Abstract
Oral tongue squamous cell carcinoma (OTSCC) is the most common type of oral cancer. Despite advances in knowledge regarding the genome-scale gene expression pattern of oral cancer, the molecular portrait of OTSCC biology has remained unclear over the last few decades. Furthermore, studies concerning OTSCC gene-expression profiles are limited or inconsistent owing to tissue heterogeneity in single-cohort studies. Consequently, the present study integrated the profile datasets of three cohorts in order to screen for differentially expressed genes (DEGs), and subsequently identified the potential candidate genes and pathways in OTSCC through gene enrichment analysis and protein-protein interaction (PPI) network construction. Using the selected Gene Expression Omnibus datasets GSE13601, GSE31056 and GSE78060, 206 DEGs (125 upregulated and 81 downregulated) were identified in OTSCC, principally associated with extracellular matrix (ECM) organization and the phosphoinositide 3-kinase/protein kinase B signaling pathway. Furthermore, 146/206 DEGs were filtered into the PPI network and 20 hub genes were sorted. Further results indicated that the two most significant modules filtered from the PPI network were associated with ECM organization and human papillomavirus infection, which are important factors affecting OTSCC pathology. Overall, a set of OTSCC-associated DEGs has been identified, including certain key candidate genes that may be of vital importance for diagnosis, therapy and prevention of this disease.
Keywords: oral tongue squamous cell carcinoma, microarray, protein-protein network, integrative bioinformatics, differentially expressed gene
Introduction
Head and neck squamous cell carcinoma (HNSCC) ranked as the sixth leading incident cancer worldwide in 2012 (1). In contrast with the slightly decreased incidence rate of general HNSCC, the occurrence of oral HNSCC has increased over the last few decades, particularly oral tongue squamous cell carcinoma (OTSCC) (2–5). OTSCC represents malignancies of the oral cavity, with a significantly increasing incidence rate reported among younger individuals from 1975 to 2007 in the USA (6). Although OTSCC cases are considered as oral squamous cell carcinoma (OSCC) or HNSCC, their distinct histological and epidemiological characteristics have been verified (7,8). Owing to the complex lymphatic network and muscular structure of the tongue, patients with OTSCC present a more aggressive phenotype compared with those with tumors affecting other parts of the body, with a higher proportion of lymph node positivity, higher recurrence and metastasis rates post-therapy, and, therefore, poorer prognosis (9,10). However, the molecular mechanisms underlying these variations remain unknown.
Gene detection techniques based on gene expression and sequence variation, including gene microarrays and sequencing, facilitate the gathering of genetic information about numerous cancer types (11–14). A large amount of functional genomic data produced by these high-throughput techniques are archived in public repositories, including the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo). Using these data, integrative analysis or re-analysis can provide valuable clues and new understanding regarding the underlying mechanism (15–17). To date, a considerable number of gene expression profiling studies on OSCC and HNSCC have been completed. However, only a few studies have focused on the transcriptome of OTSCC. The results from these independent studies are inconsistent partly due to sample heterogeneity. In the present study, two OSCC datasets and one OTSCC dataset were obtained from the GEO database and filtered prior to integrative analysis. Differentially expressed gene (DEG) screening, protein-protein interaction (PPI) network construction and gene functional annotation were performed, in order to investigate the distinct gene expression profile of patients with OTSCC.
Materials and methods
Acquisition, preprocessing and DEG screening of microarray data
The gene expression data and probe annotation files GSE13601 (18), GSE31056 (19) and GSE78060 (20) were downloaded from the GEO database for investigation. All of these datasets included microarray data of OTSCC samples. According to their anatomical definition, tongue samples were extracted from the three datasets. Raw microarray data in CEL format were processed with background correction, log2 transformation and quantile normalization using the Robust Multi-array Average (RMA) algorithm (21) in the Affy package (version 1.22.1; www.bioconductor.org/packages/2.4/bioc/html/affy.html) in R (version 3.4.3; www.r-project.org). Subsequently, the DEGs in OTSCC tissues compared with in normal tongue tissues were identified using linear models with the limma package (version 2.18.3; www.bioconductor.org/packages/2.4/bioc/html/limma.html) in R (22,23). |log2FC|≥1 (where FC is fold change) and adjusted P<0.05 were considered as the cut-off values for statistical significance. Furthermore, the intersection of the DEGs among the datasets was calculated, and the result was visualized as a Venn diagram using an online tool (bioinformatics.psb.ugent.be/webtools/Venn).
For validation, the consistency between identified DEGs from the GEO datasets and the data from The Cancer Genome Atlas (TCGA; cancergenome.nih.gov) was assessed and visualized as a Venn diagram. HNSCC gene expression data were downloaded from TCGA and a filter was applied so as to retain only the data of patients with OTSCC. Subsequently, the edgeR package (version 1.2.4; www.bioconductor.org/packages/2.4/bioc/html/edgeR.html) was used to screen for DEGs with the cut-off values of |log2FC|≥1 and adjusted P<0.05 (24).
Functional enrichment analysis
The Database for Annotation, Visualization and Integrated Discovery (DAVID) online tool (version 6.8; david.ncifcrf.gov) was applied to map candidate DEGs onto their associated biological annotation (25,26), with Gene Ontology (GO; www.geneontology.org) (27,28) and pathway analyses using the Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg) (29,30) and the Reactome (www.reactome.org) pathway databases (31,32). Adjusted P<0.05 was considered to indicate statistical significance. All significantly enriched terms were visualized in bubble chart using the ggplot2 package (version 3.1.0; docs.ggplot2.org) in R. The richness factor was calculated as the percentage of the enriched gene number relative to the background gene number for the same term.
PPI network construction
All candidate DEGs were searched in the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (version 10.5; string-db.org) and a combined score >0.4 was used as the criterion to establish the PPI network (33). All the isolated nodes were deleted from the network. The data of the PPI network were exported from the STRING website and imported into Cytoscape (version 3.5.1) software for visualization (34). Each protein in the network served as a node, and the degree and betweenness centrality were calculated using the CentiScaPe (version 2.2) plugin (35,36). The hub gene was defined as the node with a degree >10 within the top 30 betweenness centrality nodes in the present study.
Sub-network analysis
The MCODE plugin (version 1.4.2) (37) was used to identify highly interconnected regions, or clusters, in the PPI network. The degree cut-off was set to 2 and the κ-score was set to 2. The identified clusters with a score >10 were used to create a sub-network. The Cytoscape plugin ClueGO + CluePedia (version 2.5.0) (38,39), which facilitates GO and pathway enrichment analysis in a network, was applied to perform the enrichment analysis and subsequent visualization. The information from the GO and KEGG databases was combined, and the κ-coefficient threshold was set to 0.4. On the basis of the calculations, similar functional terms were marked with the same color.
