Abstract
MicroRNAs (miRNAs) have been reported to influence the occurrence and development of colorectal carcinoma (CRC). In the current research, we explored miR-140-5p function in CRC. In addition, the molecular mechanisms of miR-140-5p/SOX4 axis for CRC were investigated. The miR-140-5p and SOX4 expression levels were evaluated via RT-qPCR or western blot analysis. The MTT and Transwell assay assessed cell proliferation and invasion. Luciferase assay confirmed that miR-140-5p directly targeted SOX4. Moreover, the results showed that miR-140-5p expression was obviously declined in CRC. The low expression of miR-140-5p was related to tumor stage or metastasis. The miR-140-5p overexpression suppressed cell proliferation and invasion in CRC. Moreover, miR-140-5p was identified to directly target SOX4 and the SOX4 expression was increased in CRC. In addition, the high expression of SOX4 promoted cell proliferation and invasion in CRC. The inhibiting effect of miR-140-5p on SOX4 was identified for CRC, and it inhibited the proliferation and invasion. The novel miR-140-5p/SOX4 axis may provide a new therapy for CRC.
Keywords: colorectal carcinoma, miR-140-5p, invasion, proliferation, SOX4
Introduction
Colorectal carcinoma (CRC) is a prevalent gastrointestinal malignancy, and its morbidity ranks only second to gastric and esophageal cancer (1). The ranking of CRC patients was fifth in males and sixth in females in the most common malignant tumor deaths in China (2). CRC is second only to lung cancer in western developed countries, whereas its rate of morbidity in different countries differs by almost 60 times (3). So far, the incidence of CRC is still increasing, although various early detection methods and treatments have made great progress. Consequently, developing a new biomarker for diagnosis and clarifying the regulatory mechanism of the occurrence and progression of CRC tumors are essential.
MicroRNAs (miRNAs) regulate target gene expression through combining transcriptional mRNA (4). A previous study pointed out that miRNAs took part in multiple cancer-related signaling pathways including cell migration, invasion, proli-feration, apoptosis, and metastasis (5). Importantly, miRNAs were found to express abnormally which was relevant to CRC development. Among them, miR-17, miR-21, miR-182 and miR-203 acted as oncogenes in CRC, while miR-30a, miR-143, miR-145 and miR-195 were considered as tumor suppressors (6,7). Recently, many scholars have investigated the role of miR-140 in the process of tumor formation in various cancers, such as glioma (8), cervical cancer (9), gastric cancer (10) and breast cancer (11). Moreover, miR-140 has been considered to be involved in colorectal tumorigenesis and progression through regulating proliferation, apoptosis, differentiation, migration and invasion (12–14). However, there are no previous studies on the function of miR-140/SOX4 axis in CRC.
Sex-determining region Y-related high-mobility group box 4 (SOX4) plays a role in cancer development. Moreover, SOX4 was observed in many cancers regulated by miR-212 (15), miR-338 (16), miR-25 (17), and miR-132 (18). Vishnubalaji et al revealed that miRNA-320 suppressed CRC by targeting SOX4, FOXM1, and FOXQ1 (19). In this study, miR-140-5p expression and its clinicopathological significance in CRC were investigated. Furthermore, the role of miR-140-5p was analyzed from the perspective of cell proliferation and invasion in CRC at the same time. SOX4 directly targeted miR-140-5p. This study aimed at providing new therapeutic implications for the diagnosis of CRC.
Materials and methods
Clinical tissues
Thirty-six surgical tumor specimens and adjacent tissue samples were obtained from Shandong Provincial Third Hospital (Jinan, Shandong) after receiving written informed consent. None of the patients received treatment prior to the operation. Human tissue was frozen in liquid nitrogen and then stored at −80°C in a refrigerator for further use. This experiment was approved by the Institutional Ethics Committee of Shandong Provincial Third Hospital.
