Abstract
Colorectal cancer (CRC) is frequently diagnosed at an advanced stage of the disease, the pathogenesis of which is influenced by genetic and epigenetic events. Circulating tumor DNA (ctDNA) is extracellular DNA that is present in a number of bodily fluids, including blood, synovial fluid and cerebrospinal fluid. Compared with performing a tissue biopsy, ctDNA examination presents the advantages of minimal invasion and greater convenience. ctDNA is commonly used to identify actionable genomic alterations, monitor treatment responses, unravel therapeutic resistance and potentially detect disease progression prior to clinical and radiological confirmation. The technique can potentially serve as a non-invasive diagnostic tool in personalized medicine, as it demonstrates prognostic value in the management of patients with CRC. ctDNA detection continues to demonstrate inherent advantages compared with other methods, thus serving an increasingly important role in tumor monitoring and oncotherapy. The aim of the current review was to explore the clinical applications of ctDNA in patients with CRC, including early detection and screening, medication guidance, resistance prediction, and residual lesion and recurrence monitoring. Furthermore, several technical methods for ctDNA detection and analysis are explored, as well as other potential biomarkers.
Keywords: circulating tumor DNA, colorectal cancer, detection methods, clinical applications, biomarkers
1. Introduction
Colorectal cancer (CRC) is one of the leading causes of cancer-associated mortalities worldwide (1). Therefore, effective monitoring and therapy are particularly important in the daily management of the disease.
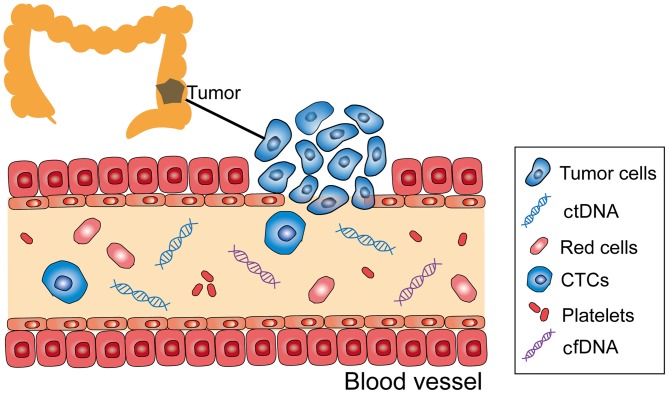
Circulating tumor DNA (ctDNA) is extracellular DNA from cancer cells that have undergone cell death, which is present in a number of bodily fluids, including blood, synovial fluid and cerebrospinal fluid; it is composed of single- or double-stranded DNA (2) (Fig. 1). ctDNA, as the first choice for liquid biopsy, has gradually evolved from research to clinical use, as its testing is non-invasive and reproducible (3), thus, it is a potential tool for detecting gene mutations.
Figure 1.
CTCs, ctDNA and cfDNA in the peripheral blood stream. ctDNA, circulating tumor DNA; CTC, circulating tumor cell; cfDNA, cell-free DNA.
CRC is a highly heterogeneous and complex disease involving various genotypes or subtypes of cells in tumors of the same histological type (4). As such, a single biopsy cannot fully demonstrate the complexity of the intra- and inter-tumor genome landscape. ctDNA presents a number of advantages over tissue biopsy in this aspect.
Traditional oncotherapy includes surgery, radiotherapy and chemotherapy. As each patient with CRC has a unique genetic and epigenetic background, patients with the same clinical and pathological characteristics of tumors may have very different therapy responses and survival rates. This phenomenon has considerably motivated research on personalized treatment. ctDNA may serve as a non-invasive diagnostic tool for individualized medicine, as it provides molecular information similar to that of invasive tumor biopsies (5).
2. Methods for ctDNA detection and analysis
Droplet digital polymerase chain reaction (ddPCR)
ddPCR is one of the most accurate and applicable tools currently available for examining genetic alterations (6). ddPCR is used to detect rare mutations, quantify copy number variations and evaluate microRNA, and the technique can be extended to clinical applications due to its relatively simple workflow. However, it can only be used to screen for known mutations.
Beads, emulsion, amplification and magnetics (BEAMing)
BEAMing is relatively sensitive and inexpensive when assessing a limited number of potential mutations (7). However, although it is fairly accurate, similar to ddPCR, BEAMing can only screen for known mutations. Furthermore, due to the complex workflow, dedicated instrumentation and the high cost of each sample, implementation of this technique in routine clinical settings is limited. Using BEAMing may be effective for detecting RAS mutations in blood (8).
Tagged-amplicon deep sequencing (TAm-Seq)
The main features of TAm-Seq include a high sequencing flux, reduced sequencing time and cost, and the ability to simultaneously sequence millions of DNA molecules, thereby enabling the analysis of the transcripts and genomes of a species in detail. Forshew et al (9) used this method to investigate the tumor protein p53 gene in patients with advanced ovarian cancer and proved the presence of metastatic mutations in multiple primary tumors; in this study, the detection rate of TAm-Seq was >2%, and its sensitivity and specificity were ~97%.
Cancer personalized profiling by deep sequencing (CAPP-Seq)
CAPP-Seq can identify multiple mutations in patients with the same type of cancer and improve the assessment of tumor heterogeneity, thereby providing more comprehensive diagnostic information. Newman et al (10) detected ctDNA by CAPP-Seq in 50% of a patient population with stage I non-small cell lung cancer and 100% of patients with the same type of cancer at stages II–IV. The study identified that CAPP-Seq could detect tumor burdens prior to medical imaging, thus indicating that this technique is valuable in monitoring residual disease. CAPP-Seq can detect all major mutation types, including single nucleotide variants, insertions, rearrangements and copy number changes (11).