Two-dimensional hierarchical clustering analysis
According to the anatomical site of samples in GSE31056, OTSCC samples and normal tongue samples were filtered into a subset of GSE31056. Following normalization, gene expression matrices of 206 DEGs from datasets GSE13601, GSE31056 and GSE78060 and the subset of GSE31056 were prepared. Unsupervised clustering was performed on the four matrices using the pheatmap package (CRAN.R-project.org/package=pheatmap) in R.
Univariate survival analysis
In order to distinguish prognostic factors for the outcome of patients with OTSCC, the 206 DEGs were subjected to overall survival (OS) analysis using the univariate Cox regression model. Owing to the unavailability of the clinical information of the samples in datasets GSE13601 and GSE78060, and the limitation in sample size of GSE31056, OTSCC gene expression data and clinical information from TCGA database were used in this analysis. Any causes of mortality were defined as events and survival was defined as a censored event. The OS analysis was performed with the R package Survival (version 2.43–3; CRAN.R-project.org/package=survival), and P<0.05 was considered to be statistically significant.
Results
Identification of DEGs in OTSCC
Following data filtering, the data of 31 OTSCC samples and 26 normal tongue samples from dataset GSE13601 were termed dataset A, 12 OTSCC samples and 39 normal tongue samples from GSE31056 were dataset B, and 26 OTSCC samples and 4 normal tongue samples of GSE78060 were dataset C. Comparing the OTSCC tissues with the normal tongue tissues, a total of 1,562, 2,584 and 1,712 DEGs were identified in datasets A, B and C, respectively. Subsequently, when the DEGs were investigated for overlap, a total of 206 consistently aberrant genes were identified (Fig. 1), comprising 125 upregulated and 81 downregulated genes (Table I). Subsequently, these DEGs were subjected to survival analysis. The results revealed that four genes, NCLN, THBS2, SPARCL1 and YKT6, were associated with the outcome of patients with OTSCC in TCGA (Table II).
Figure 1.
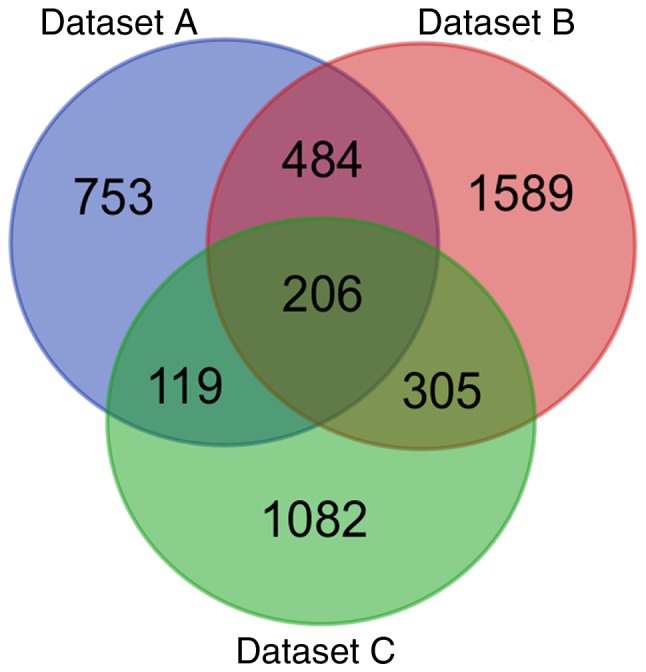
Identification of 206 common DEGs. Cohort profile datasets A, B and C were selected from datasets GSE13601, GSE31056 and GSE78060, respectively, and are indicated in different colors. The DEGs were identified using the edgeR package in R with cut-off criteria of [log2fold change] >1 and adjusted P<0.05. The overlapping areas represent the common genes. DEGs, differentially expressed genes.
Table I.
List of 206 consistently aberrant genes identified from three Gene Expression Omnibus datasets.
| Gene expression | Differentially expressed genes |
|---|---|
| Upregulated (n=125) | IFI27, CDH3, PYGL, MYO1B, MMP1, SCO2, TYMP, BNC1, COL4A1, MMP3, PTHLH, IRF6, F2RL1, COL4A2, IFI6, ACTN1, THBS2, RAB31, SLC16A1, ISG15, PRNP, KRT16, TPBG, MDFI, OSMR, PLAU, SERPINE1, PROCR, PXDN, DUSP7, ITGA6, COL1A2, SOX15, LAMB3, SHC1, NDRG1, LAMC2, ADORA2B, PDLIM4, COL5A2, GJA1, LGALS3BP, MMP13, DFNA5, IL1RAP, PDPN, RGS20, FSCN1, TPST1, STK3, SLC7A5, CTSC, ADAM10, COL7A1, UPP1, PTK7, CA2, ITGA3, GJB3, APOL1, SCG5, EIF6, PLAUR, SOX11, MMP10, COL3A1, TGFBI, MMP12, COL17A1, IRF9, ZWINT, STAT1, BPGM, PCDH7, NUP155, GNA15, POSTN, OAS1, IGFBP3, FAP, COL4A5, TUBB3, DUSP14, FST, TK1, SNAI2, FOXM1, GINS1, TRIP13, HIST1H2AE, IFIT3, PLOD2, DSG2, TGIF1, MYO10, IFI44, IFIT1, CXCL11, PRSS23, RBP1, SQLE, YKT6, KRT10, SNAPC1, BST2, HOMER3, SPP1, ENO1, DLGAP5, KIF23, OASL, COL4A6, RSAD2, CDC20, TNC, F3, FOLR3, EFNA1, PLSCR1, FN1, HIST1H2BD, GNLY, S100A3, LY6E, CCNB1 |
| Downregulated (n=81) | ADH1B, GPRASP1, MEOX2, MYRIP, CBX7, ATP6V0E2, GPR64, C7, RNASE4, ITM2A, SLC25A20, CDO1, CLDN10, MAN2A2, GNG7, SATB1, TXNIP, SERPINA5, LPIN1, ABCA3, SELENBP1, LMO2, GYPC, CXCL12, KAT2B, ZNF529, RTN1, PRELP, ANG, CFD, SSBP2, CCDC69, ENPP4, BEX4, TSPYL5, MYOC, NCLN, SYNGR1, GDF10, P2RY14, CLU, PIP5K1B, ALDH1A1, CILP, MFAP4, FRZB, IGF1, TOX3, ZBTB20, RORC, NR3C2, PTGFR, CPEB3, LGI1, SUSD5, CLGN, GAS2, LCP1, SORBS2, HLF, DPT, CX3CR1, SERPINI1, ACOX2, ASPA, PCK1, MIA, LMOD1, NFIB, SLITRK5, CRISP3, DCLK1, ANGPT1, ABCA6, FAM149A, SPARCL1, NPY1R, PTGDS, AMPD1, FBLN5, STATH |
Table II.
