.603–1.518 | 0.836 | 0.425–1.642 |
| T3-4 | 1 | 1 | ||
| Lymph node metastasis | ||||
| N0-1 | 0.685 | 0.437–1.072 | 0.626 | 0.375–1.047 |
| N2-3 | 1 | 1 | ||
| Clinical TNM staging | ||||
| I–II | 1 | 0.585–1.712 | 1.462 | 0.620–3.447 |
| III–IV | 1 | 1 | ||
| LOC541471 expression level | ||||
| Low | 0.563a | 0.360–0.881 | 0.574a | 0.365–0.904 |
| High | 1 | 1 | ||
P<0.05. HR, hazard ratio; CI, confidence interval.
Significant association of lncRNA LOC541471 with patient survival in subtypes of HNSCC. According to the anatomic neoplasm subdivision, 500 HNSCC samples from TCGA were classified as oropharyngeal, oral, laryngeal and hypopharyngeal carcinomas. On the basis of lncRNA LOC541471 expression levels, Kaplan-Meier survival analysis was performed for each subtype. The results revealed that high lncRNA LOC541471 expression was associated with poor OS compared with low lncRNA LOC541471 expression for all classified subtypes of HNSCC (Figs. 3–5).
Figure 3.
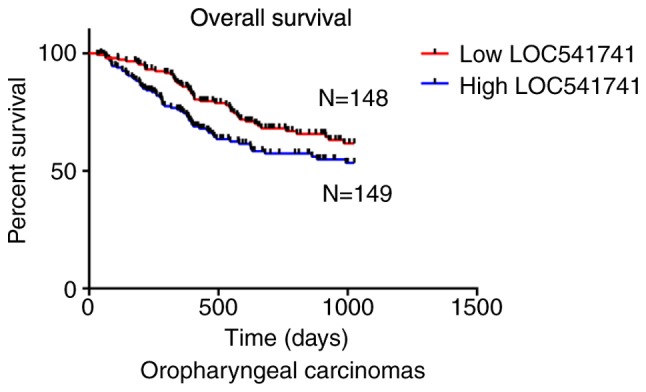
Negative association between high lncRNA LOC541471 expression and overall survival of patients with oropharyngeal carcinomas (P<0.05). LncRNA, long non-coding RNA; N, number of samples.
Figure 5.
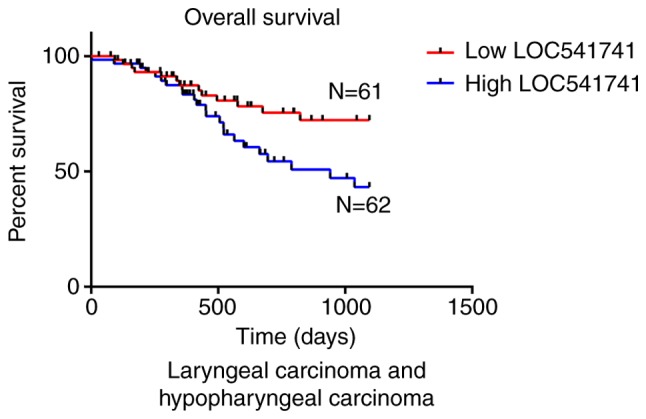
Negative association between high lncRNA LOC541471 expression and overall survival of patients with laryngeal carcinoma and hypopharyngeal carcinoma (P<0.05). LncRNA, long non-coding RNA; N, number of samples.
Discussion
Recent studies have revealed that up to 70% of the mammalian transcriptome is transcribed into lncRNAs, while coding transcripts only account for 2% of the genome (16,17). Previously, lncRNAs have been regarded as nonfunctional transcripts due to uncertainty of their function (17,18). However, recent studies have identified that lncRNAs serve an important role in tumor initiation and progression (19,20). In addition, previous studies have revealed that lncRNAs serve as vital participants in tumor cell proliferation, differentiation and apoptosis (9,21). Furthermore, lncRNAs exhibit structural stereospecificity and tissue selectivity (5,22). Therefore, dysregulated expression of lncRNAs has been identified as a biological marker in various cancer types. The lncRNAs, MALAT-1 and thymosin β-4, predict metastasis and survival in early-stage non-small cell lung cancer (23). The current study demonstrated that lncRNA LOC541471 expression was upregulated in tumor tissues and positively associated with increasing N classification and perineural invasion in patients with HNSCC. Previously, studies have revealed that perineural invasion and N classification are prognostic factors for HNSCC (15,24). Therefore, the current study analyzed the potential of lncRNA LOC541471 as a prognostic biomarker for HNSCC.
