Abstract
Background
Increasingly evidences suggest that long noncoding RNAs (lncRNAs) play important roles in various cancers. LncRNA PXN-AS1-L is recently revealed to act as on oncogene in liver cancer. However, the expression, functions, and mechanisms of action of PXN-AS-L in non-small cell lung cancer (NSCLC) remain unclear.
Methods
The expression of PXN-AS1-L in primary NSCLC tissues, NSCLC bone metastasis tissues, and cell lines was measured by quantitative real-time PCR. The correlations between PXN-AS1-L expression and clinicopathological characteristics of NSCLC patients were analyzed by Pearson Chi square test and log-rank test. The roles of PXN-AS1-L in cell viability, proliferation, apoptosis, and migration of NSCLC cells, and in vivo NSCLC tumor growth were investigated by a series of gain-of-function and loss-of-function assays. The regulatory roles of PXN-AS1-L on PXN were determined by quantitative real-time PCR and western blot.
Results
PXN-AS1-L was up-regulated in NSCLC tissues compared with noncancerous lung tissues, and PXN-AS1-L was further up-regulated in NSCLC bone metastasis tissues. Increased expression of PXN-AS1-L was positively associated with advanced TNM stages and poor prognosis. Gain-of-function and loss-of-function assays showed that PXN-AS1-L increased cell viability, promoted cell proliferation, inhibited cell apoptosis, and promoted cell migration of NSCLC cells. Xenograft assays showed that PXN-AS1-L also promoted NSCLC tumor growth in vivo. Mechanistically, we found that PXN-AS1-L, as an antisense transcript of PXN, up-regulated the expression of PXN. PXN was also up-regulated in NSCLC tissues. The expression of PXN and PXN-AS1-L was positively correlated in NSCLC tissues. Furthermore, PXN knockdown attenuated the roles of PXN-AS1-L in increasing cell viability, promoting cell proliferation, inhibiting cell apoptosis, and promoting cell migration of NSCLC cells.
Conclusions
Our data revealed that PXN-AS1-L is up-regulated and acts as an oncogene in NSCLC via up-regulating PXN. Our data suggested that PXN-AS1-L might serve as a potential prognostic biomarker and therapeutic target for NSCLC.
Keywords: Long noncoding RNA, Non-small-cell lung cancer, Proliferation, Migration, PXN
Background
Lung cancer is the most common malignancy and the leading cause of cancer death around the world, with an estimated 2,093,876 new cases and 1,761,007 deaths in 2018 worldwide [1]. The major histological subtype of lung cancer is non-small cell lung cancer (NSCLC), which mainly includes adenocarcinoma and squamous cell carcinoma, and accounts for approximate 80% of lung cancers [2]. Except the NSCLC patients which are diagnosed at early stages and could be cured by surgical resection, most NSCLC patients still have poor outcomes with the 5-year survival rate of about 20% [3]. Thus, it is of paramount importance to reveal the underlying molecular mechanisms contributing to the tumorigenesis and progression of NSCLC in order to develop novel therapeutic strategies for NSCLC [4–6].
Accumulating evidences have revealed that most of human genome is transcribed, but only about 2% of human genome encode for proteins [7]. Therefore, most of human transcripts don’t encode for proteins. Among these non-coding transcripts, long noncoding RNAs (lncRNAs) have gradually attracted people’s attention [8–12]. LncRNA is a class of non-coding transcripts with more than 200 nucleotides in length and limited protein coding potential [13–15]. Many lncRNAs are revealed to play important roles in various pathophysiological processes [16–19]. As to tumors, lncRNAs have been reported to regulate almost every aspect of biological behaviors of cancer cells, including cell proliferation, apoptosis, senescence, autophagy, migration, invasion, and so on [20–25]. Furthermore, many lncRNAs are dysregulated in various cancers and associated with early diagnosis and/or prognosis [26–28].
Several lncRNAs have been reported to have oncogenic or tumor suppressing roles in NSCLC. LncRNA MALAT1 is a positive marker of lung cancer metastasis [29]. LncRNA PVT1 is reported to promote NSCLC cell proliferation [30]. LncRNA BANCR is reported to promote NSCLC metastasis [31]. LncRNA MIR22HG is reported to inhibit cell survival [32]. LncRNA P53RRA is reported to promote ferroptosis and apoptosis [33]. LncRNA MUC5B-AS1 is reported to promote metastasis [34]. LncRNA linc00460 is reported to promote cell migration [35]. LncRNA LINC00473 is reported to promote lung cancer tumor growth [36]. Although the expression and function of these lncRNAs are studied in NSCLC, most lncRNAs transcribed from human genome are functionally unclear in NSCLC [37]. LncRNA PXN-AS1-L is a recently identified lncRNA, which is upregulated in hepatocellular carcinoma (HCC) and promoted HCC tumorigenesis via up-regulating PXN [38]. However, the expression, roles, and mechanisms of action of lncRNA PXN-AS1-L in NSCLC are still unknown.
In this study, we aimed to elucidate the expression pattern of lncRNA PXN-AS1-L in NSCLC via measuring PXN-AS1-L expression in noncancerous lung tissues, NSCLC tissues, NSCLC bone metastases tissues, normal bronchial epithelial cell line, and human NSCLC cell lines. We also investigated the biological functions of PXN-AS1-L in NSCLC using in vitro and in vivo gain-of-function and loss-of-function assays. Moreover, we explored the molecular mechanisms mediating the roles of PXN-AS1-L in NSCLC.
Methods
Cell culture and treatment
The human normal bronchial epithelial cell line 16HBE, human NSCLC cell lines NCI-H1975, A549, NCI-H1299, and SK-MES-1 were obtained from Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). 16HBE cells were cultured in Dulbecco’s Modified Eagle’s Medium (Invitrogen, Carlsbad, CA, USA). NCI-H1975 and NCI-H1299 cells were cultured in RPMI-1640 Medium (Invitrogen). A549 cells were cultured in F-12K Medium (Invitrogen). SK-MES-1 cells were cultured in Eagle’s Minimum Essential Medium (Invitrogen). All the cells were maintained in the above described medium supplemented with 10% fetal bovine serum (Invitrogen) in a humidified incubator at 37 °C with 5% CO2. Where indicated, NSCLC cells were treated with 50 µM α-amanitin (Sigma-Aldrich, Saint Louis, MO, USA) for 0–24 h as shown in the article.
Human tissue specimens
Sixty-six pairs of NSCLC tissues and adjacent noncancerous lung tissues, and ten NSCLC bone metastases tissues were acquired from NSCLC patients who underwent surgery at the General Hospital of Chinese People’s Liberation Army (Beijing, China). All the tissues were diagnosed and histologically confirmed by two pathologists. The resected specimens were immediately frozen in liquid nitrogen and stored at − 80 °C until use. The study was approved by the ethics committee of the General Hospital of Chinese People’s Liberation Army (Beijing, China) and written informed consent was obtained from all patients.
