Abstract
Background
Iron (Fe) is an essential element for plant growth and development, whereas cadmium (Cd) is non-essential and highly toxic. Previous studies showed that Fe deficiency enhanced Cd uptake and accumulation in peanuts. However, the molecular mechanism underlying the increased Cd accumulation in Fe-deficient peanut plants is poorly understood.
Results
We employed a comparative transcriptome analysis approach to identify differentially expressed genes (DEGs) in peanut roots exposed to Fe-sufficient without Cd, Fe-deficient without Cd, Fe-sufficient with Cd and Fe-deficient with Cd. Compared with the control, Fe deficiency induced 465 up-regulated and 211 down-regulated DEGs, whereas the up- and down-regulated DEGs in Cd exposed plants were 329 and 189, respectively. Under Fe-deficient conditions, Cd exposure resulted in 907 up-regulated DEGs and 953 down-regulated DEGs. In the presence of Cd, Fe deficiency induced 1042 up-regulated and 847 down-regulated genes, respectively. Based on our array data, we found that metal transporter genes such as CAX4, COPT1, IRT1, NRAMP5, OPT3, YSL3, VIT3 and VIT4 might be involved in iron homeostasis. Moreover, combined with quantitative real-time PCR, IRT1, NRAMP3, NRAMP5, OPT3, YSL3, ABCC3, ZIP1, and ZIP5 were verified to be responsible for Cd uptake and translocation in peanut plants under iron deficiency. Additionally, a larger amount of ABC transporter genes was induced or suppressed by iron deficiency under Cd exposure, indicating that this family may play important roles in Fe/Cd uptake and transport.
Conclusions
The up-regulated expression of NRAMP5 and IRT1 genes induced by iron deficiency may enhance Cd uptake in peanut roots. The decrease of Cd translocation from roots to shoots may be resulted from the down-regulation of ZIP1, ZIP5 and YSL3 under iron deficiency.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1654-9) contains supplementary material, which is available to authorized users.
Keywords: Peanut, Iron deficiency, Cadmium, Transcriptome, Gene expression
Background
Cadmium (Cd) is a non-essential and highly toxic heavy metal that is commonly released into the arable soil due to anthropogenic activities. The concentration of Cd in leaves greater than 5–10 μg g− 1 dry mass (DM) is toxic to non-tolerant crop plants [1]. For humans and animals, Cd may damage the mitochondrial and induce cell death by apoptosis and/or necrosis, leading to tissue inflammation and fibrosis [2]. Cd exposure has been shown to be associated with cancers of the prostate, lungs and testes in humans [3]. Due to its highly bioavailability, Cd is easily absorbed and accumulated in plants, and subsequently transferred to humans/animals via food chains. Thus, the presence of Cd in the arable soil can cause serious risks to human health. The minimization of Cd in edible parts of crops is an important demand especially in Cd-contaminated soil.
Iron (Fe) is an essential element that has multiple physiological functions including chlorophyll biosynthesis, photosynthesis, respiration, and redox reactions in plants. Despite its abundance in the earth’s crust, Fe often precipitates into insoluble Fe(III) oxides under aerobic conditions, especially in high-pH and calcareous soils [4]. Consequently, dissolved Fe in the soil solution is usually far lower than that required to sustain plant growth [5]. Iron deficiency causes chlorosis, growth retardation, and reduced crop productivity. It has become an important yield-limiting factor for crops growing in calcareous and alkaline soils.
Peanut (Arachis hypogaea L.) is one of the world’s fourth largest oilseed crop that is sensitive to Fe deficiency [6]. Peanut was also demonstrated to have a high capacity for accumulating Cd in both the seed and vegetative tissues depending on cultivars [7–9]. In previous studies, we have found that the uptake and accumulation of Cd in peanut plants is considerably enhanced by iron deficiency [10–13]. However, the mechanism underlying iron deficiency-induced increase of Cd accumulation in peanuts has not been well understood.
Generally, the accumulation of Cd in the shoot of plants is controlled by several biological processes, including (i) symplastic uptake by root epidermal cells, (ii) radial transport to the vascular cylinder, (iii) xylem loading, and (iv) root-to-shoot translocation [11]. Most of these processes are regulated by the transporters of several essential metals such as Fe, Mn, and Zn. During the past few years, several Fe transporters belonging to natural resistance associated macrophage proteins (Nramp), Zrt/Irt-like proteins (ZIP) and P1B-ATPases, have been identified to be responsible for the transport of Cd in plants [14]. Thomine et al. [15] found that iron deficiency could induce the expression of AtNramp3 and AtNramp4 in Arabidopsis. The increase of Cd uptake and accumulation under iron deficiency has been confirmed to be mediated by Fe2+ transporters such as OsIRT1 and OsIRT2 in rice [16]. Similarly, our results showed that the expression of AhIRT1 and AhNramp1 genes in peanut roots can be induced by iron deficiency, which is associated with Cd uptake and accumulation [13]. He et al. [17] demonstrated that Fe supply prevents Cd uptake in Arabidopsis by inhibiting AtIRT1 expression and enhancing antagonism between Fe and Cd uptake.
Taken together, we hypothesize that iron deficiency-induced expression of Fe2+ transporter genes may be responsible for the increased uptake and accumulation of Cd in iron-deficient peanut plants. To test this hypothesis, a comparative transcriptome analysis was carried out on iron-sufficient and -deficient peanut plants under Cd exposure. The aims were: (i) to obtain the gene expression patterns in the roots under iron deficiency; (ii) to identify the key genes related to Fe/Cd uptake and translocation; and (iii) to elucidate the gene regulatory network that are responsible for Cd uptake and translocation in peanut plants under iron deficiency. The results presented here will be useful for better understanding the mechanism of Cd accumulation induced by Fe deficiency and facilitate gene function studies in peanut.
Results
The accumulation and translocation of cd and Fe in plants
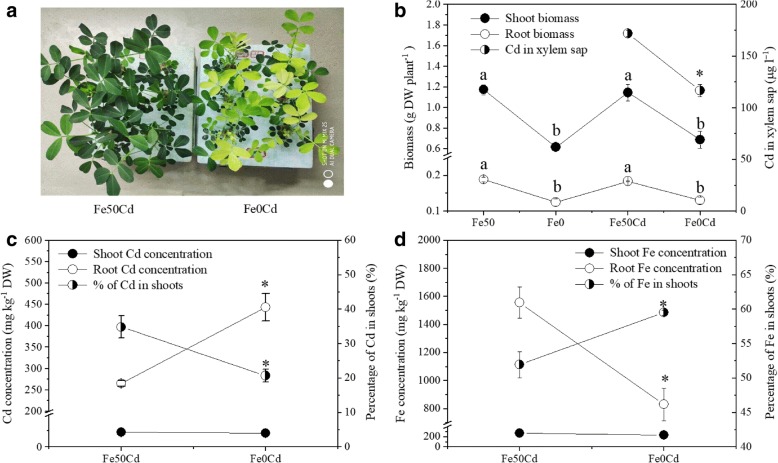
Figure 1 shows plant growth as well as the accumulation and translocation of Cd and Fe in peanut plants. Compared with normal Fe supply, Fe-deficiency inhibited plant growth and resulted in leaf chlorosis (Fig. 1 a and b), while Cd did not change plant growth in both Fe treatments (Fig. 1b). It was also observed that Fe-deficiency significantly increased root Cd concentrations but decreased Cd concentration in xylem sap and the percentage of Cd in shoots (Fig. 1 b and c), whereas Cd concentrations in the shoot were not affected (Fig. 1c). These results showed good agreement with previous findings [10–13], suggesting that Fe deficiency increased uptake but decreased root-to-shoot translocation of Cd in peanut plants. In comparison with Fe-deficient plants, Fe-sufficient plants exhibited lower root Fe concentration and higher proportion of Fe in shoots (Fig. 1d). The reverse trend in the percentage of Cd and Fe in shoots indicates that the two ions might compete with each other during the translocation from roots to shoots.
Fig. 1.
Growth (a and b) and the accumulation and translocation of Cd (c) and Fe (d) in Fe-sufficient (Fe50Cd) and Fe-deficient (Fe0Cd) peanut plants exposed to 2 μM Cd for seven days. Asterisk (*) above error bars indicate values (mean ± SE, n = 3) are significantly different between Fe-sufficient and Fe-deficient plants according to independent-samples t-test at 0.05 the level. Different letters above error bars indicate significant difference at 0.05 the level according to ANOVA followed by a Duncan multiple-range test
RNA sequencing analysis of eight cDNA libraries
To assess the global transcriptome profile of peanut roots in response to Fe-deficiency and/or Cd exposure, RNA-Seq analysis was performed on peanut roots exposed to Fe-sufficient without Cd (Fe50), Fe-deficient without Cd (Fe0), Fe-sufficient with Cd (Fe50Cd) and Fe-deficient with Cd (Fe0Cd). Two biological replicates were performed per treatment, with a total of eight cDNA libraries constructed. A total of 58,774,869, 54,456,644, 55,033,082, and 58,221,919 raw reads were produced from two biological replicate libraries of Fe50, Fe0, Fe50Cd, and Fe0Cd respectively. After removing low quality reads and reads containing adapter sequences, a total of 57,331,325, 53,124,279, 54,059,155, and 56,589,717 clean reads remained for Fe50, Fe0, Fe50Cd, and Fe0Cd, respectively. The percentage of clean reads in all eight libraries was more than 97.14%, and the percentage of Q20 exceeded 97.44% (Table 1). Pearson’s correlation coefficients for all biological replicates were 0.949–0.959 (Additional file 1: Table S1), suggesting the data were highly reproducible.
