Abstract
Background
The past few decades have witnessed a tremendous development in the field of genetics. The implementation of next generation sequencing (NGS) technologies revolutionized the field of molecular biology and made the genetic information accessible at a large scale. However, connecting a rare genetic variation to a complex phenotype remains challenging. Indeed, identifying the cause of a genetic disease requires a multidisciplinary approach, starting with the establishment of a clear phenotype with a detailed family history and ending, in some cases, with functional assays that are crucial for the validation of the pathogenicity of a mutation.
Methods
Two hundred Lebanese patients, presenting a wide spectrum of genetic disorders (neurodevelopmental, neuromuscular or metabolic disorders, etc.), sporadic or inherited, dominant or recessive, were referred, over the last three and a half years, to the Medical Genetics Unit (UGM) of Saint Joseph University (USJ). In order to identify the genetic basis of these diseases, Whole Exome Sequencing (WES), followed by a targeted analysis, was performed for each case. In order to improve the genetic diagnostic yield, WES data, generated during the first 2 years of this study, were reanalyzed for all patients who were left undiagnosed at the genetic level. Reanalysis was based on updated bioinformatics tools and novel gene discoveries.
Results
Our initial analysis allowed us to identify the specific genetic mutation causing the disease in 49.5% of the cases, in line with other international studies. Repeated WES analysis enabled us to increase the diagnostics yield to 56%.
Conclusion
The present article reports the detailed results of both analysis and pinpoints the contribution of WES data reanalysis to an efficient genetic diagnosis. Lessons learned from WES reanalysis and interpretation are also shared.
Keywords: High throughput sequencing, Exome, NGS, Mutations, Genetic heterogeneity, Genetic diagnostics, Lebanon
Background
The identification and characterization of the molecular basis of genetic disorders is crucial for the establishment of a specific diagnosis. This allows the family to benefit from an accurate genetic counseling, the patient to be aware of his disease’s prognosis and the physician to implement adequate therapeutic approaches, when possible. The last few years have seen an outstanding improvement in the field of molecular biology [1].
Indeed, the emergence of new high-throughput technologies or Next Generation Sequencing methods (NGS) such as WGS (Whole Genome Sequencing) and WES (Whole Exome Sequencing), has revolutionized the field of genetic diagnosis and made it much more efficient and affordable [2]. While WGS provides a thorough picture of the genome, WES was developed as a practical and cost effective alternative to WGS, targeting only coding regions and canonical splice sites that represent 1–1.5% of the human genome. The rationale behind this approach is that 80–85% of the mutations responsible for Mendelian diseases are located in these regions [3–5].
All these advances are driving NGS from the research field into the clinic. However, data interpretation remains the main challenge especially that thousands of variants of unknown significance are detected in each patient [6, 7].
Here we report the use of WES for genetic diagnostics purposes in 200 Lebanese patients who presented with a wide range of phenotypes, suggesting clinical and genetic heterogeneity. An initial analysis was performed followed, 2 years later, by data reanalysis in order to improve the genetic diagnostic yield. The outcome of WES in the Lebanese population is discussed and lessons learned from this study are shared.
Methods
Clinical samples
From January 2015 to June 2018, 200 patients with genetically heterogeneous disorders were included in our study. They were referred from different Lebanese areas, by private sector and academic physicians. All patients underwent a detailed review of their clinical history and a laboratory evaluation. The most common features of the 200 patients were global developmental delay/intellectual disability, encephalopathy, muscular weakness, failure to thrive and microcephaly.
Approval to conduct the study was obtained from the Ethics Committee of Saint Joseph University, Beirut, Lebanon. All patients, parents or legal guardians signed an informed consent for participation, sample collection and data publication. Peripheral blood was then collected from each individual enrolled in this study and DNA was extracted using the salting out method [8].
WES analysis
Exon capture and sequencing: The exome was captured using the SureSelect Human All Exons, reagents (Agilent Inc.® Santa Clara, CA) according to the manufacturer’s standard protocol. The concentration of each library was determined using Agilent’s QPCR NGS Library Quantification Kit (G4880A). Samples were pooled prior to sequencing with a final concentration of each sample equal to 10 nM. Sequencing was performed on the Illumina HiSeq2000 platform using TruSeq v3 chemistry.
