Abstract
Sirtuin7 (Sirt7) is a member of the Sir2 histone deacetylase family that functions in a number of physiological processes, including cellular metabolism, ageing and apoptosis. Several studies have indicated that Sirt7 may serve a vital role in promoting the development of cancer. However, to the best of our knowledge, its function in glioma progression has not been demonstrated. The present study revealed that Sirt7 expression was upregulated in human glioma tissues and that the high expression level of Sirt7 was positively associated with glioma malignancy. Further results indicated that the suppression of Sirt7 expression could inhibit the activation of phosphorylated extracellular signal-regulated kinase (p-ERK) concomitantly with decreased expression of cyclin-dependent kinase 2 in glioma cells. Phosphorylation of signal transducer and activator of transcription 3 (STAT3) inhibited when Sirt7 was downregulated by siRNA interference in glioma cell lines. The findings of the present study indicated that Sirt7 affects the malignancy of glioma cells mainly in promoting glioma proliferation and invasion through ERK and STAT3 signaling. Thus, Sirt7 may function as a valuable target for the treatment of human glioma.
Keywords: Sirtuin 7, cell proliferation, cell invasion, extracellular signal-regulated kinase, signal transducer and activator of transcription 3
Introduction
Sirtuins, which are class III histone deacetylases, are oxidized nicotinamide adenine dinucleotide (NAD+)-dependent deacetylases that are implicated in the control of a wide range of biological processes, including apoptosis, stress responses, DNA repair, cell cycling, metabolism and senescence (1,2). In total, seven sirtuins (Sirt1-7), with considerably different functions and catalytic activities, are found in mammals (3). Sirt1 has attracted considerable attention in the last decade as a result of its role in tumorigenesis in the last decade, due to its strong catalytic activity and close similarity to the yeast Sir2 protein (4).
Few studies have investigated the role of Sirtuin7 (Sirt7) in diseases (5). Sirt7 is located mainly in the nucleus, particularly the nucleolus, where it binds to rRNA. Sirt7 activity is required to resume rDNA transcription during mitosis (6). Additionally, previous studies have highlighted the role of Sirt7 in various notable cellular processes, particularly protein synthesis, chromatin remodeling, cellular survival and lipid metabolism (2,7). Of particular interest are reports concerning the high expression level of Sirt7 in certain types of cancer, indicating that Sirt7 could be considered to be an oncogene (5,8,9). However, the role of Sirt7 in glioma remains unclear, prompting this investigation into the biological characteristics of Sirt7 in human glioma.
According to the World Health Organization (WHO) clinical classification, based on the phenotype of glioma (10), >50% of glioma are glioblastoma multiform (GBM, grade IV astrocytoma). However, despite considerable therapeutic efforts, glioblastoma (GBM) remains the most common form of human brain tumor, and is associated with high morbidity, recurrence and mortality rates (11). The overall 5-year survival rate of GBM is <5%, and is even less for elderly patients (12). Thus, to understanding the mechanisms of GBM tumorigenesis, a wide range of scientific studies are required; for example, to identify a the biological marker for targeted therapy so as to decrease the recurrence rate and improve patient quality of life (13). The current study investigated the biological roles of Sirt7 in glioma tumorigenesis.
The regulation of mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK)-dependent pathways serves a role in a number of biological functions, which are particularly relevant in cancer cells, including integrin activation chemoresistance and migration (14). The Ras/Raf ERK1/2 cascade is essential for the induction of matrix metalloproteinase-9 (MMP-9) expression (15).
Signal transducer and activator of transcription 3 (STAT3) serves roles in GBM tumorigenesis and central nervous system development (16). A previous study revealed that STAT3 is activated by growth factors or extensively existed cytokines. Upon tyrosine phosphorylation by receptor-associated tyrosine kinases, STAT3 translocates to the nucleus and regulate gene transcription (17).
The present study examined Sirt7 expression in 15 clinical samples of glioma tissues and the corresponding non-tumor specimens by western blot analysis. Sirt7 was overexpressed in human glioma tissues, and its high expression level is associated with the degree of glioma malignancy. These data indicated that Sirt7 may serve as a sensitive prognostic indicator for glioma patients. In addition, Sirt7 expression was decreased in glioma cell lines using small interfering RNA (siRNA) interference. The results revealed that Sirt7 was involved in cellular proliferation, colony formation and the invasion ability of glioma cells, and that downregulation of Sirt7 decreased the expression of phosphorylated (p)-ERK, cyclin-dependent kinase 2 (CDK2) and STAT3. These results indicated that Sirt7 may be a potential target in the glioma therapy.
Materials and methods
Patient samples
Samples of glioma and matched non-tumor tissues were collected from 15 patients at Affiliated Chenggong Hospital (Xiamen, China). A total of 40% patients were female and 60% were male, the median age was 45 years, ranging from 14–74 years. Patients with malignant peripheral nerve sheath tumors or other unrelated neurologic tumors were excluded. Informed consent was obtained from all patients or family members, and the study protocol was approved by the Ethical Committee of Affiliated Chenggong Hospital. All tissues were snap-frozen and stored in liquid nitrogen prior to use. A further 20 paraffin-embedded samples obtained from Pathology Department of Affiliated Chenggong Hospital. Only samples that were diagnosed with WHO stage II and IV disease were chosen.