To further validate the hypothesis that lncRNA LOC541471 is a prognostic biomarker for HNSCC, clinical characteristics were downloaded from TCGA and analyzed. It was revealed that upregulation of lncRNA LOC541471 was associated with poor OS and poor relapse-free survival of patients. In addition, a multivariate Cox regression analysis revealed that lncRNA LOC541471 expression levels may be an independent predictor for the OS and relapse-free survival of patients. To determine whether lncRNA LOC541471 has similar prognostic potential in various subtypes of HNSCC, a total of 500 samples were classified as oropharyngeal, oral, laryngeal and hypopharyngeal. It was identified that upregulation of lncRNA LOC541471 was associated with poor survival outcomes in all subtypes of HNSCC studied. Therefore, it is proposed that lncRNA LOC541471 may have sufficient reliability and validity to serve as a prognostic biomarker for HNSCC.
The current study has indicated that LOC541471 may be considered an unfavorable prognostic factor for HNSCC. However, the specific mechanism by which LOC541471 participates in HNSCC development requires further investigation. Notably, lncRNAs can target RNA-binding proteins (RBPs) or competing endogenous RNA (ceRNA) (25). In future studies, the specific function of LOC541471, including targeting RBPs or ceRNA, may be investigated in vitro or in vivo. Identifying known genes located close to lncRNA could provide a basis for understanding its mechanism (26). Therefore, known genes located close to LOC541741 were analyzed using the University of California, Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu). A selection of these analysis results is provided in Table VI. Several of these genes have previously been identified to be associated with tumors. Low budding uninhibited by benzimidazoles 1 (BUB1) expression is an adverse prognostic marker in gastric adenocarcinoma. Tumors with low BUB1 expression have been associated with larger tumor size (27). Bcl-2-like protein 11 belongs to the Bcl-2 family, and acts as a central regulator of the intrinsic apoptotic cascade and mediates cell apoptosis (28). Proto-oncogene tyrosine-protein kinase MER inhibition combined with radiation therapy has a therapeutic effect in a subset of glioblastoma (29).
Table VI.
A selection of known genes located close to LOC541741.
| Ensembl_Gene_ID | Gene symbol | Gene_biotype |
|---|---|---|
| ENSG00000256671 | LIMS4 | protein_coding |
| ENSG00000183054 | RGPD6 | protein_coding |
| ENSG00000169679 | BUB1 | protein_coding |
| ENSG00000153093 | ACOXL | protein_coding |
| ENSG00000153094 | BCL2L11 | protein_coding |
| ENSG00000153107 | ANAPC1 | protein_coding |
| ENSG00000153208 | MERTK | protein_coding |
| ENSG00000153214 | TMEM87B | protein_coding |
| ENSG00000144152 | FBLN7 | protein_coding |
| ENSG00000144161 | ZC3H8 | protein_coding |
| ENSG00000235881 | AC114776.1 | lincRNA |
| ENSG00000236330 | RPL5P9 | processed_pseudogene |
| ENSG00000204581 | ACOXL-AS1 | antisense_RNA |
| ENSG00000230499 | AC108463.2 | lincRNA |
| ENSG00000263881 | MIR4436B2 | miRNA |
| ENSG00000266139 | MIR4435-2 | miRNA |
| ENSG00000266063 | MIR4771-2 | miRNA |
The current study preliminarily concludes that lncRNA LOC541471 is significantly associated with OS and relapse-free survival of patients and may be considered a potential unfavorable prognostic factor for HNSCC. However, information regarding the function of lncRNA LOC541471 is limited. Therefore, further studies are necessary to determine the involvement of lncRNA LOC541471 in mediating tumor biology.
Figure 4.