Genes significantly associated with overall survival in oral tongue squamous cell carcinoma.
| Gene | HR | 95% CI | P-value |
|---|---|---|---|
| NCLN | 38.678 | 1.117–6.192 | 0.0047 |
| THBS2 | 2.050 | 0.153–1.283 | 0.0127 |
| SPARCL1 | 3.333 | 0.097–2.310 | 0.0330 |
| YKT6 | 13.765 | 0.179–5.065 | 0.0354 |
HR, hazard ratio; CI, confidence interval.
Gene enrichment and functional annotation analysis of DEGs in OTSCC
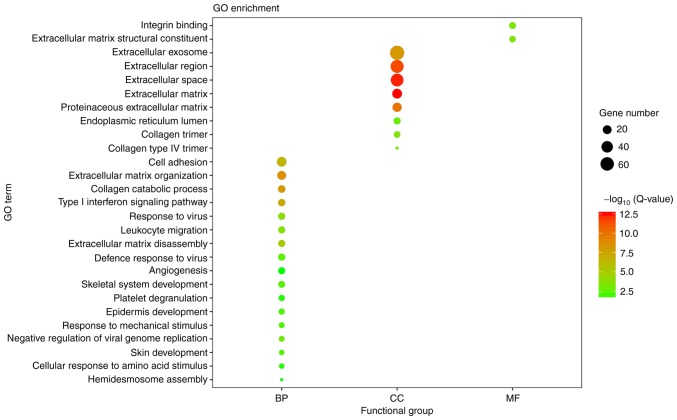
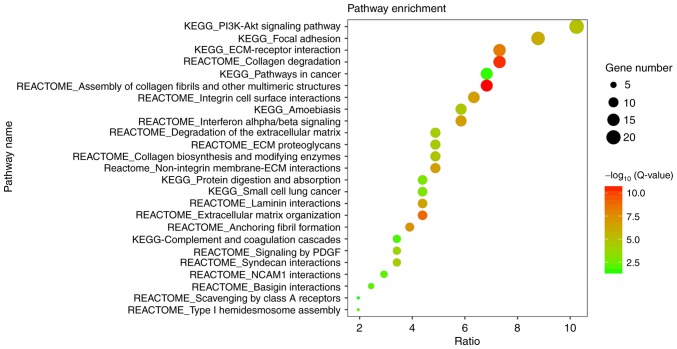
With adjusted P<0.05 as the cut-off criterion, the GO analysis was performed. A total of 206 DEGs were enriched significantly into 27 diverse GO terms, being categorized into three functional groups: Biological process (BP), cellular component (CC) and molecular function (MF) (Fig. 2). Among these terms, extracellular matrix (ECM) organization, extracellular space and ECM structural constituent were the most significant in the BP, CC and MF groups, respectively. Furthermore, the candidate genes were enriched in terms of cell adhesion, response to virus and angiogenesis. Subsequently, the pathway enrichment analysis was performed to assess the aberrant gene-associated pathways. A total of 25 significantly enriched pathways were observed (Fig. 3), a number of which are associated with ECM organization. Overall, the greatest number of genes were involved in the phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt) signaling pathway.
Figure 2.
Visualization of the GO enrichment analysis for 206 differentially expressed genes in oral tongue squamous cell carcinoma. The GO enrichment analysis was performed with the Database for Annotation, Visualization and Integrated Discovery online tool, and the detailed information is presented as a bubble chart. The y-axis represents the GO terms, the x-axis represents the BP, CC and MF functional group categorization, the size of bubbles represents the number of assigned genes, and the color of the bubbles indicates the -log10 (Q-value). The larger the number of genes associated with the term, the larger the bubble. The more significant the GO category, the higher on the color bar the bubble is. GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function; Q-value, Bonferroni-adjusted P-value.
Figure 3.
Visualization of the pathway enrichment analysis for 206 differentially expressed genes in oral tongue squamous cell carcinoma. The pathway enrichment analysis was performed with Database for Annotation, Visualization and Integrated Discovery online tool, and the detailed information is presented as a bubble chart. The y-axis represents the significantly enriched pathways, the x-axis represents the richness factor, the size of the bubbles represents the number of assigned genes, and the color of bubbles represents the -log10 (Q-value). The larger number of genes classified into the pathway, the larger the node size is. The more significant the pathway, the higher on the color bar the bubble is. KEGG, Kyoto Encyclopedia of Genes and Genomes; PI3K, phosphoinositide 3-kinase; Akt, protein kinase B; ECM, extracellular matrix; PDGF, platelet-derived growth factor; Q-value, Bonferroni-adjusted P-value.
PPI network analysis in OTSCC
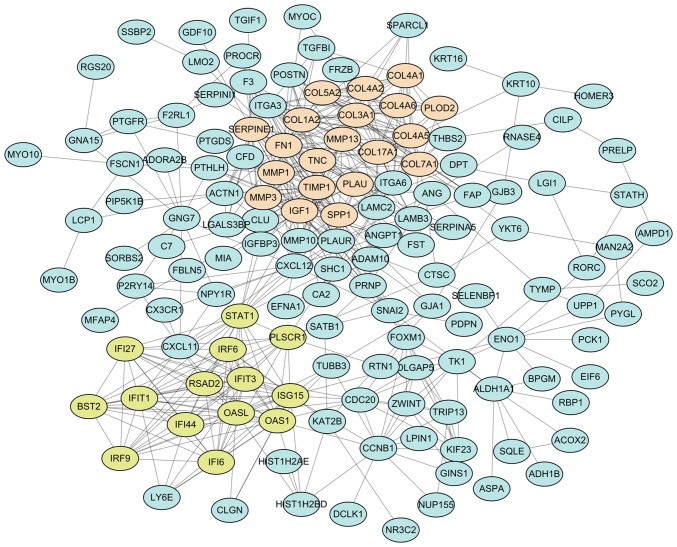
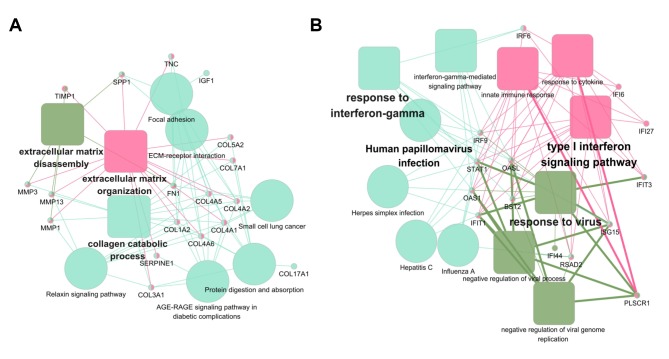
The 206 DEG-encoded proteins were searched in the STRING database, and 206 proteins in Homo sapiens matched the input. Among these, 142 proteins were filtered into the PPI network with 523 edges, whereas the remaining 64 disconnected nodes were hidden (Fig. 4). For the 142 connected nodes, the 20 central nodes were selected with the filtering criterion of degree >10 within the top 30 betweenness centrality nodes. These were FN1, IGF1, TIMP1, ISG15, STAT1, SPP1, COL17A1, SERPINE1, CXCL12, PLAU, MMP1, COL7A1, ITGA6, PLAUR, CCNB1, ACTN1, PLSCR1, CLU, CXCL11 and FOXM1, among which STAT1 and FOXM1 were identified as transcription factors. Subsequently, two modules with score >10 were identified using the MCODE plugin and marked in different colors. Module 1 consisted of 103 edges and 20 nodes in light salmon, and module 2 consisted of 85 edges and 14 nodes in yellow-green. For a further analysis, functional enrichment of these two modules was conducted using ClueGO + CluePedia plugin. The function annotation results demonstrated that module 1 was principally associated with ECM organization (Fig. 5A) and module 2 was principally associated with human papillomavirus (HPV) infection (Fig. 5B).
