Abstract
Oral cancer is a common cancer of the head and neck. Oral squamous cell carcinoma (OSCC) represents almost 90% of the total cases of head and neck cancer. Ubiquitin-specific protease 22 (USP22) is a deubiquitinating hydrolase, and it is highly expressed in various types of cancer, which also typically have a poor prognosis. Aurora-B and Survivin, which belong to the chromosomal passenger complex, are also highly expressed in a number of types of cancer. In the present study, USP22 expression and its associations with Aurora-B and Survivin, and the clinicopathological features in OSCC were explored. USP22 is highly expressed in OSCC. Overexpression of USP22 is associated with lymph node metastasis and histological grade (P<0.01). Additionally, the expression of USP22 was positively associated with Aurora-B (P<0.01), Survivin (P<0.01), and Ki-67 (P<0.01). Furthermore, USP22 small interfering RNA inhibited cell growth and reduced the expression levels of Aurora-B, Survivin and Cyclin B, together with the upregulation of cyclin-dependent kinase inhibitor 1A (p21). These data suggest that USP22, Aurora-B and Survivin promote the OSCC development and may represent novel targets for OSCC diagnosis and treatment in the future.
Keywords: oral squamous cell carcinoma, ubiquitin-specific protease 22, survivin, aurora-B
Introduction
Oral squamous cell carcinoma (OSCC) represents almost 90% of head and neck cancers (1). OSCC is the sixth most common type of malignant tumor; an estimated 50 million new cases occur annually worldwide, with data reported that the overall 5-year survival rate of OSCC has remained <50% over the past decade (2). Although there have been a large number of studies at the genetic and transcriptional levels in squamous cell carcinoma, the molecular mechanisms of carcinogenesis have not been completely elucidated. Therefore, identification of the target molecules that control the biological characteristics of OSCC would be of great clinical significance.
Deubiquitinating enzymes (DUBs) are a protease superfamily, which serve a role in the ubiquitin-proteasome system by cleaving ubiquitin chains from substrate proteins (3). Ubiquitin-specific protease 22 (USP22) is a deubiquitinating hydrolase that belongs to the DUB family (4). The USP22 gene is located on chromosome 17, and consists of 1578 base pairs that encode a protein measuring 525 amino acids long (4,5). USP22 is weakly expressed in several human tissues, including the liver, skeletal muscle and heart, and it is also strongly expressed in various types of cancer, which typically have a poor prognosis, including salivary duct carcinoma, colorectal cancer, head and neck cancer, breast cancer, and hepatocellular carcinoma (HCC) (6–12). USP22 is also considered to be a cancer stem cell marker that serves a role in the development and progression of carcinomas (4,5). USP22 can function as a subunit of the human Spt-Ada-Gcn5-acetyltransferase complex, and is involved in the transcription of target genes (5). A previous study demonstrated that USP22 could inhibit the transcription of the cyclin-dependent kinase inhibitor 1A (p21) gene by deubiquitinating the transcriptional regulator fructose-1,6-bisphosphatase (FBP1) (5). Furthermore, USP22 small interfering (si)RNA decreased the expression of Cyclin B and Survivin, and increased the expression of p21 in HCC (7). However, the precise mechanism by which USP22 affects these proteins remains unknown.
Survivin is a member of the inhibitor of apoptosis protein family and is also part of the chromosomal passenger complex (CPC), together with Aurora B kinase, inner centromere protein (INCENP) and Borealin (13). The CPC functions as an important modulator of mitosis and cytokinesis, which are known to serve a crucial role in cancer cell proliferation, resulting in more aggressive, malignant types of cancer (13,14). Several studies have suggested that Survivin is overexpressed in a variety of human cancers; however, is barely detectable in the majority of differentiated tissues (15). Additionally, it has been reported that ubiquitin carboxyl-terminal hydrolase FAF-X, a deubiquitinating enzyme, regulates chromosome alignment and segregation by regulating the dynamic association of Aurora B and Survivin to centromeres (16). Furthermore, different cullin-based ubiquitin ligase 3 adaptors regulate Aurora B during mitosis, potentially by ubiquitinating different pools of Aurora B at distinct subcellular localizations (17). Ubiquitination regulates dynamic protein-protein interactions and chromosome segregation independently of protein degradation (16). Furthermore, USP22 is associated with Survivin expression in HCC (7). Therefore, we hypothesize that the expression of Survivin and Aurora-B may be regulated by USP22 through deubiquitination.
