Abstract
Diatom diazotroph associations (DDAs) are a widespread marine planktonic symbiosis between several diatom genera and di-nitrogen (N2)-fixing bacteria. Combining single cell confocal microscopy observations and molecular genetic approaches on individual field collected cells, we determined the phylogenetic diversity, distribution and evolution of the DDAs. Confocal analyses coupled with 3-D imaging re-evaluated the cellular location of DDA symbionts. DDA diversity was resolved by paired gene sequencing (18S rRNA and rbcL genes, 16S rRNA and nifH genes). A survey using the newly acquired sequences against public databases found sequences with high similarity (99–100%) to either host (18S rRNA) or symbiont (16S rRNA) in atypical regions for DDAs (high latitudes, anoxic basin and copepod gut). Concatenated phylogenies were congruent for the host and cyanobacteria sequences and implied co-evolution. Time-calibrated trees dated the appearance of N2 fixing planktonic symbiosis from 100–50Mya and were consistent with the symbiont cellular location: symbioses with internal partners are more ancient. An ancestral state reconstruction traced the evolution of traits in DDAs and highlight that the adaptive radiation to the marine environment was likely facilitated by the symbiosis. Our results present the evolutionary nature of DDAs and provide new genetic and phenotypic information for these biogeochemically relevant populations.
Keywords: diatom, diazotroph, cyanobacteria, symbioses, evolution, distribution, confocal microscopy
Host diversity and trait based molecular dating in widely distributed N2 fixing planktonic symbioses identifies the ancestral states leading to the modern day success in DDAs.
INTRODUCTION
In the marine plankton, several diverse genera of eukaryotes live in close association (symbiosis) with cyanobacteria (Carpenter and Foster 2002; Foster, Carpenter and Bergman 2006; Foster, Collier and Carpenter 2006; Decelle, Colin and Foster 2015). Some of the cyanobacteria are N2 fixers, or diazotrophs, and therefore function as nitrogen (N) sources for their respective hosts (Foster et al.2011; Thompson et al.2012). The partnerships between heterocystous cyanobacteria Richelia intracellularis and Calothrix rhizosoleniae and a few genera of diatoms (e.g. Hemiaulus, Rhizosolenia, and Chaetoceros) are one such group. One lesser-studied symbiosis between the centric diatom Climacodium frauenfeldianum and a unicellular diazotrophic cyanobacterium also co-occurs in the plankton (Carpenter and Janson 2000). Collectively, the diatoms with N2-fixing symbionts are referred to as diatom diazotroph associations, or DDAs (Fig. S1, supporting Information).
To date, only the symbiotic partners of DDAs have been phylogenetically characterized and nothing is known about the host diatom genetic diversity (Janson et al.1999; Foster and Zehr 2006). Four draft genomes are available for the symbiotic partners, where symbiont genome size and content are related to the symbiont cellular location (Hilton et al.2013; Hilton 2014). Although it should be noted that the exact location of the R. intracellularis in Hemiaulus spp. is unknown and presumed to resemble the location in Rhizosolenia diatoms (i.e. in the diatom periplasmic space; Villareal 1992; Janson, Rai and Bergman 1995). The obligate nature in the various DDAs is also speculative and considered dependent on symbiont location (Hilton et al.2013). Whether the cellular location of the symbiont is related to other aspects of the symbiosis, e.g. how metabolites are exchanged, age of the partnerships, is also currently unknown.
Fossils for two of the DDA host diatom genera (Rhizosolenia and Hemiaulus) are reported from mid and late Cretaceous sedimentary records (100–66 million years ago, Mya) and coincides with isotopic evidence for intense N2 fixation and molecular signatures of heterocystous cyanobacteria (Sachs and Repeta 1999; Kashiyama et al.2008; Bauersachs et al. 2010). DDAs are often described as widespread in the World's oceans, yet high densities (e.g. blooms) are typically only reported in two divergent locations: seasonal summertime blooms in the North Pacific (NP) gyre (Karl et al.2012) and in areas near to large riverine inputs (e.g. Amazon, Mekong; Carpenter et al.1999; Subramaniam et al.2008; Bombar et al.2011). Moreover, a majority of abundance estimates for DDAs rely on quantitative PCR assays that quantify the symbiosis based on the symbiont nifH gene sequence (encodes nitrogenase). Methods that rely on sequences for estimating abundance are not absolute and have innate biases that complicate their use for estimating species abundances (Bonk et al.2018). Therefore, the understanding of DDA abundances, distribution and the conditions favoring their occurrence patterns is currently limited and challenging to interpret.
The primary aims of this study were four fold. First, we determined the genetic identity of field collected DDAs. For each DDA we paired 18S rRNA and rbcL genes (diatoms) and 16S rRNA and nifH genes (symbiont). Then, using the newly acquired sequences, we conducted a sequence-based survey using publicly available databases to assess the global distributions of the DDAs. Given the significance of cellular location for the symbiont genome size, content and host dependency, we re-evaluated the symbiont location using confocal microscopy. Finally, we built time-calibrated phylogenies for both partners and used them to infer the timing of the DDAs and evolution of phenotypic and life-history changes that has led to modern day DDAs.
MATERIALS AND METHODS
Sample collections
DDAs were collected from field expeditions (Fig. 1; Table S1, Supporting Information). Seawater samples (2.75–5 L) were collected from depth using a conductivity temperature depth (CTD) rosette. DDAs for microscopy observations and some phylogeny samples (see below) were filtered onto 25 or 47 mm diameter membrane filters (GE Osmonics, Fairfield, CT USA; pore sizes of 3, 5 or 8 μm) held within swinnex filter holders (Millipore, Bedford, MA USA) using a peristaltic pump or gravity filtration. The filters for microscopy were amended with 100 μL of 4% paraformaldehyde (PFA) for 1–4 h prior, rinsed three times in 50:50 solution of 0.2 μm filtered seawater (FSW): milliQ and stored dry at –20°C.
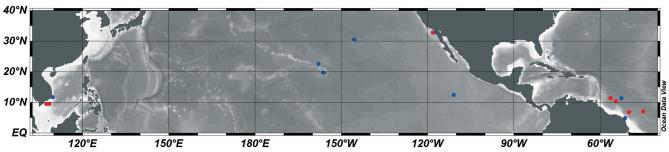
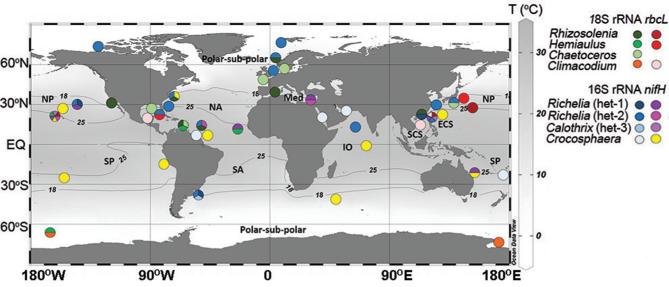
Figure 1.
Map of sampling locations for the morphological (designated red circles) and phylogeny (designated blue circles) analyses. Exact locations are summarized in Table S1 (Supporting Information).