Figure 4.
PPI network for DEGs in oral tongue squamous cell carcinoma. Using the STRING online database, a total of 142 DEG-encoded proteins were filtered into the PPI network, with the remaining 64 disconnected nodes hidden. Topology analysis was performed using the MCODE plugin and two significant sub-networks (modules), termed 1 and 2, were identified. Nodes in light salmon color represent the DEG-encoded proteins of module 1, and nodes in yellow-green color represent the DEG-encoded proteins of module 2. PPI, protein-protein interaction; DEGs, differentially expressed genes.
Figure 5.
Functional annotation of the two significant modules from the protein-protein interaction network analysis. The function annotation of the two sub-networks was performed using the ClueGO and CluePedia plugins. (A) Module 1 consists of 20 proteins that are principally associated with extracellular matrix organization. (B) Module 2 consists of 14 proteins that are principally associated with human papillomavirus infection. Solid rounded rectangles represent enriched Gene Ontology terms, solid circles represent enriched Kyoto Encyclopedia of Genes and Genomes pathways.
Validation of the distinctive expression profile in OTSCC
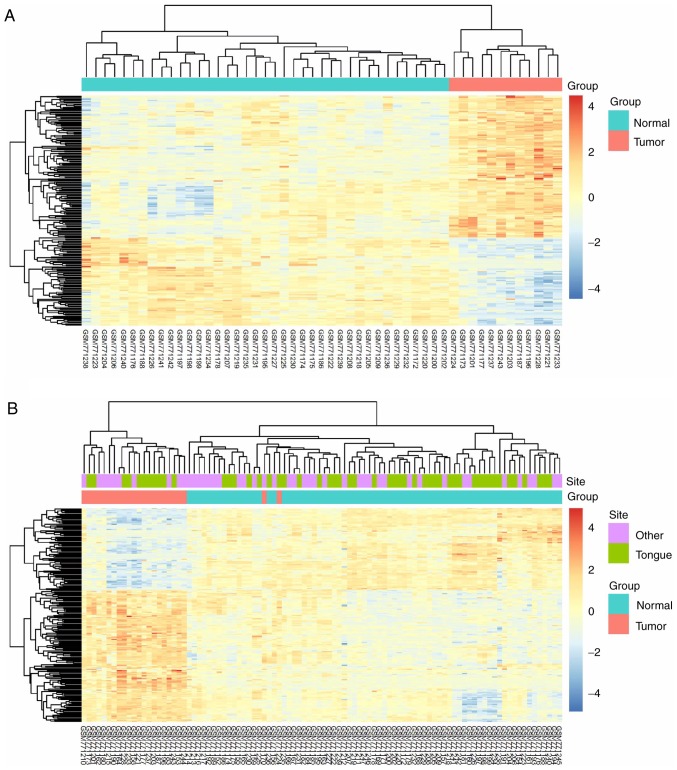
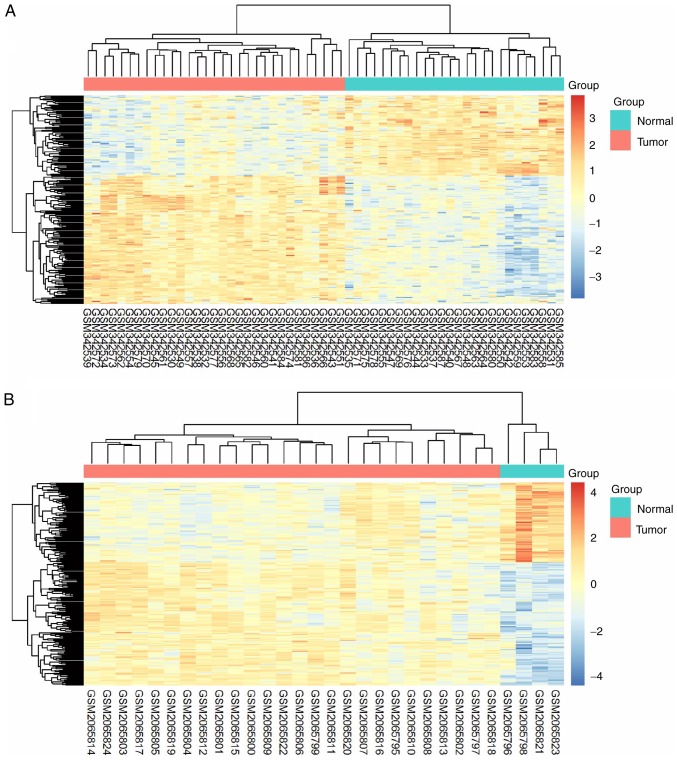
The 206 identified DEGs were subjected to a two-dimensional hierarchical clustering analysis. In dataset GSE31056, distinct clusters of tumor and normal tissues were formed for all tongue samples (Fig. 6A), whereas for all oral samples, two tumor tissues were grouped within the cluster of normal tissues (Fig. 6B). As expected, separate clusters between tongue tumors and normal tongue tissues were also observed for the samples of GSE13601 (Fig. 7A) and GSE78060 (Fig. 7B). These results revealed the differences in gene expression profiles between OSTCC and OSCC. Finally, to confirm the reliability of the identified DEGs, aberrant genes in OTSCC were screened from data of OTSCC and normal tongue tissue from TCGA database in order to investigate the overlap between the data of these two databases. In TCGA OTSCC data, 1,724 downregulated and 792 upregulated genes were identified. Although more DEGs were identified in TCGA data, a total of 119 genes (72 upregulated and 47 downregulated genes) were identified as concordant between the data of the two databases (Fig. 8) and are listed in Table III.