In the present study, USP22 expression and its correlation with Aurora-B, Survivin and the clinicopathological features in OSCC were examined. The functional associations between USP22, Aurora-B and Survivin in OSCC were also investigated.
Materials and methods
Patients and tissue samples
A total of 90 patients (66 males and 24 females), who underwent complete surgical resection between January 2009 and June 2015 at the Affiliated Hospital of Guilin Medical University (Guilin, China) were enrolled in the present study. The inclusion criteria were: i) A diagnosis of OSCC confirmed by pathology; ii) Undergone surgery for the disease. All samples were obtained following approval by the Ethics Committee of Guilin Medical University (Guilin, China). The ages of the patients ranged from 26 to 80 years (mean, 56.5 years). Histologically, 70 cases were well or moderately differentiated, 20 were poorly differentiated OSCC. Clinicopathological data, including sex, age, tumor size, lymph node metastasis and histological differentiation were recorded, the classification by the World Health Organization (WHO) were used for the degree of differentiation (18,19). Tumors from each patient were 10% formalin-fixed in room temperature for 24 h and cut into 4–5 µm sections. All the subjects provided written informed consent.
Immunohistochemical staining
The tissue sections were incubated with a primary monoclonal anti-USP22 antibody (cat. no ab71732; 1:100; Abcam, Cambridge, MA, USA), monoclonal anti-Aurora-B antibody (cat. no AMI-1; 1:200; Transduction Laboratories, San Diego, CA, USA), the anti-Ki-67 monoclonal antibody (cat. no MIB-1; 1:200, Dako; Agilent Technologies, Inc., Santa Clara, CA, USA), was used to examine antigen Ki-67 expression and the polyclonal anti-survivin antibody (1:1,000; Novus Biologicals, Littleton, CO, USA; cat. no. NB500-201). The sections were incubated with the primary antibodies at 4°C overnight following antigen retrieval for 5 min at boiling temperature (100°C) twice by microwave treatment in a citrate buffer solution (pH 6.0). Detection was performed using the avidin-biotin peroxidase complex method using a Labelled Strepavidin-biotin 2 (LSAB2) system-HRP (cat. no K0609; Dako; Agilent Technologies, Inc.), according to the manufacturer's protocol. The labeling index percentage of USP22, Aurora-B, survivin or Ki-67 was determined by examining ≥1,000 tumor cells in random 3 high-powered fields (×200 magnification; light microscope; Olympus BX53F). The expression levels of Aurora-B and survivin were divided into low expression (<20% positive cells) and high expression (≥20% positive cells) groups. The expression levels of USP22 and Ki-67 were divided into low expression (<50% positive cells) and high expression (≥50% positive cells).
Cell culture
The human OSCC cell line Ca9-22 was purchased from the Japanese Collection of Research Bioresources Cell Bank (Osaka, Japan). The cells were cultured in RPMI-1640 (Gibco, Thermo Fisher Scientific Inc.) containing penicillin (10,000 µ/ml) and streptomycin (10,000 µg/ml) (cat. no P1400; Beijing Solarbio Science & Technology Co. Ltd., Beijing, China) with 10% fetal bovine serum (FBS; Samer Feishier Technology Co., Ltd.) and maintained at 37°C in an atmosphere of 5% CO2. For the growth assay, 5×103 cells were plated onto 24-well plates, and the cells were counted at days 0, 2, 4 and 6.
Western blot analysis
The Ca9-22 cells were lysed in ice-cold RIPA buffer (cat. no. R0020; Beijing Solarbio Science & Technology Co. Ltd.) with protease inhibitor for 30 min and centrifuged at 12,000 × g for 20 min at 4°C. The protein concentration was determined by the Bradford method (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) as the standard. Each protein lysate (40 µg) were separated on 10% SDS-PAGE gels and transferred to a polyvinyl difluoride membrane (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The membrane was blocked with 5% skim milk (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) [PBS was diluted to obtain 5% milk seal solution (5 g/100 ml)] for 40 min at room temperature, and then incubated with the primary (4°C overnight) and secondary antibodies (secondary goat anti-rabbit (cat. no TA130015; 1:000) or anti-mouse antibody (cat. no TA100015; 1:000); OriGene Technologies, Inc., Rockville, MD, USA) for 1 h at room temperature. The primary antibodies were USP22 (1:1,000; Abcam; cat. no. ab71732), Survivin (cat. no. NB500-201; Novus Biologicals; 1:2,000), Aurora-B (cat. no AMI-1; 1:000; Transduction Laboratories), Cyclin B (cat. no. 610219), p21 (cat. no. clone 70) (1:1,000; Transduction Laboratories) and β-actin (1:2,000, OriGene Technologies, Inc.; cat. no TA-09).