Plasmids construction and transfection
PXN-AS1-L overexpression plasmid pcDNA3.1-PXN-AS1-L was constructed as previously described [38]. Briefly, PXN-AS1-L full-length transcript was PCR amplified by Thermo Scientific Phusion Flash High-Fidelity PCR Master Mix (Thermo-Fisher Scientific, Waltham, MA, USA) and subcloned into the BamH I and EcoR V sites of pcDNA3.1 plasmid (Invitrogen). The primers sequences were: 5′-GGTACCGAGCTCGGATCCTCGCGTTGGAGGAGCTTG-3′ (forward) and 5′-GCCACTGTGCTGGATATCCTACAAAAAAAATTTATTTAATAAAA-3′ (reverse). The cDNA oligonucleotides suppressing PXN-AS1-L or PXN expression were designed and synthesized by GenePharma (Shanghai, China). After annealing, double strand oligonucleotides were inserted to the SuperSilencing shRNA expression plasmid pGPU6/Neo (GenePharma). A scrambled shRNA was used as a negative control and designated as shControl. The shRNA sequences were: for shPXN, 5′-CACCCCTGACGAAAGAGAAGCCTAATTCAAGAGATTAGGCTTCTCTTTCGTCAGGTTTTTTG-3′ (sense), 5′-GATCCAAAAAACCTGACGAAAGAGAAGCCTAATCTCTTGAATTAGGCTTCTCTTTCGTCAGG-3′ (anti-sense); for shRXN-AS1-L, 5′-CACCGCCCAGAGGAAATCAACAAGATTCAAGAGATCTTGTTGATTTCCTCTGGGCTTTTTTG-3′ (sense), 5′-GATCCAAAAAAGCCCAGAGGAAATCAACAAGATCTCTTGAATCTTGTTGATTTCCTCTGGGC-3′ (anti-sense); for shControl, 5′-CACCGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTTG-3′ (sense), 5′-GATCCAAAAAATTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAAC-3′ (anti-sense). Transient transfection was performed using Lipofectamine 3000 (Invitrogen) in accordance with the manufacturer’s instruction.
Construction of stable cell lines
To constructing PXN-AS1-L stably overexpressed A549 cells, pcDNA3.1-PXN-AS1-L or pcDNA3.1 was transfected into A549 cells and selected with neomycin (800 µg/ml) for 4 weeks. To constructing PXN-AS1-L stably depleted NCI-H1299 cells, shPXN-AS1-L or shControl was transfected into NCI-H1299 cells and selected with neomycin (1000 µg/ml) for 4 weeks.
RNA extraction and quantitative real-time PCR analysis (qPCR)
Total RNA was extracted from indicted tissues or cells using TRIzol Reagent (Invitrogen) in accordance with the manufacturer’s protocol. Reverse transcription was performed using the M-MLV Reverse Transcriptase (Invitrogen) in accordance with the manufacturer’s protocol. Quantitative real-time PCR (qPCR) was performed in the StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using the SYBR® Premix Ex Taq™ II kit (Takara, Dalian, China) in accordance with the manufacturer’s protocols. β-actin was used as an endogenous control. The primers sequences were: for PXN-AS1-L, 5′-ACCCATCCTCAACTACCCC-3′ (forward) and 5′-ACTTCGTCTGTGCCTTCTGC-3′ (reverse); for PXN, 5′-TATCTCAGCCCTCAACACGC-3′ (forward) and 5′-GGCAGAAGGCACAGACGAA-3′ (reverse); for β-actin, 5′-GGGAAATCGTGCGTGACATTAAG-3′ (forward) and 5′-TGTGTTGGCGTACAGGTCTTTG-3′ (reverse). The relative expression of RNAs was calculated using the comparative Ct method.
Western blot analysis
Total proteins were extracted using RIPA buffer (Beyotime, Shanghai, China). Identical quantities of proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto nitrocellulose filter membranes. After being blocked with 5% not-fat milk, the membranes were incubated with primary antibodies specific for PXN (Abcam, Hong Kong, China) or β-actin (Proteintech, Rosemont, IL, USA). After being washed, the membranes were incubated with IRdye 700-conjugated goat anti-mouse IgG or IRdye 800-conjugated goat anti-rabbit IgG and were detected using an Odyssey infrared scanner (Li-Cor, Lincoln, NE, USA).
Analysis of cell proliferation and apoptosis
Cell proliferation was detected using Glo cell viability assay and Ethynyl deoxyuridine (EdU) incorporation assay. For Glo cell viability assay, 3000 indicated NSCLC cells were seeded each well in 96-well plate. At indicated time, cell viability was detected using the CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA) in accordance with the manufacturer’s protocol. EdU incorporation assay was carried out using the EdU kit (Roche, Mannheim, Germany) in accordance with the manufacturer’s protocol. The results were acquired and quantified with the Zeiss AxioPhot Photomicroscope (Carl Zeiss, Oberkochen, Germany) based on at least ten random fields. Cell apoptosis was detected using terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick end labeling (TUNEL) assay. Indicated NSCLC cells were treated with 25 ng/ml doxorubicin (Selleck, Houston, TX, USA) for 24 h. Then, cell apoptosis was detected using the TUNEL Cell Apoptosis Detection Kit (Beyotime) in accordance with the manufacturer’s protocol.
Cell migration assay
Cell migration was detected using transwell assay. 5 × 104 indicated NSCLC cells in serum-free medium were plated in the top chamber a 24-well transwell chamber (BD Biosciences, San Jose, CA, USA). Complete medium containing 10% fetal bovine serum was placed into the lower chamber. After incubation for 24 h, cells remaining on the top chamber were wiped off using a cotton swab, and the cells that had traversed the membranes were stained by crystal violet and counted.
Nude mouse xenograft assays
3 × 106 indicted NSCLC cells were subcutaneously injected into 6-week-old athymic BALB/c nude mice purchased from the Shanghai Experimental Animal Center of Chinese Academy of Sciences (Shanghai, China). Subcutaneous xenograft growth was measured weekly with a caliper, and the tumor volume was calculated as a × b2 × 0.5 (a, longest diameter; b, shortest diameter). The animal studies were approved by the ethics committee of the General Hospital of Chinese People’s Liberation Army (Beijing, China).
Immunohistochemistry (IHC)
For IHC, the subcutaneous xenografts were formalin-fixed, paraffin-embedded, and cut into 4 µm sections. The sections were incubated with primary antibodies specific for Ki67 (Abcam) or cleaved caspase-3 (Cell Signaling Technology, Boston, MA, USA). After being washed, the sections were incubated with horseradish peroxidase-conjugated second antibody (Abcam) and visualized using DAB Horseradish Peroxidase Color Development Kit (Beyotime).
Statistical analysis
All statistical analyses were performed using the GraphPad Prism Software. For comparisons, Student’s t test (two-sided), Wilcoxon signed-rank test, Mann–Whitney test, Pearson Chi square test, Log-rank test, and Pearson correlation analysis were performed as indicated. P values < 0.05 were considered as statistically significant.