Whole genome sequencing (WGS)
Complete rare tumor genome characterization shows great potential in assisting clinical decision-making and identifying unreserved treatment regimens, rare mutations and invisible oncogenes. However, several challenges limit the application of WGS in the clinical setting, including quality assurance, ethical issues and a lack of trained clinicians; it is also costly and time-consuming (12). Kim et al (13) performed WGS in ~250 untreated and 20 recurrent glioblastoma samples, and discovered that a change in the p53 pathway is a major molecular predictor of malignant glioma mutation. The results of the study thus suggested that the genetic changes in primary tumors may affect the subsequent evolution of tumor cells and the emergence of subclonal heterogeneity.
Whole exome sequencing (WES)
WES can be used to sequence the coding region of the human genome, in order to detect common or rare disease-related abnormalities. Lecomte et al (14) used WES to determine familial pancreatic cancer, and detected partner and localizer of BRCA2 as a susceptibility gene. The results of this study indicated that WES shows promising clinical utility in locating potential oncogenes and tumor suppressor genes. For example, it may provide a reference for the molecular diagnosis of tumors. Compared with traditional sequencing technology, its low cost and high yield are major advantages. However, WES currently remains in the early stages of development.
Whole genome bisulfite sequencing (WGBS-Seq)
WGBS-Seq offers single cytosine measurement resolution and high accuracy. Due to these advantages, the technique has become the gold standard in DNA methylation analysis (15). WGBS-Seq has made important contributions to the discovery of partially methylated domains in cancer cells (16). However, the method presents limitations in its operation. In particular, DNA may exist at varying degrees of degradation and the method may exhibit reduced sensitivity during detection. Table I lists the advantages and disadvantages of the various detection methods.
Table I.
Comparison of ctDNA detection methods.
| Method | Details | Detection limit, % | Target mutation | Advantage(s) | Limitation(s) | (Refs.) |
|---|---|---|---|---|---|---|
| ddPCR | Absolute quantification of the initial sample | 0.01–0.10 | Known only | High sensitivity | Only able to detect limited genomic positions in a sample | (52–54) |
| BEAMing | Bead, emulsion, amplification and magnetics | 0.01 | Known only | High sensitivity, relatively inexpensive | Only can detect known mutations | (55,56) |
| TAm-Seq | First sequencing method adapted to detect rare diagnosis mutations in cell-free DNA | 2.00 | Known and new | High sensitivity, relatively inexpensive | Less comprehensive | (9) |
| CAPP-Seq | Targeted hybrid capture | 0.01 | Known and new | High sensitivity | Less comprehensive | (10,57,58) |
| WGS | Deep sequencing of entire genome | 1.00 | Unknown | Interrogating entire genome | Low sensitivity, expensive | (59–61) |
| WES | Deep sequencing of exome | Unknown | Interrogating entire exome | Low sensitivity, expensive | (59,62) | |
| WGBS-Seq | A gold standard in DNA methylation analysis | Unknown | High accuracy and single cytosine measurement resolution | Expensive | (15) |
ctDNA, circulating tumor DNA; ddPCR, droplet digital polymerase chain reaction; BEAMing, bead, emulsion, amplification and magnetics; TAm-Seq, tagged-amplicon deep sequencing; CAPP-Seq, cancer personalized profiling by deep sequencing; WGS, whole genome sequencing; WES, whole exome sequencing; WGBS-Seq, whole genome bisulfite sequencing.
3. Clinical applications of ctDNA in CRC
Early detection and screening
An in vitro study revealed that live tumor cells could release DNA continuously, thus illustrating the feasibility of ctDNA in early-stage cancer (17). Schmiegel et al (18) recently reported that the RAS mutation status in plasma and tissue is consistent in patients with CRC. Underhill et al (19) demonstrated that the integrity of ctDNA could indicate cancer cell death, allowing the use of ctDNA fragment length as a reliable early detection marker. However, other colorectal diseases exhibit similar integrity, and whether this level of integrity could predict early cancer remains unknown. Therefore, further studies on this technique are required. The level of ctDNA in peripheral blood is closely associated with tumor initiation and progression, and patients at later stages of the disease are considered to carry more fragments than those at earlier stages (14). Furthermore, the release rate of ctDNA is associated with the location, size and vascular distribution of tumors (20). Blood tests for extensive population screening require strict specificity. Gao et al (21) indicated that ctDNA is not superior to protein biomarkers (carcinoembryonic antigen, CEA) in terms of sensitivity and specificity. As the most widely used serum markers in CRC, CEA presents no specific diagnostic value, as false-positive and false-negative results may affect early diagnosis. Therefore, using a combination of ctDNA and CEA may be a useful method for the diagnosis of early-stage CRC.
Medication guidance and resistance prediction
Anti-epidermal growth factor receptor (EGFR) monoclonal antibodies, cetuximab and panitumumab, are effective treatments for advanced CRC. These therapies function by directly blocking the EGFR pathway and enhancing the activity of chemotherapy drugs (22). The National Comprehensive Cancer Network (NCCN) 2016 guidelines state that the Kirsten rat sarcoma virus oncogene homolog (KRAS) sequence must be tested prior to administering cetuximab in CRC treatment, and only patients with KRAS wild-type tumors respond with clinical efficacy. The NCCN and the European Society for Medical Oncology suggest avoiding cetuximab and panitumumab in patients with B-raf proto-oncogene (BRAF) mutant cancer. However, even patients with KRAS wild-type tumors could have no response to anti-EGFR therapy, particularly in the event of BRAF and phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit α mutations, the overexpression of human epidermal growth factor receptor 2, MET proto-oncogene and KRAS, and the absence of phosphatase and tensin homolog expression. Tumor heterogeneity may be one of the most important reasons for the observed resistance (23). Relying on tissue biopsy to monitor resistance is unrealistic. These issues indicate the important role of ctDNA in assessing the genomic and subclone mutations repeatedly during treatment. ctDNA could also be used to monitor the clonal evolution and drug resistance of CRC in patients. ctDNA analysis could identify a second resistance mechanism not captured by single lesion biopsy (24), predicting the timing and causes of the treatment failure. Monitoring KRAS mutations in ctDNA can provide clinical guidance for the determination of treatment for patients with CRC (25). These findings indicate that ctDNA has great potential for acquired resistance monitoring.