An enrichment culture of H. hauckii-R. intracellularis (provided by AE Allen; JCVI, La Jolla, CA USA) was sampled for microscopy and phylogenetic analyses. The culture was maintained under 16:8 Light:Dark cycles in a modified RMP medium (Webb, Moffett and Waterbury 2001), at 26°C and 30–60 μmol photons m−2 s−1 with gentle shaking. Microscopy and phylogeny samples were gravity filtered onto 5 μm pore size filters (25 mm). Three microscopy samples (10 mLs) were fixed and rinsed as described above, one (10 mLs) additional microscopy sample without PFA fixation for immediate observation and two phylogeny samples (10 mLs each) were stored frozen (–80°C). The enrichment culture died shortly after the sampling.
Samples for the phylogenetic study were taken from the enrichment culture (2 samples) and the field locations using either a plankton net (80 μm mesh) or whole sea water using the CTD rosette and gravity filtration as described above (9 samples: Fig. 1; Table S1, Supporting Information). The samples collected by plankton net were towed from the stern of the research vessel during high densities of H. hauckii-R. intracellularis in the western tropical North Atlantic (WTNA; 1 sample) and R. clevei-R. intracellularis in the NP (4 samples). After collection from the CTD, the filters were placed on an oversized slide and DDAs identified using an epi-fluorescent microscope fitted with green (510–560 nm) and blue (450–490 nm) excitation filters. DDAs were isolated using a sterile micropipette onto a new sterile filter piece (5 μm pore size), and stored at –80°C. In the lab, the DDAs were re-identified, excised onto smaller filter pieces to be used directly as the template.
DNA extraction and whole genome amplification
The nucleic acids for the net samples and the positive controls (Phaeodactylum tricornutum (diatom) and Gloeothece sp. (cyanobacteria)) for the Polymerase Chain Reactions (PCRs, see below) were extracted using the DNeasy Plant Mini kit (Qiagen, Sollentuna, Sweden) with minor modifications (Foster and Zehr 2006). For DDAs isolated onto the excised filters, the filter pieces were directly added to sterile tubes and amplified using the REPLI-g Mini Kit (Qiagen, Sollentuna, Sweden) following the 50 μL protocol. Two negative controls, an excised filter containing no cells and 2 μl water, were also included. No amplifications resulted in the negative controls.
PCR, cloning and sequencing
The 11 DDA samples were processed using PCRs to amplify partial fragments: the large subunit of Ribulose-1, 5-bisphosphate (rbcL) (554 bp), the 18S rRNA (390–410 bp), the 16S rRNA (1000 bp) and nifH (359 bp). The WGA products were diluted 1:10 or 1:100. Details on the PCR primers, reaction master mixes and reaction conditions are described in the Supplementary Methods and Table S2 (Supporting Information). The PCR products were visualized on a 1.2% agarose gels, bands of appropriate size were excised, purified, ligated and cloned using commercially available kits. All details are summarized in the Supplementary Methods. Samples were sent for sequencing at Eurofins Genomics (Ebersberg, Germany) using the T7 primer (5 pmol mL −1).
Vector sequences were trimmed using Geneious® 9.1.6 software. For each library, sequences were considered different if they differed by more than one base pair (bp). Sequences determined in this study have been submitted to GenBank with accession numbers: MF536668-74 (rbcL), MF527222–30 (18S rRNA), MF536663-67 (nifH) and MF527112–16 (16S rRNA).
Microscopy observations
Two preparations for imaging and visualizing the DDAs with confocal laser scanning microscopy (CLSM) are summarized in the Supplementary Methods. Symbiotic Cells were imaged with a Zeiss LM 780 equipped with 405, 488 and 561 laser lines. The 488 and 561 laser lines were simultaneously used to visualize the chlorophyll of the host diatoms and cyanobacterial symbionts (Ex430 nm/Em680–720 nm), and the phycobilins of the cyanobacteria (Ex561 nm/Em 562–672 nm). Additionally, the 405 laser line was adopted to visualize the nuclei/nucleoids, previously stained with DAPI (Ex358 nm/Em406–568 nm). Image processing and three-dimensional reconstructions were conducted with IMARIS v.8.1 (Bitplane) using the Contour Surface tool. A minimum of 48 symbionts for each DDA type was observed and measured (Supplementary Methods).
Immunogold labeling and TEM
Cell concentrates of H. hauckii-R. intracellularis were collected from the surface with a hand towed net (70 μm mesh, 25 cm diameter), and re-suspended in a small volume of FSW (10 mLs). Small droplets (10 μL) were immobilized in 2% agar, and processed for immuno-gold staining for nitrogenase and phycoerythrin coupled to transmission electron microscopy (TEM) as previously described (Foster, Carpenter and Bergman 2006). Grids were examined with a Zeiss EM 906 at 80 kV (Zeiss).
Phylogenetic analyses
Representative sequences (57 diatom 18S rRNA and rbcL, 44 cyanobacteria 16S rRNA and nifH; Table S3, Supporting Infirmation) were downloaded from NCBI nucleotide database (https://www.ncbi.nlm.nih.gov/nucleotide/), and aligned with the sequence data generated in this study using MUSCLE v.3.831 (Edgar 2004) and concatenated in a single alignment. In order to determine the age and the timing of evolutionary events, we analyzed our data under the assumption of the presence of a molecular clock. More specifically, we addressed the known issues of heterogeneity of rates among lineages (i.e. different lineages may have different ‘clocks’), and the uncertainty in fossil calibrations, using Bayesian methods allowing for ‘relaxed clocks’ (recently reviewed in Bromham et al.2017). The output from such analyses is time calibrated phylogenetic trees, where the branching events are dated (in Myr) with a degree of uncertainty. All inferences were made using MrBayes v.2.3.7 (Ronquist et al.2012) and the fossil calibrations are summarized in Table S4 (Supporting Information). Additional details are provided in the Supplementary Methods.
Ancestral character-state reconstructions
To hypothesize about possible scenarios for trait evolution, we used statistical methods to infer ancestral character states based on the distribution of present day observations, and models of character evolution (Schluter et al.1997; Pagel 1999). A character, or trait, is then ‘mapped’ on top of a phylogenetic tree, and the outcome of such analysis is probabilistic statements about the presence of a specific character state in the ancestor (i.e. the nodes in the tree). Specifically, we applied the likelihood based ‘Re-rooting method’ (ER model; Yang, Kumar and Nei 1995), as implemented in the R-package phytools (Revell 2012). The trees from the clock-analyses were used as input, but using branch-lengths proportional to the expected number of substitutions per site. A number of both phenotypic and life-history characters were chosen (Table 1). Literature searches were used to determine the different character status for each trait (Supplementary Methods).
Table 1.
Summary of traits and character state assignments for the ancestral reconstructions presented in Figure 4 and Fig. SF4–11 (Supporting Information). Note. The character ‘Life history’ for the diatoms refers to symbiosis with cyanobacteria. N/A: Not applicable.
| Character name | Character variation | Character state | Figure(s) | |
|---|---|---|---|---|
| Cyanobacteria | Environment | non-marine | 0 | Fig. 4A; Fig. S7 |
| marine | 1 | |||
| Life History | free-living | 0 | Fig. 4A; Fig. S8 | |
| facultative symbiotic | 1 | |||
| obligate symbiotic | 2 | |||
| Location | N/A | 0 | Fig. S9 | |
| attached (epibiont) | 1 | |||
| endobiont | 2 | |||
| Heterocyst location | absent | 0 | Fig. S10 | |
| intercalary | 1 | |||
| terminal | 2 | |||
| both | 3 | |||
| Filament length | < 4μm | 0 | Fig. S11 | |
| 4μm < x < 40μm | 1 | |||
| > 40μm | 2 | |||
| Diatoms | Life History | unknown/free-living | 0 | Fig. 4B; Fig. S4 |
| facultative | 1 | |||
| obligate | 2 | |||
| Frustule silicification | poor | 0 | Fig. S5 | |
| moderate | 1 | |||
| strongly | 2 | |||
| Colony | unknown or rare | 0 | Fig. 4B; Fig. S6 | |
| forming | occasionally | 1 | ||
| frequently | 2 |
Environmental sequence survey
The sequences generated in this study (18S rRNA, rbcL, 16S rRNA and nifH) were used in BLASTn searches on NCBI, KEGG MGENES (www.genome.jp/mgenes/) and in the IMNGS database (https://www.imngs.org/) (October 2018). Sequences with at least 98% similarity and minimum coverage of 350 bp were selected and quantified for each sequence.