Analysis of Sirt7 expression using an online microarray database
Human glioma expression microarray data were downloaded from The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/), and were used to analyze difference in the expression of SIRT7 between normal brain and glioma tissues.
Cell culture and transfection
The U251 cell line was obtained from the China Center for Typical Culture Collection. Professor Xiuwu Bian (The Third Military Medical University, Chongqing, China) kindly provided the glioma CHG5 and SHG44 cell lines. U87 cell lines were purchased from the American Type Culture Collection (Manassas, VA, USA). This cell line has been reported to be misidentified/contaminated. This is not the original glioblastoma cell line established in 1968 at the University of Uppsala; as described in PubMed=27582061 (18). However, this cell line exhibits Sirt7 overexpression and meets our research requirements, it does not interfere with our experimental conclusions. Cells were cultured in DMEM (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% FBS (Gemini Bio Products, West Sacramento, CA, USA) at 37°C in a humidified atmosphere containing 5% CO2.
Owing to transfection efficiency considerations and the background high expression of Sirt7 in U87 and U251 cell lines, these two cell lines were chosen in the knockdown experiment. Two siRNAs targeting at Sirt7 (target sequence: 5′-GTGGACACTGCTTCAGAAA-3′ and 5′-CGAAGCTTTACATCGTGAA-3′) were synthesized by Guangzhou RiboBio Co., Ltd., (Guangzhou, China). Cells were seeded into a 6-well plate at 50–60% confluence, and were then transfected with oligonucleotides at a final concentration of 20 nM with the DharmaFECT reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. After 6 h, the transfection medium was replaced with 2 ml DMEM. The cells were collected for the following assays after 48 h.
Western blot analysis
Cells were harvested and lysed in radioimmunoprecipitation assay lysis buffer (50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40 and 0.5% sodium deoxycholate) The BCA Protein Assay kit was used to determine the protein concentrations according to the manufacturer's instruction (Pierce; Thermo Fisher Scientific, Inc.). Equal amounts of total proteins (25 µg) were subjected to 10% SDS-PAGE. They were then transferred to polyvinylidene difluoride membrane, incubated with 5% BSA (Sangon Biotech, Co, Ltd., Shanghai, China) blocking buffer for 1 h at room temperature and then incubated overnight with primary antibody at 4°C. The primary antibodies used were as follows: Sirt7 (cat. no. 5360), STAT3 (cat. no. 4904), p-STAT3 (cat. no. 9145), CDK2 (cat. no. 2546), MMP-9 (cat. no. 3852) (all from Cell Signaling Technology, Inc., Danvers, MA, USA, and used at a dilution of 1:1,000), and β-actin (cat. no. SC-130300, Santa Cruz Biotechnology, Inc., Dallas, TX, USA). The membranes were then incubated with the horseradish peroxidase-conjugated secondary antibody (Anti-mouse IgG, cat. no. 7076; Anti-rabbit IgG, cat. no. 7076; from Cell Signaling Technology, Inc., Danvers, MA, USA, and used at a dilution of 1:10,000). Immunoreactive bands (cat. no. WBKLS0500; Millipore Immobilon Western, ECL; EMD Millipore, Billerica, MA, USA) were quantified using ImageJ software (v.1.48; National Institutes of Health, Bethesda, MD, USA).
Immunohistochemistry (IHC)
The archived paraffin- embedded tissues were cut to a thickness of 4 µm. Immunostaining was performed using the two-step Elivision Plus kit system (Fuzhou Maixin Biotech Co., Ltd., Fuzhou, China). The sections were dewaxed in xylene, rehydrated with a series of ethanol solutions (100, 95, 80 and 70%), and then boiled in citrate buffer (pH 6.0) for 2 min in an autoclave. Next, 0.3% H2O2 was used to block endogenous peroxidase activity at room temperature for 15 min, and the sections were incubated with normal goat serum (dilution ratio 1:20) for 20 min at room temperature to reduce non-specific binding. Tissue sections were incubated with the Sirt7 antibody (cat. no. 5360s; 1:150 dilution; Santa Cruz Biotechnology, Inc.) for 2 h at room temperature. The secondary antibody was applied using the Envision Detection kit (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA). Slides were stained for 2 min with diaminobenzidine tetrahydrochloride (DAB) and then counterstained 2 min with hematoxylin at room temperature. The stained tumor cells were assessed with a Nikon microscope in 10 independent fields at magnification, ×400.
Cell viability assay
An MTS assay was used to detect cell proliferation. Cells were plated in 96-well plates at 1×104 cells/well and cultured for 4 h before each of the treatment durations (24, 36 and 48 h). A total of 20 µl MTS was added to each well and then incubated for 4 h at 37°C. The absorbance of each sample was measured at 490 nm using a microplate reader.
Colony formation assay
Cells were seeded at a density of 1,000 per well into a 6-well culture dish and incubated at 37°C. Culture medium was replaced with fresh medium every two days. After 3 weeks, cells were washed with PBS and stained with Gimesa stain for 20 min at room temperature (EMD Millipore). Only colonies with >50 cells were counted. A total of 12 random fields were counted under an upright light microscope at magnification, ×200. All experiments were performed in triplicate.