Table 1.
Overview of raw and clean reads in Fe-sufficient (Fe50) and Fe-deficient (Fe0) peanut plants exposed to 0 or 2 μM CdCl2 (Cd) for seven days
| Sample | Total raw reads | Total clean reads | Total clean bases (gb) | Clean reads q20 (%) | Clean reads q30 (%) | Clean reads in raw reads (%) | GC content (%) |
|---|---|---|---|---|---|---|---|
| Fe50_1 | 28,406,266 | 27,712,721 | 8.31 | 0.02 | 97.56 | 93.09 | 97.56 |
| Fe50_2 | 30,368,603 | 29,618,604 | 8.89 | 0.02 | 97.64 | 93.33 | 97.53 |
| Fe0_1 | 28,516,330 | 27,732,406 | 8.32 | 0.02 | 97.63 | 93.3 | 97.25 |
| Fe0_2 | 25,940,314 | 25,391,873 | 7.62 | 0.02 | 97.44 | 92.86 | 97.89 |
| Fe0Cd_1 | 28,152,785 | 27,619,944 | 8.29 | 0.02 | 97.61 | 93.23 | 98.11 |
| Fe0Cd_2 | 26,880,297 | 26,439,211 | 7.93 | 0.02 | 97.69 | 93.47 | 98.36 |
| Fe50Cd_1 | 33,142,927 | 32,227,575 | 9.67 | 0.02 | 97.68 | 93.43 | 97.24 |
| Fe50Cd_2 | 25,078,992 | 24,362,142 | 7.31 | 0.02 | 97.63 | 93.31 | 97.14 |
The high-quality clean reads were mapped to the A. duranensis genome (reference genome) using HISAT (Hierarchical indexing for spliced alignment of transcripts). Ultimately, more than 79% of the clean reads were successfully mapped for all cDNA libraries, and over 77% were observed to be unique mapped reads (Table 2), suggesting that the samples were comparable.
Table 2.
Mapping results of clean reads against the peanut genomic sequence
| sample | Total clean reads | Total mapped reads | Uniquely mapped reads | Multiple mapped reads | Spliced reads | Unspliced reads | Properly mapped reads |
|---|---|---|---|---|---|---|---|
| Fe50_1 | 55,425,442 | 43,980,460 (79.35%) | 42,937,535 (77.47%) | 1,042,925 (1.88%) | 16,275,516 (29.36%) | 26,662,019 (48.1%) | 38,544,100 (69.54%) |
| Fe50_2 | 59,237,208 | 46,911,319 (79.19%) | 45,798,117 (77.31%) | 1,113,202 (1.88%) | 17,286,768 (29.18%) | 28,511,349 (48.13%) | 41,375,852 (69.85%) |
| Fe0_1 | 55,464,812 | 45,419,922 (81.89%) | 44,410,704 (80.07%) | 1,009,218 (1.82%) | 16,915,252 (30.5%) | 27,495,452 (49.57%) | 40,481,232 (72.99%) |
| Fe0_2 | 50,783,746 | 40,804,086 (80.35%) | 39,847,134 (78.46%) | 956,952 (1.88%) | 14,885,388 (29.31%) | 24,961,746 (49.15%) | 36,103,056 (71.09%) |
| Fe0Cd_1 | 55,239,888 | 44,105,861 (79.84%) | 42,995,894 (77.83%) | 1,109,967 (2.01%) | 16,183,762 (29.3%) | 26,812,132 (48.54%) | 38,747,872 (70.14%) |
| Fe0Cd_2 | 52,878,422 | 42,298,150 (79.99%) | 41,248,604 (78.01%) | 1,049,546 (1.98%) | 15,493,356 (29.3%) | 25,755,248 (48.71%) | 37,208,590 (70.37%) |
| Fe50Cd_1 | 64,455,150 | 51,650,748 (80.13%) | 50,399,248 (78.19%) | 1,251,500 (1.94%) | 18,847,959 (29.24%) | 31,551,289 (48.95%) | 45,817,274 (71.08%) |
| Fe50Cd_2 | 48,724,284 | 38,614,623 (79.25%) | 37,716,105 (77.41%) | 898,518 (1.84%) | 13,970,730 (28.67%) | 23,745,375 (48.73%) | 34,228,200 (70.25%) |
Identification of differentially expressed genes (DEGs)
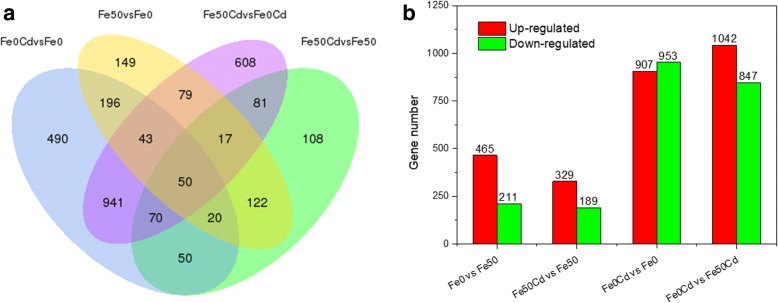
A total of 63,191 genes including 34,553 known genes and 18,030 novel genes, 8017 lncRNA, 35 misc. RNA, 580 tRNA and 1976 pseudogenes were identified in eight cDNA libraries. Pairwise comparison analysis for each gene were performed between Fe-sufficient and Fe-deficient treatments (Fe0/Fe50 and Fe0Cd/Fe50Cd), or between Cd-absent and Cd-present treatments (Fe50Cd/Fe50 and Fe0Cd/Fe0). DEGs were identified by the threshold of Padj-value < 0.05. As a result, a total of 3024 genes were differentially regulated in the four comparisons, of which 676, 1889, 518, and 1860 DEGs were identified in Fe0 vs Fe50, Fe0Cd vs Fe50Cd, Fe50Cd vs Fe50 and Fe0Cd vs Fe0, respectively (Fig. 2a). Regardless of the absence or presence of Cd, 189 genes were differentially expressed between Fe-sufficient and Fe-deficient treatments, suggesting that these genes were specifically induced or supressed by Fe deficiency. It was also found that 190 genes were specifically regulated by Cd exposure under both Fe treatments (Fig. 2a).
Fig. 2.
Gene expression profile of in Fe-sufficient (Fe50) and Fe-deficient (Fe0) peanut plants exposed to 0 or 2 μM CdCl2 (Cd) for seven days. (a) Venn diagram of DEGs. (b) The total number of up-regulated and down-regulated genes
Compared with the control (Fe50), Fe deficiency (Fe0) induced 465 up-regulated and 211 down-regulated DEGs, whereas the up- and down-regulated DEGs in Cd-exposed plants (Fe50Cd) were 329 and 189, respectively (Fig. 2b). Under Fe-deficient conditions, Cd exposure (Fe0Cd) resulted in 907 up-regulated DEGs and 953 down-regulated DEGs. In the presence of Cd, Fe deficiency (Fe0Cd) induced 1042 up-regulated and 847 down-regulated genes (Fig. 2b).
Gene ontology (GO) analysis of DEGs
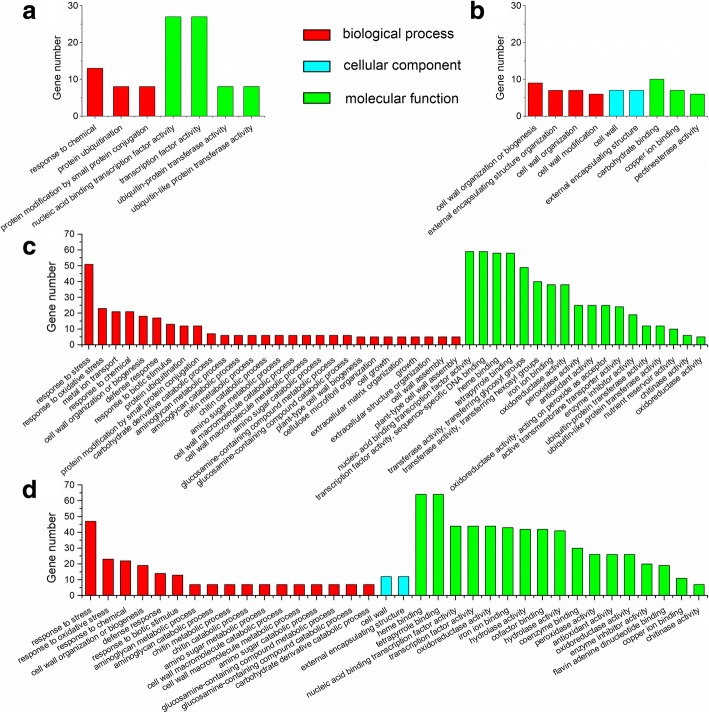
GO assignments were used to classify the functions of DEGs, and the results of significantly enriched GO terms (Padj-value < 0.05) were shown in Fig. 3. A total of 99 Fe deficiency-responsive DEGs (Fe0 vs Fe50) were assigned into 7 enriched GO terms consisting of 3 biological process (response to chemical, protein ubiquitination, protein modification by small protein conjugation) and 4 molecular function (nucleic acid binding transcription factor activity, transcription factor activity, ubiquitin-protein transferase activity, ubiquitin-like protein transferase activity) (Fig. 3a). Meanwhile, 66 Cd-responsive DEGs were assigned into 9 enriched GO terms, including 4 biological process (cell wall organization or biogenesis, external encapsulating structure organization, cell wall organization, cell wall modification), 2 cellular components (cell wall, external encapsulating structure) and 3 molecular function (carbohydrate binding, copper ion binding, pectinesterase activity) (Fig. 3b). A total of 857 DEGs between Fe0Cd and Fe0 were assigned into 46 enriched GO terms consisting of 28 biological process (response to stress, response to oxidative stress, metal ion transport, response to chemical, etc.) and 18 molecular function (nucleic acid binding transcription factor activity, transcription factor activity, heme binding, tetrapyrrole binding, etc.) subcategories (Fig. 3c), and 832 DEGs of Fe0Cd vs Fe50Cd were assigned into 36 enriched GO terms including 17 biological process (response to stress, response to oxidative stress, response to chemical, etc.), 2 cellular components (cell wall, external encapsulating structure) and 17 molecular function (heme binding, tetrapyrrole binding, etc.) subcategories (Fig. 3d).