siRNA transfection
Ca9-22 cells were transfected with Lipofectamine® 3000 Transfection reagent (Thermo Fisher Scientific, Inc.) with 150 pmol USP22 siRNA (5′-GCAGCUUCAAGGUGGACAATT-3′) and negative control siRNA (5′-UUCUCCGAACGUGUCACGUTT-3′) (Guangzhou Ribobio Co., Ltd., Guangzhou, China) in 1 ml OPTI-MEM. At 48 h following transfection, the cells were used for subsequent experiments.
Statistical analysis
The data were analyzed were using the SPSS 17.0 software package (SPPS, Inc., Chicago, IL, USA). Measurement data are presented as mean ± standard deviation. The χ2-test was used for the comparison of enumeration data. A paired student's t-test was used to compare the number of cells data between the siRNA-USP22 and the siRNA control groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of USP22, Aurora-B, Survivin and Ki-67 in OSCC tissues
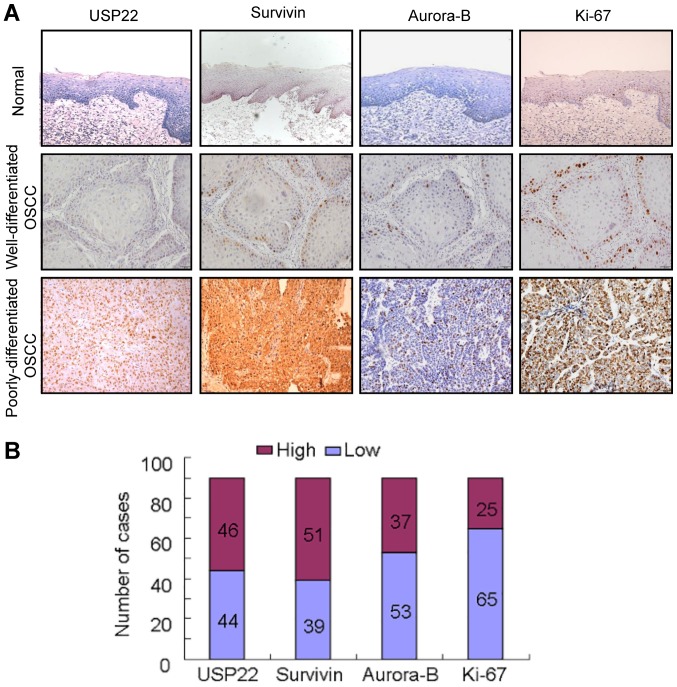
Firstly, USP22 expression was compared with Aurora-B, Survivin and Ki-67 expression in 20 normal tissues and 90 OSCC by performing immunohistochemistry. In normal colonic mucosa, USP22, Aurora-B, Survivin and Ki-67 were only distributed in the basal layers and exhibited weak staining. However, USP22, Aurora-B, Survivin and Ki-67 were strongly expressed in OSCC. Notably, the 4 proteins were more frequently expressed in poorly differentiated OSCC tissues, compared with the Well/Moderate-differentiated OSCC tissues (Fig. 1A, Tables I and II). USP22, Ki-67 and Aurora-B were primarily localized in the nuclei, while Survivin was localized to the cytoplasm and the nuclei (Fig. 1A). The number of high USP22, Survivin, Aurora-B and Ki-67 expression cases were 46/90 (51.11%), 51/90 (56.67%), 37/90 (41.11%) and 25/90 (27.78%), respectively. The number of low USP22, Survivin, Aurora-B and Ki-67 expression cases were 44/90 (48.89%), 46/90 (43.33%), 53/90 (58.89%) and 65/90 (72.22%), respectively (Fig. 1B).
Figure 1.