Cell invasion assay
An in vitro cell invasion assay was performed using a 24 well-plate transwell insert (EMD Millipore) coated with Matrigel (EMD Millipore). A total of 0.5 ml of prepared serum-free suspension of transfected cells (24 h after transfection), with a total cell count of 1×105, were added into the upper chamber (8-µm pore size), 0.5 ml cell culture medium containing 10% FBS was added to the lower chamber of the insert. After incubation for 24 h, non-invading cells in the interior of the insert were removed using a cotton swab. The invasive cells on the lower surface of the insert were treated with Gimesa stain (1:9) for 20 min at room temperature and average percentage of positive tumor cells were assayed in 10 independent fields at magnification, ×200 under an upright light microscope for research and clinical use. All experiments were performed in triplicate.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from tissues and cells using Tripure Isolation reagent (Roche Applied Science, Penzberg, Germany), according to the manufacturer's instructions. The Transcription First Strand cDNA Synthesis kit (Roche Applied Science) was used to synthesize cDNA from 1 µg total RNA. The primers sequences used are as follows: SIRT7, forward, CAGGGAGTACGTGCGGGTGT, and reverse, TCGGTCGCCGCTTCCCAGTT; GAPDH, forward, AAGTGAAGCAGGAGGGTGGAA, and reverse, CAGCCTCACCCCATTTGATG. The thermocycling conditions were as follows: Initial denaturation at 94°C for 5 min, followed by 30 cycles at 94°C for 45 sec, 59°C for 45 sec and 72°C for 45 sec, followed by 72°C for 45 sec and finally one cycle at 72°C for 10 min. Experiments were performed in triplicate with SYBR Green I Master mix (Roche Applied Science); GAPDH was used as the internal control. The results were normalized using the 2−ΔΔCq method (19).
Statistical analysis
All quantified data represented an average of at least three samples. Graph Pad Prism v.5.0 (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS v.16 (SPSS, Inc., Chicago, IL, USA) were used for statistical analysis. The data are expressed as the mean ± standard deviation. Comparisons between different groups were performed with Student's t-test and one way analysis of variance followed by a Student-Newman-Keuls post hoc test. P<0.05 was considered to indicate a statistically significant difference. TCGA statistical analysis was performed using the Mann-Whitney U-test test.
Results
Expression of Sirt7 in human glioma tissues
To examine the physiological relevance of Sirt7 in human glioma development, the expression level of Sirt7 was analyzed in clinical patient samples using a Sirt7-specific antibody. The samples were obtained from 15 patients who were classified into different grades according to their pathological diagnoses. The characteristics of the glioma patients are summarized in Table I. As shown in Fig. 1A, Sirt7 was positively expressed in tumors compared with paired adjacent non-tumor tissues. To confirm this finding, RT-qPCR and western blot analysis was performed, which revealed that Sirt7 mRNA was significantly elevated in glioma compared to the normal tissues, as was that of Sirt7 protein (Fig. 1B and C). Besides, to determine whether the levels of Sirt7 expression were associated with the grade of pathological classification, the expression level of Sirt7 was assessed in grade II and grade IV tumors. The results of this analysis revealed that Sirt7 expression was higher in grade IV than grade II gliomas (Fig. 1D).
Table I.
Characteristics of Glioma patients.
| Patient | Age, years | Sex | WHO classification | Pathological diagnosis |
|---|---|---|---|---|
| 1 | 75 | Male | III | Anaplastic astrocytoma |
| 2 | 51 | Female | IV | Glioblastoma |
| 3 | 43 | Male | II | Astroglioma |
| 4 | 38 | Male | IV | Glioblastoma |
| 5 | 14 | Female | IV | Glioblastoma |
| 6 | 41 | Male | II | Oligodendrocyte |
| 7 | 45 | Female | II | Astroglioma |
| 8 | 39 | Male | I | Pilocytic astrocytoma |
| 9 | 55 | Male | I | Pilocytic astrocytoma |
| 10 | 52 | Female | II | Oligodendrocyte |
| 11 | 52 | Male | III | Anaplastic astrocytoma |
| 12 | 44 | Male | IV | Glioblastoma |
| 13 | 58 | Female | IV | Glioblastoma |
| 14 | 53 | Male | II | Astroglioma |
| 15 | 45 | Female | IV | Glioblastoma |
WHO, World Health Organization.
Figure 1.
Sirt7 expression level in human glioma tissues. (A) Analysis of Sirt7 expression in 15 pairs of glioma tissues by western blotting. (B and C) According to the results of clinical diagnosis, the non-glioma examples were excluded. For this reason, there is a break in continuity between bands 14 and 15. Quantitative analysis revealed that Sirt7 was overexpressed in glioma tissues compared with adjacent non-tumor tissues at the protein and mRNA level. (D) The mean level of Sirt7 expression in Grade IV group was significantly higher than that in the Grade II group, presented as scatterplot graphs (*P<0.05). (E) Expression of Sirt7 was significantly higher in glioma tissues than in normal tissues in 703 samples from The Cancer Genome Analysis glioma dataset. (F) Representative immunohistochemistry images depicting the normal expression of Sirt7 in glioma adjacent non-tumor tissue (left) and the overexpression of Sirt7 in glioma tissue (right) vs. control, meanwhile pathologic gradation of glioma was assessed by hematoxylin and eosin staining (magnification, ×100). N, non-tumor; T, tumor; Sirt7, sirtuin 7.