Fig. 3.
Gene ontology classification of DEGs identified. The enriched biological process, cellular component and molecular function GO terms of DEGs between Fe-sufficient and Fe-deficient peanut roots prior to (a) or after (d) Cd exposure, and between Cd-free and Cd-treated peanut roots under Fe-sufficient (b) and Fe-deficient (c) conditions
KEEG metabolic pathway analysis of DEGs
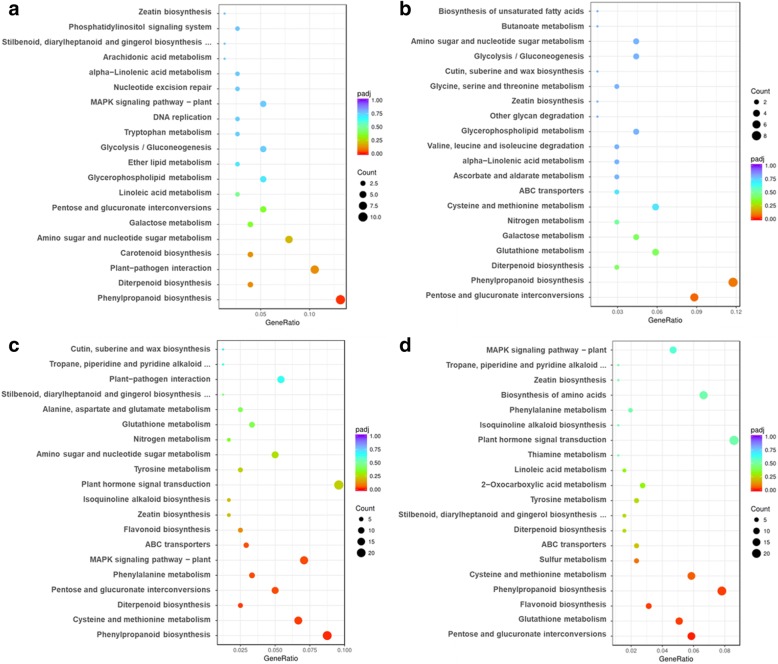
To deep insight the molecular interactions among the genes, DEGs were further annotated with Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.ad.jp/kegg/). A total of 76 (Fe0 vs Fe50), 256 (Fe0Cd vs Fe50Cd), 68 (Fe50Cd vs Fe50) and 240 (Fe0Cd vs Fe0) DEGs assigned to 48, 86, 47, and 88 pathways, respectively (Additional file 2: Table S2). The four comparisons differed from each other in metabolic pathways of DEGs (Fig. 4). The top five pathways of Fe deficiency-responsive DEGs were phenylpropanoid biosynthesis, plant-pathogen interaction, diterpenoid biosynthesis, carotenoid biosynthesis, and amino sugar and nucleotide sugar metabolism (Fig. 4a). Among these pathways, phenylpropanoid biosynthesis was identified as significantly enriched (Padj-value < 0.05). For Cd-responsive DEGs, pentose and glucuronate interconversions, phenylpropanoid biosynthesis, glutathione metabolism, galactose metabolism, and diterpenoid biosynthesis were the top five categories, and no enriched pathway was identified (Fig. 4b). A larger number of enriched pathways were found in DEGs of Fe0Cd vs Fe50Cd (pentose and glucuronate interconversions, glutathione metabolism, flavonoid biosynthesis, and phenylpropanoid biosynthesis) (Fig. 4c) and Fe0Cd vs Fe0 (phenylpropanoid biosynthesis, cysteine and methionine metabolism, diterpenoid biosynthesis, MAPK signaling pathway, pentose and glucuronate interconversions, phenylalanine metabolism, and ABC transporters) (Fig. 4d).
Fig. 4.
KEGG pathway enrichment analysis based on the differentially expressed genes between Fe-sufficient and Fe-deficient peanut roots prior to (a) or after (c) Cd exposure, and between Cd-free and Cd-treated peanut roots under Fe-sufficient (b) and Fe-deficient (d) conditions
DEGs involved in heavy metal transport
According to GO functional annotation, 54 DEGs were identified to highly similar with transporters that may be involved in the translocation of heavy metal in plants, including ATP-binding cassette transporters (ABC transporters), cation/H+ antiporters (CAXs), copper transport proteins (COPTs), Fe2+ transport proteins (IRTs), natural resistance-associated macrophage proteins (NRAMPs), metal tolerance proteins (MTPs), metal-nicotianamine transporter or yellow stripe-like transporters (YSLs), oligopeptide transporters (OTPs), vacuolar iron transporters (VITs), and zinc transporters (ZIPs) (Table 3). In the absence of Cd, ABCB19, putative ABCC15, ABCG15-like, CAX4-like, COPT1, IRT1 (107484081 and 107494754), NRAMP5 and OPT3 were up-regulated by Fe deficiency, while three genes (YSL3, VIT3 and VIT4) were down-regulated. In the presence of Cd, a total of 37 DEGs encoding metal transporters were detected between the Fe-sufficient and Fe-deficient peanut plants, including 18 up-regulated genes (ABCA2, ABCA7, ABCB19, ABCB21, ABCB11, ABCC3, ABCC8, ABCC15, ABCF1, ABCG6, ABCG11, HMA5, CAX18, Nramp3, OPT3, IRT1) and 19 down-regulated genes (ABCB11, ABCB19, ABCG5, ABCG8, ABCG32, ABCG39, COPT6, OPT7, YSL3, YSL7, VIT4, VIT3, ZIP1, ZIP5, ZIP6). Cd exposure induced seven metal transport genes in Fe-sufficient plants, including ABCB11, ABCB19, IRT1 (110279939), NRAMP5, HMA5, VIT4 (107468698) and ZIP11. In contrast, 32 transporter genes were induced or suppressed by Cd exposure under Fe deficient stress, including 19 up-regulated genes (ZIP11, ABCC4, ABCC3, ABCC8, YSL3, ABCG11, CAX20, ABCG6, NRAMP3, HMA5, OPT3, ABCB11, CAX18, ABCG11, ABCC3, ABCB19, IRT1) and 13 down-regulated genes (ABCG15, CAX4, VIT3, COPT6, COPT1, COPT2, ABCB19, ABCG5, ABCB2, ABCB11, ZIP6, ABCG5). Compared with the control, exposure of Fe-deficient plants to Cd up-regulated ABCB11, ABCB19, ABCC3, ABCC8, ABCC15, ABCF1, ABCG6, ABCG11, CAX18-like, CAX20, IRT1, NRAMP3, NRAMP5, MTP11 and OPT3, but down-regulated ABCG5, ABCG8, ABCG25, COPT1, VIT3, VIT4, ZIP1, ZIP5 and ZIP6. More importantly, it was found that three up-regulated genes (OPT3, ABCC15, IRT1) and three down-regulated genes (VIT3, VIT4, YSL3) were specifically regulated by Fe-deficiency, regardless of Cd treatments. Similarly, two genes (ZIP11 and HMA5) were found to be specifically induced by Cd in both the Fe-sufficient and Fe-deficient peanut plants.
Table 3.