Results
PXN-AS1-L was up-regulated in NSCLC and associated with poor prognosis
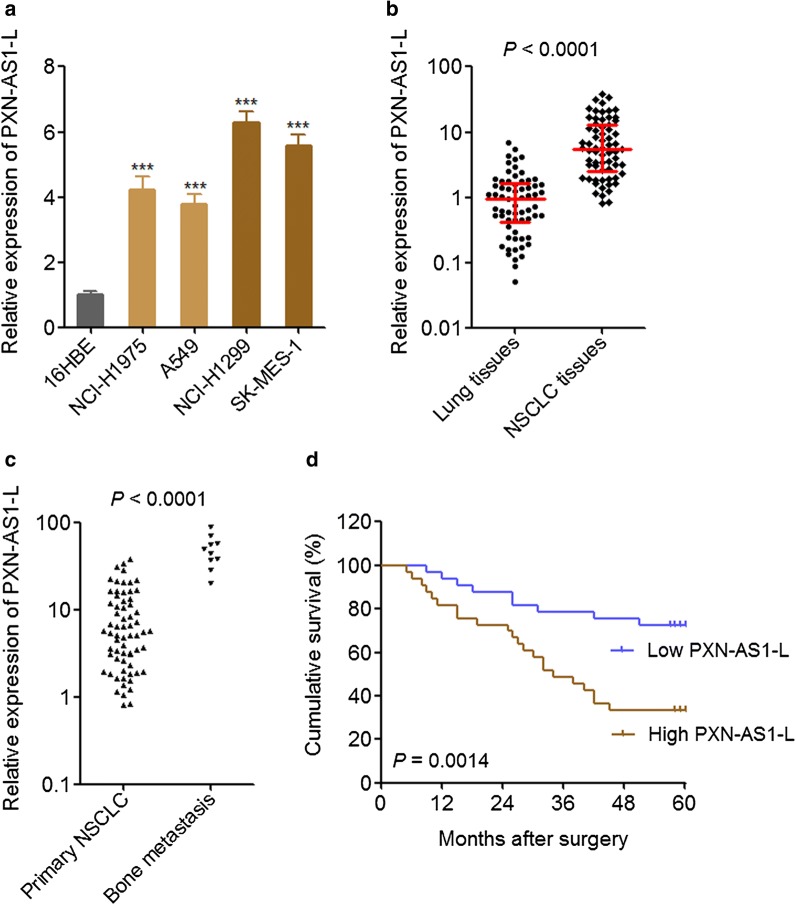
To investigate the expression pattern of PXN-AS1-L in NSCLC, we first measured the expression of PXN-AS1-L in normal bronchial epithelial cell line 16HBE and NSCLC cell lines NCI-H1975, A549, NCI-H1299, SK-MES-1. The results displayed that PXN-AS1-L was significantly up-regulated in NSCLC cell lines compared with that in normal bronchial epithelial cell line, and further up-regulated in NSCLC cell lines derived from metastatic sites (NCI-H1299 and SK-MES-1) (Fig. 1a). Then, we collected 66 pairs of NSCLC tissues and adjacent noncancerous lung tissues and measured the expression of PXN-AS1-L in these tissues. The results displayed that the expression of PXN-AS1-L was significantly higher in NSCLC tissues than that in adjacent noncancerous lung tissues (Fig. 1b). Furthermore, we collected 10 NSCLC bone metastases tissues and also measured the expression of PXN-AS1-L. The results displayed that the expression of PXN-AS1-L was further higher in bone metastases tissues than that in primary NSCLC tissues (Fig. 1c).
Fig. 1.
PXN-AS1-L was up-regulated in NSCLC and associated with poor prognosis. a The expressions of PXN-AS1-L in normal bronchial epithelial cell line 16HBE and NSCLC cell lines NCI-H1975, A549, NCI-H1299, and SK-MES-1 were detected by qPCR. Results are shown as mean ± SD of three independent experiments. ***P < 0.001 by Student’s t-test. b The expressions of PXN-AS1-L in 66 pairs of NSCLC tissues and adjacent noncancerous lung tissues were detected by qPCR. P < 0.0001 by Wilcoxon signed-rank test. c The expressions of PXN-AS1-L in 66 primary NSCLC tissues and 10 NSCLC bone metastases tissues were detected by qPCR. P < 0.0001 by Mann–Whitney test. d Kaplan–Meier survival analysis of the correlation between PXN-AS1-L expression and overall survival of these 66 NSCLC patients. The median PXN-AS1-L expression level was used as the cut-off. P < 0.0001 by Log-rank test
The correlations between the expression of PXN-AS1-L in NSCLC tissues and clinicopathological characteristics of these 66 NSCLC patients were analyzed. As shown in Table 1, increased expression of PXN-AS1-L was positively associated with larger tumor size, positive lymph nodes metastasis, and advanced TNM stages, but not associated with age, gender, and histologic subtype. Kaplan–Meier survival analysis displayed that NSCLC patients with higher PXN-AS1-L expression had worse survival than those with lower PXN-AS1-L expression (Fig. 1d). All these data together demonstrated that PXN-AS1-L was up-regulated in NSCLC, further up-regulated in metastatic NSCLC cells and tissues. Increased expression of PXN-AS1-L predicted poor outcome of NSCLC patients.
Table 1.
Correlation between PXN-AS1-L expression and clinicopathological characteristics of NSCLC patients
| Variables | Number | PXN-AS1-L | P value* | |
|---|---|---|---|---|
| Low | Higha | |||
| All cases | 66 | 33 | 33 | |
| Age | 0.620 | |||
| ≥ 60 | 37 | 20 | 17 | |
| < 60 | 29 | 13 | 16 | |
| Gender | 1.000 | |||
| Male | 48 | 24 | 24 | |
| Female | 18 | 9 | 9 | |
| Histologic subtype | 1.000 | |||
| Squamous cell carcinoma | 30 | 15 | 15 | |
| Adenocarcinoma | 36 | 18 | 18 | |
| Tumor size (cm) | 0.021 | |||
| ≤ 3 | 11 | 9 | 2 | |
| > 3 | 55 | 24 | 31 | |
| Lymph nodes metastasis | 0.003 | |||
| Positive | 26 | 7 | 19 | |
| Negative | 40 | 26 | 14 | |
| TNM stage | 0.000 | |||
| I | 28 | 23 | 5 | |
| II | 27 | 10 | 17 | |
| III | 11 | 0 | 11 | |
a The median expression level of PXN-AS1-L was used as the cut-off
* P value was acquired by Pearson Chi square test
PXN-AS1-L overexpression promoted NSCLC cell proliferation and migration
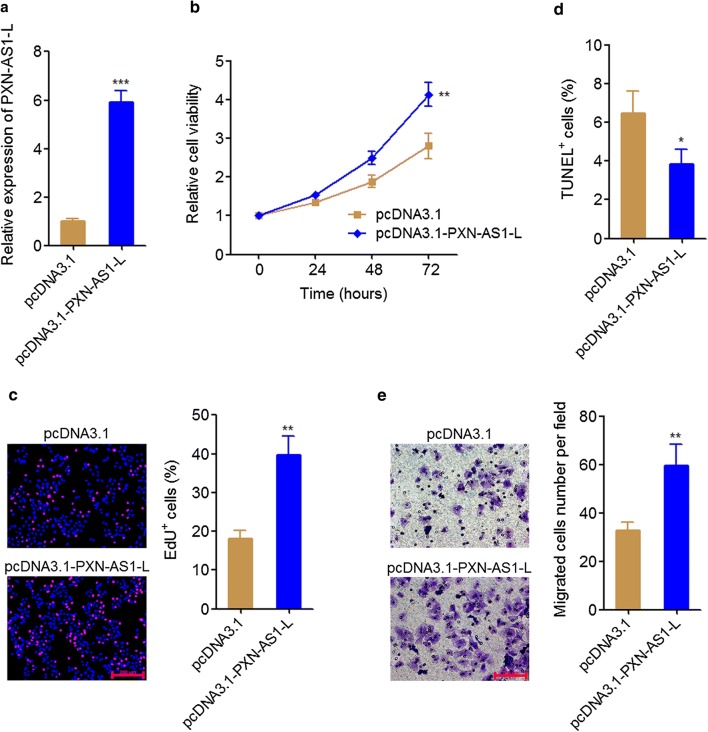
To reveal the biological effects of PXN-AS1-L on NSCLC, we stably overexpressed PXN-AS1-L in A549 cells which has a relative low expression of PXN-AS1-L among NSCLC cell lines by transfecting PXN-AS1-L overexpression plasmid (Fig. 2a). Glo cell viability assays displayed that PXN-AS1-L overexpression increased cell viability of A549 cells (Fig. 2b). EdU incorporation assays also displayed that PXN-AS1-L overexpression promoted cell proliferation of A549 cells (Fig. 2c). TUNEL assays displayed that PXN-AS1-L overexpression inhibited cell apoptosis of A549 cells (Fig. 2d). Transwell assays displayed that PXN-AS1-L overexpression promoted cell migration of A549 cells (Fig. 2e). All these data together demonstrated that PXN-AS1-L overexpression promoted cell proliferation, inhibited cell apoptosis, and promoted cell migration of NSCLC cells, suggesting that PXN-AS1-L has oncogenic roles in NSCLC.