Cell culture and transfection
The human CRC cell lines HT29 (cat. no. HTB-38), SW480 [ZK0200(XR)] and normal colorectal cell line NCM460 (cat. no. BNF-3068) were used for this experiment. All the cell lines were obtained from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All cells were seeded in DMEM supplemented by 10% fetal bovine serum (FBS) and cultured at 37°C with 5% CO2.
The miR-140-5p mimic and inhibitor, SOX4 siRNA (si-SOX4) were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China) and then they were transferred into HT29 or SW480 cells with Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA) according to the manufacturer's instructions.
RT-qPCR
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was applied for extracting total RNA containing miRNA to quantitate the miR-140-5p expression in CRC tissues and cell lines. RT-qPCR was carried out through the SYBR-Green PCR kit (Takara Bio, Inc., Otsu, Japan) on ABI 7500 Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc., Foster City, CA, USA). The reaction conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. U6 and GAPDH were used as control for miR-140-5p and SOX4. The miR-145-5p and TAGLN2 levels were analyzed using the 2−ΔΔCq method (20).
Luciferase activity assay
TargetScan (http://www.targetscan.org/) was employed to predict the biological targets of miRNAs (21). The wild or mutant type of 3′-UTR of SOX4 was inserted into the pGL3 promoter vector (Invitrogen; Thermo Fisher Scientific, Inc.) for luciferase reporter experiments. Then, wild or mutant type of 3′-UTR of SOX4 and miR-140-5p mimic were transfected into SW480 cells. After transfection for 48 h, the Dual-Luciferase Reporter Assay (Promega Corp., Madison, WI, USA) was applied to perform luciferase assays.
Cell proliferation (MTT) assay
The MTT assay was applied to measure cell proliferation. Cells (2×103) were seeded onto 96-well plates in medium containing 10% FBS. The cells containing miR-140-5p mimic or inhibitor were incubated for 0–96 h. After incubation, the cells added with MTT (Sigma-Aldrich; Merck KGaA, St. Louis, MO, USA) were incubated for 4 h at 37°C. The absorbance at 490 nm (OD=490 nm) was detected with a spectrophotometer (Molecular Devices LLC., San Jose, CA, USA).
Cell invasion assay
Transwell assay was performed to measure cell invasion. The cells were planted into the upper chambers (8 µm pore size; Corning, Inc., Corning, NY, USA) and medium with 10% FBS was added into the lower chamber. The cells were incubated for 24 h at 37°C in 5% CO2. Then the invasive cells on the lower surface were fixed with 70% ethanol and stained with crystal violet stain. Cells were counted by a light microscope (Olympus Corporation, Tokyo, Japan).
Western blot analysis
The protein samples were obtained using RIPA lysis buffer. Protein concentration was calculated using bicinchoninic acid (BCA; Beyotime Institute of Biotechnology, Shanghai, China). The 25 µl protein sample was added in the protein loaded per lane. Proteins were separated through a 12% SDS-PAGE and then incubated with 5% non-fat milk blocked membranes at room temperature for 2 h. Next we incubated the membranes overnight at 4°C with anti-SOX4 rabbit polyclonal antibody (dilution, 1:1,000; cat. no. ab80261; Abcam, Cambridge, MA, USA) anti-GAPDH mouse monoclonal antibody (dilution, 1:1,000; cat. no. 60004-1-Ig; ProteinTech, Wuhan, China) and subsequently incubated with goat anti-rabbit IgG H&L (HRP) (dilution, 1:3,000; cat. no. ab6721; Abcam) secondary antibody. Then, the protein expression levels were measured by a chemiluminescent detection system (Pierce ECL Substrate Western Blot Detection System; Pierce; Thermo Fisher Scientific, Inc., Rockford, IL, USA). Protein expression levels were quantified using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
The obtained data were shown as the mean ± SD. Enumeration data were calculated according to ANOVA with Tukey-Kramer post hoc test and χ2 test. Statistical analysis was analyzed with GraphPad Prism 6.0 and SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). It was defined as significant at P<0.05.