DEGs possibly involved in metal transport in Fe-sufficient and Fe-deficient peanut plants exposed to 0 or 2 μM CdCl2 for seven days
| Gene_ID | Gene description | Abbr. | Fe0vsFe50 | Fe50CdvsFe50 | Fe0CdvsFe50 | Fe0CdvsFe0 | Fe0CdvsFe50Cd | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| log2Fold Change | padj | log2Fold Change | padj | log2Fold Change | padj | log2Fold Change | padj | log2Fold Change | padj | |||
| 107491333 | ABC transporter A family member 2 | ABCA2 | 0.753 | 0.013 | ||||||||
| 107491332 | ABC transporter A family member 7, transcript variant × 1 | ABCA7 | 0.784 | 0.006 | ||||||||
| 107479052 | ABC transporter B family member 2-like, transcript variant X5 | ABCB2 | − 1.713 | 0.034 | ||||||||
| 107467550 | ABC transporter B family member 11 | ABCB11 | 2.600 | 0.000 | 2.761 | 0.000 | 2.126 | 0.000 | ||||
| 107495097 | ABC transporter B family member 11-like | ABCB11 | 1.352 | 0.009 | −1.652 | 0.008 | −2.400 | 0.000 | ||||
| 107471179 | ABC transporter B family member 19 | ABCB19 | 1.270 | 0.013 | 1.005 | 0.010 | −2.171 | 0.000 | −1.889 | 0.000 | ||
| 107476483 | ABC transporter B family member 19 | ABCB19 | 2.380 | 0.000 | 3.120 | 0.000 | 1.561 | 0.020 | ||||
| 107492327 | ABC transporter B family member 21-like | ABCB21 | 2.000 | 0.030 | ||||||||
| 107463694 | ABC transporter C family member 3 | ABCC3 | 1.313 | 0.000 | 1.045 | 0.004 | 1.339 | 0.000 | ||||
| 107496250 | ABC transporter C family member 3 | ABCC3 | 3.254 | 0.000 | 3.073 | 0.000 | 2.248 | 0.000 | ||||
| 107496270 | ABC transporter C family member 3-like transcript variant ×2 | ABCC3 | −2.894 | 0.044 | ||||||||
| 107475935 | ABC transporter C family member 4 | ABCC4 | 0.890 | 0.020 | ||||||||
| 107464256 | ABC transporter C family member 8, transcript variant ×2 | ABCC8 | 1.918 | 0.000 | 1.132 | 0.010 | 1.548 | 0.000 | ||||
| 107481980 | putative ABC transporter C family member 15, transcript variant X2 | ABCC15 | 2.018 | 0.000 | 2.888 | 0.000 | 2.391 | 0.000 | ||||
| 107476479 | ABC transporter F family member 1, transcript variant ×1 | ABCF1 | 1.661 | 0.000 | 1.193 | 0.007 | ||||||
| 107470452 | ABC transporter G family member 5-like | ABCG5 | −1.958 | 0.000 | −2.157 | 0.000 | −1.762 | 0.000 | ||||
| 107480222 | ABC transporter G family member 5-like | ABCG5 | −1.109 | 0.028 | −1.497 | 0.002 | −1.326 | 0.002 | ||||
| 107467717 | ABC transporter G family member 6-like | ABCG6 | 1.465 | 0.042 | 1.621 | 0.001 | 1.010 | 0.013 | ||||
| 107466837 | ABC transporter G family member 8-like | ABCG8 | −1.145 | 0.041 | − 1.331 | 0.001 | ||||||
| 107481537 | ABC transporter G family member 11 | ABCG11 | 1.012 | 0.028 | 1.305 | 0.001 | 1.107 | 0.002 | ||||
| 107481662 | ABC transporter G family member 11, transcript variant X2 | ABCG11 | 4.356 | 0.000 | 3.055 | 0.000 | ||||||
| 107492554 | ABC transporter G family member 15-like | ABCG15 | 5.706 | 0.032 | −5.572 | 0.019 | ||||||
| 107467862 | ABC transporter G family member 25 | ABCG25 | −1.042 | 0.018 | −0.948 | 0.049 | ||||||
| 107469510 | ABC transporter G family member 32 | ABCG32 | −1.273 | 0.020 | ||||||||
| 107493900 | ABC transporter G family member 39-like | ABCG39 | −2.463 | 0.017 | ||||||||
| 110273497 | Cation/H+ antiporter 4-like | CAX4 | 4.519 | 0.001 | −4.765 | 0.001 | ||||||
| 107479804 | Cation/H+ antiporter 18-like, transcript variant X2 | CAX18 | 1.903 | 0.000 | 2.838 | 0.000 | 2.334 | 0.000 | ||||
| 107491686 | Cation/H+ antiporter 20 | CAX20 | 1.106 | 0.048 | 1.366 | 0.003 | ||||||
| 107474428 | Copper transport protein ATX1 | 0.981 | 0.006 | |||||||||
| 107477078 | Copper transporter 1 | COPT1 | 1.376 | 0.036 | −1.926 | 0.002 | −3.309 | 0.000 | ||||
| 107484791 | Copper transporter 2-like, transcript variant ×1 | COPT2 | −2.964 | 0.000 | ||||||||
| 107484785 | Copper transporter 6 | COPT6 | −3.354 | 0.026 | −3.432 | 0.017 | ||||||
| 107484081 | Fe2+ transport protein 1 | IRT1 | 3.342 | 0.048 | 3.472 | 0.019 | ||||||
| 107491017 | Fe2+ transport protein 1 | IRT1 | 9.778 | 0.000 | 7.200 | 0.000 | 9.773 | 0.000 | ||||
| 107494754 | Fe2+ transport protein 1-like | IRT1 | 6.608 | 0.023 | 11.806 | 0.000 | 5.210 | 0.000 | 8.432 | 0.000 | ||
| 110279939 | Fe2+ transport protein 1-like | IRT1 | 6.658 | 0.020 | ||||||||
| 107486418 | Metal tolerance protein 11, transcript variant x2 | MTP11 | 0.890 | 0.046 | ||||||||
| 107480469 | Metal transporter Nramp3, transcript variant ×1 | Nramp3 | 2.912 | 0.000 | 1.898 | 0.000 | 3.565 | 0.000 | ||||
| 107460699 | Metal transporter Nramp5 | Nramp5 | 1.945 | 0.000 | 1.633 | 0.001 | 1.325 | 0.001 | ||||
| 107460374 | Metal-nicotianamine transporter | YSL3 | −1.920 | 0.000 | 1.230 | 0.000 | −0.648 | 0.042 | ||||
| 107489952 | Probable metal-nicotianamine transporter | YSL7 | −0.901 | 0.012 | ||||||||
| 107466086 | Oligopeptide transporter 3 | OPT3 | 1.857 | 0.000 | 4.594 | 0.000 | 2.725 | 0.000 | 4.720 | 0.000 | ||
| 107482451 | Oligopeptide transporter 7, transcript variant x1 | OPT7 | −2.576 | 0.014 | ||||||||
| 107471962 | Probable copper-transporting ATPase | HMA5 | 1.311 | 0.000 | 2.409 | 0.000 | 1.311 | 0.000 | ||||
| 107491634 | Vacuolar iron transporter homolog 3 | VIT3 | −2.675 | 0.000 | −6.523 | 0.001 | −3.851 | 0.002 | −6.562 | 0.001 | ||
| 107468698 | Vacuolar iron transporter homolog 4 | VIT4 | 2.925 | 0.032 | −5.203 | 0.001 | ||||||
| 107481738 | Vacuolar iron transporter homolog 4-like | VIT4 | −1.108 | 0.005 | −1.493 | 0.000 | ||||||
| 107481739 | Vacuolar iron transporter homolog 4-like | VIT4 | −4.145 | 0.000 | −7.910 | 0.000 | −8.370 | 0.000 | ||||
| 107461527 | Zinc transporter 1, transcript variant X1 | ZIP1 | −1.507 | 0.000 | −1.671 | 0.000 | ||||||
| 107482454 | Zinc transporter 5, transcript variant X2 | ZIP5 | −3.298 | 0.007 | ||||||||
| 107494273 | Zinc transporter 5-like | ZIP5 | −6.223 | 0.021 | ||||||||
| 107458282 | Zinc transporter 6, chloroplastic | ZIP6 | −0.933 | 0.036 | ||||||||
| 107473088 | Zinc transporter 6, chloroplastic | ZIP6 | −1.994 | 0.000 | −1.562 | 0.001 | −1.418 | 0.006 | ||||
| 107462023 | Zinc transporter 11 | ZIP11 | 0.925 | 0.009 | 0.831 | 0.034 | −0.636 | 0.036 | ||||
Data of DEGs showing a similar expression in pairwise comparisons were not shown in the table
Verification of the DEG results
To further verify the transcriptome data, ten DEGs involved in metal transport, including IRT1, NRAMP3, NRAMP5, OPT3, YSL3, CAX4, HMA5, ABCC3, ZIP1, and ZIP5, were selected for RT-qPCR analysis. The RT-qPCR results, presented in Fig. 5, showed a good agreement with the RNA-Seq data (Table 3). Under Cd-free condition, Fe deficiency up-regulated the expression of CAX4-like, NRAMP5 and OPT3, but down-regulated YSL3, ZIP1 and ZIP5 in peanut roots. For Fe-sufficient plants, the expression of IRT1, NRAMP5, HMA5 and ZIP1 was significantly induced by Cd exposure. Compared with the control, Cd exposure with Fe deficiency enhanced the expressions of OPT3, ABCC3, HMA5, and NRAMP3, but decreased those of YSL3, ZIP1 and ZIP5. Pearson’s correlation analysis showed that the relative gene expression (RT-qPCR) is significantly correlated with the foldchange of read counts (RNA-Seq) (r = 0.705, p = 0.000, n = 40). The high confirmation rate demonstrates the reliability of our RNA-Seq data.