Owing to the limited number of fresh samples, TCGA database was used to analyze the difference in expression between the normal and glioma groups. Data from 698 GBM patients and 5 control group patients were extracted from TGCA database. Expression of Sirt7 was significantly higher in the GBM patients than in the control individuals (Fig. 1E). Additionally, the pathology database was accessed and paraffin specimens (20 specimens, 9 for grade II and 11c for grade IV) were obtained. IHC assays revealed the presence of strong Sirt7 staining in the nuclei of high-grade gliomas (Fig. 1F). These results indicated that Sirt7 may have a role in the development of glioma.
Expression levels of Sirt7 in different immortalized glioma cell lines
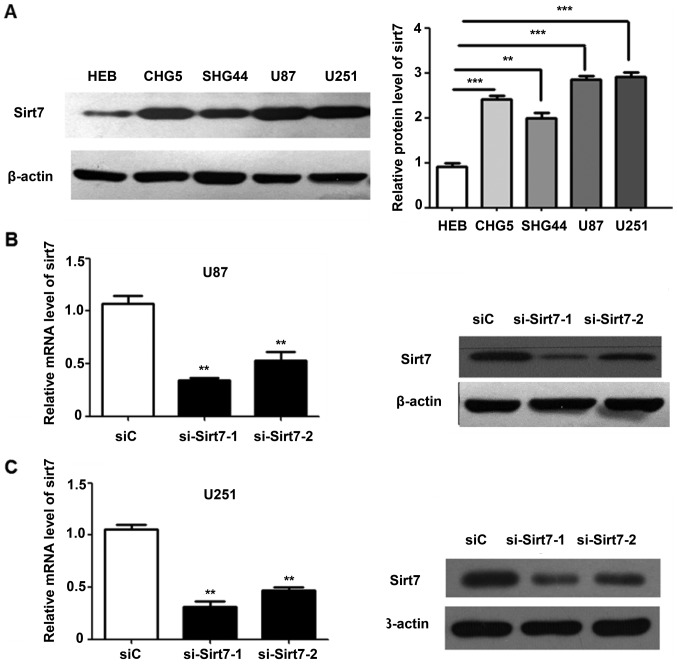
To understand the potential functions of Sirt7 in glioma cells, the expression level of Sirt7 was analyzed in the human glioma CHG5, SHG44, U87 and U251 and one normal glial HEB cell lines (20). The CHG5 and SHG44 cell lines were derived from grade II tissues, whereas U87 and U251 cell lines were obtained from grade IV tissues. Sirt7 expression was higher in glioma cells than in normal glial cells (Fig. 2A). Compared with the low-grade CHG5 and SHG44 cells, U251 and U87 cells expressed an even higher level of Sirt7, in accordance with the results from the aforementioned experiments.
Figure 2.
Sirt7 expression in glioma was suppressed following siRNA transfection. (A) Sirt7 protein level in normal glial HEB cells and glioma CHG5, SHG44, U87 and U251 cell lines (B and C) Western blotting and reverse transcription-quantitative polymerase chain reaction analysis revealed that Sirt7 was efficiently knocked down vs. control by si-Sirt7 treatment in (B) U87 and (C) U251. ***P<0.001, **P<0.01. Sirt7, sirtuin 7; siRNA, small interfering RNA; si-Sirt7, siRNA targeting Sirt7; siC, control siRNA.
As the glioma cell lines in the present study exhibited high Sirt7 expression levels, Sirt7 was not overexpressed. However, two different siRNAs targeting Sirt7 were transfected into cells, and the efficiency of Sirt7 knock down was determined by western blot analysis and RT-qPCR. Compared with the control group, the protein and mRNA expression levels of Sirt7 in the si-Sirt7 group were significantly decreased (Fig. 2B and C).
Downregulation of Sirt7 expression suppresses cell proliferation in vitro
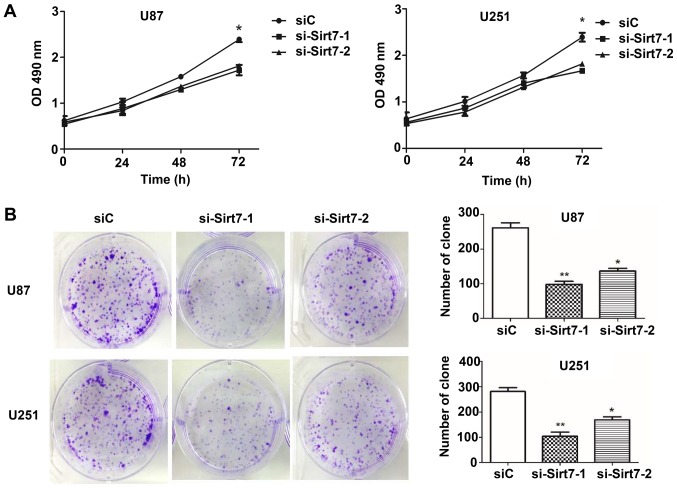
The aforementioned observations prompted the investigation of the biological function of Sirt7 in the tumorigenesis of glioma cells, which was achieved by performing MTS assays to evaluate the effect of Sirt7 on cellular proliferation. At 24 h after siRNA transfection up to 72 h, the proliferation of cells was assessed (Fig. 3A). The results demonstrated that knockdown of Sirt7 reduced the proliferation of cells in the si-Sirt7 groups when compared to the group transfected with the nonsense control siRNA. Whether Sirt7 affected the tumorigenesis of glioma cells was next assessed by detecting the colony formation ability of glioma cells following transfection. The results showed that Sirt7 knockdown significantly reduced the number of colonies formed (Fig. 3B).