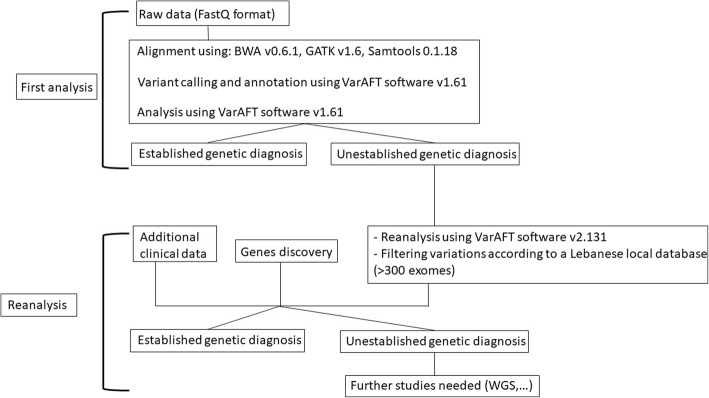
Mapping and alignment: Reads files (FASTQ) were generated from the sequencing platform via the manufacturer’s proprietary software. Reads were aligned to the hg19/b37 reference genome using the Burrows-Wheeler Aligner (BWA) package v0.6.1 [9]. Local realignment of the mapped reads around potential insertion/deletion (Indel) sites was carried out with the Genome Analysis Tool Kit (GATK) v1.6 [10]. Duplicate reads were marked using Picard v1.62. Additional BAM file manipulations were performed with Samtools 0.1.18 [11]. Base quality (Phred scale) scores were recalibrated using GATK’s covariance recalibration. SNP and Indel variants called using the GATK Unified Genotyper for each sample [12]. Variants were called using high stringency settings and annotated with VarAFT software 1.61 [13] containing information from dbSNP147 and ExAC (http://exac.broadinstitute.org/). For all patients who were left undiagnosed, reanalysis of vcf files was carried out using VarAFT2.131 and polymorphisms were further filtered using our in-house local database containing more than 300 exomes (Fig. 1).
Fig. 1.
Flowchart illustrating the methodology of WES analysis carried out in this study
In terms of functional annotation, we included only protein-altering variants, including truncating variants (stop gain/loss, start loss, or frameshift), missense variants, canonical splice-site variants, inframe indels affecting protein-coding regions, and variants within the intron–exon boundary (ten bases flanking the exonic boundaries). We focused on genotypes absent in our local control data sets. We systematically considered four different genetic models, using stratified European and African Americans in the Exon Variant Server (EVS) for minor allele frequency estimations: (a) germ-line de novo mutations, also absent in the local control populations; (b) recessive homozygous genotypes, which were heterozygous in both parents, never homozygous in controls, with a control allele frequency < 1%; (c) hemizygous X chromosome variants inherited from an unaffected heterozygous mother, with a control allele frequency < 1% and never observed in male controls or homozygous in female controls; and (d) compound heterozygous genotypes in the patient (one variant inherited from each heterozygous parent, with the two variants occurring at different genomic positions within the same gene), for which neither variant was ever homozygous in controls, and each had a control allele frequency < 1%. For the compound heterozygous genotypes, we further required study of phasing of the two variants. Genotypes meeting these criteria were referred to as “candidate genotypes,” with the genes harboring candidate genotypes referred to as “candidate genes” [14].
Variants confirmation and segregation studies by sanger sequencing
Primers were designed using Primer 3 (http://frodo.wi.mit.edu) and OLIGOS v.9.3, and checked for specificity using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). DNA sequences were obtained from UCSC and Genbank databases. Standard PCR reactions were performed using Taq DNA polymerase (Invitrogen Life Technologies, Carlsbad, CA, USA) and both strands of the resultant products were sequenced using the BigDye® Terminator v1.1 Cycle Sequencing Kit (ThermoFisher Scientific, Waltham, MA, USA) under standard conditions. The labeled products were subjected to electrophoresis on an ABI 3500 Genetic Analyzer sequencing system (ThermoFisher Scientific, Waltham, MA, USA). Electropherograms were analyzed using Sequence Analysis Software version 5.4 (ThermoFisher Scientific, Waltham, MA, USA) and compared to reference sequences using ChromasPro version 1.7.7 (Technelysium, Queensland, Australia). Nucleotide numbering reflects cDNA numbering with + 1 corresponding to the A of the ATG translation initiation codon in the reference sequences.