Figure 6.
Expression heat maps of DEGs in different samples of GSE31056. (A) In total, 206 DEGs were identified in oral tongue squamous cell carcinoma samples compared with normal tongue tissues. (B) A total of 206 DEGs in oral squamous cell carcinoma samples compared with normal oral tissues. The rows represent the genes, and the columns represent the samples. The red and blue colors indicate upregulated and downregulated genes, respectively. DEGs, differentially expressed genes.
Figure 7.
Expression heat maps of the 206 identified DEGs in oral tongue squamous cell carcinoma samples compared with normal tongue tissues in datasets (A) GSE13601 and (B) GSE78060. The rows represent the genes, and the columns represent the samples. The red and blue colors indicate upregulated and downregulated genes, respectively. DEGs, differentially expressed genes; OTSCC, oral tongue squamous cell carcinoma.
Figure 8.
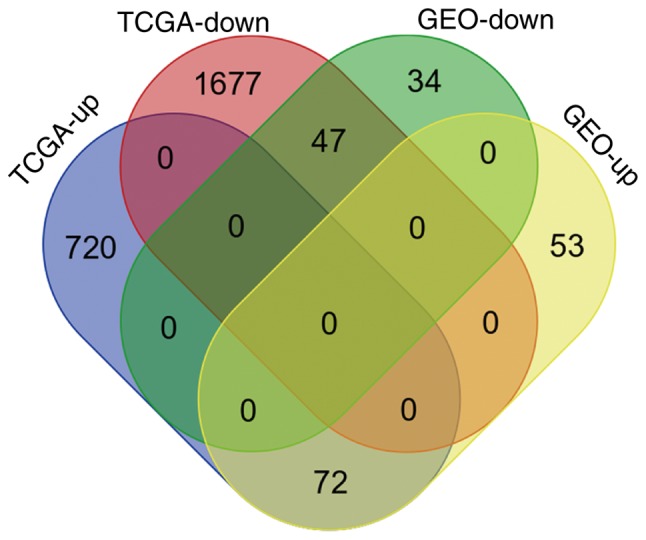
Intersection of differentially expressed genes identified from the GEO microarray datasets and a TCGA RNA sequencing dataset of oral tongue squamous cell carcinoma samples. As indicated in the Venn diagram, the results from the two databases had a total of 119 genes (72 upregulated and 47 downregulated genes) in common. TCGA, The Cancer Genome Atlas; GEO, Gene Expression Omnibus.
Table III.
List of 119 aberrant genes in oral tongue squamous cell carcinoma identified from the Gene Expression Omnibus and the cancer Genome Atlas databases.
| Gene expression | Differentially expressed genes |
|---|---|
| Upregulated (n=72) | COL4A5, CCNB1, SHC1, ITGA3, GINS1, FOXM1, PXDN, TPBG, FN1, IFI27, FST, COL5A2, SPP1, ITGA6, PLOD2, MMP1, MMP12, BNC1, KIF23, GNLY, CDH3, COL4A2, MMP3, POSTN, FSCN1, PLSCR1, DLGAP5, COL4A6, COL4A1, LAMC2, TPST1, ACTN1, COL1A2, PROCR, SLC16A1, FOLR3, IFIT3, MYO1B, PLAU, MMP13, HOMER3, PTHLH, CXCL11, MYO10, PTK7, ADAM10, CDC20, RAB31, OASL, PRNP, TRIP13, DFNA5, ISG15, PDPN, TK1, TNC, FAP, BST2, IFI6, PYGL, IFIT1, THBS2, PRSS23, SERPINE1, RSAD2, SOX11, RBP1, TGFBI, SNAI2, SCG5, IFI44, CTSC |
| Downregulated (n=47) | GDF10, CPEB3, TOX3, HLF, SORBS2, NPY1R, CLDN10, MIA, SSBP2, NR3C2, CBX7, MYOC, SLC25A20, GAS2, GNG7, RORC, PIP5K1B, LPIN1, CX3CR1, ATP6V0E2, SERPINA5, SYNGR1, CFD, RNASE4, SATB1, KAT2B, ENPP4, FAM149A, LMOD1, ASPA, AMPD1, ANG, BEX4, CRISP3, STATH, DPT, PTGDS, NFIB, SLITRK5, ALDH1A1, ITM2A, GPRASP1, ADH1B, MYRIP, FRZB, ACOX2, SELENBP1 |
Discussion
Microarrays have been extensively applied to gene expression studies of human cancer, describing the genetic profiles of the disease. In the present study, gene expression data of multiple cohorts were obtained from the GEO database for the screening of OTSCC-associated genes. Consistent with the results of previous studies on gene expression in OTSCC (40–42) and other carcinomas, including hepatocellular carcinoma, ovarian cancer and nasopharyngeal carcinoma (43–46), the present study revealed numbers of DEGs in the order of 103 in each cohort. However, the majority of previous studies were performed on a single cohort and focused on a single genetic event (40–42). Patient and sample heterogeneity in independent studies is inevitable, and consequently inconsistencies exist among these single cohorts. Furthermore, OTSCC has been classified as OSCC for investigation, thus the distinct gene expression profile underlying OTSCC remains undefined. By integrating multiple cohorts, the combination of integrative bioinformatics methods and expression datasets is an innovative way to solve these problems. Therefore, a multiple-cohort integrative analysis with a relatively stringent sample filtering was applied in the present study. The term OTSCC was used as a query to screen the candidate microarray datasets in the GEO database. First, datasets with ambiguous anatomical information were removed. Secondly, those with <30 tongue samples were removed. Thirdly, certain datasets concerning formalin-fixed paraffin-embedded samples in long-term archives were removed, owing to the poor quality of RNA. Finally, three datasets with a total of 69 tumors and 69 normal tissues of the tongue were included in the present study and an overlap of 206 DEGs was identified. Numerous studies have demonstrated that the anatomical site is one of the factors that influences progression and prognosis of OSCC (10,47). Owing to a rich blood supply and lymphatic drainage, OTSCCs are more likely to metastasize compared with other types of OSCC (48). A number of studies suggested that OTSCC possesses distinct epidemiological characteristics (7,8), and a corresponding distinct gene profile is therefore expected. From the expression data of 206 DEGs in datasets A, B and C, the distribution of the samples in two separate clusters was in agreement with their classification as tumor or normal tissue. Notably, taking all samples of GSE31056 into account, one tongue tumor sample (GSM771224) and one buccal carcinoma sample were classified within the cluster of normal tissues, which might suggest these two samples have distinct gene expression compared with the other OTSCC samples.