Fig. 2.
PXN-AS1-L overexpression promoted NSCLC cell proliferation and migration. a The expressions of PXN-AS1-L in PXN-AS1-L stably overexpressed and control A549 cells were detected by qPCR. b Cell viability of PXN-AS1-L stably overexpressed and control A549 cells was detected by Glo cell viability assays. c Cell proliferation of PXN-AS1-L stably overexpressed and control A549 cells was detected by EdU incorporation assays. The red color indicates EdU-positive cells. Scale bars = 200 μm. d Cell apoptosis of PXN-AS1-L stably overexpressed and control A549 cells was detected by TUNEL assays. e Cell migration of PXN-AS1-L stably overexpressed and control A549 cells was detected by transwell assays. Scale bars = 100 μm. Results are shown as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 by Student’s t-test
PXN-AS1-L knockdown reduced NSCLC cell proliferation and migration
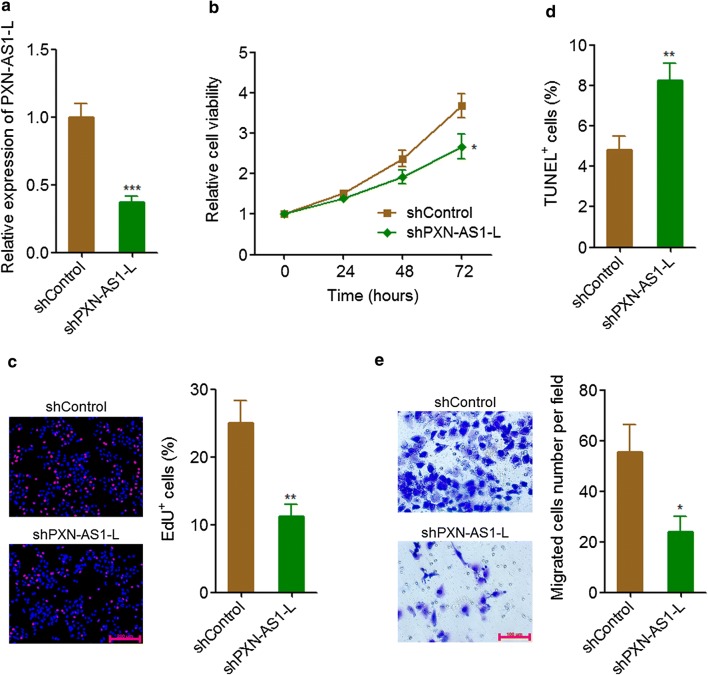
To further confirm the oncogenic roles of PXN-AS1-L on NSCLC, we stably knocked down PXN-AS1-L expression in NCI-H1299 cells which has a relative high expression of PXN-AS1-L among NSCLC cell lines by transfecting PXN-AS1-L specific shRNA (Fig. 3a). Glo cell viability assays displayed that PXN-AS1-L knockdown reduced cell viability of NCI-H1299 cells (Fig. 3b). EdU incorporation assays also displayed that PXN-AS1-L knockdown inhibited cell proliferation of NCI-H1299 cells (Fig. 3c). TUNEL assays displayed that PXN-AS1-L knockdown promoted cell apoptosis of NCI-H1299 cells (Fig. 3d). Transwell assays displayed that PXN-AS1-L knockdown inhibited cell migration of NCI-H1299 cells (Fig. 3e). All these data together demonstrated that PXN-AS1-L knockdown inhibited cell proliferation, promoted cell apoptosis, and inhibited cell migration of NSCLC cells, supporting the oncogenic roles of PXN-AS1-L in NSCLC.
Fig. 3.