RESULTS AND DISCUSSION
Cell measurements and re-evaluation of symbiont location
Wild populations were collected from diverse habitats to study the DDA morphological diversity and to make cell measurements for comparison to earlier works (Table S5, Supporting Information). The symbiont trichome length was positively correlated with host diatom cell diameter (P < 0.001, R2 = 0.2; P < 0.001, R2 = 0.4; P < 0.05, R2 = 0.4; 1, 10 and 25 meters, respectively; Table S5, Supporting Information). Despite few measurements for larger cell diameter hosts and an observed high variation, the relationships were still statistically significant. The high variation in the measurements is expected since cells are field-collected, and the growth phases are unknown and likely in a mixed state. For internal symbionts, trichome length is obviously confined to the host dimensions, while the length of the external symbionts (C. rhizosoleniae) may be unbound. Interestingly, long trichomes of C. rhizosoleniae were not observed and the phenotype-based phylogenies (see below) predict shorter trichomes as favored traits in the ancestry of DDA symbionts.
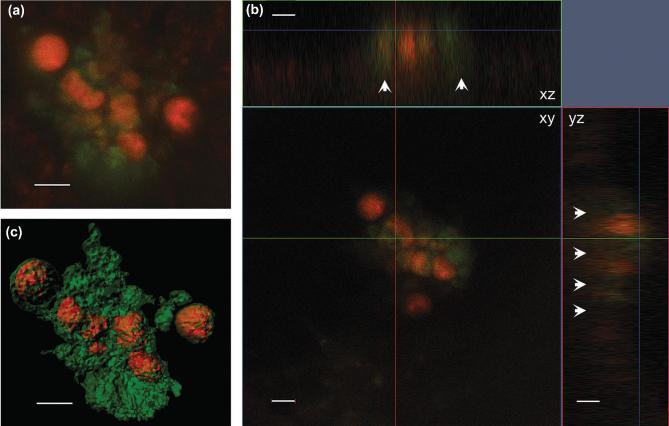
Our confocal observations confirm that symbiotic R. intracellularis resides inside the cytosol of H. hauckii. The majority (90%, or 215 of 238) of orthogonal observations (x/y, x/z, y/z projections) of symbiotic H. hauckii cells showed R. intracellularis (1–2 filaments) surrounded, partially or completely, by the auto-fluorescence derived from the host chloroplast (Fig. 2). Our observations represent the first 3-D evidence for a cytosolic location; earlier observations (e.g. light microscopy: Ferrario et al.1995; Zeev et al.2008) have all been 2-D. Observations in the chemically fixed field samples were also similar to the untreated enrichment culture suggesting that the fixation step did not influence the symbiont location (Fig. S2B and S2E, Supporting Information). The TEM coupled with immunocytochemistry also showed the vegetative cells of R. intracellularis filaments located centrally in the host (H. hauckii) cytosol (Fig. S3, Supporting Information). The cells were positively labeled with phycoerythrin and the filament surrounded by host cytoplasm (Fig. S3B and C, Supporting Information). Finally, the DAPI staining showed the symbiont located in close proximity to the host nucleus (Fig. 3). A similar location has been described for the N2 fixing cyanobacterial-like spheroid bodies of the freshwater diatom Rhopalodia gibba (Geither 1977; Prechtl et al.2004; Kneip et al.2008; Nakayama et al.2011). Close proximity to the nucleus would be strategic for energy acquisition. Although heterocystous cyanobacteria, like R. intracellularis, protect the oxygen sensitive nitrogenase enzyme by the thick envelope of the heterocyst, it is not impermeable (Walsby 1985).
Figure 2.
Laser confocal microscopy images of an H. hauckii-R. intracellularis symbiosis simultaneously excited by 488 and 561 laser lines:(a) z-stack image, (b) orthogonal views (xy, xz, yz) and (c) processed with the Contour Surface tool in IMARIS v.8.1 (Bitplane). White arrows show the fluorescence of the chloroplasts in xz and yz. Chloroplasts are shown in green, and cyanobacteria trichome (filament) in orange. Scale bar 5 μm.
Figure 3.
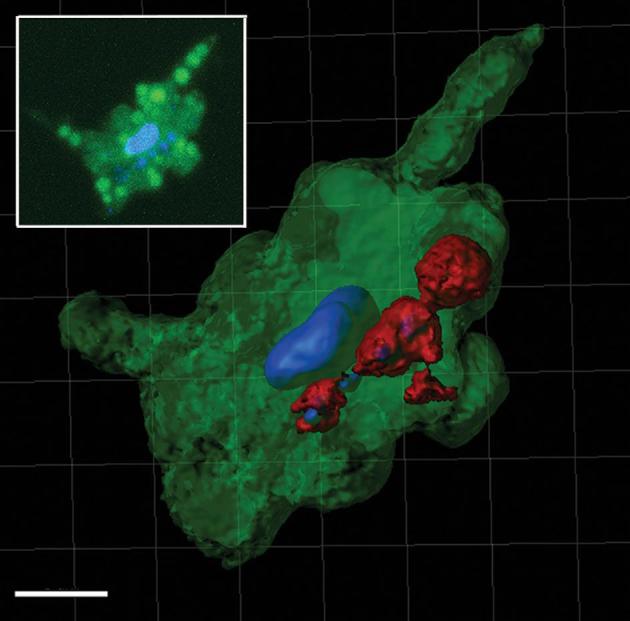
Processed confocal z-stack image using the Contour Surface tool in IMARIS v.8.1 (Bitplane) of a DAPI stained H. hauckii-R. intracellularis symbiosis showing the proximity of the symbiotic trichome (red), stained nucleoids of symbiont (blue) and the diatom stained nucleus (blue). In green the diatom chloroplasts reconstruction and includes background. Scale bar 5 μm. Inset: parallel z-stack confocal image simultaneously excited by 405 and 488 laser lines highlighting the diatom nucleus and the cyanobacteria nucleoids (in blue), and chloroplasts (brighter green circles; note, a lower degree of averaging shows some background auto-fluorescence).
As expected and consistent with earlier observations (Norris 1961), Calothrix rhizosoleniae (3–11 trichomes per chain) were observed attached externally between two Chaetoceros spp. frustules at the heterocyst (Fig. S2C and S2F, Supporting Information). Our observations for the numbers of attached epiphytes to Chaetoceros sp. chains were similar to earlier works (Gómez et al.2005; Table S5, Supporting Information) and suggested attachment space as an important factor, perhaps host-regulated. Richelia associated with Rhizosolenia (1–3 trichomes per host) was observed as previously recorded close to the diatom frustule in a fixed orientation (Sundström 1984; Villareal 1989) (Fig. S2A and S2D, Supporting Information). The number, cellular location and orientation vary in each of the symbioses, and how these characters are maintained is unknown.