GO analysis is a method used to annotate genes and provide evidence-based statements associating them with specific ontology terms (27,28), whereas pathway databases, including KEGG (29,30) and Reactome (31–32), are web resources for understanding high-level functions and interpreting pathway knowledge to support basic and clinical research. In the present study, the samples were analyzed using the DAVID online tool, which integrates a comprehensive set of functional annotation tools including the three aforementioned databases (25,26), in order to decipher the biological functions of the identified DEGs. Regarding GO annotation, the most significantly enriched terms were all associated with the ECM, in agreement with the results of a previous study (41). Certain biological ECM molecules, including fragments of glycosaminoglycan and hyaluronan, are key regulators of injury and inflammatory response during carcinogenesis (49). Certain other ECM proteins, including MMP, regulate cell motility, which may account for the high probability of distant metastasis of OTSCC. Furthermore, the appearance of cell adhesion and angiogenesis terms in OTSCC is reasonable, as these processes are associated with cancer development and metastasis. Regarding the pathway enrichment, the majority of significantly enriched terms were associated with ECM organization, in agreement with the results of the GO enrichment. The majority of detected DEGs were involved in the PI3K/Akt signaling pathway, which is a critical pathway regulating diverse cellular functions, including metabolism, growth, proliferation, survival, transcription and protein synthesis (50). Aberrant Akt signaling is the underlying defect in a number of diseases, including cancer (51). Numerous studies have demonstrated that the PI3K/Akt signaling pathway serves an essential function in the origin and progression of OTSCC (52,53). In addition, Yu et al (54) suggested that the pathway may be a key regulator of radiosensitization in patients with OSCC. Therefore, the result that the PI3K/Akt signaling pathway is affected in OTSCC is reasonable.
In order to delineate complex biological processes, including cancer initiation and progression, it is helpful to consider DEGs in the context of a complex molecular network. The STRING database is an online resource curating known and predicted PPIs for constructing functional protein association networks. Although human PPI maps represent only a fraction of the complete interaction network, their utility in interpreting complex cancer signatures has led to them being more widely used and a valuable aid in research. In the present study, a PPI network consisting of 142 nodes and 523 edges was established. Topology analysis suggested that FN1, IGF1, TIMP1, ISG15, STAT1, SPP1, COL17A1, SERPINE1, CXCL12, PLAU, MMP1, COL7A1, ITGA6, PLAUR, CCNB1, ACTN1, PLSCR1, CLU, CXCL11 and FOXM1 were the hub molecules. Among them, IGF1, FN1, SERPINE1, SPP1, COL17A1, COL7A1, MMP1, TIMP1, ITGA6 and ACTN1 are involved in the regulation of cancer cell adhesion and motility (55–58). Additionally, CCNB1, SERPINE1 and IGF1 are involved in the cellular tumor antigen p53 signaling pathway (59), and COL17A1 has been identified as a novel target of p53 with an inhibitory effect on breast cancer migration and invasion (60). Furthermore, STAT1 and FOXM1 are transcription factors. A previous study suggested that BCL10 promotes OSCC progression through activating STAT1 and ATF4 (61). Yang et al (62) provided evidence that FOXM1 is a mediator of epithelial-mesenchymal transition, facilitating OTSCC migration and invasion. Another study noted the importance of PLAU and PLAUR in complement and coagulation cascades that are linked to immune responses to tumors (63). Therefore, these molecules may represent promising candidates for molecular diagnosis and therapeutic intervention for patients with OTSCC. In further exploration, a sub-network analysis was performed and three representative modules were identified. As expected, an ECM organization-associated module was represented. Notably, the other module was associated with HPV infection, which has been identified as an emerging risk factor for OTSCC (64). Together, the results support the reliability of functional analysis of DEGs, and propose these hub genes as promising candidates for further functional experimentation.
Finally, an analysis of overlap further verified the reliability of the results of the present study. In total, 119/206 were also differentially expressed in TCGA OTSCC samples. Although the DEG lists derived from the two datasets were not identical, the disparity is explicable. First, different detecting platforms may partly account for the differences between the results from the two datasets, as neither RNA sequencing data nor microarray data cover the complete genome. Secondly, the absence of probes in certain datasets may result in fewer identified DEGs in the present analysis. Specifically, HOXD11, CDK1 and CCL15 were identified as DEGs in TCGA data. However, HOXD11, CDK1 and CCL15 are not present on the GSE13601, GSE31056 or GSE78060 arrays, respectively. Furthermore, the different genetic background of individuals and tumor heterogeneity may also be part of the reason.
In conclusion, using multiple cohort profile datasets and integrative bioinformatics analysis, the present study has identified a set of DEGs that may help in better distinguishing OTSCC from normal tongue tissue. The identified gene set may contain candidate molecular targets for disease-specific diagnosis and therapy.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by the National Natural Science Foundation of China (grant nos. 81703005 and 81201730), the Natural Science Foundation of Hunan Province (grant nos. 2017JJ3195, 2018JJ3311 and 12JJ5073), the China Postdoctoral Science Foundation (grant no. 2012M521565), the Research Project of Health and Family Planning Commission of Hunan Province (grant no. B2017098) and the Hunan Provincial Science and Technology Program (grant no. 2010FJ3078).
Availability of data and materials
The datasets analyzed during the current study are available from the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) and The Cancer Genome Atlas (cancergenome.nih.gov).