Results
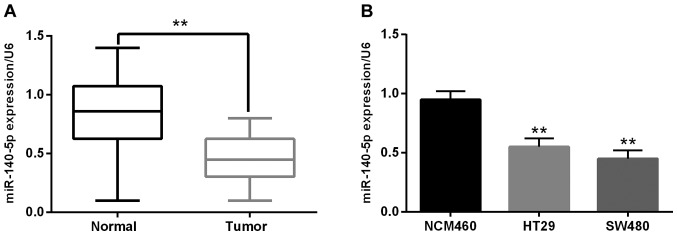
Downregulation of miR-140-5p was observed in CRC
The miR-140-5p expression was detected to verify its role in CRC carcinogenesis. Firstly, we analyzed the relationship between the clinicopathological characteristics and aberrant expression of miR-140-5p. The miR-140-5p downregulation was closely related to tumor staging or metastasis (Table I). Additionally, the RT-qPCR experiment suggested that the miR-140-5p expression was noticeably declined in CRC tissues in contrast to the adjacent normal tissues (Fig. 1A). Furthermore, we also found that miR-140-5p expression was much lower in HT29 and SW480 cell lines than that of NCM460 cells (control) (Fig. 1B). In brief, all results proved that the miR-140-5p downregulation was related to carcinogenesis of CRC.
Table I.
Clinicopathological characteristics and miR-140-5p expression in CRC.
| miR-140 expression | ||||
|---|---|---|---|---|
| Characteristics | Cases (n=36) | High | Low | P-value |
| Age (years) | 0.3173 | |||
| ≥60 | 20 | 8 | 12 | |
| <60 | 16 | 6 | 10 | |
| Tumor size (cm) | 0.6276 | |||
| ≥5 | 19 | 9 | 10 | |
| <5 | 17 | 8 | 9 | |
| Sex | 0.1213 | |||
| Male | 21 | 9 | 12 | |
| Female | 15 | 6 | 9 | |
| TNM stage | 0.0005a | |||
| I+II | 24 | 7 | 17 | |
| III+IV | 12 | 5 | 7 | |
| Lymph node metastasis | 0.005a | |||
| Yes | 23 | 6 | 17 | |
| No | 13 | 3 | 10 | |
Statistical analyses were performed by the χ2 test. CRC, colorectal carcinoma; TNM, tumor-node-metastasis.
P<0.05 was considered significant.
Figure 1.
Downregulation of miR-140-5p was observed in CRC. (A) The expression of miR-140-5p was detected via RT-qPCR in CRC tissues. (B) The expression of miR-140-5p was detected in HT29, SW480 and NCM460 cells (control). **P<0.01. CRC, colorectal carcinoma; RT-qPCR, reverse transcription-quantitative PCR.
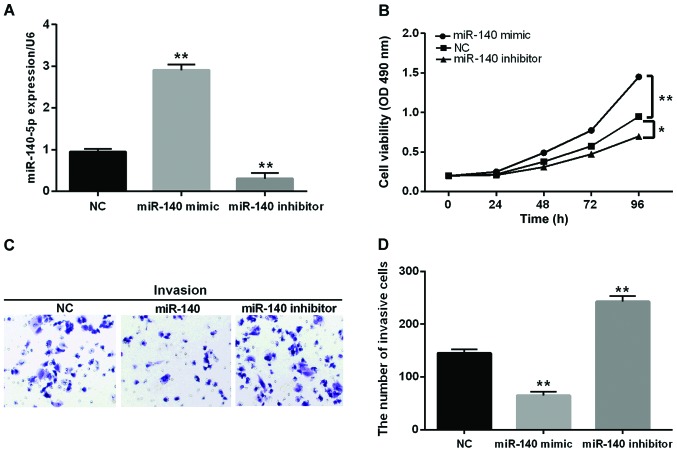
miR-140-5p inhibits CRC cell proliferation and invasion
Then, we transfected miR-140-5p mimic or inhibitor into SW480 cells to further confirm its function in CRC progression. The transfection efficiency was detected through RT-qPCR (Fig. 2A). Additionally, the cell proliferation and invasion in transfected cells was investigated by MTT and Transwell assay. The MTT analysis revealed that miR-140-5p overexpression obviously inhibited cell proliferation while the opposite effect was found in cells blocking miR-140-5p (Fig. 2B). Similarly, miR-140-5p overexpression markedly inhibited cell invasion while blocking miR-140-5p inversely regulated cell invasion (Fig. 2C and D). From the above results, we speculated that miR-140-5p had an inhibitory effect on CRC.