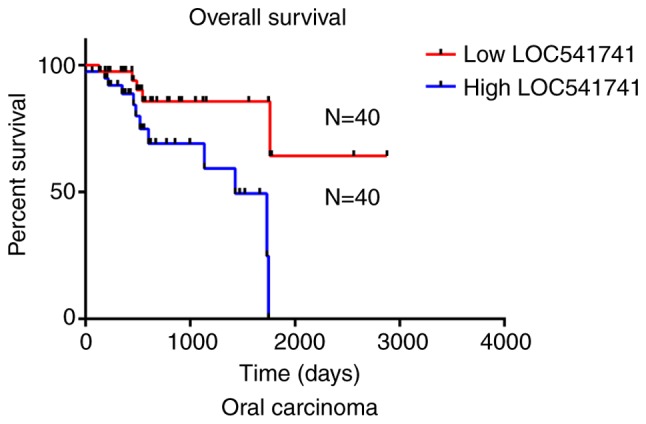
Negative association between high lncRNA LOC541471 expression and overall survival of patients with oral carcinoma (P<0.05). LncRNA, long non-coding RNA; N, number of samples.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- HNSCC
head and neck squamous cell carcinoma
- TCGA
The Cancer Genome Atlas
- lncRNA
long non-coding RNA
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 81774266).
Availability of data and materials
All the data and materials used in the present study are freely accessible in GEO (https://www.ncbi.nlm.nih.gov/geo/) and TCGA (https://cancergenome.nih.gov/).
Authors' contributions
HW performed the statistical analysis and wrote the manuscript. DY performed the statistical analysis. MW and TH designed the research, collected the data, and submitted the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Geisler S, Coller J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nohata N, Abba MC, Gutkind JS. Unraveling the oral cancer lncRNAome: Identification of novel lncRNAs associated with malignant progression and HPV infection. Oral Oncol. 2016;59:58–66. doi: 10.1016/j.oraloncology.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–761. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammerman PS, Hayes DN, Grandis JR. Therapeutic insights from genomic studies of head and neck squamous cell carcinomas. Cancer Discov. 2015;5:239–244. doi: 10.1158/2159-8290.CD-14-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. Int J Cancer. 2017;140:1955–1967. doi: 10.1002/ijc.30546. [DOI] [PubMed] [Google Scholar]
- 6.Cortez VS, Cervantes-Barragan L, Robinette ML, Bando JK, Wang Y, Geiger TL, Gilfillan S, Fuchs A, Vivier E, Sun JC, et al. Transforming growth factor-β signaling guides the differentiation of innate lymphoid cells in salivary glands. Immunity. 2016;44:1127–1139. doi: 10.1016/j.immuni.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ni W, Song E, Gong M, Li Y, Yao J, An R. Downregulation of lncRNA SDPR-AS is associated with poor prognosis in renal cell carcinoma. Onco Targets Ther. 2017;10:3039–3047. doi: 10.2147/OTT.S137641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao Y, Shi H, Ren F, Jia Y, Zhang R. Long non-coding RNA CCAT1 promotes metastasis and poor prognosis in epithelial ovarian cancer. Exp Cell Res. 2017;359:185–194. doi: 10.1016/j.yexcr.2017.07.030. [DOI] [PubMed] [Google Scholar]
- 9.Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: Current status and future perspectives. Mol Cancer. 2016;15:39. doi: 10.1186/s12943-016-0524-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenner A. Prostate cancer: PCA3 as a grade reclassification predictor. Nat Rev Urol. 2017;14:390. doi: 10.1038/nrurol.2017.70. [DOI] [PubMed] [Google Scholar]
- 11.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 12.Sacco AG, Cohen EE. Current treatment options for recurrent or metastatic head and neck squamous cell carcinoma. J Clin Oncol. 2015;33:3305–3313. doi: 10.1200/JCO.2015.62.0963. [DOI] [PubMed] [Google Scholar]
- 13.Cao W, Liu JN, Liu Z, Wang X, Han ZG, Ji T, Chen WT, Zou X. A three-lncRNA signature derived from the Atlas of ncRNA in cancer (TANRIC) database predicts the survival of patients with head and neck squamous cell carcinoma. Oral Oncol. 2017;65:94–101. doi: 10.1016/j.oraloncology.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 14.Kyzas PA, Loizou KT, Ioannidis JP. Selective reporting biases in cancer prognostic factor studies. J Natl Cancer Inst. 2005;97:1043–1055. doi: 10.1093/jnci/dji184. [DOI] [PubMed] [Google Scholar]