Residual lesion and recurrence monitoring
Patients with locally advanced rectal cancer generally receive neoadjuvant chemotherapy followed by radical surgery. Sensitive monitoring of neoadjuvant chemoradiotherapy (nCRT) or post-surgical recurrence is important for patients with CRC, as early detection of recurrence is associated with increased survival times (26). Early-stage recurrence is mainly due to incomplete resectional treatments or the existence of unknown metastasis (27). A relapse rate of 50–60% was previously observed in patients with stage III cancer following complete resectional treatments (28). In general, ~80% of the recurrence occurred in the first 2 years after surgery (29,30). Carpinetti et al (31) compared ctDNA levels with clinical, radiological and pathological responses to nCRT, and identified that ctDNA could be used to detect disease recurrence and monitor treatment responses to nCRT, preceding an increase in CEA levels and radiological diagnosis. Diehl et al (32) detected independent gene mutations in patients with CRC and identified 16 cases of postoperative ctDNA in patients with recurrence and 4 cases of non-detectable ctDNA in patients with no lesions. A previous study revealed that, compared with conventional follow-up, ctDNA monitoring could identify the recurrence of CRC progression 10 months earlier than radiological reports (29), as tumor progression is accompanied by an accumulation of mutations (27). Tie et al (33) performed large-scale assays to determine the feasibility of using ctDNA in detecting residual disease among patients who had not been treated with adjuvant chemotherapy; ctDNA was detected postoperatively in 14 (7.9%) of 178 patients, 11 (79%) of whom exhibited recurrence at a median follow-up time of 27 months, whereas recurrence occurred in only 16 (9.8%) of the remaining 164 patients with negative ctDNA. While these findings suggest that ctDNA detection may provide direct evidence of residual disease, Habr-Gama et al (26) came to contrasting conclusions and suggested that whether ctDNA levels are proportional to systemic tumor burden is not clear since no independent method is yet available to detect this burden. Clinical applications of ctDNA detection in patients with CRC are summarized in Table II. Besides CRC, ctDNA detection is widely used in the detection of numerous other types of cancer (Table III). Although ctDNA has great prospects in monitoring the responses of patients post-surgery, a number of obstacles remain, including early risk prediction, real-time tracking of tumor progression and mutation detection of treatment resistance (34).
Table II.
Clinical applications of ctDNA detection in patients with colorectal cancer.
| Author, year | Patients, n | Potential clinical utility | Biomarker(s) | Detection method(s) | (Refs.) |
|---|---|---|---|---|---|
| Church et al, 2014 | 7,941 | Screening | Methylated SEPT9 | qPCR | (63) |
| Diehl et al, 2008 | 18 | Prognosis | APC, KRAS, PIK3CA, TP53 | BEAMing, qPCR | (32) |
| Cabel et al, 2017 | 15 | Immunotherapy monitoring | KRAS, BRAF, EGFR, TP53 | bi-PAP, NGS, ddPCR | (64) |
| Xu et al, 2017 | 32 | Resistance | PIK3CA, KRAS, AKT1 | Sanger sequencing | (65) |
| Vidal et al, 2017 | 115 | Diagnosis and treatment monitoring | KRAS, NRAS | BEAMing | (66) |
| Grasselli et al, 2017 | 146 | Treatment selection | KRAS, NRAS | BEAMing, qPCR | (67) |
| Kakizawa et al, 2017 | 16 | Treatment monitoring | KRAS | ddPCR | (68) |
| Nq et al, 2017 | 44 | Early detection of metastasis | KRAS | Multiplex-PCR amplicon sequencing | (69) |
| Garrigou et al, 2016 | 240 | Monitoring mutations | MettDNA (WIF1, NPY, PENK) | ddPCR | (70) |
| Siravegna et al, 2015 | 100 | Monitoring clonal evolution and resistance | KRAS, MAP2K1, NRAS, MET, FLT3, ERBB2, EGFR | BEAMing, ddPCR, qPCR | (71) |
| Mouliere et al, 2011 | 12 | Detection of tumor weight | KRAR, ACTB | qPCR | (72) |
qPCR, quantitative polymerase chain reaction; BEAMing, bead, emulsion, amplification and magnetics; bi-PAP, bidirectional pyrophosphorolysis-activated polymerization; NGS, next generation sequencing; ddPCR, droplet digital polymerase chain reaction.
Table III.