PXN-AS1-L knockdown inhibited NSCLC cell proliferation and migration. a The expressions of PXN-AS1-L in PXN-AS1-L stably knocked down and control NCI-H1299 cells were detected by qPCR. b Cell viability of PXN-AS1-L stably knocked down and control NCI-H1299 cells was detected by Glo cell viability assays. c Cell proliferation of PXN-AS1-L stably knocked down and control NCI-H1299 cells was detected by EdU incorporation assays. The red color indicates EdU-positive cells. Scale bars = 200 μm. d Cell apoptosis of PXN-AS1-L stably knocked down and control NCI-H1299 cells was detected by TUNEL assays. e Cell migration of PXN-AS1-L stably knocked down and control NCI-H1299 cells was detected by transwell assays. Scale bars = 100 μm. Results are shown as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 by Student’s t-test
PXN-AS1-L promoted the growth of NSCLC xenograft in vivo
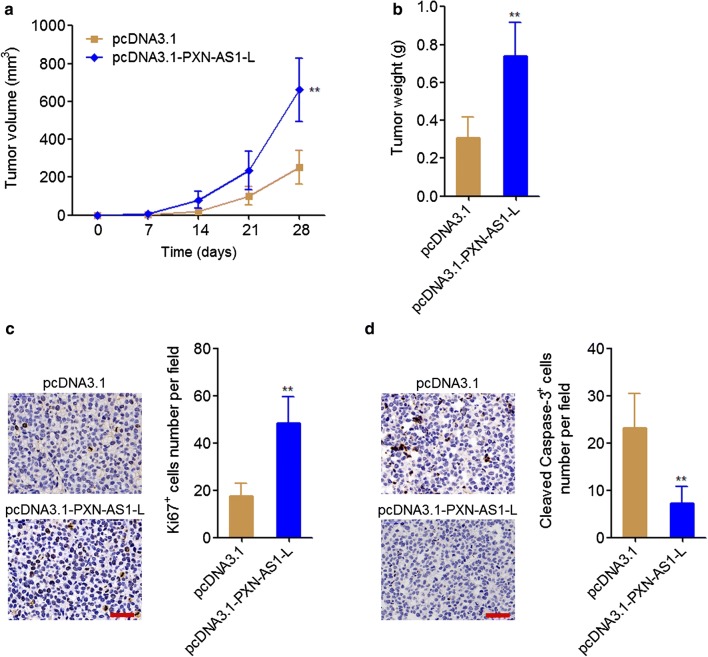
To explore whether PXN-AS1-L also have oncogenic roles in vivo, PXN-AS1-L stably overexpressed and control A549 cells were subcutaneously injected into nude mice. Xenografts growth rates were measured every 7 days. At the 28th day, the xenografts were excised and weighed. The results displayed that PXN-AS1-L overexpression markedly promoted xenograft growth in vivo, and PXN-AS1-L overexpressed A549 cells formed much larger tumor than those formed by control A549 cells (Fig. 4a, b). Ki67 staining of xenograft displayed that PXN-AS1-L overexpression promoted A549 cell proliferation in vivo (Fig. 4c). Cleaved caspase-3 staining of xenograft displayed that PXN-AS1-L overexpression inhibited A549 cell apoptosis in vivo (Fig. 4d). All these data together demonstrated that PXN-AS1-L also had oncogenic roles in vivo.
Fig. 4.
PXN-AS1-L promoted NSCLC xenograft growth in vivo. a Tumor volumes of subcutaneous xenografts derived from PXN-AS1-L stably overexpressed and control A549 cells were measured every 7 days. b Tumor weights of subcutaneous xenografts derived from PXN-AS1-L stably overexpressed and control A549 cells at the 28th day after injection. c In vivo cell proliferation of PXN-AS1-L stably overexpressed and control A549 cells was evaluated using Ki67 immunohistochemistry staining of subcutaneous xenografts. Scale bars = 50 µm. d In vivo cell apoptosis of PXN-AS1-L stably overexpressed and control A549 cells was evaluated using cleaved caspase-3 immunohistochemistry staining of subcutaneous xenografts. Scale bars = 50 µm. Results are shown as mean ± SD of six mice in each group. **P < 0.01 by Mann–Whitney test
PXN-AS1-L up-regulated PXN expression
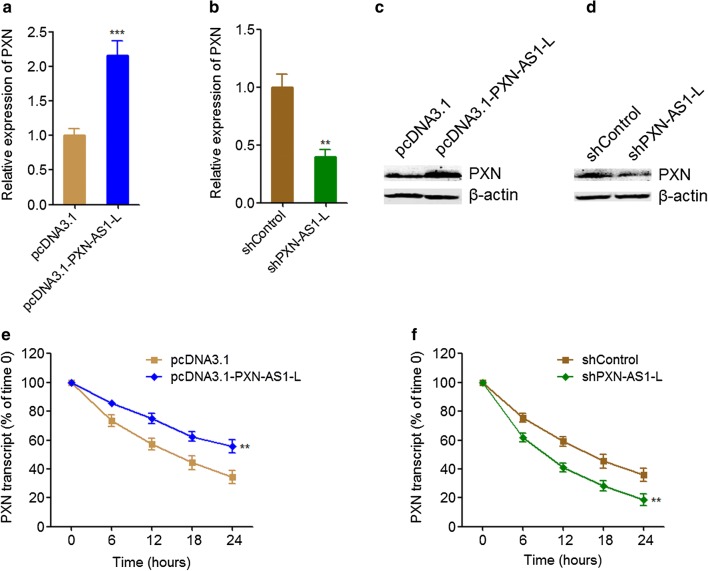
PXN-AS1-L is previously reported to up-regulate PXN expression in HCC cells [38]. PXN has been revealed to play oncogenic roles in NSCLC in several reports [39–41]. Therefore, we first investigated whether PXN-AS1-L also regulates PXN expression in NSCLC and whether the oncogenic roles of PXN-AS1-L in NSCLC are dependent on the regulation of PXN. The expressions of PXN in PXN-AS1-L stably overexpressed and control A549 cells, and PXN-AS1-L stably depleted and control NCI-H1299 cells were measured by qRT-PCR and western blot. The results displayed that PXN-AS1-L overexpression up-regulated PXN mRNA level (Fig. 5a). Conversely, PXN-AS1-L knockdown reduced PXN mRNA level (Fig. 5b). Western blot assays also displayed that PXN-AS1-L overexpression up-regulated PXN protein level and while PXN-AS1-L knockdown reduced PXN protein level (Fig. 5c, d). PXN-AS1-L is reported to decrease PXN mRNA degradation and increase PXN mRNA stability in HCC cells [38]. To investigate whether the same mechanism was employed by PXN-AS1-L in NSCLC, we next determined the effects of PXN-AS1-L on PXN mRNA stability in NSCLC cell. After transiently overexpressing PXN-AS1-L in A549 cells or depleting PXN-AS1-L in NCI-H1299 cells, the cells were treated with α-amanitin to block new RNA synthesis. Next, the loss of PXN mRNA was measured. As shown in Fig. 5e, f, PXN-AS1-L overexpression elongated the half-life of PXN mRNA, and conversely, PXN-AS1-L knockdown shortened the half-life of PXN mRNA, which suggested that PXN-AS1-L also increase PXN mRNA stability in NSCLC cells. All these data together demonstrated that PXN-AS1-L up-regulated PXN expression in NSCLC.
Fig. 5.