Results
Over a 42-month period, 200 cases with heterogeneous genetic disorders were enrolled in this study. Among these patients, around 80% (159 samples) were younger than 18 years of age. From the latter, 150 cases were children, including the case of a fetus whose specimen was collected after a terminated pregnancy.
Patients were referred by physicians for presenting with (Table 1): neurodevelopmental disorders (in 39.5% of the cases), neuromuscular disorders (10%), metabolic and mitochondrial disorders (9.5%), renal disorders (5%), hearing disorders (3.5%), isolated epilepsy (3%), bone diseases (2%), leukodystrophy (2%), visual disorders (1%) and other rare diseases referred as “others” in all tables (24.5%). WES was performed on these patients for diagnosis purposes.
Table 1.
Distribution of cases between disease groups with the corresponding success rate
| Pathologies | Number of patients presenting the pathology | Number of patients with established molecular diagnosis | Percentage of patients with established molecular diagnosis (%) |
|---|---|---|---|
| Neurodevelopmental disorders | 79 | 24 | 30.4 |
| Neuromuscular disorders | 20 | 17 | 85 |
| Metabolic and mitochondrial disorders | 19 | 16 | 84.2 |
| Renal disorders | 10 | 6 | 60 |
| Hearing disorders | 7 | 7 | 100 |
| Epilepsy | 6 | 4 | 66.7 |
| Bone diseases | 4 | 3 | 75 |
| Leukodystrophy | 4 | 3 | 75 |
| Visual disorders | 2 | 2 | 100 |
| Others | 49 | 30 | 61.2 |
| Total | 200 | 112 | 56 |
Our first analysis yielded a success rate of 49.5%. In other words, genetic diagnosis was established in 99 patients, after the first analysis (Table 2). In order to improve the diagnosis yield, reanalysis of WES data, generated during the first 2 years of this study, was carried out. Reanalysis was performed using new bioinformatics algorithms and was based on the newly established local database, new genes discoveries and additional clinical information for each of the undiagnosed patients. The second analysis allowed the identification of the pathogenic mutations in 13 additional cases (Table 3) corresponding to 6.5% increase in the genetic diagnosis rate, thus leading to an overall success rate of 56%. The 112 patients with established genetic diagnosis included 54 patients with autosomal dominant diseases, 49 with autosomal recessive diseases, and 9 with X-linked diseases. Different types of mutations were detected including small frameshift, nonsense, splice sites, and missense mutations.
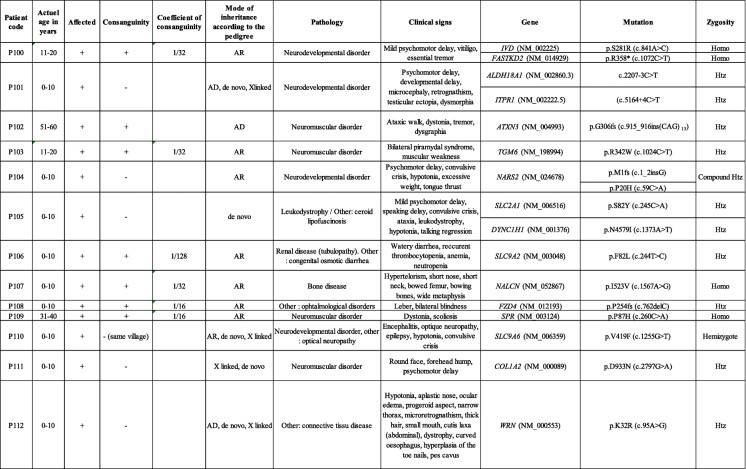
Table 2.