Use of circulating tumor DNA detection in various cancer types.
| Cancer | Diagnostic biomarker(s) | Sample size, n | Detection method(s) | (Refs.) |
|---|---|---|---|---|
| Head and neck squamous cell carcinoma | TP53 | 6 | ddPCR | (73) |
| Non-small cell lung cancer | EGFR, T790M | 10 | ddPCR | (74) |
| TP53, KRAS, EGFR | 68 | NGS | (75) | |
| EGFR, KRAS, PIK3CA, TP53 | 58 | Targeted sequencing | (76) | |
| Renal cell carcinoma | TP53, VHL, EGFR, NF1, ARID1A | 220 | NGS | (77) |
| MET | Immunohistochemistry, direct DNA sequencing, qPCR | (78) | ||
| Breast cancer | TP53 | 46 | ddPCR | (79) |
| TP53 | 40 | Microfluidic digital PCR | (80) | |
| ESR1, ERBB2, PIK3CA, AKT1 E17K | 96 | mdPCR | (81) |
ddPCR, droplet digital polymerase chain reaction; NGS, next generation sequencing; mdPCR, multiplex digital PCR.
ctDNA and DNA methylation (DNAme)
DNAme is an important epigenetic modification that refers to a reversible and heritable approach for regulating genomic functions; it mainly occurs in 5′-cytosine-guanine-3′ (CpG) dinucleotide sites in the G/C nucleotide-intensive sequences (CpG islands) (35). With further understanding of DNA methylation mechanisms and advances in methylation chip technology, DNAme has demonstrated great value in early tumor screening, prognostic evaluation and chemosensitivity prediction (36–38). Detection of tumor-specific DNAme alterations in ctDNA may assist in monitoring the tumor burden and treatment responses of patients with breast or hepatocellular cancer (39). Differentially methylated regions of ctDNA were tested in healthy subjects and patients with colon cancer, and it was demonstrated that the DNAme detection method exhibits high sensitivity and specificity for CRC (40).
4. Other potential predictive biomarkers
Circulating tumor cells (CTCs)
CTCs refer to various types of tumor cells with the antigenicity and genetic characteristics of a specific tumor type, found in the peripheral blood (41). The CellSearch® CTC test (Menarini Silicon Biosystems, Inc., Huntington Valley, PA, USA) is the only Food and Drug Administration-approved in vitro CTC diagnostic tool with positive counts associated with overall survival (OS) and progression-free survival (PFS) in patients with CRC (42). Lu et al (43) revealed that patients with detectable CTCs following adjuvant chemotherapy have higher recurrence rates than those without. Krebs et al (44) also suggested that patients with elevated CTC blood levels (≥3 cells per 7.5 ml) are more likely to benefit from intensive chemotherapy regimens than those with lower CTC levels, indicating that CTC levels may aid in guiding the adjustment of the treatment protocols of patients with CRC. In addition, CTCs are advantageous in other areas, for example, while complex chromosomal rearrangements, including translocation, cannot be easily detected using ctDNA, it is possible with CTC detection (45). The genetic and phenotypic profiles of CTCs are usually different from those of primary tumors; therefore, these biomarkers may be used to determine the most effective targeted therapy.
Cell-free DNA (cfDNA)
Patients with CRC have higher total cfDNA levels (which include ctDNA) than healthy individuals (46,47), implying that cfDNA levels may distinguish patients with cancer from those without. The level of degradation of cfDNA may be a sensitive indicator of therapeutic effect and disease progression (48,49), similar to the ctDNA. Lin et al (50) confirmed that patients with lower cfDNA levels have improved 5-year OS rates compared with those with higher levels. Kitahara et al (51) evaluated the levels and integrity (as determined by the ratio of long/short fragments) of cfDNA in plasma samples collected from 93 patients with CRC prior to immunotherapy and drew similar conclusions that cfDNA integrity could be a predictive biomarker of immunotherapy efficacy. Higher cfDNA integrity tends to indicate excessive tumor necrosis factors and a high likelihood of tumor rupture; it also predicts immunosuppressive resistance and shorter PFS time in patients with CRC (49).
5. Conclusions and perspectives
With the development of novel molecular targeted agents and the application of individualized treatment, the survival rates of patients with metastatic CRC have significantly improved. There is a close association between targeted therapy and gene detection. Compared with tissue biopsy, ctDNA examination presents the advantages of minimal invasion, convenience and no contact with radioactive material. The analysis of ctDNA can therefore be used to guide immediate management, clarify drug resistance mechanisms and detect minimal residual diseases or recurrence prior to imaging. Tissue biopsy is a powerful method to obtain static information on the cancer, while ctDNA detection is more timely and accurate. The main advantage of ctDNA analysis is its high specificity, as mutations in ctDNA are consistent with those in the tumor DNA. Although ctDNA has great prospects in monitoring the responses of patients post-surgery, a number of obstacles, including early risk prediction, real-time tracking of tumor progression and mutation detection of treatment resistance, have been encountered (34). Additionally, operating procedures during the extraction of cfDNA have not been standardized, with the cost and practicality of the associated technical methods being important issues hindering its wider clinical applications. Despite these shortcomings, ctDNA detection presents inherent advantages over other methods, indicating that it may serve an increasingly important role in tumor monitoring and oncotherapy. As the technique is gradually adopted in clinical applications, a standardized known-marker detection database could be established and a standard list of ctDNA markers across different tumor types could be made available. In addition, more detection kits accompanying diagnostic reagent standards should continue to be issued. In the future, the combination of ctDNA analysis and clinical immunotherapy could be attempted.
Acknowledgements
The authors would like to thank Dr Leilei Zhou, Dr Xia Zhao and Dr Fan Fan of the Research Center of Clinical Oncology (Nanjing, China) for their technical assistance and useful discussions.
Funding
The present study was supported by the Science Foundation of Jiangsu Province (grant no. BE2016795).
Availability of data and materials
Not applicable.