Authors' contributions
YL, YZ and XZ conceived and designed the study; YL, RW, HW, BZ, SD, QD, MP, XS, JY and RH performed data analysis; YL wrote the manuscript; YL, XZ, HW, and YZ edited the manuscript prior to acceptance. All authors reviewed the manuscript and approved the final version.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Harris SL, Kimple RJ, Hayes DN, Couch ME, Rosenman JG. Never-smokers, never-drinkers: Unique clinical subgroup of young patients with head and neck squamous cell cancers. Head Neck. 2010;32:499–503. doi: 10.1002/hed.21220. [DOI] [PubMed] [Google Scholar]
- 3.Chaturvedi AK, Engels EA, Anderson WF, Gillison ML. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. J Clin Oncol. 2008;26:612–619. doi: 10.1200/JCO.2007.14.1713. [DOI] [PubMed] [Google Scholar]
- 4.Shiboski CH, Schmidt BL, Jordan RC. Tongue and tonsil carcinoma: Increasing trends in the U.S. population ages 20–44 years. Cancer. 2005;103:1843–1849. doi: 10.1002/cncr.20998. [DOI] [PubMed] [Google Scholar]
- 5.Tota JE, Anderson WF, Coffey C, Califano J, Cozen W, Ferris RL, St John M, Cohen EE, Chaturvedi AK. Rising incidence of oral tongue cancer among white men and women in the United States, 1973–2012. Oral Oncol. 2017;67:146–152. doi: 10.1016/j.oraloncology.2017.02.019. [DOI] [PubMed] [Google Scholar]
- 6.Patel SC, Carpenter WR, Tyree S, Couch ME, Weissler M, Hackman T, Hayes DN, Shores C, Chera BS. Increasing incidence of oral tongue squamous cell carcinoma in young white women, age 18 to 44 years. J Clin Oncol. 2011;29:1488–1494. doi: 10.1200/JCO.2010.31.7883. [DOI] [PubMed] [Google Scholar]
- 7.Li R, Koch WM, Fakhry C, Gourin CG. Distinct epidemiologic characteristics of oral tongue cancer patients. Otolaryngol Head Neck Surg. 2013;148:792–796. doi: 10.1177/0194599813477992. [DOI] [PubMed] [Google Scholar]
- 8.Bello IO, Soini Y, Salo T. Prognostic evaluation of oral tongue cancer: Means, markers and perspectives (I) Oral Oncol. 2010;46:630–635. doi: 10.1016/j.oraloncology.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Sano D, Myers JN. Metastasis of squamous cell carcinoma of the oral tongue. Cancer Metastasis Rev. 2007;26:645–662. doi: 10.1007/s10555-007-9082-y. [DOI] [PubMed] [Google Scholar]
- 10.Jansen L, Buttmann-Schweiger N, Listl S, Ressing M, Holleczek B, Katalinic A, Luttmann S, Kraywinkel K, Brenner H. GEKID Cancer Survival Working Group: Differences in incidence and survival of oral cavity and pharyngeal cancers between Germany and the United States depend on the HPV-association of the cancer site. Oral Oncol. 2018;76:8–15. doi: 10.1016/j.oraloncology.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mikhail S, Faltas B, Salem ME, Bekaii-Saab T. Application of next-generation sequencing in gastrointestinal and liver tumors. Cancer Lett. 2016;374:187–191. doi: 10.1016/j.canlet.2016.02.029. [DOI] [PubMed] [Google Scholar]
- 13.Ou J, Wu T, Sijmons R, Ni D, Xu W, Upur H. Prevalence of BRCA1 and BRCA2 germline mutations in breast cancer women of multiple ethnic region in northwest China. J Breast Cancer. 2013;16:50–54. doi: 10.4048/jbc.2013.16.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu F, Zhang Z, Zhang Y, Chen Y, Yang X, Li J, Zhao J. Genetic polymorphisms in the telomere length-related gene ACYP2 are associated with the risk of colorectal cancer in a Chinese Han population. Oncotarget. 2017;8:9849–9857. doi: 10.18632/oncotarget.14219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y, Bao Y, Ma M, Yang W. Identification of key candidate genes and pathways in colorectal cancer by integrated bioinformatical analysis. Int J Mol Sci. 2017;18:722. doi: 10.3390/ijms18040722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao P, Hu W, Wang H, Yu S, Li C, Bai J, Gui S, Zhang Y. Identification of differentially expressed genes in pituitary adenomas by integrating analysis of microarray data. Int J Endocrinol. 2015;2015:164087. doi: 10.1155/2015/164087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li WX, He K, Tang L, Dai SX, Li GH, Lv WW, Guo YC, An SQ, Wu GY, Liu D, Huang JF. Comprehensive tissue-specific gene set enrichment analysis and transcription factor analysis of breast cancer by integrating 14 gene expression datasets. Oncotarget. 2017;8:6775–6786. doi: 10.18632/oncotarget.14286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estilo CL, O-charoenrat P, Talbot S, Socci ND, Carlson DL, Ghossein R, Williams T, Yonekawa Y, Ramanathan Y, Boyle JO, et al. Oral tongue cancer gene expression profiling: Identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer. 2009;9:11. doi: 10.1186/1471-2407-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reis PP, Waldron L, Perez-Ordonez B, Pintilie M, Galloni NN, Xuan Y, Cervigne NK, Warner GC, Makitie AA, Simpson C, et al. A gene signature in histologically normal surgical margins is predictive of oral carcinoma recurrence. BMC Cancer. 2011;11:437. doi: 10.1186/1471-2407-11-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enokida T, Fujii S, Takahashi M, Higuchi Y, Nomura S, Wakasugi T, Yamazaki T, Hayashi R, Ohtsu A, Tahara M. Gene expression profiling to predict recurrence of advanced squamous cell carcinoma of the tongue: Discovery and external validation. Oncotarget. 2017;8:61786–61799. doi: 10.18632/oncotarget.18692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 22.Diboun I, Wernisch L, Orengo CA, Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics. 2006;7:252. doi: 10.1186/1471-2164-7-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Gene Ontology Consortium: Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331–D338. doi: 10.1093/nar/gkw1108. (D1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472–D477. doi: 10.1093/nar/gkt1102. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabregat A, Jupe S, Matthews L, Sidiropoulos K, Gillespie M, Garapati P, Haw R, Jassal B, Korninger F, May B, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. (D1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scardoni G, Petterlini M, Laudanna C. Analyzing biological network parameters with CentiScaPe. Bioinformatics. 2009;25:2857–2859. doi: 10.1093/bioinformatics/btp517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scardoni G, Tosadori G, Faizan M, Spoto F, Fabbri F, Laudanna C. Biological network analysis with CentiScaPe: Centralities and experimental dataset integration. Version 2. F1000Res. 2014;3:139. doi: 10.12688/f1000research.4477.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bindea G, Galon J, Mlecnik B. CluePedia cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: A cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun W, Qiu Z, Huang W, Cao M. Gene expression profiles and protein-protein interaction networks during tongue carcinogenesis in the tumor microenvironment. Mol Med Rep. 2018;17:165–171. doi: 10.3892/mmr.2017.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, Liu J, Fu X, Yang A. Identification of key genes and pathways in tongue squamous cell carcinoma using bioinformatics analysis. Med Sci Monit. 2017;23:5924–5932. doi: 10.12659/MSM.905035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carinci F, Lo Muzio L, Piattelli A, Rubini C, Chiesa F, Ionna F, Palmieri A, Maiorano E, Pastore A, Laino G, et al. Potential markers of tongue tumor progression selected by cDNA microarray. Int J Immunopathol Pharmacol. 2005;18:513–524. doi: 10.1177/039463200501800311. [DOI] [PubMed] [Google Scholar]