USP22 expression and its association with Survivin, Aurora-B and Ki-67 in OSCC. (A) Expression of USP22, Survivin, Aurora-B and Ki-67 was examined by immunohistochemistry in normal oral mucosa and OSCC tissues. In normal oral mucosa, USP22 were only distributed in the basal and parabasal layers and exhibited weak staining. In well-differentiated OSCC cases, USP22-positive cells were observed predominantly in the periphery of the tumor nests, while in poorly differentiated OSCC cases, USP22-positive cells were present throughout the tumor nests. The expression patterns of Survivin, Aurora-B and Ki-67 appear similar to USP22 in the same cases. (B) The quantity of high or low expression of USP22, Survivin, Aurora-B and Ki-67 in 90 OSCC cases. The quantity of high USP22, Survivin, Aurora-B and Ki-67 expression cases were 46/90, 51/90, 37/90 and 25/90 respectively. The quantity of low USP22, Survivin, Aurora-B and Ki-67 expression cases were 44/90, 46/90, 53/90 and 65/90 respectively. USP22, ubiquitin-specific protease 22; Ki-67, antigen Ki-67; OSCC, oral squamous cell carcinoma.
Table I.
USP22 expression and its association with clinicopathological features in oral squamous cell carcinoma.
| USP22 expression | |||
|---|---|---|---|
| Clinicopathological features | Low | High | P-value |
| Tissue type | |||
| Normal | 20 | 0 | |
| OSCC | 44 | 46 | |
| Age, years | |||
| ≥50 | 34 | 39 | 0.363 |
| <50 | 10 | 7 | |
| Sex | |||
| Male | 30 | 36 | 0.280 |
| Female | 14 | 10 | |
| Tumor size, mm | |||
| ≥15 | 20 | 18 | 0.544 |
| <15 | 24 | 28 | |
| Histological differentiation | |||
| Poor | 3 | 17 | <0.001 |
| Well/Moderate | 41 | 29 | |
| Lymph node metastasis | |||
| Negative | 41 | 31 | 0.002 |
| Positive | 3 | 15 | |
USP22, ubiquitin-specific protease 22; OSCC, oral squamous cell carcinoma.
Table II.
Survivin and Aurora-B expression and their associations with clinicopathological features in oral squamous cell carcinoma.
| Survivin expression | Aurora-B expression | |||||
|---|---|---|---|---|---|---|
| Clinicopathological features | Low | High | P-value | Low | High | P-value |
| Tissue type | ||||||
| Normal | 20 | 0 | 20 | 0 | ||
| OSCC | 39 | 51 | 53 | 37 | ||
| Age, years | ||||||
| ≥50 | 33 | 40 | 0.458 | 45 | 28 | 0.271 |
| <50 | 6 | 11 | 8 | 9 | ||
| Sex | ||||||
| Male | 17 | 43 | 0.0011 | 34 | 32 | 0.018 |
| Female | 16 | 8 | 19 | 5 | ||
| Tumor size, mm | ||||||
| ≥15 | 16 | 22 | 0.841 | 20 | 18 | 0.302 |
| <15 | 23 | 29 | 33 | 19 | ||
| Histological differentiation | ||||||
| Poor | 2 | 18 | 0.0006 | 4 | 16 | <0.001 |
| Well/Moderate | 37 | 33 | 49 | 21 | ||
| Lymph node metastasis | ||||||
| Negative | 36 | 35 | 0.0064 | 47 | 24 | 0.007 |
| Positive | 3 | 16 | 6 | 13 | ||
OSCC, oral squamous cell carcinoma.
Association between USP22, Aurora-B, Survivin and the clinicopathological features of OSCC
Next, the associations between the clinicopathological features of OSCCC and USP22, Aurora-B, Survivin expression and the clinicopathological features of OSCC were examined. High expression of USP22 was associated with lymph node metastasis (P<0.01) and histological grade (P<0.01), however, not with age, sex and tumor size in OSCC (Table I). The expression of Aurora-B and Survivin were associated with sex (P<0.05 and P<0.01, respectively), lymph node metastasis (P<0.001) and histological differentiation (P<0.01); however, not with tumor size and age in OSCC (Table II).
