Abstract
Previous studies have reported that microRNAs regulate gene expression and transcription. miR-21 have been identified to play a role in many types of cancer. KLF5 functions as a tumor inhibitor in certain cancers. However, the role of KLF5 plays in hepatocellular carcinoma (HCC), especially concerning the relationship between miR-21 and the KLF5 gene remains to be determined. Reverse transcription-quantitative PCR (RT-qPCR), western blot analysis, as well as luciferase reporter and Transwell assays were used to determine the expression of miR-21 and KLF5 in Huh 7, SK-HEP-1, LO-2, and HCC tissues. In HCC cells and tissues, the upregulation of miR-21 was identified. HCC cell migratory and invasive abilities significantly increased because of miR-21 overexpression. KLF5 expression was inhibited by miR-21 by targeting its 3′-UTR. KLF5 overexpression alleviated the effect induced by miR-21 on the migratory and invasive ability of the Huh 7 cells. The results therefore show that, HCC cell migration and invasion is significantly suppressed by miR-21 via targeting KLF5. The newly identified miR-21/KLF5 axis provides a useful therapeutic biomarker for HCC treatment.
Keywords: miR-21, invasion, migration, Kruppel-like family of transcription factor 5, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is ranked second worldwide with regard to tumor-related death (1). In addition, chronic infection through the hepatitis B virus (HBV) is strongly associated with death (2). Surgical resection or transplantation is used as a form of treatment of HCC, when the disease is detected early. However, in most cases, patients are diagnosed with HCC at an advanced stage. Consequently, doctors have limited treatment options because of lack of effective therapies. Activation of proto-oncogenes and deactivation of cancer-inhibitor genes play a pivotal role in tumorigenesis (3).
MicroRNAs can suppress the expression of related genes or induce their degradation at the post-transcriptional level (4–6). Approximately 1–5% of animal genes have been predicted to comprise miRNAs (7). Existing miRNAs have been identified in a number of organisms. Cell proliferation, differentiation, and apoptosis are regulated in the post-transcriptional regulation mechanism by various miRNAs (8,9). Therefore, human cancer cells developing from normal cells through a series of disorders are caused by the aberrant regulation of miRNAs (10).
The dysregulated expression of many microRNAs, including miR-21 in HCC, has been reported, which were identified in genomic analysis (11,12). The up- or downregulation of miR-21 were revealed in abundant malignancies, often overexpressed in various types of cancer, such as multiple myeloma pathogenesis, lung cancer (13–15). It has been confirmed that programmed cell death 4 (PDCD4) as a tumor suppressor may be downregulated by miR-21 (16). The tumorigenesis of colorectal carcinoma may also be stimulated by miR-21 at the post-transcriptional level (17). In addition, miR-21 controlled breast cancer cell progression by targeting the PDCD4 factor (18). The expression of long non-coding RNA (IncRNA) growth arrest-specific 5 (GAS5) can be inhibited by miR-21. Of note, GAS5 is capable of suppressing miR-21 expression (19). In HCC, HBV X protein (HBx) downregulates PDCD4 expression via miR-21 (20). The function of miR-21 was also present in pancreatic, gastric and lung cancer (21–23). The Kruppel-like family of transcription factors (KLFs), are considered tumor suppressors. The cell growth of breast cancer was inhibited via KLF5 (24). In addition, the cell growth, cell cycle regulation, and angiogenesis can be influenced by KLF5. In human bladder cancer, Gao et al found that cell proliferation and tumorigenesis were promoted by KLF5 (25). Downregulation of KLF5 is often detected in prostate cancer (26).
However, whether the regulation of migration and invasion in HCC is induced via miR-21 through KLF5 remains to be elucidated. To analyze the function of miR-21, miR-21 upregulation in HCC was identified in tumor tissues and cell line in this study using RT-qPCR. The results showed that in HCC cells, miR-21 overexpression promoted metastasis and invasion. Furthermore, KLF5 as a downstream target of miR-21 was inhibited by miR-21. The present findings demonstrate a valuable therapeutic biomarker for HCC differentiation and treatment.