Authors' contributions
HL and CJ carried out the collection and reading of the reference, and HL participated in writing of the manuscript. JW, JN, HS and XX participated in the analysis and interpretation of the references. YD, RL, SD and JF participated in the study design of the review. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Oke S, Martin A. Insights into the role of the intestinal microbiota in colon cancer. Therap Adv Gastroenterol. 2017;10:417–428. doi: 10.1177/1756283X17694832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng F, Su L, Qian C. Circulating tumor DNA: A promising biomarker in the liquid biopsy of cancer. Oncotarget. 2016;7:48832–48841. doi: 10.18632/oncotarget.9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 4.Myint NNM, Verma AM, Fernandez-Garcia D, Sarmah P, Tarpey PS, Al-Aqbi SS, Cai H, Trigg R, West K, Howells LM, et al. Circulating tumor DNA in patients with colorectal adenomas: Assessment of detectability and genetic heterogeneity. Cell Death Dis. 2018;9:894. doi: 10.1038/s41419-018-0934-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger AW, Schwerdel D, Welz H, Marienfeld R, Schmidt SA, Kleger A, Ettrich TJ, Seufferlein T. Treatment monitoring in metastatic colorectal cancer patients by quantification and KRAS genotyping of circulating cell-free DNA. PLoS One. 2017;12:e0174308. doi: 10.1371/journal.pone.0174308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang BO, Xu CW, Shao Y, Wang HT, Wu YF, Song YY, Li XB, Zhang Z, Wang WJ, Li LQ, Cai CL. Comparison of droplet digital PCR and conventional quantitative PCR for measuring EGFR gene mutation. ExpTher Med. 2015;9:1383–1388. doi: 10.3892/etm.2015.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. [PubMed] [Google Scholar]
- 8.Garcia-Foncillas J, Alba E, Aranda E, Díaz-Rubio E, López-López R, Tabernero J, Vivancos A. Incorporating BEAMing technology as a liquid biopsy into clinical practice for the management of colorectal cancer patients: An expert taskforce review. Ann Oncol. 2017;28:2943–2949. doi: 10.1093/annonc/mdx501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, Dawson SJ, Piskorz AM, Jimenez-Linan M, Bentley D, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 10.Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, Liu CL, Neal JW, Wakelee HA, Merritt RE, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seidman AD, Fornier MN, Esteva FJ, Tan L, Kaptain S, Bach A, Panageas KS, Arroyo C, Valero V, Currie V, et al. Weekly trastuzumab and paclitaxel therapy for metastatic breast cancer with analysis of efficacy by HER2 immunophenotype and gene amplification. J Clin Oncol. 2001;19:2587–2595. doi: 10.1200/JCO.2001.19.10.2587. [DOI] [PubMed] [Google Scholar]
- 12.McGuire AL, Caulfield T, Cho MK. Research ethics and the challenge of whole-genome sequencing. Nat Rev Genet. 2008;9:152–156. doi: 10.1038/nrg2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H, Zheng S, Amini SS, Virk SM, Mikkelsen T, Brat DJ, Grimsby J, Sougnez C, Muller F, Hu J, et al. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015;25:316–327. doi: 10.1101/gr.180612.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lecomte T, Berger A, Zinzindohoué F, Micard S, Landi B, Blons H, Beaune P, Cugnenc PH, Laurent-Puig P. Detection of free-circulating tumor-associated DNA in plasma of colorectal cancer patients and its association with prognosis. Int J Cancer. 2002;100:542–548. doi: 10.1002/ijc.10526. [DOI] [PubMed] [Google Scholar]
- 15.Wardenaar R, Liu H, Colot V, Colomé-Tatché M, Johannes F. Evaluation of MeDIP-chip in the context of whole-genome bisulfite sequencing (WGBS-seq) in Arabidopsis. Methods Mol Biol. 2013;1067:203–224. doi: 10.1007/978-1-62703-607-8_13. [DOI] [PubMed] [Google Scholar]
- 16.Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22:246–258. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Vaart M, Pretorius PJ. The origin of circulating free DNA. Clin Chem. 2007;53:2215. doi: 10.1373/clinchem.2007.092734. [DOI] [PubMed] [Google Scholar]
- 18.Schmiegel W, Scott RJ, Dooley S, Lewis W, Meldrum CJ, Pockney P, Draganic B, Smith S, Hewitt C, Philimore H, et al. Blood-based detection of RAS mutations to guide anti-EGFR therapy in colorectal cancer patients: Concordance of results from circulating tumor DNA and tissue-based RAS testing. Mol Oncol. 2017;11:208–219. doi: 10.1002/1878-0261.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Underhill HR, Kitzman JO, Hellwig S, Welker NC, Daza R, Baker DN, Gligorich KM, Rostomily RC, Bronner MP, Shendure J. Fragment length of circulating tumor DNA. PLoS Genet. 2016;12:e1006162. doi: 10.1371/journal.pgen.1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haber DA, Velculescu VE. Blood-based analyses of cancer: Circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014;4:650–661. doi: 10.1158/2159-8290.CD-13-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Y, Zhang K, Xi H, Cai A, Wu X, Cui J, Li J, Qiao Z, Wei B, Chen L. Diagnostic and prognostic value of circulating tumor DNA in gastric cancer: A meta-analysis. Oncotarget. 2017;8:6330–6340. doi: 10.18632/oncotarget.14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sameen S, Barbuti R, Milazzo P, Cerone A, Del Re M, Danesi R. Mathematical modeling of drug resistance due to KRAS mutation in colorectal cancer. J Theor Biol. 2016;389:263–273. doi: 10.1016/j.jtbi.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 23.Zhai Z, Yu X, Yang B, Zhang Y, Zhang L, Li X, Sun H. Colorectal cancer heterogeneity and targeted therapy: Clinical implications, challenges and solutions for treatment resistance. Semin Cell Dev Biol. 2017;64:107–115. doi: 10.1016/j.semcdb.2016.08.033. [DOI] [PubMed] [Google Scholar]