Figure 3.
Effects of Sirt7 knockdown on cell proliferation and colony formation ability. (A) MTS assays on U87 and U251 cells following transfection with si-Sirt7 or negative control siRNA. (B) Downregulation of Sirt7 inhibited glioma cell line colony formation ability in U87 and U251 cell lines. **P<0.01, *P<0.05. Sirt7, sirtuin 7; siRNA, small interfering RNA; si-Sirt7, siRNA targeting Sirt7; siC, control siRNA.
Sirt7 knockdown weakens cell invasion
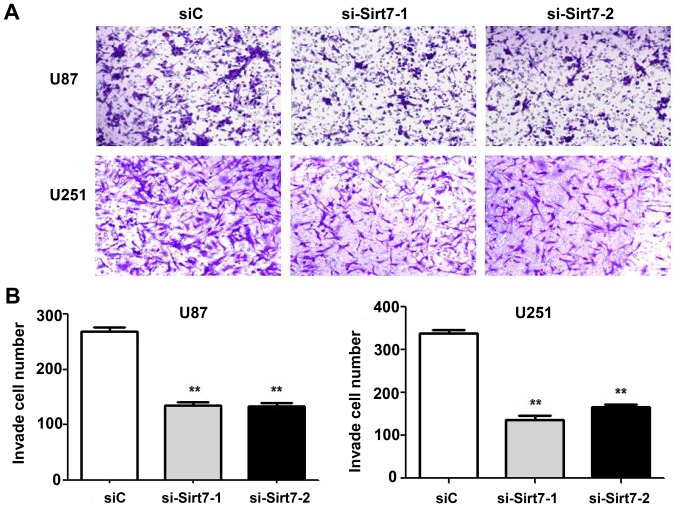
Invasive ability is one of the features that best reflects the malignancy of glioma cells. Thus, the effects of Sirt7 on cell invasion was assessed by Matrigel invasion assays. Sirt7 was knocked down in the U87 and U251 cell lines by transfection with si-Sirt7. The si-Sirt7 group exhibited reduced invasion into the lower chamber compared to the control group (Fig. 4A and B; P<0.01). Depletion of Sirt7 significantly reduced cell invasion, indicating that Sirt7 may participate in the progression of glioma.
Figure 4.
Sirt7 knockdown inhibited cell invasion. (A) Downregulation of Sirt7 following siRNA treatment inhibited cell invasion in U87 and U251 cell lines. (B) Quantification analysis showed that Sirt7 knockdown could suppress cell invasion in U87 and U251 cell lines (**P<0.01). Sirt7, sirtuin 7; siRNA, small interfering RNA; si-Sirt7, siRNA targeting Sirt7; siC, control siRNA.
Sirt7 knockdown reduces the phosphorylation level of ERK and STAT3, and inhibits the expression of MMP9 and CDK2
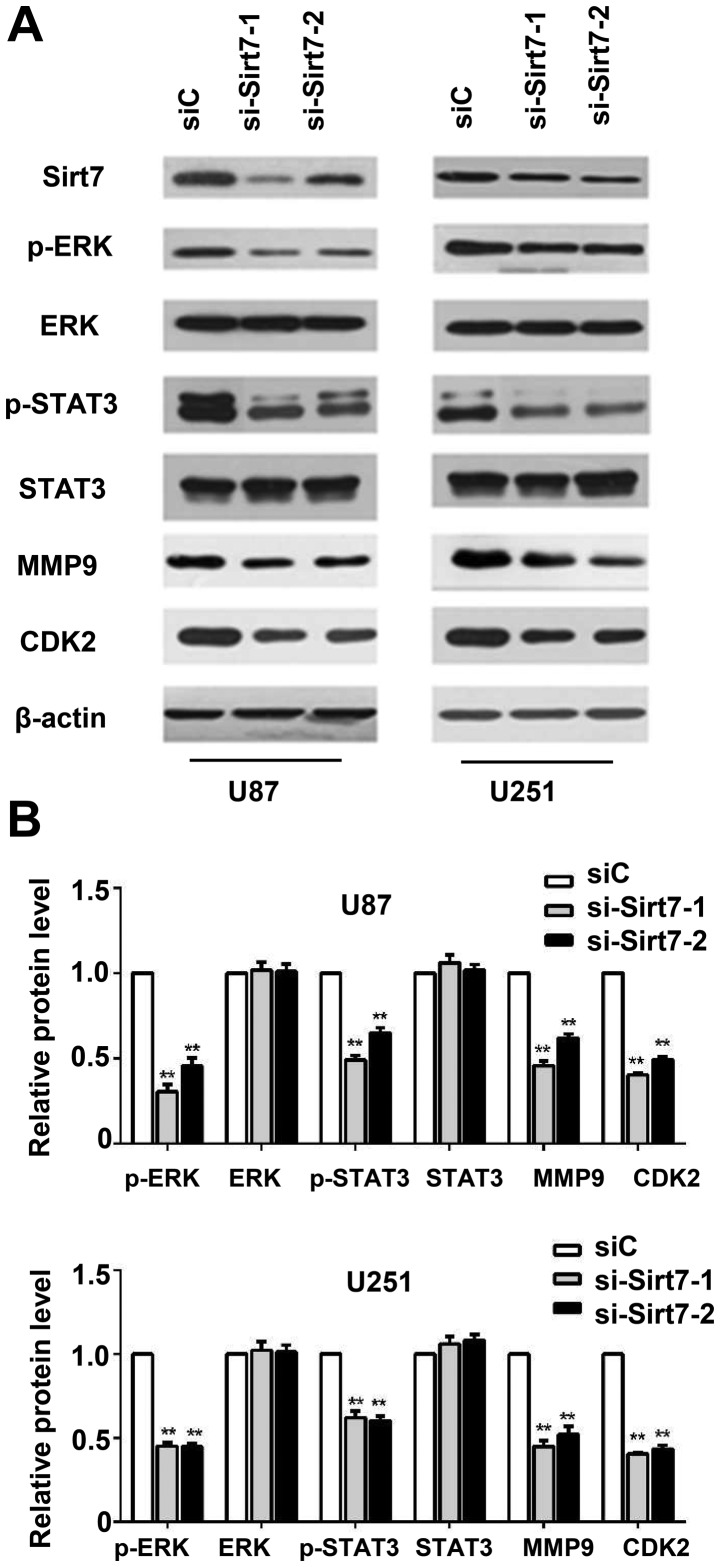
To investigate the molecular mechanism by which downregulation of Sirt7 inhibited glioma cell proliferation and invasion, whether knockdown of Sirt7 influenced ERK signaling activation was examined. The level of p-ERK decreased when Sirt7 expression was suppressed (Fig. 5A and B). Compared with the control group, the si-Sirt7 group exhibited a decrease in the levels of p-STAT3 and MMP9. To determine the function of Sirt7 in cell cycle progression, Sirt7 downregulation was performed, which decreased the expression of CDK2 (Fig. 5A and B), indicating that Sirt7 may participate in cell cycle regulation, which could further influence the glioma tumorigenesis.
Figure 5.