Association between USP22, Aurora-B, Survivin and Ki-67 expression in OSCC
In order to understand the role of USP22, the present study examined the association between USP22, Aurora-B, Survivin and Ki-67 in OSCC. Among the 90 OSCC cases, 46 cases exhibited the high USP22 expression, and 44 displayed a low USP22 expression in OSCC tissues (Table III). Of the 46 cases of high USP22 expression, the number of high Survivin, Aurora-B and Ki-67 expression cases were 33/46 (71.74%), 27/46 (58.70%) and 20/46 (43.48%), respectively (Table III). Of the 44 cases with a low USP22 expression, the number of low Survivin, Aurora-B and Ki-67 expression cases were 26/44 (59.09%), 34/44 (77.27%) and 39/44 (88.64%), respectively (Table III). USP22 expression was positively associated with Aurora-B, Survivin and Ki-67 (P<0.01, Table III). In addition, the clinicopathological findings were compared with the co-expression of USP22, Aurora-B and Survivin in OSCC. OSCC cases with a high expression of USP22, Aurora-B and Survivin exhibited increased lymph node metastasis (P<0.01) and poor differentiation (P=0.00006) when compared with OSCC cases with low expression of USP22, Aurora-B and Survivin (Table IV). These data indicate that USP22 promotes the development of OSCC together with Aurora-B and Survivin. These data indicate that USP22 promotes the development of OSCC together with Survivin and Aurora-B.
Table III.
Association between USP22 and survivin, between USP22 and Aurora-B expression and between USP22 and Ki-67 in oral squamous cell carcinoma.
| USP22 expression | ||||
|---|---|---|---|---|
| Protein expression | Low (n=44) | High (n=46) | Total | P-value |
| Survivin | ||||
| Low | 26 | 13 | 39 | 0.003 |
| High | 18 | 33 | 51 | |
| Aurora-B | ||||
| Low | 34 | 19 | 53 | 0.001 |
| High | 10 | 27 | 37 | |
| Ki-67 | ||||
| Low | 39 | 26 | 65 | 0.001 |
| High | 5 | 20 | 25 | |
USP22, ubiquitin-specific protease 22; Ki-67, antigen Ki-67.
Table IV.
USP22 and survivin, and USP22 and Aurora-B expression and their associations with clinicopathological features in oral squamous cell carcinoma.
| USP22/survivin/Aurora-B expression | ||||
|---|---|---|---|---|
| Clinicopathological features | All high (n=26) | Other (n=38) | All low (n=26) | P-value |
| Age, years | ||||
| ≥50 | 21 | 31 | 21 | 0.995 |
| <50 | 5 | 7 | 5 | |
| Sex | ||||
| Male | 22 | 30 | 14 | 0.025 |
| Female | 4 | 8 | 12 | |
| Tumor size, mm | ||||
| ≥15 | 9 | 20 | 9 | 0.232 |
| <15 | 17 | 18 | 17 | |
| Histological differentiation | ||||
| Poor | 13 | 7 | 0 | <0.001 |
| Well/Moderate | 13 | 31 | 26 | |
| Lymph node metastasis | ||||
| Negative | 16 | 29 | 26 | 0.003 |
| Positive | 10 | 9 | 0 | |
Suppression of cell growth by USP22 knockdown in OSCC cells
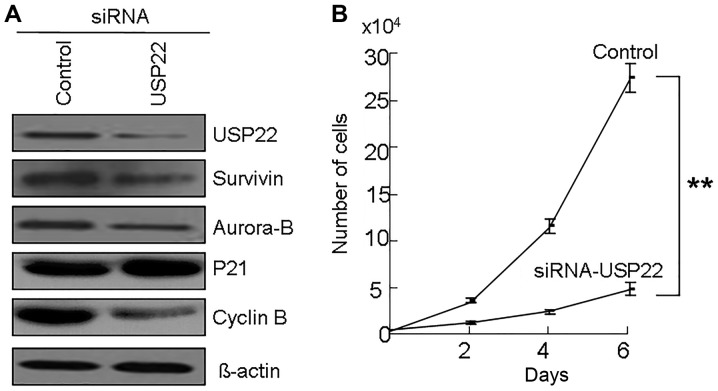
To understand the role of USP22 in the development of OSCC, USP22 was silenced by siRNA in OSCC cells. It was identified that USP22 siRNA decreased the expression of USP22 protein (Fig. 2A) and also inhibited cell growth in OSCC cells (Fig. 2B). Moreover, USP22 siRNA increased the p21 protein levels and decreased the Cyclin B protein levels in OSCC cells (Fig. 2A). Thus, we have found a possible relationship between USP22, Aurora-B and Survivin in OSCC tissues (Fig. 1 and Table III). Notably, USP22 siRNA reduced the protein levels of Aurora-B and Survivin in OSCC cells (Fig. 2B). These results suggest that the expression of Aurora-B and Survivin may be regulated by USP22.