- 24.Tie J, Kinde I, Wang Y, Wong HL, Roebert J, Christie M, Tacey M, Wong R, Singh M, Karapetis CS, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol. 2015;26:1715–1722. doi: 10.1093/annonc/mdv177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takayama Y, Suzuki K, Muto Y, Ichida K, Fukui T, Kakizawa N, Ishikawa H, Watanabe F, Hasegawa F, Saito M, et al. Monitoring circulating tumor DNA revealed dynamic changes in KRAS status in patients with metastatic colorectal cancer. Oncotarget. 2018;9:24398–24413. doi: 10.18632/oncotarget.25309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habr-Gama A, Gama-Rodrigues J, São Julião GP, Proscurshim I, Sabbagh C, Lynn PB, Perez RO. Local recurrence after complete clinical response and watch and wait in rectal cancer after neoadjuvant chemoradiation: Impact of salvage therapy on local disease control. Int J Radiat Oncol Biol Phys. 2014;88:822–828. doi: 10.1016/j.ijrobp.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 27.Zhou J, Chang L, Guan Y, Yang L, Xia X, Cui L, Yi X, Lin G. Application of circulating tumor DNA as a non-invasive tool for monitoring the progression of colorectal cancer. PLoS One. 2016;11:e0159708. doi: 10.1371/journal.pone.0159708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zippi M, De Toma G, Minervini G, Cassieri C, Pica R, Colarusso D, Stock S, Crispino P. Desmoplasia influenced recurrence of disease and mortality in stage III colorectal cancer within five years after surgery and adjuvant therapy. Saudi J Gastroenterol. 2017;23:39–44. doi: 10.4103/1319-3767.199114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinert T, Schøler LV, Thomsen R, Tobiasen H, Vang S, Nordentoft I, Lamy P, Kannerup AS, Mortensen FV, Stribolt K, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut. 2016;65:625–634. doi: 10.1136/gutjnl-2014-308859. [DOI] [PubMed] [Google Scholar]
- 30.Scholefield JH, Steele RJ. British Society For Gastroenterology; Association of Coloproctology for Great Britain and Ireland: Guidelines for follow up after resection of colorectal cancer. Gut. 2002;51(Suppl 5):V3–V5. doi: 10.1136/gut.51.suppl_5.v3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carpinetti P, Donnard E, Bettoni F, Asprino P, Koyama F, Rozanski A, Sabbaga J, Habr-Gama A, Parmigiani RB, Galante PA, et al. The use of personalized biomarkers and liquid biopsies to monitor treatment response and disease recurrence in locally advanced rectal cancer after neoadjuvant chemoradiation. Oncotarget. 2015;6:38360–38371. doi: 10.18632/oncotarget.5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, Silliman N, Tacey M, Wong HL, Christie M, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92. doi: 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pantel K, Alix-Panabières C. Liquid biopsy: Potential and challenges. Mol Oncol. 2016;10:371–373. doi: 10.1016/j.molonc.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadiyska T, Nossikoff A. Stool DNA methylation assays in colorectal cancer screening. World J Gastroenterol. 2015;21:10057–10061. doi: 10.3748/wjg.v21.i35.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuo CC, Shih YL, Su HY, Yan MD, Hsieh CB, Liu CY, Huang WT, Yu MH, Lin YW. Methylation of IRAK3 is a novel prognostic marker in hepatocellular carcinoma. World J Gastroenterol. 2015;21:3960–3969. doi: 10.3748/wjg.v21.i13.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takai D. Aberrant methylation of circulating DNA for prediction of chemo-sensitivity of non-small cell lung cancer. Gan To Kagaku Ryoho. 2010;37:189–193. (In Japanese) [PubMed] [Google Scholar]
- 39.Fackler MJ, Lopez Bujanda Z, Umbricht C, Teo WW, Cho S, Zhang Z, Visvanathan K, Jeter S, Argani P, Wang C, et al. Novel methylated biomarkers and a robust assay to detect circulating tumor DNA in metastatic breast cancer. Cancer Res. 2014;74:2160–2170. doi: 10.1158/0008-5472.CAN-13-3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitchell SM, Ho T, Brown GS, Baker RT, Thomas ML, McEvoy A, Xu ZZ, Ross JP, Lockett TJ, Young GP, et al. Evaluation of methylation biomarkers for detection of circulating tumor DNA and application to colorectal cancer. Genes. 2016;7(pii):E125. doi: 10.3390/genes7120125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Racila E, Euhus D, Weiss AJ, Rao C, McConnell J, Terstappen LW, Uhr JW. Detection and characterization of carcinoma cells in the blood. Proc Natl Acad Sci USA. 1998;95:4589–4594. doi: 10.1073/pnas.95.8.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harouaka R, Kang Z, Zheng SY, Cao L. Circulating tumor cells: Advances in isolation and analysis, and challenges for clinical applications. Pharmacol Ther. 2014;141:209–221. doi: 10.1016/j.pharmthera.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu CY, Tsai HL, Uen YH, Hu HM, Chen CW, Cheng TL, Lin SR, Wang JY. Circulating tumor cells as a surrogate marker for determining clinical outcome to mFOLFOX chemotherapy in patients with stage III colon cancer. Br J Cancer. 2013;108:791–797. doi: 10.1038/bjc.2012.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krebs MG, Renehan AG, Backen A, Gollins S, Chau I, Hasan J, Valle JW, Morris K, Beech J, Ashcroft L, et al. Circulating tumor cell enumeration in a phase II trial of a four-drug regimen in advanced colorectal cancer. Clin Colorectal Cancer. 2015;14:115–122. doi: 10.1016/j.clcc.2014.12.006. e1-e2. [DOI] [PubMed] [Google Scholar]