Clinical and genetic characteristics of families with established genetic diagnosis following the first analysis
AD Autosomal dominant, AR Autosomal recessive, Htz Heterozygous, Homoz Homozygous
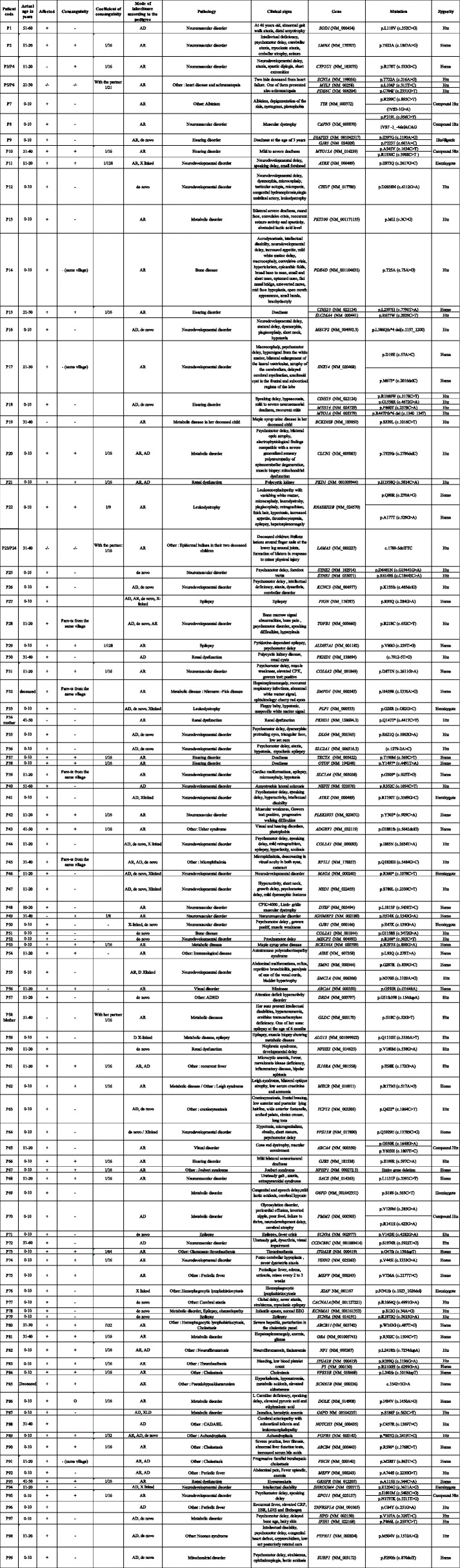
Table 3.
Clinical and genetic characteristics of the 13 additional cases with established genetic diagnosis after reanalysis
AD Autosomal dominant, AR Autosomal recessive, Htz Heterozygous, Homoz Homozygous.
There was a trend towards an association between the rate of positive diagnosis and the disease group. Indeed, the highest diagnosis success rate corresponds to the group of hearing and visual disorders (100%), followed by neuromuscular disorders (85%), metabolic and mitochondrial disorders (84.2%), bone diseases and leukodystrophy (75%), epilepsy (66.7%), others (61.2%), renal disorders (60%) and neurodevelopmental disorders (30.4%) (Table 1).
Of the 136 identified mutated alleles, 66.9% were novel variants at the time of diagnosis.
Discussion
WES is a relevant, efficient and cost effective genetic diagnostic tool that allows the detection of a wide range of genetic variants including point mutations (missense and nonsense mutations), small deletions and insertions in addition to canonical splicing mutations [15]. However, linking genetic variations to diseases still represents a big challenge in many cases.
Here we report the first study on WES outcome in the Lebanese population. Exome sequencing performed in 200 cases included in our study first yielded a 49.5% overall success rate, which is concordant with other international studies [16]. This rate was variable depending on the disease group. It barely reached 30.4% in some cases such as in neurodevelopmental diseases including intellectual disabilities (ID) and autism spectrum disorders (ASD) but was extended to 100% in the case of some diseases with well-established molecular mechanisms, such as visual and hearing disorders.