Materials and methods
Tissue samples
Between March, 2013 and May, 2016, 80 patients selected from People's Hospital of Linyi (Linyi, China), were included in this study. Following surgical resection, tumor tissues obtained from the patients, were quickly frozen in liquid nitrogen and then stored at −80°C until further use. None of the patients participating in the study underwent chemo- or radiation therapy.
The present study was approved by the Ethics Committee of People's Hospital of Linyi. Informed consent was obtained from all the patients whose tissues were used in this study.
Cell culture
Three cell lines (Huh 7 and SK-HEP-1) and normal human hepatocytes (LO-2) were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and Sciencell (San Diego, CA, USA), respectively. These cell lines were cultured with DMEM medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), added with 10% FBS (Biowest, South America Origin, Paris, France), 100 IU/ml penicillin and 100 µg/ml streptomycin, and placed in a humidity incubator at 37°C and 5% CO2.
Plasmid construction
In our study, the psiCHECK-2 plasmid (Promega, Madison, WI, USA) was selected for the dual luciferase reporter assay. The partial KLF5 gene served as the gene from which the 3′-untranslated regions (UTRs) were taken and cloned for the appropriate miR-21-binding site. Then, it was linked into the downstream of the luciferase reporter gene of psiCHECK-2. The plasmids containing miR-21 or LNA anti-miR-21 were constructed. In order to knock down the KLF5 gene, KLF5 siRNA was produced.
Transfection
We purchased miR-21 mimic and inhibitor from GenePharma Co., Ltd. (Suzhou, China) and KLF5 vector from Shanghai Genechem Co., Ltd. (Shanghai, China). Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) and RNAiMAX (Invitrogen; Thermo Fisher Scientific, Inc.) were used to transfect miR-21 mimic, miR-21 inhibitor, KLF5 vector or both KLF5 vector and miR-21 mimic, respectively. For transfection, Huh 7 or SK-HEP-1 cells were seeded in 24- or 6-well plates and then transfected with the indicated plasmids for 48 h using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) following the manufacturer's protocol. The cells were collected for the following experiments after 48 h of transfection.
RNA extraction and reverse transcription-quantitative PCR
TRIzol reagent (Takara, Dalian, China) was used to isolate total RNA from HCC tissues and three cell lines Huh 7, SK-HEP-1 and LO-2 cell. The SYBR PrimeScript RT-PCR kit (Takara) was applied for reverse-transcription of RNA to detect miR-21 and KLF5. The Roche Light Cycler 480 instrument (Roche, Basel, Switzerland) was used to perform reverse transcription-quantitative PCR (RT-qPCR). The snRNA U6 or GAPDH were identified as the internal control for miRNA or KLF5, respectively. The miR-21 and KLF5 relative expression ratio was determined using the 2−∆∆Cq algorithm (27). PCR primers used were as follows: miR-21, forward: 5′-TAGCTTATCAGACTGATG-3′ and reverse: 5′-TGGTGTCGTGGAGTCG-3′; U6, forward: 5′-CTCGCTTCGGCAGCACA-3′ and reverse: 5′-AACGCTTCACGAATTTGCGT-3′; KLF5 forward:, 5′-ATCGAGATGTTCGCTCGTGC-3′; and reverse,: 5′-TTTAAAGGCAGACACTGAGTCAG-3′; GAPDH forward: 5′-ATCGTCCACCGCAAATGCTTCTA-3′; and reverse: 5′-AGCCATGCCAATCTCATCTTGTT-3′. All the RT-qPCR experiments were repeated three times.
Western blot analysis
Tissue powder (200 mg) stored at −80°C were lysed in lysis buffer (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) for protein extraction. The concentration of the extracted protein was analyzed using BCA kit (Beyotime Institute of Biotechnology, Haimen, China). A 20 µg protein sample was loaded in each lane, and gel electrophoresis was conducted using 12% SDS-PAGE to separate the extracted proteins, which were subsequently transferred on nitrocellulose membranes that blocked overnight in Tris-buffered saline including 0.1% Tween-20 and 5% skimmed milk powder at 4°C. The membranes were then incubated with the primary antibody: rabbit monoclonal anti-KLF5 (ab 137676; 1:1,000; Abcam, Cambridge, MA, USA) at 4°C for 8 h. Rabbit monoclonal to GAPDH (EPR16891; 1:5000; Abcam) was chosen as the internal reference. Then, they were incubated with secondary antibody goat anti-rabbit IgG-HRP (sc-2004; 1:3,000; Santa Cruz Biotechnology, Inc. Santa Cruz, CA, USA) at room temperature for 1 h. The proteins of interest were detected by the enhanced chemiluminescence (ECL) detection system (Sea Biotech, Shanghai, China). Finally, the extracted protein was quantified using the Bio-Rad GelDoc 2000 instrument (Bio-Rad, Hercules, CA, USA).