PXN-AS1-L up-regulated PXN expression. a PXN mRNA levels in PXN-AS1-L stably overexpressed and control A549 cells were detected by qPCR. b PXN mRNA levels in PXN-AS1-L stably depleted and control NCI-H1299 cells were detected by qPCR. c PXN protein levels in PXN-AS1-L stably overexpressed and control A549 cells were detected by western blot. d PXN protein levels in PXN-AS1-L stably depleted and control NCI-H1299 cells were detected by western blot. e After transient overexpressing PXN-AS1-L in A549 cells, the stability of PXN mRNA over time was determined after blocking new RNA synthesis with α-amanitin (50 µM) and normalized to 18S rRNA (a product of RNA polymerase I that is unchanged by α-amanitin). f After transient depleting PXN-AS1-L in NCI-H1299 cells, the stability of PXN mRNA over time was determined after blocking new RNA synthesis with α-amanitin (50 µM) and normalized to 18S rRNA. Results are shown as mean ± SD of three independent experiments. **P < 0.01, ***P < 0.001 by Student’s t-test
PXN was up-regulated in NSCLC and positively correlated with PXN-AS1-L expression
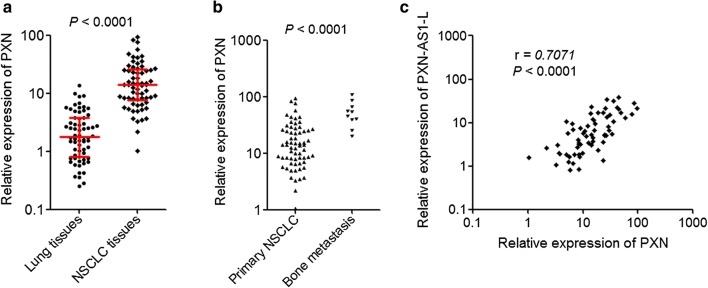
To explore whether the regulation of PXN by PXN-AS1-L also exist in vivo, we measured PXN expression in the same 66 pairs of NSCLC tissues and adjacent noncancerous lung tissues used in Fig. 1b. The results displayed that the expression of PXN was also significantly higher in NSCLC tissues than that in adjacent noncancerous lung tissues (Fig. 6a). Furthermore, our results also displayed that the expression of PXN was further higher in bone metastases tissues than that in primary NSCLC tissues (Fig. 6b). Correlation analysis displayed that the expression level of PXN-AS1-L was positively correlated with that of PXN in these 66 NSCLC tissues (Fig. 6c), supporting the positive regulation of PXN by PXN-AS1-L.
Fig. 6.
PXN was up-regulated in NSCLC and positively correlated with PXN-AS1-L expression. a The expressions of PXN in 66 pairs of NSCLC tissues and adjacent noncancerous lung tissues were detected by qPCR. P < 0.0001 by Wilcoxon signed-rank test. b The expressions of PXN-AS1-L in 66 primary NSCLC tissues and 10 NSCLC bone metastases tissues were detected by qPCR. P < 0.0001 by Mann–Whitney test. c The correlation between the expression level of PXN-AS1-L and PXN was analyzed in these 66 NSCLC tissues. r = 0.7071, P < 0.0001 by Pearson correlation analysis
PXN knockdown attenuated the oncogenic roles of PXN-AS1-L in NSCLC
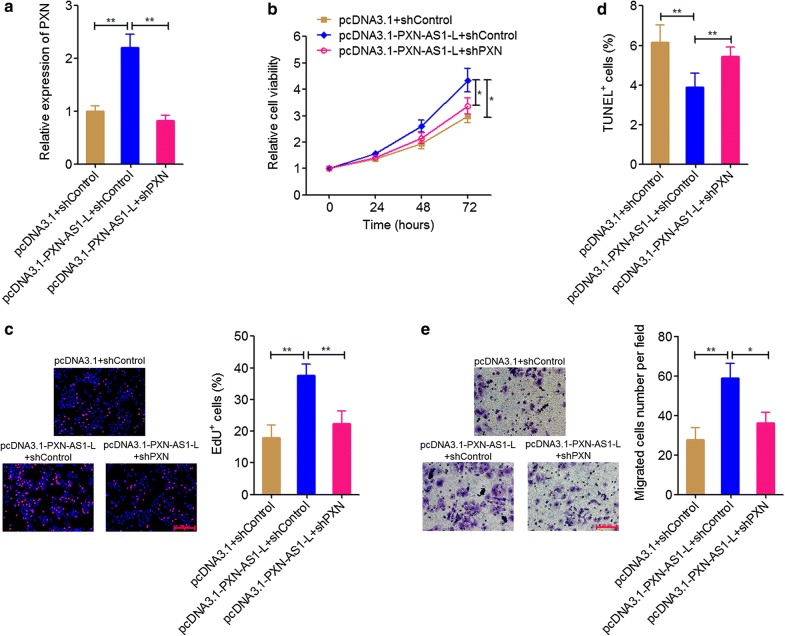
To explore whether the oncogenic roles of PXN-AS1-L in NSCLC are dependent on the positive regulation of PXN, we knocked down PXN in PXN-AS1-L stably overexpressed A549 cells by transient transfection of PXN specific shRNA (Fig. 7a). Glo cell viability assays displayed that PXN knockdown attenuated the increasing of cell viability induced by PXN-AS1-L overexpression (Fig. 7b). EdU incorporation assays also displayed that PXN knockdown attenuated the pro-proliferative roles of PXN-AS1-L (Fig. 7c). TUNEL assays displayed that PXN knockdown attenuated the cell apoptosis repressive roles of PXN-AS1-L (Fig. 7d). Transwell assays displayed that PXN knockdown attenuated the increasing of cell migration induced by PXN-AS1-L overexpression (Fig. 7e). All these data together demonstrated that PXN knockdown attenuated the roles of PXN-AS1-L in promoting cell proliferation, inhibiting cell apoptosis, and promoting cell migration of NSCLC cells. These data also suggested that the oncogenic roles of PXN-AS1-L in NSCLC were at least partially dependent on the positive regulation of PXN.
Fig. 7.
PXN knockdown attenuated the oncogenic roles of PXN-AS1-L in NSCLC. a The expressions of PXN in PXN-AS1-L stably overexpressed and control A549 cells after transient transfection of PXN specific shRNA were detected by qPCR. b Cell viability of PXN-AS1-L stably overexpressed and control A549 cells after transient transfection of PXN specific shRNA was detected by Glo cell viability assays. c Cell proliferation of PXN-AS1-L stably overexpressed and control A549 cells after transient transfection of PXN specific shRNA was detected by EdU incorporation assays. The red color indicates EdU-positive cells. Scale bars = 200 μm. d Cell apoptosis of PXN-AS1-L stably overexpressed and control A549 cells after transient transfection of PXN specific shRNA was detected by TUNEL assays. e Cell migration of PXN-AS1-L stably overexpressed and control A549 cells after transient transfection of PXN specific shRNA was detected by transwell assays. Scale bars = 100 μm. Results are shown as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 by Student’s t-test
Discussion
LncRNA PXN-AS1-L has 863 nucleotides in length. The gene encoding PXN-AS1-L locates at chromosome 12q24.23 and is reverse complementary to PXN. PXN-AS1-L is recently identified to have oncogenic roles in HCC [38]. In this study, we further studied the expression, roles, and mechanisms of action of PXN-AS1-L in NSCLC.
First, we found that PXN-AS1-L is up-regulated in NSCLC cell lines compared with normal bronchial epithelial cell line. Moreover, PXN-AS1-L is further up-regulated in NSCLC cell lines derived from metastatic sites (NCI-H1299 and SK-MES-1) compared with that derived from primary sites (NCI-H1975 and A549). In clinical tissues specimens, we also found that PXN-AS1-L is up-regulated in NSCLC tissues compared with noncancerous lung tissues and is further up-regulated in NSCLC bone metastases tissues. Analyses of the correlation between PXN-AS1-L expression and clinicopathological characteristics revealed that PXN-AS1-L was positively associated with tumor size, lymph nodes metastasis, advanced TNM stages, and poor prognosis. These data confirmed that PXN-AS1-L is up-regulated in NSCLC and implied that PXN-AS1-L may be involved in the progression of NSCLC. Whether the up-regulation of PXN-AS1-L in cancer is lung and liver cancer specific or cancer-popular need further investigation. Furthermore, multi-center prospective study investigating the correlation between the expression of PXN-AS1-L and prognosis of NSCLC patients would be more significant for the application of PXN-AS1-L for clinical outcome prediction.