In order to improve the diagnostic yield, WES raw data that was generated over the first 2 years were reanalyzed. Repeated WES analysis enabled us to establish the genetic diagnosis, for 13 additional patients who were left undiagnosed, corresponding to around 7% increase in the diagnosis rate. This positive outcome is due to many factors, among which: the development of new annotation and filtering tools (including new bioinformatics variant effect prediction logarithms), the continuous revelation of new genes in addition to the implementation at our research unit of a Lebanese WES database that allowed us to filter out all “private” Lebanese polymorphisms. Apart from these factors, the 2 years of experience in WES analysis were an asset that helped us improve our performance in data interpretation. Here we report some of the lessons learned from our study:
Two monogenic diseases can masquerade as a single entity: the first molecular investigation of both parents (P5 and P6) of two deceased patients presenting with heart failure and achromatopsia did not lead to any success. However, reanalysis helped to identify, in each parent, two heterozygous mutations in MYL3 and PDE6C, responsible for two different conditions: heart defect (OMIM #608751) and achromatopsia (OMIM #613093), respectively. The observation, in a single patient, of multiple “hits” leading to a unique clinical presentation is nowadays becoming more and more acceptable.
Molecular geneticists need to be aware of filtering out homozygous mutations classified, by bioinformatics logarithms, as “heterozygous uncertain” due to poor read depth: A homozygous mutation in PET100 in the patient P13, classified as heterozygous uncertain by our logarithm was missed in our first WES analysis. Reanalysis allowed the detection of this candidate mutation and Sanger sequencing enabled its confirmation.
Patients from consanguineous families can still present with compound heterozygous mutations: Patients P10 and P95, issued form consanguineous Lebanese families, were for instance shown to carry respectively two compound heterozygous mutations in MYO15A responsible for an autosomal recessive type of deafness (OMIM #600316) and in SPG11 responsible for spastic paraplegia type 11 (OMIM #604360).
De-novo mutations linked to autosomal dominant conditions can occur in patients from consanguineous families: A de novo mutation in GJB3 (p.S199R) involved in hearing loss was detected in patient P66 who is issued from a consanguineous Lebanese family.
Variable Expressivity and incomplete penetrance of some diseases might mislead the choice of the filtering strategy: Patient P94 who was expected to carry a de novo mutation, was found to have a hemizygous mutation (p.E1204G) in SHROOM4, inherited from his mother who shares with him milder clinical signs.
Pathogenic mutations with a high frequency in a population exist! Filtering out common variants can be risky in the case of genetic disorders that are frequent in a population: In patient P87 presenting with G6PD deficiency, a disease frequently seen in the Lebanese population, WES reanalysis led to the identification of a pathogenic mutation in G6PD (p.S188F) that is present in high frequency (greater than 1%) in several arab populations [17].
WES has technical limitations: Large deletions encompassing candidate genes were detected by CGH arrays in two patients (not included in the study), who were left undiagnosed by WES.
WES coverage data might, in some cases, enable the detection of gene deletion: NPHP1 gene coverage was for instance equal to zero in patient P67 who presented with Joubert syndrome. This gene was fully covered in other patients run on the same sequencing chip. Owing to the potential involvement of the gene in the disease, CGH array was performed and a homozygous deletion encompassing the entire NPHP1 sequence was confirmed, thus explaining the clinical presentation.
Last but not least,
Communication between clinicians and geneticists is the ultimate key for genetic diagnostics success: In our first WES analysis, genetic diagnostic was not established for the patient P106 who was referred to our center as presenting with Bartter syndrome. While following up on all undiagnosed cases during our WES reanalysis, a pathogenic mutation in SLC9A2 (p.F82 L) was highlighted and communicated to the physician who re-evaluated the patient and modified the initial diagnosis to congenital chloride diarrhea, known to result from mutations in SLC9A2 [18].
On the other hand, failure of genetic diagnostics in the remaining patients can be due to several factors: i) the clinical and genetic heterogeneity of some entities: More than 1000 genes are for instance linked to ASD, thus making genetic diagnostics very challenging (https://gene.sfari.org/database/human-gene/); ii) the detection in isolated cases of private mutations whose pathogenicity is hard to be demonstrated in addition to iii) purely technical limitations associated with WES: Some bioinformatics pipelines are for example unable to detect CNVs (Copy Number Variation), rearrangements or triplet expansions. Homopolymers can for instance be miscalled, thus generating false positive variants. Furthermore, lack of coverage of coding regions (inefficient capture of regions rich in GC or bad read depth) and misalignment of reads to the reference sequence are all factors that can occur and lead to false negatives [19, 20].