Luciferase reporter analysis
We strived to seek antisense matches of KLF5 3′-UTR sequences against miR-21 by means of TargetScan. In order to perform luciferase reporter analysis, 100 ng of the negative control or miR-21 mimic and psiCHECK-2-KLF5-3′-UTR-WT, or psiCHECK-2-KLF5-3′-UTR-MT were co-transfected into Huh 7 cell line. At 48 h after transfection, the cells were gathered and analyzed via dual-luciferase reporter assay system (Promega, Madison, WI, USA). Transfections were performed three times.
Transwell migration and invasion assays
The 24-well Transwell chambers (Millipore Corporation, Billerica, MA, USA) were used for the cell migration assay according to the manufacturer's protocol. DMEM medium containing 5% fetal bovine serum (FBS) was added to the lower chamber as a chemoattractant. After incubation for 24 h, the cells adhering to the upper membrane were removed with cotton wool, whereas the cells that had migrated or invaded through the membrane were fixed with methanol and stained with 0.1% crystal violet in 5% CO2 at 37°C for 15 min. For the invasion assay, the procedure was similar to the cell migration assay, except that membranes of the upper chambers were pre-coated with 100 ml Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). The cells were photographed under a phase-contrast microscope (Olympus, Tokyo, Japan) and counted in five randomly chosen fields. Each experiment was performed in triplicate.
Statistical analysis
The results are presented as the mean ± standard deviation (SD). Statistical analysis was conducted using SPSS 17.0 (SPSS, Inc., Chicago, IL, USA). Each experiment was repeated at least three times. The Student's t-test or Tukey-Kramer post hoc test after one-way analysis of variance (ANOVA) in SPSS were used to analyze the differences between the groups. Correlation between mRNA and miRNA were estimated using the Spearman's correlation method. P<0.05 was considered to indicate a significant difference. We generated graphs using GraphPad Prism 6 Software (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Upregulation of miR-21 in the HCC tissues
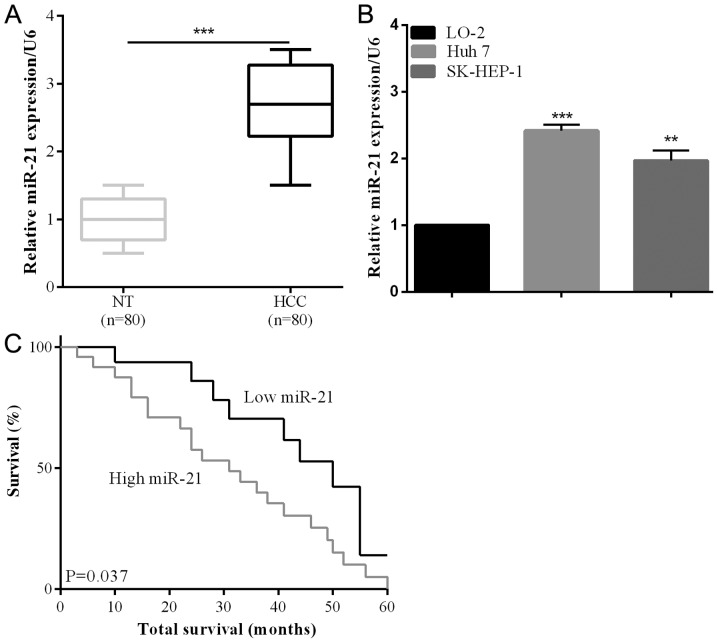
The upregulation of miR-21 was identified in HCC cancer tissues, as in a previous study (28). Eighty collected human HCC tissues and their adjacent non-tumorous samples were analyzed using RT-qPCR to investigate miR-21 expression in this study. miR-21 expression was significantly increased compared with non-tumorous in HCC tissues (P<0.001) (Fig. 1A). According to the RT-qPCR results, miR-121 expression was increased in HCC cell lines (Huh 7 and SK-HEP-1) compared with LO-2 normal liver cells (Fig. 1B). The correlation between clinicopathological characteristics and the miR-21 expression level was also detected. We observed that the lager tumor size was associated with an increase of miR-21 expression (P=0.0042) (Table I). Additionally, poor 5-year survival rate was significantly associated with miR-21 overexpression (P=0.037) (Fig. 1C), suggesting a relationship between the prognostic and miR-21 expression level.