Host and symbiont sequence diversity
The DDAs are considered to be highly host specific, (Janson et al.1999; Foster and Zehr 2006), yet the genetic identity of the hosts has never been investigated. For each DDA, we report paired genetic sequences, such that one individual DDA resulted in 18S rRNA and rbcL sequences for the hosts, and 16S rRNA and nifH sequences for the symbionts. In one DDA, however, the host diatom (C. compressus) failed to amplify the rbcL gene. Eight to twelve identical clones were retrieved for each library, except for a few samples that showed more than one phylotype (sample IDs 10010, 404, 5239; Supplementary Results and Table S6, Supporting Information). Prior to the phylogenetic analysis, host and symbiont sequences were characterized using the BLASTn tool on NCBI.
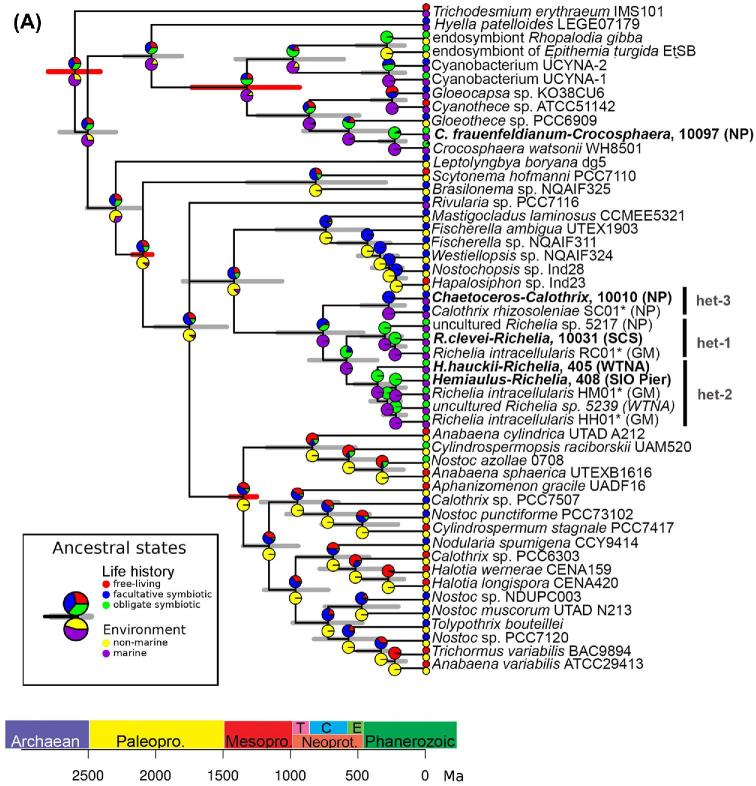
Consistent with previous work (Foster and Zehr 2006), our symbiont sequences clustered as expected and diverged into three well-supported clades (het-1, -2 and -3; Fig. 4A). The average sequence diversity amongst the heterocystous symbionts was similar to values reported earlier (Foster and Zehr 2006) for both 16S rRNA (97.5% vs. 98.2%) and nifH (91.8% vs. 91.1%) (Table S7A, Supporting Information). The symbiont sequence similarity was also independent of geographic location (Janson et al.1999; Foster and Zehr 2006) since highly similar sequences were derived from DDA cells collected from very distant oceans. Smaller sequence diversity might be expected amongst internal symbionts given that an internal cellular location would be more stable and perhaps limit genetic drift in a small population (Giovannoni, Thrash and Temperton 2014).
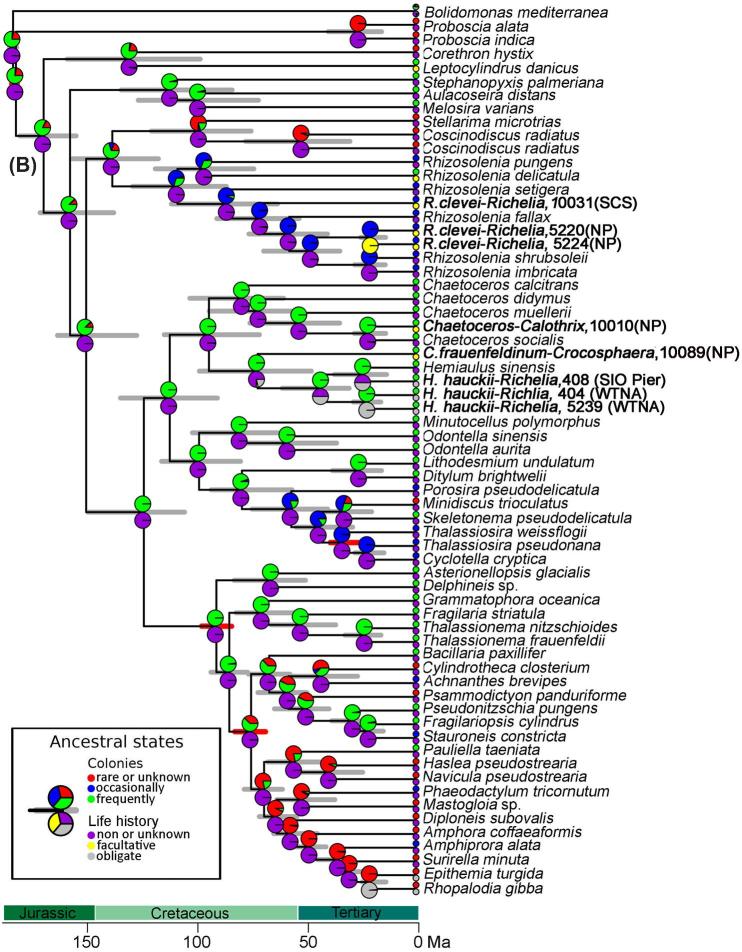
Figure 4.
(A) Chronogram based on concatenated sequences (16S rRNA and nifH) depicting relationships among cyanobacteria taxa. Branch lengths are proportional to time (Mya). The 95% highest posterior density intervals for node ages are indicated by gray bars. Red bars indicate calibrated nodes defined in Table S4 (Supporting Information). Two characters are displayed: life history (upper pie-chart), and environment (lower pie-chart); the states assigned for each character are color-coded and shown in the figure legend. The small circles at the tips represent the observed data (present day); the pie-charts on the internal nodes are the probability distributions of the ancestral character state. The position of the circles (i.e. upper or lower) is consistent with the position of the pie-chart. Accession numbers for species in tree are provided in Table S3 (Supporting Information), and the sequences generated from our study are shown in bold.(B) Chronogram based on concatenated sequences (18S rRNA and rbcL) depicting relationships among diatom taxa. Branch lengths are proportional to time (Ma). The 95% highest posterior density intervals for node ages are indicated by gray bars. Red bars indicate calibrated nodes defined in Table S4 (Supporting Information). Two characters are displayed: colonies (upper pie-chart), and life history (lower pie-chart); the states assigned for each character are color-coded and shown in the figure legend. The small circles at the tips represent the observed data (present day); the pie-charts on the internal nodes are the probability distributions of the ancestral character state. The lower pie-charts at each node correspond to the lower circle at the tip (and vice-versa for the upper pie-charts). Accession numbers for species in tree are provided in Table S3 (Supporting Information), and sequences generated in our study are shown in bold.