- 43.Yin F, Shu L, Liu X, Li T, Peng T, Nan Y, Li S, Zeng X, Qiu X. Microarray-based identification of genes associated with cancer progression and prognosis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2016;35:127. doi: 10.1186/s13046-016-0403-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li S, Li H, Xu Y, Lv X. Identification of candidate biomarkers for epithelial ovarian cancer metastasis using microarray data. Oncol Lett. 2017;14:3967–3974. doi: 10.3892/ol.2017.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duffy MJ. Use of biomarkers in screening for cancer. Adv Exp Med Biol. 2015;867:27–39. doi: 10.1007/978-94-017-7215-0_3. [DOI] [PubMed] [Google Scholar]
- 46.Wen X, Tang X, Li Y, Ren X, He Q, Yang X, Zhang J, Wang Y, Ma J, Liu N. Microarray expression profiling of long non-coding RNAs involved in nasopharyngeal carcinoma metastasis. Int J Mol Sci. 2016;17(pii):E1956. doi: 10.3390/ijms17111956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chi AC, Day TA, Neville BW. Oral cavity and oropharyngeal squamous cell carcinoma-an update. CA Cancer J Clin. 2015;65:401–421. doi: 10.3322/caac.21293. [DOI] [PubMed] [Google Scholar]
- 48.Ho CM, Lam KH, Wei WI, Lau SK, Lam LK. Occult lymph node metastasis in small oral tongue cancers. Head Neck. 1992;14:359–363. doi: 10.1002/hed.2880140504. [DOI] [PubMed] [Google Scholar]
- 49.Misra S, Hascall VC, Markwald RR, Ghatak S. Interactions between hyaluronan and its receptors (CD44, RHAMM) regulate the activities of inflammation and cancer. Front Immunol. 2015;6:201. doi: 10.3389/fimmu.2015.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carnero A, Blanco-Aparicio C, Renner O, Link W, Leal JF. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets. 2008;8:187–198. doi: 10.2174/156800908784293659. [DOI] [PubMed] [Google Scholar]
- 51.Tran NT, Su H, Khodadadi-Jamayran A, Lin S, Zhang L, Zhou D, Pawlik KM, Townes TM, Chen Y, Mulloy JC, Zhao X. The AS-RBM15 lncRNA enhances RBM15 protein translation during megakaryocyte differentiation. EMBO Rep. 2016;17:887–900. doi: 10.15252/embr.201541970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Massarelli E, Liu DD, Lee JJ, El-Naggar AK, Lo Muzio L, Staibano S, De Placido S, Myers JN, Papadimitrakopoulou VA. Akt activation correlates with adverse outcome in tongue cancer. Cancer. 2005;104:2430–2436. doi: 10.1002/cncr.21476. [DOI] [PubMed] [Google Scholar]
- 53.Wang K, Lin C, Wang C, Shao Q, Gao W, Song B, Wang L, Song X, Qu X, Wei F. Silencing Kif2a induces apoptosis in squamous cell carcinoma of the oral tongue through inhibition of the PI3K/Akt signaling pathway. Mol Med Rep. 2014;9:273–278. doi: 10.3892/mmr.2013.1804. [DOI] [PubMed] [Google Scholar]
- 54.Yu CC, Hung SK, Lin HY, Chiou WY, Lee MS, Liao HF, Huang HB, Ho HC, Su YC. Targeting the PI3K/AKT/mTOR signaling pathway as an effectively radiosensitizing strategy for treating human oral squamous cell carcinoma in vitro and in vivo. Oncotarget. 2017;8:68641–68653. doi: 10.18632/oncotarget.19817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Novak M, Leonard MK, Yang XH, Kowluru A, Belkin AM, Kaetzel DM. Metastasis suppressor NME1 regulates melanoma cell morphology, self-adhesion and motility via induction of fibronectin expression. Exp Dermatol. 2015;24:455–461. doi: 10.1111/exd.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu YC, Chang JT, Liao CT, Kang CJ, Huang SF, Chen IH, Huang CC, Huang YC, Chen WH, Tsai CY, et al. OncomiR-196 promotes an invasive phenotype in oral cancer through the NME4-JNK-TIMP1-MMP signaling pathway. Mol Cancer. 2014;13:218. doi: 10.1186/1476-4598-13-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moilanen JM, Löffek S, Kokkonen N, Salo S, Väyrynen JP, Hurskainen T, Manninen A, Riihilä P, Heljasvaara R, Franzke CW, et al. Significant role of collagen XVII and integrin beta4 in migration and invasion of the less aggressive squamous cell carcinoma cells. Sci Rep. 2017;7:45057. doi: 10.1038/srep45057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brooks DL, Schwab LP, Krutilina R, Parke DN, Sethuraman A, Hoogewijs D, Schörg A, Gotwald L, Fan M, Wenger RH, Seagroves TN. ITGA6 is directly regulated by hypoxia-inducible factors and enriches for cancer stem cell activity and invasion in metastatic breast cancer models. Mol Cancer. 2016;15:26. doi: 10.1186/s12943-016-0510-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Joerger AC, Fersht AR. The p53 pathway: Origins, inactivation in cancer, and emerging therapeutic approaches. Annu Rev Biochem. 2016;85:375–404. doi: 10.1146/annurev-biochem-060815-014710. [DOI] [PubMed] [Google Scholar]
- 60.Yodsurang V, Tanikawa C, Miyamoto T, Lo PHY, Hirata M, Matsuda K. Identification of a novel p53 target, COL17A1, that inhibits breast cancer cell migration and invasion. Oncotarget. 2017;8:55790–55803. doi: 10.18632/oncotarget.18433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu TS, Tan CT, Chang CC, Lin BR, Lai WT, Chen ST, Yen-Ping Kuo M, Rau CL, Jaw FS, Chang HH. B-cell lymphoma/leukemia 10 promotes oral cancer progression through STAT1/ATF4/S100P signaling pathway. Oncogene. 2017;36:5440. doi: 10.1038/onc.2017.250. [DOI] [PubMed] [Google Scholar]
- 62.Yang H, Wen L, Wen M, Liu T, Zhao L, Wu B, Yun Y, Liu W, Wang H, Wang Y, Wen N. FoxM1 promotes epithelial-mesenchymal transition, invasion and migration of tongue squamous cell carcinoma cells through a c-Met/AKT-dependent positive feedback loop. Anticancer Drugs. 2018;29:216–226. doi: 10.1097/CAD.0000000000000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li D, Wei P, Peng Z, Huang C, Tang H, Jia Z, Cui J, Le X, Huang S, Xie K. The critical role of dysregulated FOXM1-PLAUR signaling in human colon cancer progression and metastasis. Clin Cancer Res. 2013;19:62–72. doi: 10.1158/1078-0432.CCR-12-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zumsteg ZS, Cook-Wiens G, Yoshida E, Shiao SL, Lee NY, Mita A, Jeon C, Goodman MT, Ho AS. Incidence of oropharyngeal cancer among elderly patients in the united states. JAMA Oncol. 2016;2:1617–1623. doi: 10.1001/jamaoncol.2016.1804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analyzed during the current study are available from the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) and The Cancer Genome Atlas (cancergenome.nih.gov).