Figure 1.
miR-21 was significantly upregulated in HCC tissues. (A) The expression levels of the miR-21 in HCC and theirs corresponding tissues were tested using RT-qPCR. The expression levels were calculated using the miR21/U6 expression ratio (2−∆∆Cq). (B) The miR-12 expression in HCC cell lines (Huh 7 and SK-HEP-1) and the LO-2 normal liver cells were tested using RT-qPCR. (C) The overall survival rate of the patients associated with the expression level of miR-21. **P<0.01, ***P<0.001.
Table I.
miR-21 expression and clinicopathological characteristics in HCC patients.
| miR-21 expression | ||||
|---|---|---|---|---|
| Variables | No. | Low n (%) | High n (%) | P-valuea |
| Age (year) | 0.768 | |||
| <50 | 39 | 18 (46.2) | 21 (53.8) | |
| ≥50 | 61 | 30 (49.2) | 31 (50.8) | |
| Sex | 0.519 | |||
| Male | 52 | 25 (48.1) | 27 (51.9) | |
| Female | 48 | 20 (41.7) | 28 (58.3) | |
| Tumor size (cm) | 0.0042 | |||
| <5 | 31 | 15 (48.4) | 16 (51.6) | |
| ≥5 | 69 | 14 (20.3) | 55 (79.7) | |
| Tumor differentiation | 0.732 | |||
| I + II | 55 | 25 (45.5) | 30 (54.5) | |
| III + IV | 45 | 22 (48.9) | 23 (51.1) | |
| Metastasis | 0.658 | |||
| No | 41 | 19 (46.3) | 22 (53.7) | |
| Yes | 59 | 25 (42.4) | 24 (57.6) | |
P-values are calculated with Chi-square test.
miR-21 overexpression increases the number of invasive and migrated cells in HCC
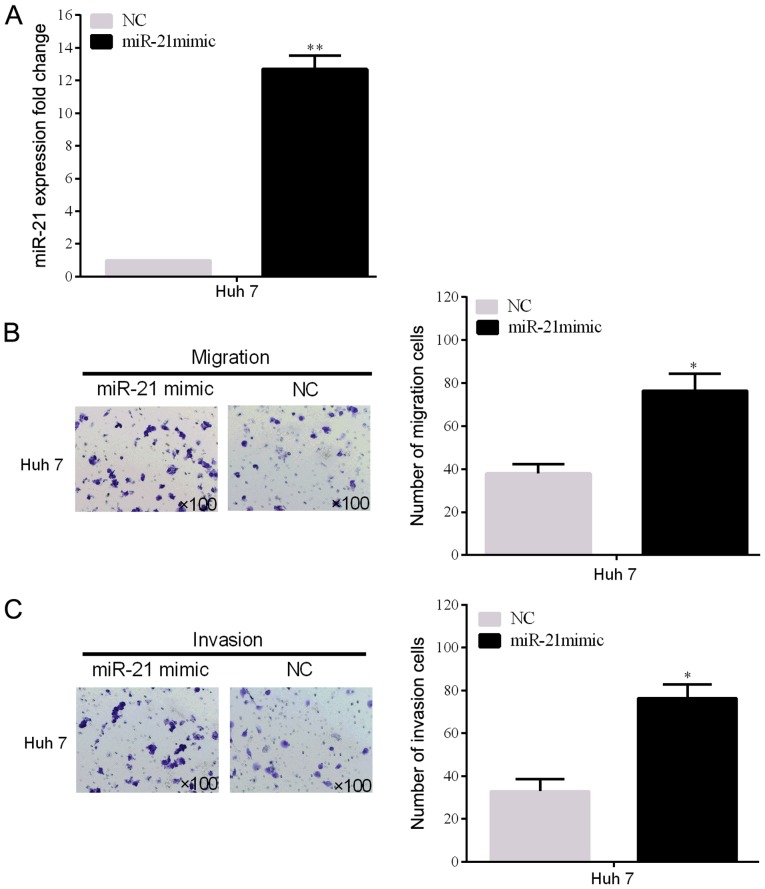
To prove the miR-21 biological influence of cell invasion and migration, miR-21 expression levels of the Huh 7 cell line transfected with miR-21 mimic plasmid was detected (P<0.05) (Fig. 2A). In addition, the invasion and migration rates of the two HCC cell lines (SMMC-7721) were remarkably promoted by miR-21 overexpression, which were demonstrated by the Transwell assays (Fig. 2B and C).