Figure 2.
miR-140-5p inhibits CRC cell proliferation and invasion. (A) miR-140-5p expression was examined in HT29 cells that contained miR-140-5p mimics or inhibitor via RT-qPCR. (B) The cell proliferation was measured in cells containing miR-140-5p mimics or inhibitor via MTT. (C and D) The cell invasion was measured in cells containing miR-140-5p mimics or inhibitor via Transwell analysis. *P<0.05; **P<0.01. CRC, colorectal carcinoma; RT-qPCR, reverse transcription-quantitative PCR.
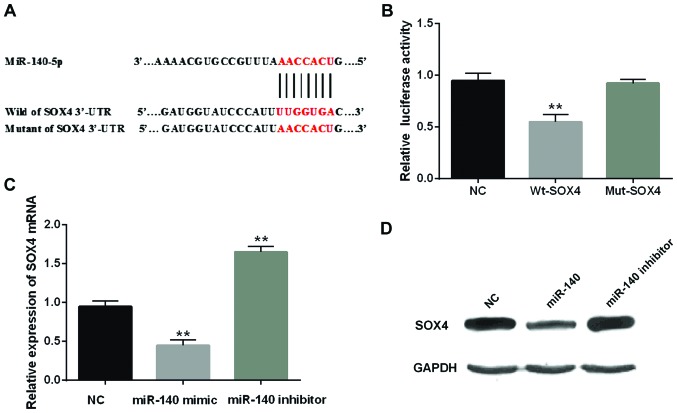
miR-140-5p directly targets SOX4 in CRC cells
As a target gene, SOX4 was analyzed to elucidate the regulated mechanisms in CRC. Among them, SOX4 as an oncogene demonstrated that overexpression of SOX4 was strongly correlated with the malignant diffusion of tumor in CRC (22). We presumed that miR-140-5p targeted SOX4 (Fig. 3A). In order to verify the above conclusion, we transfected Wt-SOX4 or Mut-SOX4 into SW480 cells containing miR-140-5p mimic. We found that miR-140-5p overexpression obviously reduced the luciferase activity of Wt-SOX4 while there was almost no change in Mut-SOX4 (Fig. 3B). In addition, the overexpression of miR-140-5p reduced the expression of mRNA and protein of SOX4. Conversely, the knockout of miR-140-5p promoted the SOX4 expression (Fig. 3C and D). Hence, miR-140-5p would directly target SOX4.
Figure 3.
miR-140-5p directly targets SOX4 in CRC cells. (A) The binding sites of miR-140-5p on the 3′-UTR of SOX4. (B) Luciferase reporter assay. (C and D) The mRNA and protein expression of SOX4 was analyzed in cells containing miR-140-5p mimic or inhibitor. **P<0.01. CRC, colorectal carcinoma.