Figure 2.
The effects of USP22 knockdown in OSCC cells. (A) USP22 siRNA were transfected into Ca9-22 cells. At 48 h following transfection, cells were collected and the expression of Survivin, Aurora-B, p21 and cyclin B was examined by western blot analysis. β-actin was used as a control. (B) Cell growth of siRNA treated Ca9-22 cells. At 48 h following USP22 siRNA treatment, 5,000 cells were plated on 24-well plates. At 24 h, the cell number was counted as 0 day. The cell number was subsequently counted at days 2, 4, and 6. **P<0.01. USP22, ubiquitin-specific protease 22; OSCC, oral squamous cell carcinoma; siRNA, small interfering RNA; p21, cyclin-dependent kinase inhibitor 1A.
Discussion
The putative cancer stem cell marker USP22 has been demonstrated to be overexpressed in a number of types of cancer (6–12). The present study identified that USP22 was highly expressed in OSCC, particularly in poorly differentiated cancers, and that high USP22 expression was significantly associated with the malignant behaviors of OSCC, including lymph node metastasis and poor differentiation. Previous findings demonstrated that a high expression of USP22 was associated with a poor prognosis in various types of cancer, such as salivary duct carcinoma, colorectal cancer, head and neck cancer, breast cancer, and HCC (7–12). The results of the present study also demonstrated that USP22 expression was positively associated with the expression of the cell proliferation marker Ki-67. Additionally, it was observed that cell growth was suppressed by USP22 siRNA in OSCC cells. USP22 siRNA also increased the expression of p21 and reduced the expression of Cyclin B in OSCC cells. USP22 is a positive regulator of tumor growth and depletion of USP22 leads to cell cycle arrest at the G1 phase (5). USP22, a member of the ubiquitin hydrolase family, promotes the occurrence and development of tumors by blocking ubiquitin-mediated protein degradation, including FBP1, p21, p53, c-Myc and BMI-1 (the Polycomb repressor complex 1), thereby enhancing the stability of certain cancer genes (4,17,20). Previous studies have demonstrated that USP22 deubiquitinates histones H2A and H2B and regulates cell growth, cell-cycle regulation and signal transduction (4,5,17). Furthermore, USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator, FBP1 (5). USP22 may also alter the expression level of multiple tumor-associated regulatory factors, such as c-Myc, BMI-1, p53, p21 and Cyclin D (14,20).
CPC contains Survivin, Borealin, Aurora-B and INCENP, and acts as a critical mitotic regulator that controls the cell cycle and serves a crucial role in the expansion of tumor cells (13,14). In the present study, the expression of USP22 was identified to be positively associated with Aurora-B and Survivin in OSCC. In addition, high USP22, Aurora-B, or Survivin expression is associated with a poor prognosis in OSCC, especially when all are highly expressed concomitantly. Survivin inhibits apoptosis, regulates chromosome separation and cell division (13–15), and is highly expressed in various types of cancer, which typically have a poor prognosis, including colorectal cancer, head and neck cancer, endometrial carcinoma and hepatocellular carcinoma (7,21–26). Aurora-B regulates cytokinesis and chromosome segregation together with Survivin, Borealin and INCENP (7,8,23,24). Aurora B is highly expressed in a number of types of cancer, including head and neck, colon, liver and breast cancer, and is associated with malignancy indicators, including the histological differentiation and lymph node metastasis (23–26). USP22, Aurora-B and Survivin are highly expressed in OSCC, and serve an important role in the tumorigenesis of oral cancer most likely by disrupting cell cycle progression (23,26,27). USP22, Aurora-B and Survivin expression may be promising markers for predicting the malignant behaviors of OSCC. These observations are supported by previous data suggesting that high levels of Survivin and Aurora-B were associated with more malignant phenotypes and that they were independent prognostic indicators for multifarious cancers (9,22–26).