Figure 2.
Overexpression of miR-21 promotes HCC cell line invasion and migration in vitro. (A) The successful overexpression of miR-21 in Huh 7 was confirmed by RT-qPCR. (B and C) Cell line invasion and migration were determined using the Transwell assays in the two HCC cell lines (Huh 7). *P<0.05, **P<0.01.
miR-21 inhibits the KLF5 expression and binds to the 3′-UTR of the KLF5
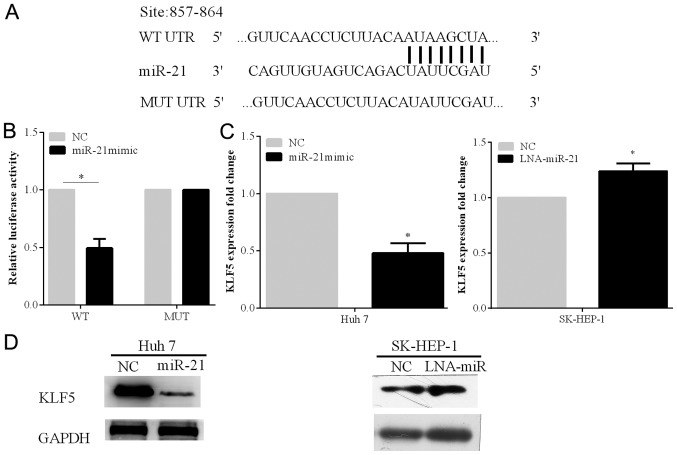
To identify the potential downstream targets of miR-21, bioinformatics analysis was carried out on the website TargetScan (http://www.targetscan.org/). By the online prediction, KLF5 was selected as the target gene of miR-21. The prediction showed that KLF5 gene has one binding site to miR-21 at 857–864 bp in the 3′-UTR (Fig. 3A). In order to verify the prediction, the luciferase reporter analysis was carried out using the Huh 7 cell line.
Figure 3.
The expression of KLF5 can be suppressed by miR-21, which binds to 3′-UTR. (A) The binding site of KLF for miR-21 was predicted using the online TargetScan tool. (B) Huh 7 cells were transfected with the Renilla luciferase psiCHECK-2 plasmid, and psiCHECK-2 plasmid which had either wide-type or mutant KLF5 3′-UTR, and the miR-21 mimic. (C) Huh 7 cell lines were transfected with NC or the miR-21 mimic. The SK-HEP-1 cell line was transfected with NC or LNA-miR-21. The expression of the KLF5 was tested by RT-qPCR. (D) The protein expression of KLF5 was determined using western blot analysis in the Huh 7 and SK-HEP-1 cell lines. *P<0.05.
The relative luciferase activity of the cells transfected with the miR-21 mimic and the psi-KLF5-3′-UTR-WT plasmid was significantly decreased. On the other hand, when the miR-21 mimic and psi-KLF5-3′-UTR-MUT plasmid containing mutant sites at the 3′-UTR of KLF5 were transfected into the cell line, the reduction extent of the luciferase activity was alleviated (P<0.05) (Fig. 3B).
Furthermore, we found that the protein and mRNA expression levels of KLF5 were markedly inhibited by miR-21 overexpression in Huh 7 cells. The protein and mRNA expression levels of KLF5 were increased following with the silenced miR-21 in SK-HEP-1 cells (P<0.05) (Fig. 3C and D).