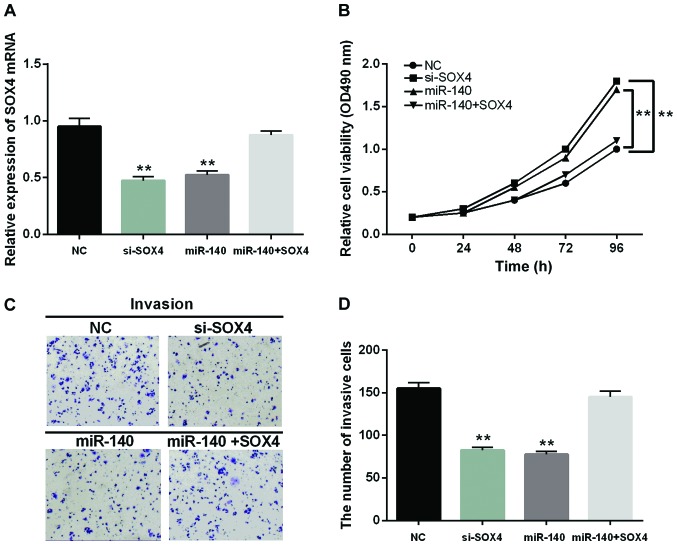
miR-140-5p regulates the progression of CRC through affecting SOX4 expression
For better understanding the interaction between miR-140-5p and SOX4, the si-SOX4 was applied to suppress the expression of SOX4 so as to identify its function in CRC. Moreover, the negative control or SOX4 expression vector was transfected into SW480 cells, which overexpressed miR-140-5p to further explore their function. The expression of SOX4 in the transfected cells were measured by RT-qPCR (Fig. 4A). Consistent with the expected result, the decrease of SOX4 expression suppressed cell proliferation and invasion (Fig. 4B-D). A similar result was identified in the cells containing miR-140-5p mimic as well. Furthermore, the SOX4 overexpression could conversely change the inhibition of cell proliferation and invasion induced by miR-140-5p (Fig. 4B-D). Thus, miR-140-5p inhibited the proliferation and invasion of CRC cells by regulating SOX4 expression.
Figure 4.
miR-140-5p regulates the progression of CRC through affecting SOX4 expression. (A) The mRNA expression of SOX4 was measured in cells containing si-SOX4 or SOX4 plasmid with miR-140-5p. (B) The cell proliferation in cells containing si-SOX4 or SOX4 plasmid with miR-140-5p. (C and D) Transwell analysis of SW480 cells with si-SOX4 or SOX4 plasmid with miR-140-5p. **P<0.01. CRC, colorectal carcinoma; si-SOX4, SOX4 siRNA.
Discussion
CRC patients still suffer a diagnosis in late stage and a high mortality, although the treatment for CRC has made great progress. Hence, exploiting novel biomarkers and applying them to early diagnosis, as well as improved cure rate will be the main tasks for us in the future. Moreover, CRC tumors occur through multiple steps to activate the oncogene or inactivate tumor suppressor that impact different regulatory pathways in CRC tumors (14). Opportunely, miRNAs have been found to take part in various biological processes in different tumors, which regulate mRNA stability or prevent mRNA translation by combining with its target gene (23). In addition, miRNA has been observed as an oncogene or a suppressor in previous studies. Among them, the different roles of miR-140-5p were identified in various cancers (8–12). Nevertheless, there were no sufficient investigations about the effect of miR-140-5p on the proliferation and invasion of CRC cells.
In this study, miR-140-5p inhibited the proliferation and invasion of CRC cells. In addition, the decreased expression of miR-140-5p was found in CRC which was consistent with other cancers (8–11). It proved that miR-140-5p had an inhibiting effect on tumor progression. Furthermore, we confirmed that low miR-140-5p expression was closely related to advanced clinical stage and lymph node metastasis which was similar to the previous study (12). These experimental results indicated that overexpression of miR-140-5p suppressed the tumorige-nesis and progression of CRC.
In addition, we paid more attention to SOX4 based on the prediction software databases in CRC. Previous studies reported that abnormal expression of many transcription factors, such as CBFB, SMARCC1 and SOX4, were all related to the occurrence of CRC (24). In this study, we silenced the SOX4 gene in SW480 cells and further explored its function in CRC progression. We found that the SOX4 overexpression promoted cell proliferation and invasion in CRC. The same result for cell proliferation and invasion was identified in esophageal tumor as well (25). Moreover, the SOX4 overexpression has been found to enhance cell migrated and invasive abilities in renal cell carcinoma (26). Some research indicated that SOX4 could accelerate malignant progression of CRC. Moreover, the interactions of miR-140-5p and SOX4 were investigated thoroughly in this study and miR-140-5p overexpression inversely regulated the SOX4 expression. Therefore, the miR-140-5p/SOX4 axis was considered to provide an effective biomarker for CRC diagnosis.