Conclusion
In conclusion, WES allowed a rapid and cost effective identification of the molecular bases of heterogeneous genetic disorders in the Lebanese population. Our study yielded an overall success rate of 56%, of which around 7% is due to data reanalysis, thus pinpointing the utility of WES reanalysis that should take into consideration the updates in bioinformatics logarithms/annotation tools, novel genetic findings in addition to the occurrence in patients of new clinical manifestations.
Undiagnosed patients need to undergo further molecular investigation. For these cases, Whole Genome Sequencing (WGS) can be performed in order to enhance coverage performance and to detect non-coding mutations (variants modifying gene expression or affecting cryptic splice sites) and structural variations including big insertions and deletions. However, this approach remains laborious and very costly.
On the other hand, current genetic diagnosis studies focuses on the protein-coding regions and ignores the vast majority of non-coding regulating elements such us non-coding RNAs. We believe that the recent advances in the fields of computational biology and experimental technology will allow a better characterization of these elements, which might enable their integration in similar studies in the future.
Altogether, the outstanding improvement in high-throughput sequencing techniques will enable the establishment of low-cost genetic diagnostics, the identification of novel genes and the elucidation of physiological mechanisms, all driving towards personalized medicine.
Acknowledgements
We express our deepest gratitude and sympathy to the families for their full cooperation throughout the study.
Funding
This work (samples collection, analysis, and interpretation of data) was supported by grants from Saint Joseph University, Beirut, Lebanon.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author upon a reasonable request.
Abbreviations
- AD
Autosomal dominant
- AR
Autosomal recessive
- CGH
Comparative Genome Hybridization
- CNVs
Copy number variations
- NGS
Next-generation sequencing
- WES
Whole-exome sequencing
- WGS
Whole-genome sequencing
Authors’ contributions
NJ, CyM and EC conceived, designed the study, performed data interpretation and wrote the manuscript. SC, ZH, NH, JAG, RG and NS conducted the experiments and performed data analysis. EF, JPD and VD conducted acquisition and bioinformatics analysis of the raw data. AF, CDK, MR, ChM, AdM, SR, BG, HM, MB, RD, DB and AnM performed the recruitment and referral of patients, their clinical investigation and the correlation between genotype – phenotype. All authors have read and approved the manuscript.
Ethics approval and consent to participate
Approval to conduct the study was obtained from the Ethics Committee of Saint Joseph University, Beirut, Lebanon (Reference: FM327). All patients, parents or legal guardians signed an informed consent for participation and sample collection.
Consent for publication
All patients, parents or legal guardians signed an informed consent for data publication.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nadine Jalkh, Email: nadine.jalkh@usj.edu.lb.
Sandra Corbani, Email: sandra.corbani@usj.edu.lb.
Zahraa Haidar, Email: zahraa.haidar@net.usj.edu.lb.
Nadine Hamdan, Email: nadine.hamdan@net.usj.edu.lb.
Elias Farah, Email: elias.farah@usj.edu.lb.
Joelle Abou Ghoch, Email: joelle.aboughoch@usj.edu.lb.
Rouba Ghosn, Email: rouba.ghosn@usj.edu.lb.
Nabiha Salem, Email: nabiha.salem@usj.edu.lb.
Ali Fawaz, Email: dralifawaz@hotmail.com.
Claudia Djambas Khayat, Email: claudiakhayat@yahoo.fr.
Mariam Rajab, Email: drmariam1@hotmail.com.
Chebl Mourani, Email: cheblmourani@gmail.com.
Adib Moukarzel, Email: amoukarzel@pol.net.
Simon Rassi, Email: srassi@hotmail.com.
Bernard Gerbaka, Email: bernard.gerbaka@usj.edu.lb.
Hicham Mansour, Email: hicham.mansour@gmail.com.