Previous to our study, the phylogenetic identity for the unicells of C. frauenfeldianum was limited to one partial 16S rRNA sequence (Carpenter and Janson 2000), and the host sequence identity was unknown. The 16S rRNA sequence (sample ID:10097) reported here was 99% similar to the previously published sequence (Accession no. AF193247), and 99% similar to both 16S rRNA and nifH sequences of C. watsonii WH8501 (Accession no. CGA000167195) (Table S7A, Supporting Information). Hence the phylogenetic inference robustly places the symbiont of C. frauenfeldianum with C. watsonii WH8501 (Fig. 4A). An unusual high genetic conservation is reported for C. watsonii isolates, despite possessing diverse phenotypes (cell size, presence/absence of extracellular polysaccharides) and life histories (colonial, free-living) (Zehr et al.2007; Bench et al. 2011, 2013). It would be interesting to confirm if the C. frauenfeldianum symbionts share similar genome conservation or contain unique genome information for a symbiotic life history.
The concatenated phylogeny based on the host sequences was congruent with the symbionts. Host sequences clustered according to the genus they belong to (Rhizosolenia, Hemiaulus, Chaetoceros) (Table S7B, Supporting Information). More specifically, the symbiotic Rhizosolenia sequences (sample IDs 5220, 5244 and 10031) clustered within the Rhizosolenia lineage, together with R. delicatula, R. fallax, R. imbricata, R. pungens, R. setigera, and R. shrubsoleii (Fig. 4B). The replicate bloom samples from the NP gyre (sample IDs 5221, 5220, 5244) were more distant (86% similar 18S rRNA, 93% rbcL) to the sample from the SCS (sample ID 10031), suggesting broader host diversity. However the SCS sample has only been identified to genus, while the NP bloom was R. clevei. Similarly, the 2 Hemiaulus samples (sample IDs 404/405, 5239) collected during separate H. hauckii blooms in the WTNA (2003, 2010) clustered together, and clustered with the symbiotic H. hauckii collected at SIO Pier (sample ID 407/408) and collectively symbiotic H. hauckii sequences clustered with the only culture representative of the genus, i.e. H. sinensis (Fig. 4B). Sister to this clade was the sequences derived from diatom-host C. frauenfeldianum (Sample ID 10089). The Climacodium genus belongs to the Hemiaulaceae family. Noteworthy, the sequences for C. frauenfeldianum represent the first genetic characterization of the genus Climacodium Grunow (1868).
The phylogenetic reconstruction placed the symbiotic Chaetoceros compressus (sample ID 10010) within the Chaetoceros clade, composed by C. calcitrans, C. didymus, C. muelleri and C socialis. Our 18S rRNA sequence was most similar (99%) to C. socialis (Accession no. JQ217339) (Table S7B, Supporting Information); unfortunately we were unable to resolve the rbcL sequence for C. compressus since the sample failed to amplify.
Environmental distribution of DDAs
The newly acquired paired sequences were used to estimate the distribution of the DDAs by Blast searches in public databases (i.e. NCBI, KEGG MGENES and IMNGS; Fig. 5; Table S8, Supporting Information). Our results confirm earlier works with DDAs widely distributed across much of the oligotrophic gyres, while also showing evidence for an extended range to cooler waters (Fig. 5). The number of sequences with high identity (99–100%) to the host/symbiont sequences retrieved was low (329 sequences: 31 rbcL; 24 18S rRNA; 196 nifH; 78 16S rRNA sequences) and related to the limitations in the sequence databases (e.g. different molecular markers than sequenced here; limited or under sampling (Caputo et al.2018); comparing a partial sequence).
Figure 5.
Global distribution of sequences with similarity to DDA partners (sequence identity ≥ 98%; minimum coverage ≥ 350 bp). The host (18S rRNA, rbcL) and cyanobacterial sequences (nifH, 16S rRNA) generated in this study were used in BLASTn searches on NCBI, KEGG MGENES and IMNGS databases, and are color-coded. The presence of more than one sequence at the same geographic location is represented by a pie-chart. If only one sequence was retrieved, it is shown by a solid colored circle. The latitudinal 18°C and 25°C isotherms (average values of yearly means from the World Ocean Atlas 2013: NOAA’s National Oceanographic Data Center—Ocean Climate Laboratory) and ocean basin are designated.
Despite the limitations, there were a few notable exceptions (Table S8, Supporting Information; Fig. 5). For example, we identified sequences with high similarity (99–100%) to the 18S rRNA sequences from 3 DDA hosts and the 16S rRNA sequence from the Richelia symbiont of H. hauckii in the high latitudes (i.e. polar and sub-polar regions). The average temperature in these regions (−2°C to 2°C) is dramatically colder than regions where DDAs thrive (ca. 25°C). Considering that diatoms are known to grow in polar waters (Miettinen 2018), and other heterocystous cyanobacteria prefer cooler temperatures (Staal, Meysman and Stal 2003; Stal 2009), sub-polar regions could potentially favor the DDAs. The R. clevei host 18S rRNA sequence was similar to a sample collected in the anoxic zone of the Cariaco Basin, while the H. hauckii host 18SrRNA sequence matched a clonal sequence from a copepod gut, hence perhaps an important role of the DDAs also resides in the trophic chain (Conroy et al.2017). To date, DDAs have largely been reported in the upper euphotic zone in tropical/subtropical waters, and in a few older and recent reports from moored sediment traps (Scharek et al.1999; Scharek, Tupas and Karl 1999; Caffin et al.2018; Spungin et al.2018). Thus, we identified new distributions for the DDA partners and symbioses. Our results also concur with recent reports detecting another N2 fixing symbiont, UCYN-A, in Arctic waters (Farnelid et al.2016; Martínez-Pérez et al.2016; Shiozaki et al.2017; Harding et al. 2018).
Evolution of DDAs
Our new sequence data also allowed for the direct estimation of the timing and conditions, which has led to the emergence of DDAs. Using the newly generated sequences in a relax clock model, we estimated both phylogeny and divergence times for both symbiotic partners (Drummond et al.2006), and included two other eukaryotic-N2 fixing symbiosis: the marine prymnesiophyte-UCYN-A symbiosis and two freshwater DDAs (E. turgida and R. gibba) (Fig. 4).
Our data suggests an evolutionary progression where symbionts gradually move inside their hosts and have progressively smaller genomes. For example, the symbionts of the oldest partnerships (e.g. Hemiaulus, Rhizosolenia) are more integrated into the host cell and possess more eroded (draft) genomes (e.g. 2.21–5.4 Mbp; lack N-related transporters and reductases), while the most recent symbiosis (Chaetoceros sp.) has external/attached symbionts with a larger genome (5.97 Mbp) (Hilton et al.2013; 2014). The appearance of the C. frauenfeldianum-Crocosphaera symbiosis, for which symbiont location is presumed internal (Carpenter and Janson 2000), was also estimated between 100–50 Mya (Fig. 4B). Given the trend followed by the other DDAs, we would expect the symbiotic Crocosphaera to be internal and to possess similar pattern of genome reduction.