- 45.Pailler E, Adam J, Barthélémy A, Oulhen M, Auger N, Valent A, Borget I, Planchard D, Taylor M, André F, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol. 2013;31:2273–2281. doi: 10.1200/JCO.2012.44.5932. [DOI] [PubMed] [Google Scholar]
- 46.Lan YT, Chen MH, Fang WL, Hsieh CC, Lin CH, Jhang FY, Yang SH, Lin JK, Chen WS, Jiang JK, et al. Clinical relevance of cell-free DNA in gastrointestinal tract malignancy. Oncotarget. 2017;8:3009–3017. doi: 10.18632/oncotarget.13821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boni L, Cassinotti E, Canziani M, Dionigi G, Rovera F, Dionigi R. Free circulating DNA as possible tumour marker in colorectal cancer. Surg Oncol. 2007;16(Suppl 1):S29–S31. doi: 10.1016/j.suronc.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 48.Yu D, An G, Xu L. Investigation of efficacy evaluation comparison of cfDNA and CEA in colorectal cancer. Clin La. 2016;62:1947–1953. doi: 10.7754/Clin.Lab.2016.160215. [DOI] [PubMed] [Google Scholar]
- 49.Seifert L, Werba G, Tiwari S, Giao Ly NN, Alothman S, Alqunaibit D, Avanzi A, Barilla R, Daley D, Greco SH, et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature. 2016;532:245–249. doi: 10.1038/nature17403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin JK, Lin PC, Lin CH, Jiang JK, Yang SH, Liang WY, Chen WS, Chang SC. Clinical relevance of alterations in quantity and quality of plasma DNA in colorectal cancer patients: Based on the mutation spectra detected in primary tumors. Ann Surg Oncol. 2014;21(Suppl 4):S680–S686. doi: 10.1245/s10434-014-3804-5. [DOI] [PubMed] [Google Scholar]
- 51.Kitahara M, Hazama S, Tsunedomi R, Takenouchi H, Kanekiyo S, Inoue Y, Nakajima M, Tomochika S, Tokuhisa Y, Iida M, et al. Prediction of the efficacy of immunotherapy by measuring the integrity of cell-free DNA in plasma in colorectal cancer. Cancer Sci. 2016;107:1825–1829. doi: 10.1111/cas.13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Isobe K, Hata Y, Tochigi N, Kaburaki K, Kobayashi H, Makino T, Otsuka H, Ishida F, Hirota N, Sano G, et al. Usefulness of nanofluidic digital PCR arrays to quantify T790M mutation in EGFR-mutant lung adenocarcinoma. Cancer Genomics Proteomics. 2015;12:31–37. [PubMed] [Google Scholar]
- 54.Link-Lenczowska D, Pallisgaard N, Cordua S, Zawada M, Czekalska S, Krochmalczyk D, Kanduła Z, Sacha T. A comparison of qPCR and ddPCR used for quantification of the JAK2 V617F allele burden in Ph negative MPNs. Ann Hematol. 2018 Jul 28; doi: 10.1007/s00277-018-3451-1. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci USA. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakamura K. Circulating tumor DNA (ctDNA) detection using BEAMing and its clinical significance. Rinshobyori. Rinsho Byori. 2016;64:400–406. (In Japanese) [PubMed] [Google Scholar]
- 57.Bratman SV, Newman AM, Alizadeh AA, Diehn M. Potential clinical utility of ultrasensitive circulating tumor DNA detection with CAPP-Seq. Expert Rev Mol Diagn. 2015;15:715–719. doi: 10.1586/14737159.2015.1019476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Przybyl J, Chabon JJ, Spans L, Ganjoo KN, Vennam S, Newman AM, Forgó E, Varma S, Zhu S, Debiec-Rychter M, et al. Combination approach for detecting different types of alterations in circulating tumor DNA in leiomyosarcoma. Clin Cancer Res. 2018;24:2688–2699. doi: 10.1158/1078-0432.CCR-17-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heitzer E, Ulz P, Belic J, Gutschi S, Quehenberger F, Fischereder K, Benezeder T, Auer M, Pischler C, Mannweiler S, et al. Tumor-associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5:30. doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Casuscelli J, Weinhold N, Gundem G, Wang L, Zabor EC, Drill E, Wang PI, Nanjangud GJ, Redzematovic A, Nargund AM, et al. Genomic landscape and evolution of metastatic chromophobe renal cell carcinoma. JCI Insight. 2017;2(pii):92688. doi: 10.1172/jci.insight.92688. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dolzhenko E, van Vugt JJFA, Shaw RJ, Bekritsky MA, van Blitterswijk M, Narzisi G, Ajay SS, Rajan V, Lajoie BR, Johnson NH, et al. Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. 2017;27:1895–1903. doi: 10.1101/gr.225672.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hintzsche JD, Robinson WA, Tan AC. A survey of computational tools to analyze and interpret whole exome sequencing data. Int J Genomics. 2016;2016:7983236. doi: 10.1155/2016/7983236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR, Castaños-Vélez E, Blumenstein BA, Rösch T, Osborn N, et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63:317–325. doi: 10.1136/gutjnl-2012-304149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cabel L, Riva F, Servois V, Livartowski A, Daniel C, Rampanou A, Lantz O, Romano E, Milder M, Buecher B, et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: A proof-of-concept study. Ann Oncol. 2017;28:1996–2001. doi: 10.1093/annonc/mdx212. [DOI] [PubMed] [Google Scholar]