Malek Baassiri, Email: malek-baassiri-med@hotmail.com.
Rawane Dagher, Email: rawanedagher@gmail.com.
David Breich, Email: drdavidbr@hotmail.com.
André Mégarbané, Email: jean-pierre.desvignes@univ-amu.fr.
Jean Pierre Desvignes, Email: valerie.delague@univ-amu.fr.
Valérie Delague, Email: valerie.delague@univ-amu.fr.
Cybel Mehawej, Email: cybel.mehawej@usj.edu.lb.
Eliane Chouery, Phone: 00 961 1 421 630, Email: eliane.chouery@usj.edu.lb.
References
- 1.Green ED, Guyer MS. National Human Genome Research Institute. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–213. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 2.Soden SE, Saunders CJ, Willig LK, Farrow EG, Smith LD, Petrikin JE, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med. 2014;6:265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonnefond A, Durand E, Sand O, De Graeve F, Gallina S, Busiah K, et al. Molecular diagnosis of neonatal diabetes mellitus using next-generation sequencing of the whole exome. PLoS One. 2010;5:e13630. doi: 10.1371/journal.pone.0013630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon-Salazar TJ, Silhavy JL, Udpa N, Schroth J, Bielas S, Schaffer AE, et al. Exome sequencing can improve diagnosis and alter patient management. Sci Transl Med. 2012;4:138ra78. doi: 10.1126/scitranslmed.3003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maddalena A, Bale S, Das S, Grody W, Richards S, ACMG Laboratory Quality Assurance Committee Technical standards and guidelines: molecular genetic testing for ultra-rare disorders. Genet Med. 2005;7:571–583. doi: 10.1097/01.GIM.0000182738.95726.ca. [DOI] [PubMed] [Google Scholar]
- 7.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 8.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinforma Oxf Engl. 2009;25:2744–2750. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinforma Oxf Engl. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desvignes J-P, Bartoli M, Delague V, Krahn M, Miltgen M, Béroud C, et al. VarAFT: a variant annotation and filtration system for human next generation sequencing data. Nucleic Acids Res. 2018;46(W1):W545-W553. [DOI] [PMC free article] [PubMed]
- 14.Zhu X, Petrovski S, Xie P, Ruzzo EK, Lu Y-F, McSweeney KM, et al. Whole-exome sequencing in undiagnosed genetic diseases: interpreting 119 trios. Genet Med. 2015;17:774–781. doi: 10.1038/gim.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kryukov GV, Shpunt A, Stamatoyannopoulos JA, Sunyaev SR. Power of deep, all-exon resequencing for discovery of human trait genes. Proc Natl Acad Sci U S A. 2009;106:3871–3876. doi: 10.1073/pnas.0812824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yavarna T, Al-Dewik N, Al-Mureikhi M, Ali R, Al-Mesaifri F, Mahmoud L, et al. High diagnostic yield of clinical exome sequencing in middle eastern patients with Mendelian disorders. Hum Genet. 2015;134:967–980. doi: 10.1007/s00439-015-1575-0. [DOI] [PubMed] [Google Scholar]
- 17.Doss CGP, Alasmar DR, Bux RI, Sneha P, Bakhsh FD, Al-Azwani I, et al. Genetic epidemiology of Glucose-6-phosphate dehydrogenase deficiency in the Arab world. Sci Rep. 2016;6:37284. doi: 10.1038/srep37284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiagarajah JR, Donowitz M, Verkman AS. Secretory diarrhoea: mechanisms and emerging therapies. Nat Rev Gastroenterol Hepatol. 2015;12:446–457. doi: 10.1038/nrgastro.2015.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ku C-S, Cooper DN, Polychronakos C, Naidoo N, Wu M, Soong R. Exome sequencing: dual role as a discovery and diagnostic tool. Ann Neurol. 2012;71:5–14. doi: 10.1002/ana.22647. [DOI] [PubMed] [Google Scholar]
- 20.Singleton AB. Exome sequencing: a transformative technology. Lancet Neurol. 2011;10:942–946. doi: 10.1016/S1474-4422(11)70196-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author upon a reasonable request.