In conclusion, this study first revealed that miR-140-5p inhibited the SOX4 expression and contributed to the proli-feration and invasion of CRC cells. The miR-140-5p/SOX4 axis may provide new therapeutic implications for CRC. miR-140-5p performed as a tumor suppressor through downregulating SOX4 which can be developed as an effective therapeutic path for the treatment of CRC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
ZZ contributed significantly to the data analysis and wrote the manuscript. WL performed the data analyses. JL contributed to the conception of the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Shandong Provincial Third Hospital (Jinan, Shandong). Signed informed consents were obtained from the patients or guardians.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 2.Zheng ZX, Zheng RS, Zhang SW, Chen WQ. Colorectal cancer incidence and mortality in China, 2010. Asian Pac J Cancer Prev. 2014;15:8455–8460. doi: 10.7314/APJCP.2014.15.19.8455. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris TJ, McCormick F. The molecular pathology of cancer. Nat Rev Clin Oncol. 2010;7:251–265. doi: 10.1038/nrclinonc.2010.41. [DOI] [PubMed] [Google Scholar]
- 6.Meng WJ, Yang L, Ma Q, Zhang H, Adell G, Arbman G, Wang ZQ, Li Y, Zhou ZG, Sun XF. MicroRNA expression profile reveals miR-17-92 and miR-143-145 cluster in synchronous colorectal cancer. Medicine (Baltimore) 2015;94:e1297. doi: 10.1097/MD.0000000000001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun G, Cheng YW, Lai L, Huang TC, Wang J, Wu X, Wang Y, Huang Y, Wang J, Zhang K, et al. Signature miRNAs in colorectal cancers were revealed using a bias reduction small RNA deep sequencing protocol. Oncotarget. 2016;7:3857–3872. doi: 10.18632/oncotarget.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Y, Li Y, Wu C, Zhou L, Han X, Wang Q, Xie X, Zhou Y, Du Z. MicroRNA-140-5p inhibits cell proliferation and invasion by regulating VEGFA/MMP2 signaling in glioma. Tumour Biol. 2017;39:1010428317697558. doi: 10.1177/1010428317697558. [DOI] [PubMed] [Google Scholar]
- 9.Su Y, Xiong J, Hu J, Wei X, Zhang X, Rao L. MicroRNA-140-5p targets insulin like growth factor 2 mRNA binding protein 1 (IGF2BP1) to suppress cervical cancer growth and metastasis. Oncotarget. 2016;7:68397–68411. doi: 10.18632/oncotarget.11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou J, Xu Y. MicroRNA-140 inhibits cell proliferation in gastric cancer cell line HGC-27 by suppressing SOX4. Med Sci Monit. 2016;22:2243–2252. doi: 10.12659/MSM.896633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu Y, Qin T, Li J, Wang L, Zhang Q, Jiang Z, Mao J. MicroRNA-140-5p inhibits invasion and angiogenesis through targeting VEGF-A in breast cancer. Cancer Gene Ther. 2017;24:386–392. doi: 10.1038/cgt.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang W, Zou C, Pan L, Xu Y, Qi W, Ma G, Hou Y, Jiang P. MicroRNA-140-5p inhibits the progression of colorectal cancer by targeting VEGFA. Cell Physiol Biochem. 2015;37:1123–1133. doi: 10.1159/000430237. [DOI] [PubMed] [Google Scholar]