Overexpression of KLF5 alleviates the miR-21-driven promoting effect on cell line invasion and migration in vitro
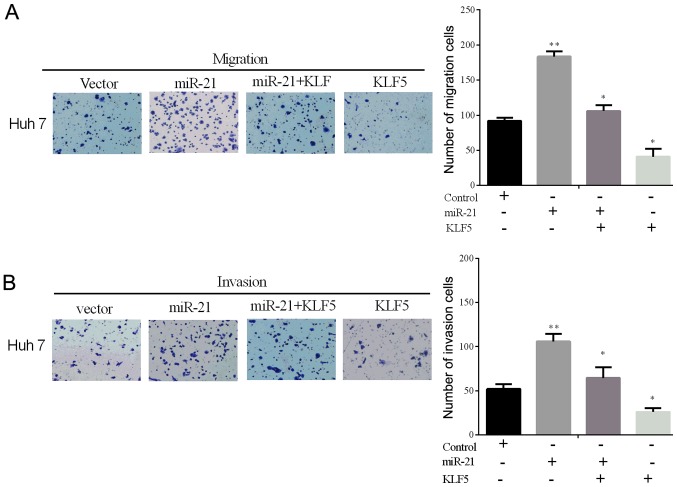
To confirm whether KLF5 overexpression could reverse the miR-21-driven promoting effect on the cell invasive and migratory ability in vitro, the expressing plasmid containing the KLF5 without the 3′-UTR was constructed. The invasion and migration rates were tested using the Transwell assays in the selected cell line (Huh 7) transfected with the miR-21 mimic and the KLF5 lacking the 3′-UTR (Fig. 4A and B). From the results, we could identify that the overexpression of KLF5 alleviates the miR-21-driven promoting role of cell invasion and migration in vitro.
Figure 4.
Overexpression of KLF5 alleviates the miR-21-driving promoting effect of cell line migration and invasion in vitro. (A and B) Transwell assays were used to determine the Huh 7 cell line migration and invasion following transfection with the miR-21 mimic and KLF5 without its 3′-UTR or control plasmid. *P<0.05, **P<0.01.
Discussion
A number of onco-miRNAs have been previously identified, especially miR-21 (28). The function of miR-21 which is frequently upregulated has been identified in numerous cancers including colorectal cancer (29), breast carcinoma (18), pancreatic carcinoma (21), gastric carcinoma (22), lung carcinoma (30,31), colorectal carcinoma (32) and hepatocellular carcinoma (33). In the present study, it was found that the miR-21 expression level was enhanced in HCC tumor tissues compared to the corresponding non-tumorous tissues. In addition, the invasive and migratory ability of the hepatoma cell line in vitro could be promoted by miR-21 re-expression. At the post-transcriptional level, we first demonstrated that miR-21 negatively modulated KLF5 by combining with the 3′-UTR of the tumor suppressor in this study.
Previous findings have shown that, PDCD4 expression was downregulated via HBx, which was partially modulated by miR-21 (20). In the present study, we did not employ HBV and HBx. However, HBV-infected hepatic cells are involved in the formation of HCC. Therefore, identifying the correlation between HBx and KLF5 in future investigations is imperative. miR-21 also has a great impact on the pathogenesis of atrial fibrosis. In addition, miR-21 inhibition induced by the antagomir-21 could suppress the fibrosis of atrial fibrillation post-myocardium infarction (34). This confirms that miR-21 has various different functions in different cancer types.
As a member of the Kruppel-like family, KLF5 is involved in the modulation of various physiological and pathological processes, KLF5 has been reported to regulate embryonic development, stress response, cell proliferation and differentiation, carcinogenesis and cardiovascular remolding through various regulatory mechanisms, as indicated by previous findings (35,36). In addition, KLF5 contributes to bladder cancer angiogenesis via the VEGFA pathway. The GC boxes and CACCC elements of VEGFA combine with KLF5 to regulate tumorigenesis. The silence of KLF5 can remarkably inhibit the expression of VEGFA (25). In our study, miR-21 regulated KLF5 by binding to its 3′-UTR. Additionally, HCC cell invasion and migration can be promoted through the knockdown of KLF5 by siRNA in vitro.
miR-153 reduced KLF5 expression by combining with its 3′-UTR, as is evident from our results. In addition, the expression of KLF5, suppressed by miR-153, was verified by the luciferase reporter assay (37). ERβ is a nuclear receptor for estrogen (E2). E2 biphasically regulated KLF5 transcription through ERβ to modulate prostate tumor progression, which showed that the KLF5 expression was associated with the progression of prostate tumor (38). Thus, we may develop or identify effective drugs to prevent the progression of HCC.