The location of the UCYN-A is still debated, however many consider it an internal symbiont (Hagino et al.2013), and its’ genome is extremely reduced (1.44 Mbp; Tripp et al.2010). The draft genomes of the internal R. intracellularis symbionts are similarly reduced, but less so than UCYN-A. The UCYN-A appearance followed a similar trend as the internal DDA symbionts, emerging as the oldest partnership. The symbionts of the freshwater DDAs are also internal and have similarly small genomes as UCYN-A (E. turgida, 2.7 Mbp; Nakayama et al.2014). However, our analysis predicts their appearance rather recently (ca 25 Mya), and actually coincides with the external Chaetoceros bearing hosts. It seems that the marine DDA symbionts are undergoing a slower co-evolutionary process compared to the UCYN-A, perhaps related to a larger starting genome (e.g. the heterocystous Anabaena variabilis ATCC 29413, 7.1 Mbp vs. the unicell Cyanothece ATCC 51142, 5.4 Mbp) (Welsh et al.2008; Thiel et al.2014). A combination of higher host dependency (e.g. UCYN-A dependence on host for carbon; Thompson et al.2012) and environment (e.g. oligotrophic conditions favor genome streamlining) could also control and accelerate UCYN-A genome reduction (Giovannoni, Thrash and Temperton. 2014).
Besides determining the age and order of the various planktonic symbiosis, we evaluated the conditions at the time of the DDA appearances and speculated whether these influenced the formation of partnerships. The surface ocean during the late Cretaceous was highly stratified, low in dissolved nutrients, and the surface temperatures were particularly high due to an increase of atmospheric CO2 (Erbacher et al.2001; Wilson and Norris 2001). These conditions reflect where DDAs are found in modern oceans: warm low nutrient, open ocean. The later appearances of symbiotic Chaetoceros and the freshwater DDAs (ca. 25 Mya) were at a time of an intense diversification of the Chaetoceros genus characterized during the Oligocene-Miocene transition (OMT; Suto 2006) (Fig. 4B). During the OMT, the surface ocean was cooling down, and most geological records report diatoms in resting spore stages (Suto, 2006). The unfavorable conditions during the OMT might have led C. compressus to a different survival strategy, such as evolving a symbiosis with an N2 fixing partner and taking refuge in the open sea.
Ancestral state of DDAs
We used an ancestral state reconstruction to trace the evolution of phenotypic and life-history traits in both partners that has led to the DDAs in the modern ocean (Table 1; Fig. 4; Fig. S4–11, Supporting Information). In such analysis, a particular character trait is scored for each tip in the tree (for the example, ‘life-history’: zero free-living diatoms, one for facultative symbiotic diatoms and two for obligate symbiotic diatoms (Table 1; Fig. 4B)). Then, given the tree, the branch-lengths and the model of character evolution, we can make probabilistic statements of the possible state for the ancestor (internal nodes in the tree). It would then be possible to trace, for example, the transition from free-living to symbiotic. The ancestral transitions from asymbiotic diatoms to obligate symbioses were congruent with the molecular phylogenies. For example, more ancient symbioses have a deeper ancestral lineage leading to obligate symbioses (e.g. 3rd diatom ancestor Hemiaulus was asymbiotic) (Fig. 4B, Fig. S4, Supporting Information).
Several of the ancestral traits are shared between symbiotic and asymbiotic diatoms, while some traits appear as adaptive phenotypes for aiding the diatom-cyanobacteria bond (Fig. S4–S11, Supporting Information). For example, the ancestor of the symbiotic diatoms (freshwater, R. gibba and marine DDAs) and the majority of asymbiotic diatoms possessed either a strong silicified frustule (i.e. Hemiaulus and Climacodium ancestors) or a moderately silicified frustule (i.e. Rhizosolenia and Chaetoceros ancestors) (Fig. S5, Supporting Information). A recent study suggested that a thick frustule was crucial for diatom success (Jewson and Harwood 2017). A strong frustule was perhaps also a necessary trait for not just free-living diatoms, but for symbiotic diatoms considering that it provides physical protection, and perhaps a means for keeping an internal symbiont more secure. All DDAs form colonies or chains when blooming, hence we used colony as a trait to trace the host diatom ancestor. Our analyses predicted a colony-forming ancestor (Fig. 4B; Fig. S6, Supporting Information), likely a strategic life history trait to also maintain buoyancy and enhance light capture.
The evolutionary reconstruction of the cyanobacterial partner highlighted that the prokaryotic ancestor was likely a filamentous non-marine species living first in a facultative symbiotic state (Fig. 4A; Fig. S7 and S8, Supporting Information). The transition to a symbiotic life-style appeared to happen gradually through an intermediate ancestor which generated two separate lineages: 1) episymbiotic strains (i.e. C. rhizosoleniae) and 2) endosymbiotic strains (i.e. R. intracellularis) (Fig. S9, Supporting Information). The adaptive radiation to the marine environment also occurred later, likely facilitated by the symbiosis. Our results were also consistent with recent studies on cyanobacteria evolution, which dates the appearance of marine planktonic species starting from the late Proterozoic (ca 500 Mya) and describes the precursor of the heterocystous cyanobacteria as non-marine cyanobacterium (Uyeda, Harmon and Blank 2016; Sánchez-Baracaldo et al.2017). Finally, in contrast to many of the terrestrial symbiotic plants with heterocystous cyanobacteria, which have maintained long trichomes with intercalary heterocysts, the DDA symbionts evolved short trichomes with terminal heterocysts (Fig. S10 and S11, Supporting Information). The transition to shorter filaments with one terminal heterocyst was likely independent of symbiont location (e.g. external symbionts followed the same transition), but perhaps related to space for residing in/on a diatom host and the preferred phenotype for providing sufficient N to both partners.
CONCLUSIONS
Our confocal observations resolved an internal cellular location for R. intracellularis in H. hauckii. The DDAs are thus a unique system defined by a continuum of internal, partial and truly external symbionts. The cellular location is related to the age of the DDA partnerships: older symbionts are more internal. Paired genetic information for each DDAs is now linked back to a specific consorting pair. Given that DDAs are often estimated exclusively by the symbiont presence, the new genetic information is applicable for developing other methods to verify the new distribution of the DDAs (cooler waters). The ancestral state reconstructions predicted short filaments and terminal heterocysts as favored traits in DDA symbionts. The DDAs are amongst the oldest planktonic partnerships, emerging during a time of elevated sea surface temperatures and high CO2 (Erbacher et al.2001; Wilson and Norris, 2001). These conditions are reminiscent of future climate scenarios and perhaps will influence the numbers and types of consortia.
Supplementary Material
Acknowledgements
We thank the captain and crew of the R/V Knorr and PIs of the ANACONDA project. Samples from SCS were collected in collaboration with Drs J Montoya (Georgia Tech, GA, USA), Ajit Subramaniam (Columbia U, NY, USA) and Maren Voss (IOW, Rostock, Germany) by Andreas Novotny (Stockholm University) on board the R/V Falkor. A special acknowledgement to Director Mary A Raven, and Dr. Geoff Lewis of University of California Santa Barbara, CA USA and Dr. Stina Höglund from SU for assistance in the confocal image analyses. Confocal microscopy was done at the Image Facility of SU. We acknowledge Dr. AE Allen and H Zheng from UC San Diego, CA for providing the H. hauckii-R. intracellularis enrichment culture. We thank Drs Sandra Lage and Sara Rydberg from SU for providing the P. tricornutum culture, Dr. Denis Warshan and Mariana P. Braga for assistance with the statistical analysis and other members of the Foster lab for support. And finally we thank the four anonymous reviewers of our manuscript, each have provided invaluable and insightful suggestions that have greatly improved the presentation of our manuscript. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project SNIC 2017/7–309.