Sirt7 knockdown decreased the phosphorylation of ERK, STAT3, and inhibited expression of MMP9 and CDK2 in U87 and U251 glioma cells. (A) Western blot was used to detect the key signal molecules, with β-actin used as an internal control. (B) Quantification of the results in (A) Data are expressed as the mean ± standard deviation of triplicate experiments. **P<0.01 indicates significantly different from the siC group. ERK, extracellular signal-regulated kinase; STAT3, signal transducer and activator of transcription 3; MMP9, matrix metallopeptidase 9; CDK2, cyclin-dependent kinase 2; siRNA, small interfering RNA; si-Sirt7, siRNA targeting Sirt7; siC, control siRNA.
Discussion
The mammalian sirtuin family has attracted considerable attention as regulators of numerous cellular processes, including metabolism, aging, cell cycle progression and cell survival under stress conditions (4,21). Sirt7, the newest member of the sirtuin family, is a nuclear protein that interacts with RNA polymerase I. Depletion of Sirt7 halts cellular proliferation and triggers apoptosis (22). The dysregulated expression of Sirt7 has been detected in several malignancies, including colorectal cancer, gastric cancer, breast cancer and hepatocellular carcinoma (8,23). Ford et al (22) revealed that Sirt7 is associated with transcriptionally active rRNA genes, by interacting with RNA polymerase I (Pol I) and histones. Knockdown of Sirt7 resulted in decreased association of Pol I with rDNA and a reduction of Pol I transcription. Sirt7 is an NAD1-dependent histone 3 acetyl Lys18 deacetylase that stabilizes the transformed state of cancer cells (24).
However, to the best of our knowledge, no intensive study of the biological functions of Sirt7 in human glioma has been conducted. The present study observed that significantly higher Sirt7 expression levels were observed in glioma tissues than in adjacent non-tumor tissues. Furthermore, the results of the present study demonstrated that Sirt7 upregulation was significantly associated with higher WHO grades, indicating that Sirt7 may participate in the progression of glioma. More notably, in functional experiments, Sirt7 knockdown markedly inhibited cell colony formation and invasion in vitro. Additionally, Sirt7 inactivation suppressed MMP9 and CDK2 expression, indicating that Sirt7 may lead to mitotic stimulation and uncontrolled cell growth during glioma progression. This finding was consistent with the results of a study of human hepatocellular carcinoma tumorigenesis, which indicated that Sirt7-knockdown was associated with G1 phase cell cycle arrest (8). However, further research into the mechanism of suppression is required.