Our previous research demonstrated that USP22 can positively regulate the expression of Survivin in HCC (7); however, it is unclear whether USP22 regulates the expression of Aurora-B or Survivin in OSCC. In the present study, the results demonstrated that the expression of USP22 was positively associated with Aurora-B and Survivin in OSCC. A previous study reported that BMI-1 induces repressive epigenetic controlling of the Survivin promoter (16). USP22 also upregulates BMI-1 and may upregulate Survivin via BMI-1 overexpression (20). Previous studies have indicated that Aurora-B is degraded by the ubiquitin/proteasome pathway in the final stage of cell division (28,29). Indeed, the present study identified that USP22 siRNA decreased Aurora-B and Survivin expression in OSCC cells. These data indicate that the expression of Survivin and Aurora-B may be regulated by USP22. However, the mechanism of USP22 regulating Survivin and Aurora-B is still unclear and needs further investigations.
In conclusion, the results of the present study suggest that USP22 may be involved in the progression of OSCC, in cooperation with Aurora-B and Survivin. USP22, Aurora-B and Survivin are markedly associated with the occurrence and development of OSCC and are novel potential targets for the diagnosis and treatment of OSCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Natural Science Foundation of Guangxi (grant nos. 2015GXNSF, AA139110 and 2017GxNSFDA198022); and The National Natural Science Foundation of China (grant nos. 81460411 and 81660450).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
TL, JL, GQ and SZ designed the present research. TL, YK and GQ analyzed and explained the patient data, and were major contributors in writing the manuscript. QC and SJ performed the histological examination of the samples. SM, WS, YK analyzed and interpreted the patient data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All steps involved in the study involving human participants were according to the ethical standards of the institutional and national research committee and the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The present study was conducted following approval by the Ethics Committee of Guilin Medical University (Guilin, China). Written informed consent was provided by all individual participants included in the work.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Wikner J, Gröbe A, Pantel K, Riethdorf S. Squamous cell carcinoma of the oral cavity and circulating tumor cells. World J Clin Oncol. 2014;5:114–124. doi: 10.5306/wjco.v5.i2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mascitti M, Orsini G, Tosco V, Monterubbianesi R, Balercia A, Putignano A, Procaccini M, Santarelli A. An overview on current non-invasive diagnostic devices in oral oncology. Front Physiol. 2018;9:1510. doi: 10.3389/fphys.2018.01510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Zhang XY, Pfeiffer HK, Thorne AW, McMahon SB. USP22, an hSAGA subunit and potential cancer stem cell marker, reverses the polycomb-catalyzed ubiquitylation of histone H2A. Cell Cycle. 2008;7:1522–1524. doi: 10.4161/cc.7.11.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jordan CT. Cancer stem cell biology: From leukemia to solid tumors. Curr Opin Cell Biol. 2004;16:708–712. doi: 10.1016/j.ceb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Tang B, Liang X, Tang F, Zhang J, Zeng S, Jin S, Zhou L, Kudo Y, Qi G. Expression of USP22 and survivin is an indicator of malignant behavior in hepatocellular carcinoma. Int J Oncol. 2015;47:2208–2216. doi: 10.3892/ijo.2015.3214. [DOI] [PubMed] [Google Scholar]
- 8.Lee HJ, Kim MS, Shin JM, Park TJ, Chung HM, Baek KH. The expression patterns of deubiquitinating enzymes, USP22 and Usp22. Gene Expr Patterns. 2006;6:277–284. doi: 10.1016/j.modgep.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lv L, Xiao XY, Gu ZH, Zeng FQ, Huang LQ, Jiang GS. Silencing USP22 by asymmetric structure of interfering RNA inhibits proliferation and induces cell cycle arrest in bladder cancer cells. Mol Cell Biochem. 2011;346:11–21. doi: 10.1007/s11010-010-0585-4. [DOI] [PubMed] [Google Scholar]
- 11.Liu YL, Yang YM, Xu H, Dong XS. Aberrant expression of USP22 is associated with liver metastasis and poor prognosis of colorectal cancer. J Surg Oncol. 2011;103:283–289. doi: 10.1002/jso.21802. [DOI] [PubMed] [Google Scholar]