Fig. 5.
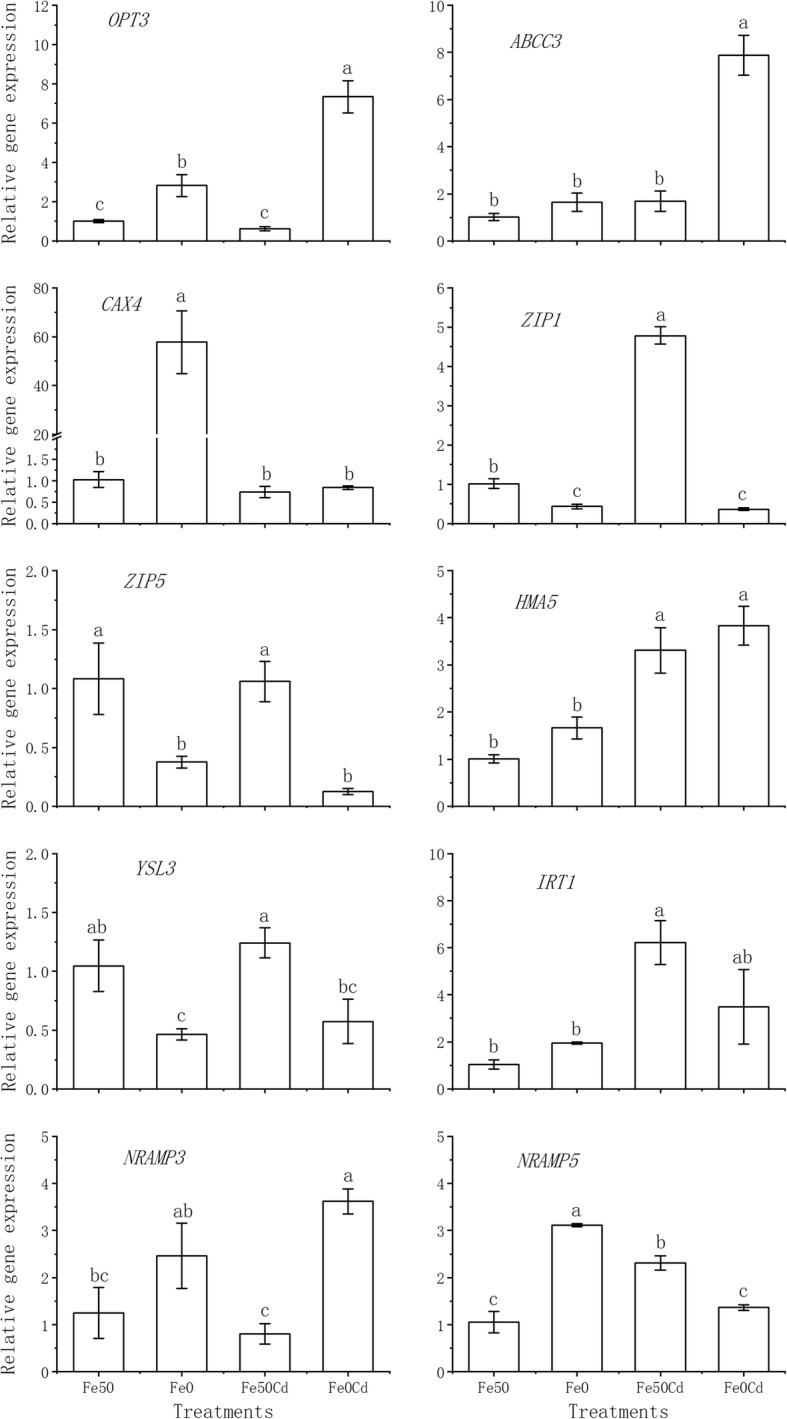
The qRT-PCR analysis of metal transport-related genes in peanut roots exposed to 0 or 2 μM Cd under Fe-sufficient (Fe50) and Fe-deficient (Fe0) conditions. The relative expression of each gene was calculated as the 2−ΔΔCT value and normalized by geometric mean of three stably expressed reference genes. Data are presented as means with SE (n = 3). Different letters above error bars indicate that values are significantly different (P < 0.05) according to Duncan’s multiple range test after one-way analysis of variance
Discussion
Although iron deficiency has been demonstrated to considerably enhance the uptake and accumulation of Cd in peanut plants [10–13], limited information is available about the physiological and molecular mechanisms underlying iron deficiency-induced increase of Cd accumulation in peanut. RNA-seq analysis has been used for revealing the molecular mechanisms of Cd uptake, translocation and accumulation in many plant species [18–20]. The complete genome sequencing of A. duranensis and A. ipaensis, the diploid ancestors of cultivated peanut [21], has shed light to genomic studies on cultivated peanut. However, sequence information of peanut in response to iron deficiency and Cd exposure is scarce. In this study, we obtained 57,331,325, 53,124,279, 54,059,155, and 56,589,717 clean reads from the peanut roots treated with Fe50, Fe0, Fe50Cd, and Fe0Cd respectively. More than 79% of clean reads for all cDNA libraries were functionally annotated in the A. duranensis genome [21], and more than 77% were unique mapped reads (Table 2). A total of 3024 genes were identified to be DEGs in pairwise comparisons: Fe0 vs Fe50 (676), Fe0Cd vs Fe50Cd (1889), Fe50Cd vs Fe50 (518), and Fe0Cd vs Fe0 (1860) (Fig. 2a). Of them, 54 DEGs were identified to highly similar with transporters that may be involved in the uptake and translocation of Fe/Cd in plants. These results provide clues to mechanisms underpinning Cd uptake and accumulation in Fe-deficient peanut plants.
Genes involved in Fe uptake and translocation were greatly induced by Fe deficiency in peanut roots. As an Fe-efficient plant that develops strategy I mechanism in response to Fe deficiency, peanut takes up Fe2+ by increasing Fe2+ transporter coupled with an increase of ferric reductase activity and rhizosphere acidification by releasing protons from the roots under Fe-deficient conditions [6]. We found that both the Fe2+ transport gene IRT1 and ferric reductase FRO1 were up-regulated by Fe deficiency in peanut roots (Table 3, Fig. 5). The result indicates that IRT1/FRO1 system constitutes the major pathway for Fe entry into root epidermal cells, and induction of these genes might improve Fe nutrition under Fe-deficient stress. A transporter gene of NRAMP family, NRAMP5 (NRAMP1) [13], was also highly induced in roots under Fe-deficient conditions (Table 3, Fig. 5). The NRAMP5 (NRAMP1) have been shown to function in Fe transport in several plants such as Arabidopsis [22], Noccaea caerulescens [23], Malus baccata [24], peanut [25] and rice [26]. The induction of NRAMP5 (NRAMP1) under Fe-deficiency indicates that this gene is involved in the uptake of Fe by peanut roots. Interestingly, both the IRT1 and NRAMP5 are shown to be involved in Cd transport into root cells across membrane [13, 16, 27]. The current results confirmed that iron deficiency-induced expression of IRT1 and NRAMP5 may be responsible for the increase of Cd uptake and accumulation in Fe-deficient peanut plants [13].
We also found that large number of ABC transporter genes were significantly induced or suppressed by Fe deficiency and/or Cd exposure (Table 3). Although the detailed functions of ABC transporter genes is poorly understood, some members have been verified to play roles in the uptake and translocation of Fe/Cd in plants [28]. ABCB19, an auxin transporter that mediates long-distance polar auxin transport in stems and roots [29, 30], was induced by Fe deficiency and/or Cd exposure (Table 3). Fe deficiency can increase the levels of auxin in the roots, which may promote the formation of root hair [31]. ABCC3 (MRP3), like most ABCC transporters such as such as ABCC2 and ABCC3, is vacuolar membrane-localized protein involved in the vacuolar transport of PC-Cd complexes [32]. The induction of ABCC3 in peanut roots by Fe deficiency with Cd exposure (Table 3, Fig. 5) may contribute to vacuolar Cd sequestration, enhancing Cd detoxification and reducing root-to-shoot translocation of Cd. ABCC8 homologous to MRP6 in Arabidopsis, which was shown to be part of a cluster (AtMRP6, AtMRP3 and AtMRP7, as well as SAT3) involved in metal tolerance [33].
Besides ABC transporters, another peptide transporter OPT3 was significantly induced under Fe-deficient conditions (Table 3, Fig. 5). In Arabidopsis, OPT3 was shown to load iron into the phloem, facilitate iron recirculation from the xylem to the phloem, and regulate both shoot-to-root iron signaling and iron redistribution from mature to developing tissues [34, 35]. Reduced expression of OPT3 induces an over accumulation of Fe in roots and leaves, partially due to an elevated expression of the IRT1 [36]. Therefore, the strong induction of OPT3 in the root of Fe-deficient peanut seedlings suggests that the gene play a role in the redistribution of Fe between vegetative tissues. Moreover, OPT3 was also found to have an impact on the uptake and translocation of Cd in Arabidopsis [34, 35]. Mendoza-Cózatl et al. [35] observed that an OPT3 mutant of Arabidopsis, opt3–2, over-accumulates Cd in seeds and roots, but under-accumulates Cd in leaves. However, Zhai et al. [34] demonstrated that OPT3 is not involved in the phloem-based remobilization of Cd from source to sink tissues, despite it can mediate Cd transport in vitro. They speculated that increased Cd in seeds of the mutant is due to the impact of reduced expression of OPT3 on other transporters such as YSL1 or YSL3 [34]. YSL3 is a plasma-localized transporter delivering a broad range of nicotianamine-metal complexes. SnYSL3 gene is shown to be involved in the translocation and detoxification of Cd in Solanum nigrum, and its overexpression in Arabidopsis increased the translocation of Cd and Fe from roots to shoots [37]. In the present study, YSL3 was down-regulated in peanut roots under Fe-deficient stress (Table 3, Fig. 5), which may be partially responsible for the reduced root-to-shoot translocation of Cd in Fe-deficient plants.