Second, we investigated the in vitro and in vivo roles of PXN-AS1-L in NSCLC using gain-of-function and loss-of-function assays. Our results revealed that PXN-AS1-L overexpression increases NSCLC cell viability, promotes NSCLC cell proliferation, inhibits NSCLC cell apoptosis, and promotes NSCLC cell migration. Conversely, PXN-AS1-L knockdown decreases NSCLC cell viability, inhibits NSCLC cell proliferation, promotes NSCLC cell apoptosis, and represses NSCLC cell migration. Therefore, our data demonstrated that PXN-AS1-L also acts as an oncogene in NSCLC, similar to the roles of PXN-AS1-L in HCC. Whether the oncogenic role of PXN-AS1-L is lung and liver cancer specific or cancer-popular also need further investigation. Nevertheless, the up-regulation of PXN-AS1-L and oncogenic roles of PXN-AS1-L in NSCLC suggested that PXN-AS1-L may be a potential therapeutic target for NSCLC. Increasing evidences have shown that noncoding RNAs could be targeted by chemically modified complementary oligonucleotides, which are revealed to be effective treatment in animal models and clinical trials involving humans [42, 43]. Therefore, developing chemically modified complementary oligonucleotides targeting PXN-AS1-L would be potential therapeutic strategy for NSCLC.
The focal adhesion protein PXN mediates critical signal transduction and plays important roles in cell survival and migration [44, 45]. Due to the reverse complementation between PXN-AS1-L and PXN, we investigated whether PXN-AS1-L regulates PXN and whether PXN is the mediator of the oncogenic roles of PXN-AS1-L in NSCLC. In this study, we found that PXN-AS1-L up-regulated PXN expression. Similar to the expression pattern of PXN-AS1-L in NSCLC, PXN is also up-regulated in NSCLC tissues compared with noncancerous lung tissues and is further up-regulated in NSCLC bone metastases tissues. The expression of PXN-AS1-L is positively associated with that of PXN in NSCLC tissues. Furthermore, knockdown of PXN attenuated the oncogenic roles of PXN-AS1-L in NSCLC. All these data support the positive regulation of PXN by PXN-AS1-L and the importance of PXN in the oncogenic roles of PXN-AS1-L in NSCLC.
Conclusions
In conclusion, our data showed that PXN-AS1-L is up-regulated in NSCLC, predicts poor outcome of NSCLC patients, and has oncogenic roles in NSCLC via up-regulating PXN. Our data also implied that PXN-AS1-L may be a potential prognostic biomarker and therapeutic target for NSCLC.
Authors’ contributions
XZ, WH, and ZZ designed the study; ZZ, ZP, JC, JW, YH, and KS carried out the experiments; XZ, WH, ZZ, ZP, and JC collected and analyzed the data. XZ, WH, and ZZ wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The data used and analyzed during this study are available from the corresponding author on request.
Consent for publication
The authors agree for publication.
Ethics approval and consent to participate
Use of patient tissue samples and nude mice were approved by the ethics committee of the General Hospital of Chinese People’s Liberation Army (Beijing, China).
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81371911) and Beijing science and technique star foundation (No. 2010B80).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- NSCLC
non-small cell lung cancer
- lncRNAs
long noncoding RNAs
- HCC
hepatocellular carcinoma
- qPCR
quantitative real-time PCR
- EdU
ethynyl deoxyuridine
- TUNEL
terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick end labeling
- IHC
immunohistochemistry
Contributor Information
Zhifa Zhang, Email: zhangzhifa301@sina.com.
Zhaohui Peng, Email: zhaohuipeng_R@163.com.
Junying Cao, Email: shenzongchaosheng@163.com.
Jiaqi Wang, Email: 1327623@qq.com.
Yongyu Hao, Email: kyluakylua@163.com.
Kai Song, Email: songkai301@sina.com.
Yan Wang, Email: wangyan301@sina.com.
Wei Hu, Email: huweicj@163.com.
Xuesong Zhang, Email: zhangxuesong301@sina.com.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018 doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14:535–546. doi: 10.1038/nrc3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reck M, Heigener DF, Mok T, Soria JC, Rabe KF. Management of non-small-cell lung cancer: recent developments. Lancet. 2013;382:709–719. doi: 10.1016/S0140-6736(13)61502-0. [DOI] [PubMed] [Google Scholar]
- 4.Sekimoto N, Suzuki Y, Sugano S. Decreased KPNB1 expression is induced by PLK1 inhibition and leads to apoptosis in lung adenocarcinoma. J Cancer. 2017;8:4125–4140. doi: 10.7150/jca.21802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samulin Erdem J, Skaug V, Haugen A, Zienolddiny S. Loss of MKK3 and MK2 copy numbers in non-small cell lung cancer. J Cancer. 2016;7:512–515. doi: 10.7150/jca.13651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boldrini L, Giordano M, Niccoli C, Melfi F, Lucchi M, Mussi A, Fontanini G. Role of microRNA-33a in regulating the expression of PD-1 in lung adenocarcinoma. Cancer Cell Int. 2017;17:105. doi: 10.1186/s12935-017-0474-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan X, Hu Z, Feng Y, Hu X, Yuan J, Zhao SD, Zhang Y, Yang L, Shan W, He Q, et al. Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell. 2015;28:529–540. doi: 10.1016/j.ccell.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 11.Battaglin F, Naseem M, Puccini A, Lenz HJ. Molecular biomarkers in gastro-esophageal cancer: recent developments, current trends and future directions. Cancer Cell Int. 2018;18:99. doi: 10.1186/s12935-018-0594-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivanovic RF, Viana NI, Morais DR, Silva IA, Leite KR, Pontes-Junior J, Inoue G, Nahas WC, Srougi M, Reis ST. miR-29b enhances prostate cancer cell invasion independently of MMP-2 expression. Cancer Cell Int. 2018;18:18. doi: 10.1186/s12935-018-0516-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 15.Flintoft L. Non-coding RNA: structure and function for lncRNAs. Nat Rev Genet. 2013;14:598. doi: 10.1038/nrg3561. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, Xiao ZD, Han L, Zhang J, Lee SW, Wang W, Lee H, Zhuang L, Chen J, Lin HK, et al. LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol. 2016;18:431–442. doi: 10.1038/ncb3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu D, Yang F, Yuan JH, Zhang L, Bi HS, Zhou CC, Liu F, Wang F, Sun SH. Long noncoding RNAs associated with liver regeneration 1 accelerates hepatocyte proliferation during liver regeneration by activating Wnt/beta-catenin signaling. Hepatology. 2013;58:739–751. doi: 10.1002/hep.26361. [DOI] [PubMed] [Google Scholar]