- 15.Dunne AA, Müller HH, Eisele DW, Kessel K, Moll R, Werner JA. Meta-analysis of the prognostic significance of perinodal spread in head and neck squamous cell carcinomas (HNSCC) patients. Eur J Cancer. 2006;42:1863–1868. doi: 10.1016/j.ejca.2006.01.062. [DOI] [PubMed] [Google Scholar]
- 16.Xu S, Wang P, You Z, Meng H, Mu G, Bai X, Zhang G, Zhang J, Pang D. The long non-coding RNA EPB41L4A-AS2 inhibits tumor proliferation and is associated with favorable prognoses in breast cancer and other solid tumors. Oncotarget. 2016;7:20704–20717. doi: 10.18632/oncotarget.8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou AE, Zheng H, Saad MA, Rahimy M, Ku J, Kuo SZ, Honda TK, Wang-Rodriguez J, Xuan Y, Korrapati A, et al. The non-coding landscape of head and neck squamous cell carcinoma. Oncotarget. 2016;7:51211–51222. doi: 10.18632/oncotarget.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu JW, Kong Y, Sun X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol. 2016;142:1571–1579. doi: 10.1007/s00432-016-2152-1. [DOI] [PubMed] [Google Scholar]
- 19.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Lena PG, Paz-Gallardo A, Paramio JM, Garcia-Escudero R. Clusterization in head and neck squamous carcinomas based on lncRNA expression: Molecular and clinical correlates. Clin Epigenetics. 2017;9:36. doi: 10.1186/s13148-017-0334-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: A new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Salvo TG. Evolving targeted therapies for right ventricular failure. Expert Opin Biol Ther. 2015;15:1263–1283. doi: 10.1517/14712598.2015.1054277. [DOI] [PubMed] [Google Scholar]
- 23.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 24.Fanelli MF, Oliveira TB, Braun AC, Corassa M, Abdallah EA, Nicolau UR, da Silva Alves V, Garcia D, Calsavara VF, Kowalski LP, Chinen LTD. Evaluation of incidence, significance, and prognostic role of circulating tumor microemboli and transforming growth factor-β receptor I in head and neck cancer. Head Neck. 2017;39:2283–2292. doi: 10.1002/hed.24899. [DOI] [PubMed] [Google Scholar]
- 25.Huang T, Huang W, Lu H, Zhang BY, Ma J, Zhao D, Wang YJ, Yu DH, He X. Identification and validation a TGF-β-associated long non-coding RNA of head and neck squamous cell carcinoma by bioinformatics method. J Transl Med. 2018;16:46. doi: 10.1186/s12967-018-1418-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du M, Huang T, Wu J, Gu JJ, Zhang N, Ding K, Qian LX, Lu ZW, Zhang WJ, Tian XK, et al. Long non-coding RNA n326322 promotes the proliferation and invasion in nasopharyngeal carcinoma. Oncotarget. 2017;9:1843–1851. doi: 10.18632/oncotarget.22828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stahl D, Braun M, Gentles AJ, Lingohr P, Walter A, Kristiansen G, Gütgemann I. Low BUB1 expression is an adverse prognostic marker in gastric adenocarcinoma. Oncotarget. 2017;8:76329–76339. doi: 10.18632/oncotarget.19357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Duan J, Qu Y, Deng T, Liu R, Zhang L, Bai M, Li J, Ning T, Ge S, et al. Onco-miR-24 regulates cell growth and apoptosis by targeting BCL2L11 in gastric cancer. Protein Cell. 2016;7:141–151. doi: 10.1007/s13238-015-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J, Frady LN, Bash RE, Cohen SM, Schorzman AN, Su YT, Irvin DM, Zamboni WC, Wang X, Frye SV, et al. MerTK as a therapeutic target in glioblastoma. Neuro Oncol. 2018;20:92–102. doi: 10.1093/neuonc/nox111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data and materials used in the present study are freely accessible in GEO (https://www.ncbi.nlm.nih.gov/geo/) and TCGA (https://cancergenome.nih.gov/).