In summary, results of the present study show that, the invasion and migration of HCC cells may be significantly suppressed by miR-21 by targeting the tumor suppressor KLF5. Accordingly, the miR-21/KLF5 axis may open up a novel way for treating HCC, such as tumor invasion and metastasis.
Acknowledgements
Not applicable.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
JW and YC contributed to the conception of the study; MX (3rd author) contributed significantly to data analysis and manuscript preparation; XZ performed the data analysis and wrote the manuscript; YZ and MX (last author) helped perform the analysis with constructive discussions. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Approval for the study was obtained from the Ethics Committee of People's Hospital of Linyi (Linyi, China). Informed consent was obtained from all the patients whose tissues were used in this study.
Patient consent for publication
Not applicable.
Competing interest
The authors declare that they have no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Tan A, Yeh SH, Liu CJ, Cheung C, Chen PJ. Viral hepatocarcinogenesis: from infection to cancer. Liver Int. 2008;28:175–188. doi: 10.1111/j.1478-3231.2007.01652.x. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Rana TM. Illuminating the silence: Understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 5.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 6.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 8.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 9.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 10.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 11.Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, Schmittgen TD. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang WY, Zhang HF, Wang L, Ma YP, Gao F, Zhang SJ, Wang LC. miR-21 expression predicts prognosis in hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2014;38:715–719. doi: 10.1016/j.clinre.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, Taccioli C, Zanesi N, Alder H, Hagan JP, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci USA. 2008;105:12885–12890. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia H, Zhang W, Zhang B, Zhao Y, Zhao Y, Li S, Liu Y. miR-21 modulates the effect of EZH2 on the biological behavior of human lung cancer stem cells in vitro. Oncotarget. 2017;8:85442–85451. doi: 10.18632/oncotarget.20006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Q, Wang Z, Hu Y, Li J, Li X, Zhou L, Huang Y. miR-21 promotes migration and invasion by the miR-21-PDCD4-AP-1 feedback loop in human hepatocellular carcinoma. Oncol Rep. 2012;27:1660–1668. doi: 10.3892/or.2012.1682. [DOI] [PubMed] [Google Scholar]
- 17.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 18.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, Wu F, Mo YY. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013;20:1558–1568. doi: 10.1038/cdd.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu X, Dong S, Qiao F, Lu S, Song Y, Lao Y, Li Y, Zeng T, Hu J, Zhang L, et al. HBx-mediated miR-21 upregulation represses tumor-suppressor function of PDCD4 in hepatocellular carcinoma. Oncogene. 2013;32:3296–3305. doi: 10.1038/onc.2013.150. [DOI] [PubMed] [Google Scholar]
- 21.Sicard F, Gayral M, Lulka H, Buscail L, Cordelier P. Targeting miR-21 for the therapy of pancreatic cancer. Mol Ther. 2013;21:986–994. doi: 10.1038/mt.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang SM, Huang C, Li XF, Yu MZ, He Y, Li J. miR-21 confers cisplatin resistance in gastric cancer cells by regulating PTEN. Toxicology. 2013;306:162–168. doi: 10.1016/j.tox.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ, Wang YK, Zeng F, Zhou JH, Zhang YK. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol. 2012;29:618–626. doi: 10.1007/s12032-011-9923-y. [DOI] [PubMed] [Google Scholar]