- 12.Luo Y, Zeng FQ, Gu ZH, Wang L, Wang ZY, Jiang GS, Xiao XY. Quantitative analysis of the putative cancer stem cell marker USP22 mRNA in the transitional cell carcinoma of the bladder and the relationship between USP22 and the grading of tumor. J Clin Urol. 2009;24:140–144. [Google Scholar]
- 13.Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13:789–803. doi: 10.1038/nrm3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van der Waal MS, Hengeveld RC, van der Horst A, Lens SM. Cell division control by the chromosomal passenger complex. Exp Cell Res. 2012;318:1407–1420. doi: 10.1016/j.yexcr.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 15.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. doi: 10.1038/nrc968. [DOI] [PubMed] [Google Scholar]
- 16.Vong QP, Cao K, Li HY, Iglesias PA, Zheng Y. Chromosome alignment and segregation regulated by ubiquitination of survivin. Science. 2005;310:1499–1504. doi: 10.1126/science.1120160. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 18.Sun Z, Hu S, Luo Q, Ye D, Hu D, Chen F. Overexpression of SENP3 in oral squamous cell carcinoma and its association with differentiation. Oncol Rep. 2013;29:1701–1706. doi: 10.3892/or.2013.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnes L, Eveson JW, Reichart P, Sidransky D. World Health Organization Classification of Tumours. IARC Press; 2005. Pathology and genetics of head and neck tumours; pp. 168–175. [Google Scholar]
- 20.Liu YL, Zheng J, Tang LJ, Han W, Wang JM, Liu DW, Tian QB. The deubiquitinating enzyme activity of USP22 is necessary for regulating HeLa cell growth. Gene. 2015;572:49–56. doi: 10.1016/j.gene.2015.06.075. [DOI] [PubMed] [Google Scholar]
- 21.Maerki S, Olma MH, Staubli T, Steigemann P, Gerlich DW, Quadroni M, Sumara I, Peter M. The Cul3-KLHL21 E3 ubiquitin ligase targets aurora B to midzone microtubules in anaphase and is required for cytokinesis. J Cell Biol. 2009;187:791–800. doi: 10.1083/jcb.200906117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi G, Tuncel H, Aoki E, Tanaka S, Oka S, Kaneko I, Okamoto M, Tatsuka M, Nakai S, Shimamoto F. Intracellular localization of survivin determines biological behavior in colorectal cancer. Oncol Rep. 2009;22:557–562. doi: 10.3892/or_00000471. [DOI] [PubMed] [Google Scholar]
- 23.Qi G, Kudo Y, Ando T, Tsunematsu T, Shimizu N, Siriwardena SB, Yoshida M, Keikhaee MR, Ogawa I, Takata T. Nuclear Survivin expression is correlated with malignant behaviors of head and neck cancer together with Aurora-B. Oral Oncol. 2010;46:263–270. doi: 10.1016/j.oraloncology.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 24.Tuncel H, Shimamoto F, Kaneko Guangying Qi H, Aoki E, Jikihara H, Nakai S, Takata T, Tatsuka M. Nuclear Aurora B and cytoplasmic Survivin expression is involved in lymph node metastasis of colorectal cancer. Oncol Lett. 2012;3:1109–1114. doi: 10.3892/ol.2012.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hori M, Miki T, Okamoto M, Yazama F, Konishi H, Kaneko H, Shimamoto F, Ota T, Temme A, Tatsuka M. The detergent-soluble cytoplasmic pool of survivin suppresses anoikis and its expression is associated with metastatic disease of human colon cancer. PLoS One. 2013;8:e55710. doi: 10.1371/journal.pone.0055710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi G, Ogawa I, Kudo Y, Miyauchi M, Siriwardena BS, Shimamoto F, Tatsuka M, Takata T. Aurora-B expression and its correlation with cell proliferation and metastasis in oral cancer. Virchows Arch. 2007;450:297–302. doi: 10.1007/s00428-006-0360-9. [DOI] [PubMed] [Google Scholar]
- 27.Qi G, Mi S, Jin S, Shao W, Liu J, Liu T, Zeng S, Lu H. The role of the cancer stem cell marker USP22 in tumors. Stem Cell Trans Invest. 2016;3:e1219. [Google Scholar]
- 28.Kurai M, Shiozawa T, Shih HC, Miyamoto T, Feng YZ, Kashima H, Suzuki A, Konishi I. Expression of Aurora kinases A and B in normal, hyperplastic, and malignant human endometrium: Aurora B as a predictor for poor prognosis in endometrial carcinoma. Hum Pathol. 2005;36:1281–1288. doi: 10.1016/j.humpath.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen HG, Chinnappan D, Urano T, Ravid K. Mechanism of Aurora-B degradation and its dependency on intact KEN and A-boxes: Identification of an aneuploidy-promoting property. Mol Cell Biol. 2005;25:4977–4992. doi: 10.1128/MCB.25.12.4977-4992.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.