FUNDING
This work was funded by the Knut and Alice Wallenberg Foundation to RAF. Earlier sample collections in the WTNA were funded by NSF grant (OCE 092 9015 to RAF).
Conflict of Interest. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest
REFERENCES
- Bauersachs T, Speelman EN, Hopmans EC et al. . Fossilized glycolipids reveal past oceanic N2 fixation by heterocystous cyanobacteria. Proc Natl Acad Sci 2010;107:19190–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bench SR, Heller P, Frank I et al. . Whole genome comparison of six Crocosphaera watsonii strains with differing phenotypes. J Phycol 2013;49:786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bench SR, Ilikchyan IN, Tripp HJ et al. . Two Strains of Crocosphaera watsonii with highly conserved genomes are distinguished by strain-specific features. Front Microbio 2011;2:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombar D, Moisander PH, Dippner JW et al. . Distribution of diazotrophic microorganisms and nifH gene expression in the Mekong River plume during intermonsoon. Mar Ecol Prog Ser 2011;424:39–52. [Google Scholar]
- Bonk F, Popp D, Harms H et al. . PCR-based quantification of taxa-specific abundances in microbial communities: quantifying and avoiding common pitfalls. J Microbiol Methods 2018;153:139–47. [DOI] [PubMed] [Google Scholar]
- Bromham L, Duchêne S, Hua X et al. . Bayesian molecular dating: opening up the black box. Biol Rev 2018;93:1165–91. [DOI] [PubMed] [Google Scholar]
- Caffin M, Moutin T, Foster RA et al. . N2 fixation as a dominant new N source in the western tropical South Pacific Ocean (OUTPACE cruise). Biogeosciences 2018;15:2565–85. [Google Scholar]
- Caputo AC, Stenegren M, Pernice MC et al. . A Short comparison of two marine planktonic diazotrophic symbioses highlights an un-quantified disparity. Front Mar Sci 2018;5:1–8.29552559 [Google Scholar]
- Carpenter EJ, Foster RA. Marine cyanobacterial symbioses. : Rai AN, Bergman B, Rasmussen U (eds). Cyanobacteria in Symbiosis. Dordrecht: Springer, 2002, 11–7 [Google Scholar]
- Carpenter EJ, Janson S. Intracellular cyanobacterial symbionts in the marine diatom Climacodium frauenfeldianum (Bacillariophyceae). J Phycol 2000;36:540–4. [DOI] [PubMed] [Google Scholar]
- Carpenter EJ, Montoya JP, Burns J et al. . Extensive bloom of a N2-fixing diatom/cyanobacterial association in the tropical Atlantic Ocean. Mar Ecol Prog Ser 1999;185:273–83. [Google Scholar]
- Conroy BJ, Steinberg DK, Song B et al. . Mesozooplankton graze on cyanobacteria in the Amazon River Plume and Western Tropical North Atlantic. Front Microbiol 2017;8:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decelle J, Colin S, Foster RA. Photosymbiosis in marine planktonic protists. : Ohtsuka S, Suzaki T, Horiguchi T et al. (eds). Marine Protists. Tokyo: Springer, 2015, 465–500. [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ et al. . Relaxed phylogenetics and dating with confidence. PLoS Biol 2006;4:699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004;32:1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbacher J, Huber BT, Norris RD et al. . Increased thermohaline stratification as a possible cause for an ocean anoxic event in the Cretaceous period. Nature 2001;409:325–7. [DOI] [PubMed] [Google Scholar]
- Farnelid H, Turk-Kubo K, Munoz-Marın M et al. . New insights into the ecology of the globally significant uncultured nitrogen-fixing symbiont UCYN-A. 2016. Aquat Microb Ecol 2016;77:125–38. [Google Scholar]
- Ferrario ME, Villafane V, Helbling W et al. . The occurrence of the symbiont Richelia in Rhizosolenia and Hemiaulus in North Pacific. Rev Bras Bio 1995;55:439–43. [Google Scholar]
- Foster RA, Carpenter EJ, Bergman B. Unicellular cyanobionts in open ocean dinoflagellates, radiolarians, and tintinnids: ultrastructural characterization and immuno-localization of phycoerythrin and nitrogenase. J Phycol 2006;42:453–63. [Google Scholar]
- Foster RA, Collier JL, Carpenter EJ. Reverse transcription PCR amplification of cyanobacterial symbiont 16S rRNA sequences from single non-photosynthetic eukaryotic marine planktonic host cells. J Phycol 2006;42:243–50. [Google Scholar]
- Foster RA, Zehr JP. Characterization of diatom–cyanobacteria symbioses on the basis of nifH, hetR and 16S rRNA sequences. Environ Microbiol 2006;8:1913–25. [DOI] [PubMed] [Google Scholar]
- Foster RA, Kuypers MMM, Vagner T et al. . Nitrogen fixation and transfer in open ocean diatom–cyanobacterial symbioses. ISME J 2011;5:1484–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geither L. Zur Entwicklungsgeschichte der Epithemiaceen Epithemia, Rhopalodia und Denticula (Diatomophyceae) und ihre vermutlich symbiotischen Spharoidkorper. Pl Syst Evol 1977;128:259–75. [Google Scholar]
- Giovannoni SJ, Thrash JC, Temperton B. Implications of streamlining theory for microbial ecology. ISME J 2014;8:1553–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez F, Furuya KEN, Takeda S et al. . Distribution of the cyanobacterium Richelia intracellularis as an epiphyte of the diatom Chaetoceros compressus in the western Pacific Ocean. J Plankton Res 2005;27:323–30. [Google Scholar]
- Hagino K, Onuma R, Kawachi M et al. . Discovery of an endosymbiotic nitrogen-fixing cyanobacterium UCYN-A in Braarudosphaera bigelowii (Prymnesiophyceae). PLoS One 2013;8:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding K, Turk-Kubo KA, Sipler RE et al. . Symbiotic unicellular cyanobacteria fix nitrogen in the Arctic Ocean. 2018;52:13371–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton JA, Foster RA, Tripp HJ et al. . Genomic deletions disrupt nitrogen metabolism pathways of a cyanobacterial diatom symbiont. Nat Commun 2013;4:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton JA. Ecology and evolution of diatom-associated cyanobacteria through genetic analyses. Ph.D. Thesis. University of California, Santa Cruz, CA USA, 2014. [Google Scholar]
- Janson S, Rai AN, Bergman B. Intracellular cyanobiont Richelia intracellularis: ultrastructure and immuno-localisation of phycoerythrin, nitrogenase, Rubisco and glutamine synthetase. Mar Biol 1995;124:1–8. [Google Scholar]
- Janson S, Woulters J, Bergman B et al. . Host specificity in the Richelia-diatom symbiosis revealed by hetR gene sequence analysis. Environ Microbiol 1999;1:431–8. [DOI] [PubMed] [Google Scholar]
- Jewson DH, Harwood DM. Diatom life cycles and ecology in the Cretaceous. J Phycol 2017;53:616–28. [DOI] [PubMed] [Google Scholar]
- Karl DM, Church MJ, Dore JE et al. . Predictable and efficient carbon sequestration in the North Pacific Ocean supported by symbiotic nitrogen fixation. Proc Natl Acad Sci 2012;109:1842–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiyama Y, Ogawa NO, Kuroda J et al. . Diazotrophic cyanobacteria as the major photoautotrophs during mid-Cretaceous oceanic anoxic events: Nitrogen and carbon isotopic evidence from sedimentary porphyrin. Org Geochem 2008;39:532–49. [Google Scholar]