ERKs are required for cell growth, and are reported to exert a diversity of biological functions in differentiation, apoptosis, inflammation, oncogenesis and angiogenesis in numerous tumor types (25–27). ERK signaling is activated in the majority of gliomas (28). In glioma cell lines and stem cell-enriched primary cultures of glioblastoma, constitutive activation of ERK kinases has been observed (29). ERK phosphorylation is a major upstream activator of MMP9 (30). STAT3, which was originally identified to be a mediator of inflammatory response to interleukin-6 (IL-6) (31), is required for brain tumor formation, as well as for maintenance of multipotency in GBM stem cells (32,33). We hypothesized that Sirt7 is important to this activation in two pathways; knockdown of Sirt7 expression decreased the phosphorylation level of ERK and STAT3, which indicated that STAT3 and ERK may be downstream effectors of Sirt7. Nevertheless, the identity of the specific signaling pathway in which Sirt7 is involved remains unclear. Therefore, additional research is required to confirm the findings of the present study.
The results of the present study demonstrated that the targeted knockdown of Sirt7 inhibited glioma cell proliferation and invasion in vitro. Sirt7 selectively modulated the cell cycle and invasion-associated protein synthesis, partly through the ERK/MMP-9 signaling axis. In addition, interference of Sirt7 expression disrupted STAT3 signaling via inhibition of STAT3 phosphorylation, which is required for proliferation and the maintenance of multipotency in glioblastoma stem cells (34). Unfortunately, phosphatase inhibitors were not added to the lysis buffer during preparation of the patient samples; as a result, the phosphorylation status of STAT3 and ERK1/2 in tumor and adjacent normal tissues could not be detected. Finally, a reduction in CDK2 levels was observed in U87 and U251 cell lines when Sirt7 expression was suppressed, indicating that the suppression of Sirt7 may cause cell cycle arrest and inhibit cell growth. Thus, the findings of the present study demonstrated that Sirt7 could represent a potential therapeutic target in glioma. These data identified the potential clinical value of quantifying Sirt7 expression, which may aid assessment of the clinical prognosis. Future studies should investigate the detailed association between STAT3, ERK1/2, and Sirt7. A number of research topics remain to be studied, which include whether activating p-ERK and p-STAT3 directly or indirectly can antagonize the induction of Sirt7-inhibited cell proliferation and invasion.
Acknowledgements
The authors would like to thank Dr. Kathryn Williams (University of Florida, Gainesville, FL, USA) for writing assistance, and Professor Xiuwu Bian (Third Military Medical University, Chongqing, China) for the gift of the glioma cell lines.
Glossary
Abbreviations
- p-ERK
phosphorylated extracellular signal- regulated kinase
- CDK2
cyclin-dependent kinase 2
- STAT3
signal transducer and activator of transcription 3
- siRNA
small interfering RNA
Funding
The present study was supported by grants from the National Natural Science Foundation of China (grant no. 81402064), Science and Technology Project of Xiamen (grant nos. 3502Z20164043, 3502Z20154039 and 3502Z20124019).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
PM, KL, QL, WS and TJ conceived the study, participated in its design and drafted the manuscript. DL participated in the cell experiments. WY and ZL performed sample collection and IHC staining. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The research was approved by the Ethics Committee of the Chinese People's Liberation Army 174 Hospital (Affiliated Chenggong Hospital, Xiamen University). Informed consent was provided by all patients or their family members.
Patient consent for publication
Informed permission for publication was acquired from all patients.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kiran S, Anwar T, Kiran M, Ramakrishna G. Sirtuin 7 in cell proliferation, stress and disease. Rise of the Seventh Sirtuin! Cell Signal. 2015;27:673–682. doi: 10.1016/j.cellsig.2014.11.026. [DOI] [PubMed] [Google Scholar]
- 3.North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haigis MC, Sinclair DA. Mammalian sirtuins: Biological insights and disease relevance. Ann Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang S, Chen P, Huang Z, Hu X, Chen M, Hu S, Hu Y, Cai T. Sirt7 promotes gastric cancer growth and inhibits apoptosis by epigenetically inhibiting miR-34a. Sci Rep. 2015;5 doi: 10.1038/srep09787. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Grob A, Roussel P, Wright JE, McStay B, Hernandez-Verdun D, Sirri V. Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J Cell Sci. 2009;122:489–498. doi: 10.1242/jcs.042382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshizawa T, Karim MF, Sato Y, Senokuchi T, Miyata K, Fukuda T, Go C, Tasaki M, Uchimura K, Kadomatsu T, et al. SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metab. 2014;19:712–721. doi: 10.1016/j.cmet.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 8.Kim JK, Noh JH, Jung KH, Eun JW, Bae HJ, Kim MG, Chang YG, Shen Q, Park WS, Lee JY, et al. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57:1055–1067. doi: 10.1002/hep.26101. [DOI] [PubMed] [Google Scholar]