Several other transporter genes such as CAX4-like, COPT1, VIT3 and VIT4, were highly induced or suppressed by Fe deficiency (Table 3, Fig. 5). CAX4 is a cation/H+ antiporter that plays a key role in mediating cations, such as Ca2+ and Cd2+, influx into the vacuole [38]. Thus, the induction of CAX4-like by Fe deficiency in peanut roots may enhance vacuole sequestration of Cd and consequently, reducing root-to-shoot Cd translocation. COPT1 participates in copper acquisition and accumulation and regulates root elongation and pollen development [39]. In the current study, we found that COPT1 was up-regulated by Fe deficiency, but down-regulated in Fe-deficient seedlings exposed to Cd. We inferred that COPT1 indirectly effect Fe and/or Cd acquisition by regulating root elongation. Regardless of Cd exposure, the vacuolar iron transporters, VIT3 and VIT4, were suppressed under Fe deficiency condition. These alterations could reduce Fe sequestration in the vacuolar of root cells, and therefore enhance Fe transport from roots to shoots. NRAMP3 localizes at the vacuolar membrane, and is able to release Fe and Cd from the vacuolar under limited Fe conditions [40, 41]. The induction of NRAMP3 may improve the remobilization of Fe from vacuoles of root cells in Fe-deficient peanut plants particularly under Cd stress.
Apart from abovementioned genes, three genes belonging to ZIP family, ZIP1, ZIP5, and MTP11, differentially expressed between the control and Cd exposure with Fe deficiency (Table 3, Fig. 5). ZIP1 shows a high degree of homology to the AtZIP2 from Arabidopsis [42] and OsZIP1 from rice [43], while ZIP5 is homologous to AtZIP1 [42]. Both the AtZIP2 and OsZIP1 show a broad substrate specificity for divalent cations including Cd [42, 43]. In Arabidopsis, AtZIP2 is a plasma membrane localized transporter that may contribute to Mn/Zn movement in the stele to the xylem parenchyma, for subsequent xylem loading and transport to the shoot [44]. Similarly, OsZIP1 transcripts were localized to the epidermis and stele of roots of zinc-deprived plants, suggesting the involvement of this transporter in zinc absorption or transfer from the vascular tissue [43]. AtZIP1 is a vacuolar transporter that may play a role in remobilizing Mn from the vacuole to the cytoplasm in root stellar cells, and may contribute to radial movement to the xylem parenchyma [44]. Thus, the reduced expression of ZIP1 and ZIP5 in the roots of Fe-deficient peanut plants under Cd stress may inhibit root-to-shoot Cd translocation. In the case of MTP11, it was shown to function in Mn transport and tolerance by sequestering Mn into the pre-vacuolar compartments in Arabidopsis [45]. Whether MTP11 is associated with Fe and/or Cd transport need to be further investigated.
Conclusions
In conclusion, the current comparative study revealed that CAX4, COPT1, IRT1, NRAMP3, NRAMP5, OPT3, YSL3, VIT3 and VIT4 might be involved in iron homeostasis in Fe-deficient peanut plants. More importantly, some genes such as IRT1, NRAMP3, NRAMP5, OPT3, YSL3, ABCC3, ZIP1, and ZIP5, were identified to be responsible for the uptake, distribution, and translocation of Cd in peanut plants under iron deficiency. Based on our array data, we proposed a model to explain why iron deficiency induced an increase of Cd uptake but a decrease of Cd translocation from roots to shoots in peanut plants. The up-regulated expression of NRAMP5 and IRT1 genes induced by iron deficiency may enhance the uptake of cadmium by peanut roots. Iron deficiency-induced down-regulation of ZIP1, ZIP5 and YSL3 might result in a decrease of Cd xylem (or phloem) loading in root stele, and consequently, reducing root-to-shoot Cd translocation in peanut plants. Additionally, although detailed information is still unclear, larger amounts of ABC transporter genes were induced or suppressed by iron deficiency under Cd exposure, indicating that further study of this family would be helpful to understand the mechanism of Fe/Cd uptake and transport.
Methods
Plant growth and treatment
Peanut plants (Arachis hypogaea cv. Fenghua 1) were cultured as the conditions previously reported by Chen et al. [13] in a growth chamber of the Huaibei Normal University. Seeds (obtained commercially from the Peanut Institute of Shandong Province, Qingdao) were sterilized with 1% sodium hypochlorite for 10 min, and then they were rinsed with tap water for 24 h and germinated in well-washed sand. After 5 days of emergence, the uniform sized seedlings were selected and transferred to polyethylene pot (six plants per pot) containing 3.5 L of nutrient solution (pH 5.8) [11]. Seven-day-old seedlings were pretreated with (Fe50) or without (Fe0) 50 μM FeEDTA for 7 d in basal nutrient solution. Thereafter, 0 or 2 μM CdCl2 were added in nutrient solution for each Fe treatment, and continuously cultured for one week. The Cd level (2 μM) was arranged according to the Cd concentration in the soil solution of a Cd-contaminated farmland in China (soil moisture at 60% of field capacity) [7]. The experiment was arranged as a completely random design with nine replications (pots). During the growing period, the nutrient solution was renewed twice a week, and pots were randomly arranged and moved daily to minimize position effects.
Root samples for RNA-seq (two biological replicates) and qRT-PCR (three biological replicates) analysis were collected separately from Fe-sufficient and Fe-deficient plants after Cd treatment. Each biological replicate contains a pool of six different plants growing in each pot. All samples were immediately frozen in liquid nitrogen and stored at − 80 °C.
Determination of cd and Fe in plants
Three pots of plants for each Fe treatment under Cd exposure were used for collecting xylem sap excluded from the cut surface as the method described by Uraguchi et al. [46]. After weighing, the collected sap was used for determining the Cd concentration by graphite furnace atomic absorption spectrometry (GF-AAS). Roots and shoots were separated and rinsed three times with deionized water. Thereafter, plant samples were oven-dried at 70 °C for constant weight. The dried tissues were weighed and ground into powder. The concentration of Cd and Fe in plant samples was measured by flame AAS after digested with HNO3–HClO4 (3:1, v/v).
The translocation of Cd and Fe from roots to shoots was indicated as the percentage of metal in shoots, which was calculated as follows [7]:
cDNA library construction and RNA sequencing
Total RNA was extracted by using Trizol® Reagent (Invitrogen) and purified using the RNeasy Plant Mini kit (Qiagen) according to the manufacturer’s instructions. The purity and integrity of RNA were analyzed using NanoPhotometer® spectrophotometer (IMPLEN, CA, USA) and Agilent 2100 Bioanalyzer (Agilent, USA), respectively. Eight cDNA libraries named Fe50_1, Fe50_2, Fe0_1, Fe0_2, Fe50Cd _1, Fe50Cd _2, Fe0Cd_1 and Fe0Cd_2 were constructed as the method previously described by Hu et al. [47]. The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq™ 2500 platform and 125 bp/150 bp paired-end reads were generated. All library construction and sequencing were done at the Novogene Bioinformatics Institute (Beijing, China).
Bioinformatics analysis
Data filtering and mapping
To obtain high-quality clean reads, the raw reads from RNA-seq were filtered by removing the adaptor sequences, ambiguous ‘N’ nucleotides and low-quality sequences. Clean reads were aligned to the peanut reference genome (NCBI) using HISAT2 (v2.2.4, http://www.ccb.jhu.edu/software/hisat) with default parameters. The mapped reads of each sample were assembled by StringTie (v1.3.3b) [48] in a reference-based approach.
Identification of DEGs
The numbers of reads mapped to the reference were counted using featureCounts v1.5.0-p3 [49]. The gene expression levels were represented by the expected number of Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced (FPKM), which was calculated on the basis of the length of the gene and reads count mapped to this gene. Differential expression analysis of two groups was performed using the DESeq2 R package (1.16.1) according to the method described by Love et al. [50]. The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted Padj-value < 0.05 found by DESeq2 were assigned as differentially expressed.
Gene ontology (GO) and pathway analysis of DEGs
Gene Ontology (GO) enrichment analysis of differentially expressed genes was implemented by the clusterProfiler R package, in which gene length bias was corrected.