- Kneip C, Voss C, Lockhart PJ et al. . The cyanobacterial endosymbiont of the unicellular algae Rhopalodia gibba shows reductive genome evolution. BMC Evol Biol 2008;8:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Pérez C, Mohr W, Löscher CR et al. . The small unicellular diazotrophic symbiont, UCYN-A, is a key player in the marine nitrogen cycle. Nat Microbiol 2016;1:1–7. [DOI] [PubMed] [Google Scholar]
- Miettinen A. Diatoms in arctic regions: potential tools to decipher environmental changes. Polar Sci 2018;18:220–6. [Google Scholar]
- Nakayama T, Ikegami Y, Nakayama T et al. . Spheroid bodies in rhopalodiacean diatoms were derived from a single endosymbiotic cyanobacterium. J Plant Res 2011;124:93–7. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Kamikawa R, Tanifuji G et al. . Complete genome of a nonphotosynthetic cyanobacterium in a diatom reveals recent adaptations to an intracellular lifestyle. Proc Natl Acad Sci USA: 2014;111:11407–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris RE. Observations on phytoplankton organisms collected on the NZOI Pacific cruise, September, 1958. NZ J Sci 1961;4:162–88. [Google Scholar]
- Pagel M. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Syst Biol 1999;48:612–22. [Google Scholar]
- Prechtl J, Kneip C, Lockhart P et al. . Intracellular spheroid bodies of Rhopalodia gibba have nitrogen-fixing apparatus of cyanobacterial origin. Mol Biol Evol 2004;21:1477–81. [DOI] [PubMed] [Google Scholar]
- Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 2012;3:217–23. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P et al. . MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol 2012;61:539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs JP, Repeta DJ. Oligotrophy and nitrogen fixation during eastern Mediterranean sapropel events. Science 1999;286:2485–8. [DOI] [PubMed] [Google Scholar]
- Sánchez-Baracaldo P, Raven JA, Pisani D et al. . Early photosynthetic eukaryotes inhabited low-salinity habitats. Proc Natl Acad Sci USA 2017;114:E7737–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharek R, Latasa M, Karl DM et al. . Temporal variations in diatom abundance and downward vertical flux in the oligotrophic North Pacific gyre. Deep Sea Res Part I 1999;46:1051–75. [Google Scholar]
- Scharek R, Tupas LM, Karl DM. Diatom fluxes to the deep sea in the oligotrophic North Pacific gyre at station ALOHA. Mar Ecol Prog Ser 1999;182:55–67. [Google Scholar]
- Schluter D, Price T, Mooers AO et al. . Likelihood of ancestor states in adaptive radiation. Evolution 1997;51:1699–711. [DOI] [PubMed] [Google Scholar]
- Shiozaki T, Bombar D, Riemann L et al. . Basin scale variability of active diazotrophs and nitrogen fixation in the North Pacific, from the tropics to the subarctic Bering Sea. Global Biogeochem Cycles 2017;31:996–1009. [Google Scholar]
- Shiozaki T, Fujiwara A, Ljichi M et al. . Diazotroph community structure and the role of nitrogen fixation in the nitrogen cycle in the Chukchi Sea (western Arctic Ocean). Limnol Oceanogr 2018;63:2191–205. [Google Scholar]
- Spungin D, Belkin N, Foster RA et al. . Programmed cell death in diazotrophs and the fate of organic matter in the western tropical South Pacific Ocean during the OUTPACE cruise. Biogeosciences 2018;15:3893–908. [Google Scholar]
- Staal M, Meysman FJ, Stal LJ. Temperature excludes N2-fixing heterocystous cyanobacteria in the tropical oceans. Nature 2003;425:504–7. [DOI] [PubMed] [Google Scholar]
- Stal LJ. Is the distribution of nitrogen-fixing cyanobacteria in the oceans related to temperature?. Environ Microbiol 2009;11:1632–45. [DOI] [PubMed] [Google Scholar]
- Subramaniam A, Yager PL, Carpenter EJ et al. . Amazon River enhances diazotrophy and carbon sequestration in the tropical North Atlantic Ocean. Proc Natl Acad Sci 2008;105:10460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundström BG. Observations on Rhizosolenia clevei Ostenfeld (Bacillariophyceae) and Richelia intracellularis Schmidt (Cyanophyceae). Bot Mar 1984;27:345–55. [Google Scholar]
- Suto I. The explosive diversification of the diatom genus Chaetoceros across the Eocene/Oligocene and Oligocene/Miocene boundaries in the Norwegian Sea. Mar Micropaleontol 2006;58:259–69. [Google Scholar]
- Thiel T, Pratte BS, Zong J et al. . Complete genome sequence of Anabaena variabilis ATCC 29413. Stand Genomic Sci 2014;9:562–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AW, Foster RA, Krupke A et al. . Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 2012;337:1546–50. [DOI] [PubMed] [Google Scholar]
- Tripp HJ, Bench SR, Turk KA et al. . Metabolic streamlining in an open-ocean nitrogen-fixing cyanobacterium. Nature 2010;464:90–4. [DOI] [PubMed] [Google Scholar]
- Uyeda JC, Harmon LJ, Blank CE. A Comprehensive study of cyanobacterial morphological and ecological evolutionary dynamics through deep geologic time. PLoS One 2016;11:1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villareal TA. cycles in the nitrogen-fixing Rhizosolenia (Bacillariophyceae)-Richelia (Nostocaceae) symbiosis. British Phycolo J 1989;24:357–65. [Google Scholar]
- Villareal TA. Marine nitrogen-fixing diatom-cyanobacteria symbioses. : Carpenter EJ, Capone DG, Reuter JG (eds). Marine Pelagic Cyanobacteria: Trichodesmium and other Diazotrophs. Dordecht: Kluwer, 1992, 163–75. [Google Scholar]
- Walsby AE. The permeability of heterocysts to the gases nitrogen and oxygen. Proc R Soc Lond B 1985;226:345–66. [Google Scholar]
- Webb EA, Moffett JW, Waterbury JB. Iron stress in open-ocean cyanobacteria (Synechococcus, Trichodesmium, and Crocosphaera spp.): identification of the IdiA protein. Appl Environ Microbiol 2001;67:5444–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh EA, Liberton M, Stöckel J et al. . The genome of Cyanothece 51142, a unicellular diazotrophic cyanobacterium important in the marine nitrogen cycle. Proc Natl Acad Sci 2008; 105: 15094–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson PA, Norris RD. Warm tropical ocean surface and global anoxia during the mid-Cretaceous period. Nature 2001;412:425–9. [DOI] [PubMed] [Google Scholar]
- Yang Z, Kumar S, Nei M. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 1995;141:1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeev EB, Yogev T, Man-Aharonovich D et al. . Seasonal dynamics of the endosymbiotic, nitrogen-fixing cyanobacterium Richelia intracellularis in the eastern Mediterranean Sea. ISME J 2008;2:911–23. [DOI] [PubMed] [Google Scholar]
- Zehr JP, Bench SA, Mondragon EA et al. . Low genomic diversity in tropical oceanic N2-fixing cyanobacteria. Proc Natl Acad Sci 2007;104:17807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.