- 24.Chen C, Bhalala HV, Qiao H, Dong JT. A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer. Oncogene. 2002;21:6567–6572. doi: 10.1038/sj.onc.1205817. [DOI] [PubMed] [Google Scholar]
- 25.Gao Y, Wu K, Chen Y, Zhou J, Du C, Shi Q, Xu S, Jia J, Tang X, Li F, et al. Beyond proliferation: KLF5 promotes angiogenesis of bladder cancer through directly regulating VEGFA transcription. Oncotarget. 2015;6:43791–43805. doi: 10.18632/oncotarget.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C, Bhalala HV, Vessella RL, Dong JT. KLF5 is frequently deleted and down-regulated but rarely mutated in prostate cancer. Prostate. 2003;55:81–88. doi: 10.1002/pros.10205. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Wagenaar TR, Zabludoff S, Ahn SM, Allerson C, Arlt H, Baffa R, Cao H, Davis S, Garcia-Echeverria C, Gaur R, et al. Anti-miR-21 suppresses hepatocellular carcinoma growth via broad transcriptional network deregulation. Mol Cancer Res. 2015;13:1009–1021. doi: 10.1158/1541-7786.MCR-14-0703. [DOI] [PubMed] [Google Scholar]
- 29.Sazanov AA, Kiselyova EV, Zakharenko AA, Romanov MN, Zaraysky MI. Plasma and saliva miR-21 expression in colorectal cancer patients. J Appl Genet. 2017;58:231–237. doi: 10.1007/s13353-016-0386-x. [DOI] [PubMed] [Google Scholar]
- 30.Lasithiotaki I, Tsitoura E, Koutsopoulos A, Lagoudaki E, Koutoulaki C, Pitsidianakis G, Spandidos DA, Siafakas NM, Sourvinos G, Antoniou KM. Aberrant expression of miR-21, miR-376c and miR-145 and their target host genes in Merkel cell polyomavirus-positive non-small cell lung cancer. Oncotarget. 2016;8:112371–112383. doi: 10.18632/oncotarget.11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charkiewicz R, Pilz L, Sulewska A, Kozlowski M, Niklinska W, Moniuszko M, Reszec J, Manegold C, Niklinski J. Validation for histology-driven diagnosis in non-small cell lung cancer using hsa-miR-205 and hsa-miR-21 expression by two different normalization strategies. Int J Cancer. 2016;138:689–697. doi: 10.1002/ijc.29816. [DOI] [PubMed] [Google Scholar]
- 32.Yu Y, Nangia-Makker P, Farhana L, G Rajendra S, Levi E, Majumdar AP. miR-21 and miR-145 cooperation in regulation of colon cancer stem cells. Mol Cancer. 2015;14:98. doi: 10.1186/s12943-015-0372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li ZB, Li ZZ, Li L, Chu HT, Jia M. MiR-21 and miR-183 can simultaneously target SOCS6 and modulate growth and invasion of hepatocellular carcinoma (HCC) cells. Eur Rev Med Pharmacol Sci. 2015;19:3208–3217. [PubMed] [Google Scholar]
- 34.Adam O, Löhfelm B, Thum T, Gupta SK, Puhl SL, Schäfers HJ, Böhm M, Laufs U. Role of miR-21 in the pathogenesis of atrial fibrosis. Basic Res Cardiol. 2012;107:278. doi: 10.1007/s00395-012-0278-0. [DOI] [PubMed] [Google Scholar]
- 35.Tetreault MP, Yang Y, Katz JP. Krüppel-like factors in cancer. Nat Rev Cancer. 2013;13:701–713. doi: 10.1038/nrc3582. [DOI] [PubMed] [Google Scholar]
- 36.Dong JT, Chen C. Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell Mol Life Sci. 2009;66:2691–2706. doi: 10.1007/s00018-009-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu R, Shi P, Nie Z, Liang H, Zhou Z, Chen W, Chen H, Dong C, Yang R, Liu S, et al. Mifepristone suppresses basal triple-negative breast cancer stem cells by down-regulating KLF5 expression. Theranostics. 2016;6:533–544. doi: 10.7150/thno.14315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakajima Y, Osakabe A, Waku T, Suzuki T, Akaogi K, Fujimura T, Homma Y, Inoue S, Yanagisawa J. Estrogen exhibits a biphasic effect on prostate tumor growth through the estrogen receptor β-KLF5 pathway. Mol Cell Biol. 2015;36:144–156. doi: 10.1128/MCB.00625-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.