GO terms with corrected P value less than 0.05 were considered significantly enriched by differential expressed genes. KEGG pathways were retrieved from KEGG web server (http://www.genome.jp/kegg/). The clusterProfiler R package was used to test the statistical enrichment of differential expression genes in KEGG pathways.
qRT-PCR validation
Ten DEGs were randomly selected for qRT-PCR validation. Primer sequences of these genes as well as reference genes are listed in Additional file 3: Table S3. Total RNA (0.5 μg) from each root sample was reverse transcribed into cDNA using Prime Script® RT reagent Kit (Takara, Dalian, China) and random primers following manufacturer’s instructions. Quantitative PCR reactions were performed in 20 μl reaction volumes using a SYBR Premix EX Taq Kit (Takara) according to the manufacturer’s instructions. Reactions were carried out on an ABI7300 (Applied Biosystems, CA, USA). Each biological replicate was technically replicated three times. The relative expression levels of the selected genes were calculated using the 2-ΔΔCT method and normalized by geometric mean of three stably expressed housekeeping genes (AhADH3, Ah60S and Ahactin) [51, 52].
Additional files
Table S1. Pearson correlation between samples. (DOCX 13 kb)
Table S2. Overview of KEGG pathways for DEGs in four comparisons. (XLSX 21 kb)
Table S3. The primers used in RT-qPCR analysis. (DOCX 15 kb)
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the Natural Science Foundation of China (No. 31671599) and the Innovation Team of Scientific Research Platform of Anhui Province (No. KJ2015TD001). The funding body had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Availability of data and materials
The RNA sequence dataset supporting the results of this article is available in the repository of NCBI Sequence Read Archive (SRA) with the GenBank accession No.: PRJNA436766 (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA436766).
Abbreviations
- ABC
ATP-binding cassette transporter protein
- CAX
Cation/H+ exchanger
- Cd
Cadmium
- COPT
Copper transporter
- DEG
Differentially expressed gene
- GO
Gene ontology
- HMA
Cadmium/zinc/copper-transporting ATPase (heavy metal-associated domain)
- KEGG
Kyoto encyclopedia of genes and genomes
- MTP
Metal tolerance protein
- NA
Nicotianamine
- NRAMP
Natural resistance-associated macrophage protein
- OPT
Oligopeptide transporter
- PC
Phytochelatin
- qRT-PCR
Quantitative Real-time PCR
- YSL
Yellow stripe-like transporter
- ZIP
Zinc-regulated transporter/Iron-regulated transporter-like Protein
Authors’ contributions
SG designed the experiments. LJ, QJ, YR and CQ performed the peanut cultivation and sample collection. CC, LJ, QJ, and CQ performed the experiments. SG and CC wrote and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests for this research.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chu Chen, Email: nwangyuan@hotmail.com.
Qiqi Cao, Email: 993359663@qq.com.
Qun Jiang, Email: 1472032988@qq.com.
Jin Li, Email: 2071272849@qq.com.
Rugang Yu, Email: yurugang@126.com.
Gangrong Shi, Email: swsgr@126.com.
References
- 1.White PJ, Brown PH. Plant nutrition for sustainable development and global health. Ann Bot. 2010;105(7):1073–1080. doi: 10.1093/aob/mcq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thijssen S, Cuypers A, Maringwa J, Smeets K, Horemans N, Lambrichts I, Van Kerkhove E. Low cadmium exposure triggers a biphasic oxidative stress response in mice kidneys. Toxicology. 2007;236(1):29–41. doi: 10.1016/j.tox.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 3.Bertin G, Averbeck D. Cadmium: cellular effects, modifications of biomolecules, modulation of DNA repair and genotoxic consequences (a review) Biochimie. 2006;88(11):1549–1559. doi: 10.1016/j.biochi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Lindsay WL, Schwab AP. The chemistry of iron in soils and its availability to plants. J Plant Nutr. 1982;5(4–7):821–840. doi: 10.1080/01904168209363012. [DOI] [Google Scholar]
- 5.Eroglu S, Meier B, von Wirén N, Peiter E. The vacuolar manganese transporter MTP8 determines tolerance to iron deficiency-induced chlorosis in Arabidopsis. Plant Physiol. 2016;170(2):1030–1045. doi: 10.1104/pp.15.01194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su Y, Zhang Z, Su G, Liu J, Liu C, Shi G. Genotypic differences in spectral and photosynthetic response of peanut to iron deficiency. J Plant Nutr. 2015;38(1):145–160. doi: 10.1080/01904167.2014.920392. [DOI] [Google Scholar]
- 7.Shi G, Su G, Lu Z, Liu C, Wang X. Relationship between biomass, seed components and seed cd concentration in various peanut (Arachis hypogaea L.) cultivars grown on cd-contaminated soils. Ecotoxicol Environ Saf. 2014;110:174–181. doi: 10.1016/j.ecoenv.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Liu C, Yu R, Shi G. Effects of drought on the accumulation and redistribution of cadmium in peanuts at different developmental stages. Arch Agr Soil Sci. 2016;63(8):1049–1057. doi: 10.1080/03650340.2016.1271120. [DOI] [Google Scholar]
- 9.Mclaughlin MJ, Bell MJ, Wright GC, Cozens GD. Uptake and partitioning of cadmium by cultivars of peanut (Arachis hypogaea L.) Plant Soil. 2000;222(1–2):51–58. doi: 10.1023/A:1004771712840. [DOI] [Google Scholar]
- 10.Su Y, Liu J, Lu Z, Wang X, Zhang Z, Shi G. Effects of iron deficiency on subcellular distribution and chemical forms of cadmium in peanut roots in relation to its translocation. Environ Exp Bot. 2014;97:40–48. doi: 10.1016/j.envexpbot.2013.10.001. [DOI] [Google Scholar]
- 11.Su Y, Wang X, Liu C, Shi G. Variation in cadmium accumulation and translocation among peanut cultivars as affected by iron deficiency. Plant Soil. 2013;363(1–2):201–213. doi: 10.1007/s11104-012-1310-8. [DOI] [Google Scholar]
- 12.Shi G, Sun L, Wang X, Liu C. Leaf responses to iron nutrition and low cadmium in peanut: anatomical properties in relation to gas exchange. Plant Soil. 2014;375(1):99–111. doi: 10.1007/s11104-013-1953-0. [DOI] [Google Scholar]
- 13.Chen C, Xia S, Deng R, Liu C, Shi G. AhIRT1 and AhNRAMP1 metal transporter expression correlates with cd uptake in peanuts under iron deficiency. PLoS One. 2017;12(10):e0185144. doi: 10.1371/journal.pone.0185144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao L, Chang J, Chen R, Li H, Lu H, Tao L, Xiong J. Comparison on cellular mechanisms of iron and cadmium accumulation in rice: prospects for cultivating Fe-rich but cd-free rice. Rice. 2016;9(1):39. doi: 10.1186/s12284-016-0112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc Natl Acad Sci U S A. 2000;97(9):4991–4996. doi: 10.1073/pnas.97.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakanishi H, Ogawa I, Ishimaru Y, Mori S, Nishizawa NK. Iron deficiency enhances cadmium uptake and translocation mediated by the Fe2+ transporters OsIRT1 and OsIRT2 in rice. Soil Sci Plant Nutr. 2006;52(4):464–469. doi: 10.1111/j.1747-0765.2006.00055.x. [DOI] [Google Scholar]
- 17.He XL, Fan SK, Zhu J, Guan MY, Liu XX, Zhang YS, Jin CW. Iron supply prevents cd uptake in Arabidopsis by inhibiting IRT1 expression and favoring competition between Fe and cd uptake. Plant Soil. 2017;416(1):453–462. doi: 10.1007/s11104-017-3232-y. [DOI] [Google Scholar]
- 18.Yu R, Li D, Du X, Xia S, Liu C, Shi G. Comparative transcriptome analysis reveals key cadmium transport-related genes in roots of two pak choi (Brassica rapa L. ssp. chinensis) cultivars. BMC Genomics. 2017;18(1):587. doi: 10.1186/s12864-017-3973-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang YY, Shen C, Chen JX, He CT, Zhou Q, Tan X, Yuan JG, Yang ZY. Comparative transcriptome analysis of two ipomoea aquatica forsk. Cultivars targeted to explore possible mechanism of genotype-dependent accumulation of cadmium. J Agr Food Chem. 2016;64(25):5241–5250. doi: 10.1021/acs.jafc.6b01267. [DOI] [PubMed] [Google Scholar]
- 20.Xu J, Sun J, Du L, Liu X. Comparative transcriptome analysis of cadmium responses in Solanum nigrum and Solanum torvum. New Phytol. 2012;196(1):110–124. doi: 10.1111/j.1469-8137.2012.04235.x. [DOI] [PubMed] [Google Scholar]
- 21.Bertioli DJ, Cannon SB, Froenicke L, Huang G, Farmer AD, Cannon EKS, Liu X, Gao D, Clevenger J, Dash S, Ren L, Moretzsohn MC, Shirasawa K, Huang W, Vidigal B, Abernathy B, Chu Y, Niederhuth CE, Umale P, Araújo ACG, Kozik A, Kim KD, Burow MD, Varshney RK, Wang X, Zhang X, Barkley N, Guimarães PM, Isobe S, Guo B, Liao B, Stalker HT, Schmitz RJ, Scheffler BE, Leal-Bertioli SCM, Xun X, Jackson SA, Michelmore R, Ozias-Akins P. The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat Genet. 2016;48(4):438–446. doi: 10.1038/ng.3517. [DOI] [PubMed] [Google Scholar]