- 9.Geng Q, Peng H, Chen F, Luo R, Li R. High expression of Sirt7 served as a predictor of adverse outcome in breast cancer. Int J Clin Exp Pathol. 2015;8:1938–1945. [PMC free article] [PubMed] [Google Scholar]
- 10.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. New Eng J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 12.Chen J, McKay RM, Parada LF. Malignant glioma: Lessons from genomics, mouse models, and stem cells. Cell. 2012;149:36–47. doi: 10.1016/j.cell.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: Pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:829–848. doi: 10.1007/s00401-015-1432-1. [DOI] [PubMed] [Google Scholar]
- 14.Eckert A, Böck BC, Tagscherer KE, Haas TL, Grund K, Sykora J, Herold-Mende C, Ehemann V, Hollstein M, Chneiweiss H, et al. The PEA-15/PED protein protects glioblastoma cells from glucose deprivation-induced apoptosis via the ERK/MAP kinase pathway. Oncogene. 2008;27:1155–1166. doi: 10.1038/sj.onc.1210732. [DOI] [PubMed] [Google Scholar]
- 15.Zeigler ME, Chi Y, Schmidt T, Varani J. Role of ERK and JNK pathways in regulating cell motility and matrix metalloproteinase 9 production in growth factor-stimulated human epidermal keratinocytes. J Cell Physiol. 1999;180:271–284. doi: 10.1002/(SICI)1097-4652(199908)180:2<271::AID-JCP15>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 16.Levy DE, Darnell JE., Jr Stats: Transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 17.Iwamaru A, Szymanski S, Iwado E, Aoki H, Yokoyama T, Fokt I, Hess K, Conrad C, Madden T, Sawaya R, et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene. 2007;26:2435–2444. doi: 10.1038/sj.onc.1210031. [DOI] [PubMed] [Google Scholar]
- 18.Allen M, Bjerke M, Edlund H, Nelander S, Westermark B. Origin of the U87MG glioma cell line: Good news and bad news. Sci Transl Med. 2016;8:354re353. doi: 10.1126/scitranslmed.aaf6853. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Dai Z, Wu J, Chen F, Cheng Q, Zhang M, Wang Y, Guo Y, Song T. CXCL5 promotes the proliferation and migration of glioma cells in autocrine- and paracrine-dependent manners. Oncol Rep. 2016;36:3303–3310. doi: 10.3892/or.2016.5155. [DOI] [PubMed] [Google Scholar]
- 21.Liu T, Liu PY, Marshall GM. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res. 2009;69:1702–1705. doi: 10.1158/0008-5472.CAN-08-3365. [DOI] [PubMed] [Google Scholar]
- 22.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075–1080. doi: 10.1101/gad.1399706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashraf N, Zino S, Macintyre A, Kingsmore D, Payne AP, George WD, Shiels PG. Altered sirtuin expression is associated with node-positive breast cancer. Br J Cancer. 2006;95:1056–1061. doi: 10.1038/sj.bjc.6603384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Mansour SJ, Matten WT, Hermann AS, Candia JM, Rong S, Fukasawa K, Vande Woude GF, Ahn NG. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 26.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- 27.Chen D, Zuo D, Luan C, Liu M, Na M, Ran L, Sun Y, Persson A, Englund E, Salford LG, et al. Glioma cell proliferation controlled by ERK activity-dependent surface expression of PDGFRA. PLoS One. 2014;9:e87281. doi: 10.1371/journal.pone.0087281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Guo Q, Wang R, Xu G, Li P, Sun Y, She X, Liu Q, Chen Q, Yu Z, et al. The D Domain of LRRC4 anchors ERK1/2 in the cytoplasm and competitively inhibits MEK/ERK activation in glioma cells. J Hematol Oncol. 2016;9:130. doi: 10.1186/s13045-016-0355-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glassmann A, Reichmann K, Scheffler B, Glas M, Veit N, Probstmeier R. Pharmacological targeting of the constitutively activated MEK/MAPK-dependent signaling pathway in glioma cells inhibits cell proliferation and migration. Int J Oncol. 2011;39:1567–1575. doi: 10.3892/ijo.2011.1165. [DOI] [PubMed] [Google Scholar]
- 30.Lin F, Chengyao X, Qingchang L, Qianze D, Enhua W, Yan W. CRKL promotes lung cancer cell invasion through ERK-MMP9 pathway. Mol Carcinog. 2015;54:E35–E44. doi: 10.1002/mc.22148. [DOI] [PubMed] [Google Scholar]
- 31.Justicia C, Gabriel C, Planas AM. Activation of the JAK/STAT pathway following transient focal cerebral ischemia: Signaling through Jak1 and Stat3 in astrocytes. Glia. 2000;30:253–270. doi: 10.1002/(SICI)1098-1136(200005)30:3<253::AID-GLIA5>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 32.Dasgupta A, Raychaudhuri B, Haqqi T, Prayson R, Van Meir EG, Vogelbaum M, Haque SJ. Stat3 activation is required for the growth of U87 cell-derived tumours in mice. Eur J Cancer. 2009;45:677–684. doi: 10.1016/j.ejca.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu H, Jove R. The STATs of cancer-new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 34.Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem cells. 2009;27:2383–2392. doi: 10.1002/stem.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.