- 13.Zhai H, Fesler A, Ba Y, Wu S, Ju J. Inhibition of colorectal cancer stem cell survival and invasive potential by hsa-miR-140-5p mediated suppression of Smad2 and autophagy. Oncotarget. 2015;6:19735–19746. doi: 10.18632/oncotarget.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu L, Lu Y, Han X, Zhao W, Li J, Mao J, Wang B, Shen J, Fan S, Wang L, et al. microRNA-140-5p inhibits colorectal cancer invasion and metastasis by targeting ADAMTS5 and IGFBP5. Stem Cell Res Ther. 2016;7:180. doi: 10.1186/s13287-016-0438-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Jiang C, Wang H, Zhou L, Jiang T, Xu Y, Xia L. MicroRNA-212 inhibits the metastasis of nasopharyngeal carcinoma by targeting SOX4. Oncol Rep. 2017;38:82–88. doi: 10.3892/or.2017.5641. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Jin Y, Zhao M, Xie Q, Zhang H, Wang Q, Ma Q. MicroRNA-338-3p functions as tumor suppressor in breast cancer by targeting SOX4. Int J Oncol. 2015;47:1594–1602. doi: 10.3892/ijo.2015.3114. [DOI] [PubMed] [Google Scholar]
- 17.Chen B, Liu J, Qu J, Song Y, Li Y, Pan S. MicroRNA-25 suppresses proliferation, migration, and invasion of osteosarcoma by targeting SOX4. Tumour Biol. 2017;39:1010428317703841. doi: 10.1177/1010428317703841. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Zu L, Wang Y, Wang M, Chen P, Zhou Q. miR-132 inhibits lung cancer cell migration and invasion by targeting SOX4. J Thorac Dis. 2015;7:1563–1569. doi: 10.3978/j.issn.2072-1439.2015.09.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vishnubalaji R, Hamam R, Yue S, Al-Obeed O, Kassem M, Liu FF, Aldahmash A, Alajez NM. MicroRNA-320 suppresses colorectal cancer by targeting SOX4, FOXM1, and FOXQ1. Oncotarget. 2016;7:35789–35802. doi: 10.18632/oncotarget.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Volinia S, Visone R, Galasso M, Rossi E, Croce CM. Identification of microRNA activity by Targets' Reverse EXpression. Bioinformatics. 2010;26:91–97. doi: 10.1093/bioinformatics/btp598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu F, Min J, Cao X, Liu L, Ge Z, Hu J, Li X. MiR-363-3p inhibits the epithelial-to-mesenchymal transition and suppresses metastasis in colorectal cancer by targeting Sox4. Biochem Biophys Res Commun. 2016;474:35–42. doi: 10.1016/j.bbrc.2016.04.055. [DOI] [PubMed] [Google Scholar]
- 23.Smits M, Nilsson J, Mir SE, van der Stoop PM, Hulleman E, Niers JM, de Witt Hamer PC, Marquez VE, Cloos J, Krichevsky AM, et al. miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget. 2010;1:710–720. doi: 10.18632/oncotarget.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen CL, Christensen LL, Thorsen K, Schepeler T, Sørensen FB, Verspaget HW, Simon R, Kruhøffer M, Aaltonen LA, Laurberg S, et al. Dysregulation of the transcription factors SOX4, CBFB and SMARCC1 correlates with outcome of colorectal cancer. Br J Cancer. 2009;100:511–523. doi: 10.1038/sj.bjc.6604884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koumangoye RB, Andl T, Taubenslag KJ, Zilberman ST, Taylor CJ, Loomans HA, Andl CD. SOX4 interacts with EZH2 and HDAC3 to suppress microRNA-31 in invasive esophageal cancer cells. Mol Cancer. 2015;14:24. doi: 10.1186/s12943-014-0284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ruan H, Yang H, Wei H, Xiao W, Lou N, Qiu B, Xu G, Song Z, Xiao H, Liu L, et al. Overexpression of SOX4 promotes cell migration and invasion of renal cell carcinoma by inducing epithelial-mesenchymal transition. Int J Oncol. 2017;51:336–346. doi: 10.3892/ijo.2017.4010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.