- 22.Curie C, Alonso JM, Le Jean M, Ecker JR, Briat JF. Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem J. 2000;347(Pt3):749–755. doi: 10.1042/bj3470749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milner MJ, Mitani-Ueno N, Yamaji N, Yokosho K, Craft E, Fei Z, Ebbs S, Clemencia ZM, Ma JF, Kochian LV. Root and shoot transcriptome analysis of two ecotypes of Noccaea caerulescens uncovers the role of NcNramp1 in cd hyperaccumulation. Plant J. 2014;78(3):398–410. doi: 10.1111/tpj.12480. [DOI] [PubMed] [Google Scholar]
- 24.Xiao H, Yin L, Xu X, Li T, Han Z. The iron-regulated transporter, MbNRAMP1, isolated from malus baccata is involved in Fe, Mn and cd trafficking. Ann Bot. 2008;102(6):881–889. doi: 10.1093/aob/mcn178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiong H, Kobayashi T, Kakei Y, Senoura T, Nakazono M, Takahashi H, Nakanishi H, Shen H, Duan P, Guo X, Nishizawa NK, Zuo Y. AhNRAMP1 iron transporter is involved in iron acquisition in peanut. J Exp Bot. 2012;63(12):4437–4446. doi: 10.1093/jxb/ers117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi R, Ishimaru Y, Senoura T, Shimo H, Ishikawa S, Arao T, Nakanishi H, Nishizawa NK. The OsNRAMP1 iron transporter is involved in cd accumulation in rice. J Exp Bot. 2011;62(14):4843–4850. doi: 10.1093/jxb/err136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat J-F, Curie C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 2002;14(6):1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briat J-F, Curie C, Gaymard F. Iron utilization and metabolism in plants. Curr Opin Plant Biol. 2007;10(3):276–282. doi: 10.1016/j.pbi.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Wu G, Cameron JN, Ljung K, Spalding EP. A role for ABCB19-mediated polar auxin transport in seedling photomorphogenesis mediated by cryptochrome 1 and phytochrome B. Plant J. 2010;62(2):179–191. doi: 10.1111/j.1365-313X.2010.04137.x. [DOI] [PubMed] [Google Scholar]
- 30.Lewis DR, Wu G, Ljung K, Spalding EP. Auxin transport into cotyledons and cotyledon growth depend similarly on the ABCB19 multidrug resistance-like transporter. Plant J. 2009;60(1):91–101. doi: 10.1111/j.1365-313X.2009.03941.x. [DOI] [PubMed] [Google Scholar]
- 31.Romera FJ, Lucena C, Alcàntara E. Plant hormones influencing Iron uptake in plants. In: Barton LL, Abadia J, editors. Iron nutrition in plants and Rhizospheric microorganisms. Dordrecht: Springer Netherlands; 2006. pp. 251–278. [Google Scholar]
- 32.Brunetti P, Zanella L, De Paolis A, Di Litta D, Cecchetti V, Falasca G, Barbieri M, Altamura MM, Costantino P, Cardarelli M. Cadmium-inducible expression of the ABC-type transporter AtABCC3 increases phytochelatin-mediated cadmium tolerance in Arabidopsis. J Exp Bot. 2015;66(13):3815–3829. doi: 10.1093/jxb/erv185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gaillard S, Jacquet H, Vavasseur A, Leonhardt N, Forestier C. AtMRP6/AtABCC6 , an ATP-binding cassette transporter gene expressed during early steps of seedling development and up-regulated by cadmium in Arabidopsis thaliana. BMC Plant Biol. 2008;8(1):22. doi: 10.1186/1471-2229-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhai Z, Gayomba SR, Jung H, Vimalakumari NK, Piñeros M, Craft E, Rutzke MA, Danku J, Lahner B, Punshon T, Guerinot ML, Salt DE, Kochian LV, Vatamaniuk OK. OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and cadmium in Arabidopsis. Plant Cell. 2014;26(5):2249–2264. doi: 10.1105/tpc.114.123737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendoza-Cózatl DG, Xie Q, Akmakjian GZ, Jobe TO, Patel A, Stacey MG, Song L, Demoin DW, Jurisson SS, Stacey G, Schroedera JI. OPT3 is a component of the iron-signaling network between leaves and roots and misregulation of OPT3 leads to an over-accumulation of cadmium in seeds. Mol Plant. 2014;7(9):1455–1469. doi: 10.1093/mp/ssu067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan MA, Castro-Guerrero NA, Mcinturf SA, Nguyen NT, Dame AN, Wang J, Bindbeutel RK, Joshi T, Jurisson SS, Nusinow DA. Changes in iron availability in Arabidopsis are rapidly sensed in the leaf vasculature and impaired sensing leads to opposite transcriptional programs in leaves and roots. Plant Cell Environ. 2018;41(10):2263–2276. doi: 10.1111/pce.13192. [DOI] [PubMed] [Google Scholar]
- 37.Feng S, Tan J, Zhang Y, Liang S, Xiang S, Wang H, Chai T. Isolation and characterization of a novel cadmium-regulated yellow stripe-like transporter (SnYSL3) in Solanum nigrum. Plant Cell Rep. 2017;36(2):281–296. doi: 10.1007/s00299-016-2079-7. [DOI] [PubMed] [Google Scholar]
- 38.Mei H, Cheng NH, Zhao J, Park S, Escareno RA, Pittman JK, Hirschi KD. Root development under metal stress in Arabidopsis thaliana requires the H+/cation antiporter CAX4. New Phytol. 2009;183(1):95–105. doi: 10.1111/j.1469-8137.2009.02831.x. [DOI] [PubMed] [Google Scholar]
- 39.Sancenón V, Puig S, Mateu-Andrés I, Dorcey E, Thiele DJ, Peñarrubia L. The Arabidopsis copper transporter COPT1 functions in root elongation and pollen development. J Biol Chem. 2004;279(15):15348–15355. doi: 10.1074/jbc.M313321200. [DOI] [PubMed] [Google Scholar]
- 40.Oomen RJFJ, Wu J, Lelièvre F, Blanchet S, Richaud P, Barbier-Brygoo H, Aarts MGM, Thomine S. Functional characterization of NRAMP3 and NRAMP4 from the metal hyperaccumulator Thlaspi caerulescens. New Phytol. 2009;181(3):637–650. doi: 10.1111/j.1469-8137.2008.02694.x. [DOI] [PubMed] [Google Scholar]
- 41.Mary V, Schnell Ramos M, Gillet C, Socha AL, Giraudat J, Agorio A, Merlot S, Clairet C, Kim SA, Punshon T, Guerinot ML, Thomine S. Bypassing iron storage in endodermal vacuoles rescues the iron mobilization defect in the natural resistance associated-macrophage protein3natural resistance associated-macrophage protein4 double mutant. Plant Physiol. 2015;169(1):748–759. doi: 10.1104/pp.15.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grotz N, Fox T, Connolly E, Park W, Guerinot ML, Eide D. Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc Natl Acad Sci U S A. 1998;95(12):7220–7224. doi: 10.1073/pnas.95.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramesh SA, Shin R, Eide DJ, Schachtman DP. Differential metal selectivity and gene expression of two zinc transporters from rice. Plant Physiol. 2003;133(1):126–134. doi: 10.1104/pp.103.026815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milner MJ, Seamon J, Craft E, Kochian LV. Transport properties of members of the ZIP family in plants and their role in Zn and Mn homeostasis. J Exp Bot. 2013;64(1):369–381. doi: 10.1093/jxb/ers315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delhaize E, Gruber BD, Pittman JK, White RG, Leung H, Miao Y, Jiang L, Ryan PR, Richardson AE. A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J. 2007;51(2):198–210. doi: 10.1111/j.1365-313X.2007.03138.x. [DOI] [PubMed] [Google Scholar]
- 46.Uraguchi S, Mori S, Kuramata M, Kawasaki A, Arao T, Ishikawa S. Root-to-shoot cd translocation via the xylem is the major process determining shoot and grain cadmium accumulation in rice. J Exp Bot. 2009;60(9):2677–2688. doi: 10.1093/jxb/erp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu L, Li H, Chen L, Lou Y, Amombo E, Fu J. RNA-seq for gene identification and transcript profiling in relation to root growth of bermudagrass (Cynodon dactylon) under salinity stress. BMC Genomics. 2015;16(1):575. doi: 10.1186/s12864-015-1799-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao Y, Smyth GK, Shi W. featureCounts. An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genom Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genom Biol. 2002;3(7):research0034.0031. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brand Y, Hovav R. Identification of suitable internal control genes for quantitative real-time PCR expression analyses in peanut (Arachis hypogaea) Peanut Sci. 2010;37(1):12–19. doi: 10.3146/PS09-014.1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Pearson correlation between samples. (DOCX 13 kb)
Table S2. Overview of KEGG pathways for DEGs in four comparisons. (XLSX 21 kb)
Table S3. The primers used in RT-qPCR analysis. (DOCX 15 kb)
Data Availability Statement
The RNA sequence dataset supporting the results of this article is available in the repository of NCBI Sequence Read Archive (SRA) with the GenBank accession No.: PRJNA436766 (https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA436766).