- 65.Xu JM, Wang Y, Wang YL, Wang Y, Liu T, Ni M, Li MS, Lin L, Ge FJ, Gong C, et al. PIK3CA mutations contribute to acquired cetuximab resistance in patients with metastatic colorectal cancer. Clin Cancer Res. 2017;23:4602–4616. doi: 10.1158/1078-0432.CCR-16-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vidal J, Muinelo L, Dalmases A, Jones F, Edelstein D, Iglesias M, Orrillo M, Abalo A, Rodríguez C, Brozos E, et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol. 2017;28:1325–1332. doi: 10.1093/annonc/mdx125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grasselli J, Elez E, Caratù G, Matito J, Santos C, Macarulla T, Vidal J, Garcia M, Viéitez JM, Paéz D, et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol. 2017;28:1294–1301. doi: 10.1093/annonc/mdx112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kakizawa N, Suzuki K, Fukui T, Takayama Y, Ichida K, Muto Y, Hasegawa F, Watanabe F, Kikugawa R, Tsujinaka S, et al. Clinical and molecular assessment of regorafenib monotherapy. Oncol Rep. 2017;37:2506–2512. doi: 10.3892/or.2017.5456. [DOI] [PubMed] [Google Scholar]
- 69.Ng SB, Chua C, Ng M, Gan A, Poon PS, Teo M, Fu C, Leow WQ, Lim KH, Chung A, et al. Individualised multiplexed circulating tumour DNA assays for monitoring of tumour presence in patients after colorectal cancer surgery. Sci Rep. 2017;7:40737. doi: 10.1038/srep40737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garrigou S, Perkins G, Garlan F, Normand C, Didelot A, Le Corre D, Peyvandi S, Mulot C, Niarra R, Aucouturier P, et al. A study of hypermethylated circulating tumor DNA as a universal colorectal cancer biomarker. Clin Chem. 2016;62:1129–1139. doi: 10.1373/clinchem.2015.253609. [DOI] [PubMed] [Google Scholar]
- 71.Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, Ponzetti A, Cremolini C, Amatu A, Lauricella C, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21:827. doi: 10.1038/nm.3870. [DOI] [PubMed] [Google Scholar]
- 72.Mouliere F, Robert B, ArnauPeyrotte E, Del Rio M, Ychou M, Molina F, Gongora C, Thierry AR. High fragmentation characterizes tumour-derived circulating DNA. PLoS One. 2011;6:e23418. doi: 10.1371/journal.pone.0023418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Ginkel JH, Huibers MMH, van Es RJJ, de Bree R, Willems SM. Droplet digital PCR for detection and quantification of circulating tumor DNA in plasma of head and neck cancer patients. BMC Cancer. 2017;17:428. doi: 10.1186/s12885-017-3424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Husain H, Melnikova VO, Kosco K, Woodward B, More S, Pingle SC, Weihe E, Park BH, Tewari M, Erlander MG, et al. Monitoring daily dynamics of early tumor response to targeted therapy by detecting circulating tumor DNA in urine. Clin Cancer Res. 2017;23:4716–4723. doi: 10.1158/1078-0432.CCR-17-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Villaflor V, Won B, Nagy R, Banks K, Lanman RB, Talasaz A, Salgia R. Biopsy-free circulating tumor DNA assay identifies actionable mutations in lung cancer. Oncotarget. 2016;7:66880–66891. doi: 10.18632/oncotarget.11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen KZ, Lou F, Yang F, Zhang JB, Ye H, Chen W, Guan T, Zhao MY, Su XX, Shi R, et al. Circulating Tumor DNA Detection in early-stage non-small cell lung cancer patients by targeted sequencing. Sci Rep. 2016;6:31985. doi: 10.1038/srep31985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pal SK, Sonpavde G, Agarwal N, Vogelzang NJ, Srinivas S, Haas NB, Signoretti S, McGregor BA, Jones J, Lanman RB, et al. Evolution of circulating tumor DNA profile from first-line to subsequent therapy in metastatic renal cell carcinoma. Eur Urol. 2017;72:557–564. doi: 10.1016/j.eururo.2017.03.046. [DOI] [PubMed] [Google Scholar]
- 78.Zhao H, Nolley R, Chan AMW, Rankin EB, Peehl DM. Cabozantinib inhibits tumor growth and metastasis of a patient-derived xenograft model of papillary renal cell carcinoma with MET mutation. Cancer Biol Ther. 2017;18:863–871. doi: 10.1080/15384047.2016.1219816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Riva F, Bidard FC, Houy A, Saliou A, Madic J, Rampanou A, Hego C, Milder M, Cottu P, Sablin MP, et al. Patient-specific circulating tumor DNA detection during neoadjuvant chemotherapy in triple-negative breast cancer. Clin Chem. 2017;63:691–699. doi: 10.1373/clinchem.2016.262337. [DOI] [PubMed] [Google Scholar]
- 80.Parkinson CA, Gale D, Piskorz AM, Biggs H, Hodgkin C, Addley H, Freeman S, Moyle P, Sala E, Sayal K, et al. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: A retrospective study. PLoS Med. 2016;13:e1002198. doi: 10.1371/journal.pmed.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hrebien S, O'Leary B, Beaney M, Schiavon G, Fribbens C, Bhambra A, Johnson R, Garcia-Murillas I, Turner N. Reproducibility of digital PCR assays for circulating tumor DNA analysis in advanced breast cancer. PLoS One. 2016;11:e0165023. doi: 10.1371/journal.pone.0165023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.