- 19.Zhang C, Yuan J, Hu H, Chen W, Liu M, Zhang J, Sun S, Guo Z. Long non-coding RNA CHCHD4P4 promotes epithelial-mesenchymal transition and inhibits cell proliferation in calcium oxalate-induced kidney damage. Braz J Med Biol Res. 2017;51:e6536. doi: 10.1590/1414-431X20176536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin A, Hu Q, Li C, Xing Z, Ma G, Wang C, Li J, Ye Y, Yao J, Liang K, et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat Cell Biol. 2017;19:238–251. doi: 10.1038/ncb3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, Wang C, Hawke DH, Wang S, Zhang Y, et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu XT, Yuan JH, Zhu TT, Li YY, Cheng XY. Long noncoding RNA glypican 3 (GPC3) antisense transcript 1 promotes hepatocellular carcinoma progression via epigenetically activating GPC3. FEBS J. 2016;283:3739–3754. doi: 10.1111/febs.13839. [DOI] [PubMed] [Google Scholar]
- 23.Li JK, Chen C, Liu JY, Shi JZ, Liu SP, Liu B, Wu DS, Fang ZY, Bao Y, Jiang MM, et al. Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol Cancer. 2017;16:111. doi: 10.1186/s12943-017-0681-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parasramka M, Yan IK, Wang X, Nguyen P, Matsuda A, Maji S, Foye C, Asmann Y, Patel T. BAP1 dependent expression of long non-coding RNA NEAT-1 contributes to sensitivity to gemcitabine in cholangiocarcinoma. Mol Cancer. 2017;16:22. doi: 10.1186/s12943-017-0587-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernando TR, Contreras JR, Zampini M, Rodriguez-Malave NI, Alberti MO, Anguiano J, Tran TM, Palanichamy JK, Gajeton J, Ung NM, et al. The lncRNA CASC15 regulates SOX4 expression in RUNX1-rearranged acute leukemia. Mol Cancer. 2017;16:126. doi: 10.1186/s12943-017-0692-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizrahi I, Mazeh H, Grinbaum R, Beglaibter N, Wilschanski M, Pavlov V, Adileh M, Stojadinovic A, Avital I, Gure AO, et al. Colon cancer associated transcript-1 (CCAT1) expression in adenocarcinoma of the stomach. J Cancer. 2015;6:105–110. doi: 10.7150/jca.10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Yang F, Yuan JH, Yuan SX, Zhou WP, Huo XS, Xu D, Bi HS, Wang F, Sun SH. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013;34:577–586. doi: 10.1093/carcin/bgs381. [DOI] [PubMed] [Google Scholar]
- 28.Hosseini ES, Meryet-Figuiere M, Sabzalipoor H, Kashani HH, Nikzad H, Asemi Z. Dysregulated expression of long noncoding RNAs in gynecologic cancers. Mol Cancer. 2017;16:107. doi: 10.1186/s12943-017-0671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan L, Sun M, Liu GJ, Wei CC, Zhang EB, Kong R, Xu TP, Huang MD, Wang ZX. Long noncoding RNA PVT1 promotes non-small cell lung cancer cell proliferation through epigenetically regulating LATS2 expression. Mol Cancer Ther. 2016;15:1082–1094. doi: 10.1158/1535-7163.MCT-15-0707. [DOI] [PubMed] [Google Scholar]
- 31.Sun M, Liu XH, Wang KM, Nie FQ, Kong R, Yang JS, Xia R, Xu TP, Jin FY, Liu ZJ, et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer. 2014;13:68. doi: 10.1186/1476-4598-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su W, Feng S, Chen X, Yang X, Mao R, Guo C, Wang Z, Thomas DG, Lin J, Reddy RM, et al. Silencing of long non-coding RNA MIR22HG triggers cell survival/death signaling via oncogenes YBX1, MET, and p21 in lung cancer. Cancer Res. 2018;78:3207–3219. doi: 10.1158/1538-7445.AM2018-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao C, Wang X, Liu Y, Wang M, Yan B, Jiang Y, Shi Y, Shen Y, Liu X, Liai W, et al. A G3BP1-interacting lncRNA promotes ferroptosis and apoptosis in cancer via nuclear sequestration of p53. Cancer Res. 2018;78:3484–3496. doi: 10.1158/1538-7445.AM2018-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan S, Liu Q, Hu Z, Zhou Z, Wang G, Li C, Xie W, Meng G, Xiang Y, Wu N, et al. Long non-coding RNA MUC5B-AS1 promotes metastasis through mutually regulating MUC5B expression in lung adenocarcinoma. Cell Death Dis. 2018;9:450. doi: 10.1038/s41419-018-0472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li K, Sun D, Gou Q, Ke X, Gong Y, Zuo Y, Zhou JK, Guo C, Xia Z, Liu L, et al. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018;420:80–90. doi: 10.1016/j.canlet.2018.01.060. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z, Li JL, Lin S, Cao C, Gimbrone NT, Yang R, Fu DA, Carper MB, Haura EB, Schabath MB, et al. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J Clin Invest. 2016;126:2267–2279. doi: 10.1172/JCI85250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 2014;15:429. doi: 10.1186/s13059-014-0429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan JH, Liu XN, Wang TT, Pan W, Tao QF, Zhou WP, Wang F, Sun SH. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 2017;19:820–832. doi: 10.1038/ncb3538. [DOI] [PubMed] [Google Scholar]
- 39.Jiang H, Zhang H, Hu X, Li W. Knockdown of long non-coding RNA XIST inhibits cell viability and invasion by regulating miR-137/PXN axis in non-small cell lung cancer. Int J Biol Macromol. 2018;111:623–631. doi: 10.1016/j.ijbiomac.2018.01.022. [DOI] [PubMed] [Google Scholar]
- 40.Wu DW, Chen CY, Chu CL, Lee H. Paxillin confers resistance to tyrosine kinase inhibitors in EGFR-mutant lung cancers via modulating BIM and Mcl-1 protein stability. Oncogene. 2016;35:621–630. doi: 10.1038/onc.2015.120. [DOI] [PubMed] [Google Scholar]
- 41.Bi Y, Han Y, Bi H, Gao F, Wang X. miR-137 impairs the proliferative and migratory capacity of human non-small cell lung cancer cells by targeting paxillin. Hum Cell. 2014;27:95–102. doi: 10.1007/s13577-013-0085-4. [DOI] [PubMed] [Google Scholar]
- 42.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 43.Slack FJ. Tackling tumors with small RNAs derived from transfer RNA. N Engl J Med. 2018;378:1842–1843. doi: 10.1056/NEJMcibr1716989. [DOI] [PubMed] [Google Scholar]
- 44.Huang C, Rajfur Z, Borchers C, Schaller MD, Jacobson K. JNK phosphorylates paxillin and regulates cell migration. Nature. 2003;424:219–223. doi: 10.1038/nature01745. [DOI] [PubMed] [Google Scholar]
- 45.Liu S, Thomas SM, Woodside DG, Rose DM, Kiosses WB, Pfaff M, Ginsberg MH. Binding of paxillin to alpha4 integrins modifies integrin-dependent biological responses. Nature. 1999;402:676–681. doi: 10.1038/45264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used and analyzed during this study are available from the corresponding author on request.