Abstract
MicroRNAs serve crucial functions in cancer progression by inhibiting the translation of target genes and causing mRNA degradation. However, the underlying regulatory mechanism of Let-7b in osteosarcoma (OS) has not, to the best of our knowledge, been comprehensively elucidated. The aim of the present study was to investigate the function of Let-7b in OS and clarify the regulation of insulin-like growth factor 1 receptor (IGF1R) by Let-7b. It was observed that Let-7b was significantly downregulated in OS tissues and cell lines compared with the matched adjacent non-tumorous tissues and human normal osteoblastic cell line hFOB 1.19. Overexpression of Let-7b significantly inhibited the proliferation and invasion of U2OS and SAOS-2 cells. A luciferase reporter assay validated that IGF1R was a downstream and functional target of Let-7b. Let-7b was also able to decrease the expression levels of IGF1R protein. Functional studies revealed that the antitumor effect of Let-7b was probably due to targeting and suppressing IGF1R expression. Furthermore, in OS tissues, IGF1R was identified to be significantly upregulated and negatively correlated with Let-7b levels. In conclusion, the results of the present study indicated that Let-7b suppresses OS cellular proliferation and invasion via targeting IGF1R. A novel candidate prognostic factor was identified and it is suggested that the Let-7b/IGF1R axis may represent a novel anti-metastasis therapeutic target in OS.
Keywords: Let-7b, insulin-like growth factor 1 receptor, osteosarcoma, proliferation, invasion
Introduction
Osteosarcoma (OS) is one of the most common primary malignant tumors of bone and the third most frequent cancer in young adolescents and children (1). In the last several decades, the treatments for OS have included surgical resection, radiotherapy and combinatorial chemotherapy, and, although the survival rates of patients with OS have improved, aggressive invasion into local tissue and a markedly high metastatic rate remain the primary causes of tumor-associated mortality (2,3). Therefore, identification of novel therapeutic approaches against specific targets to systematically combat OS is an urgent requirement (4).
MicroRNAs (miRNAs) are a group of small non-coding RNA molecules between 19 and 25 nucleotides in length that have highly conserved sequences in plants and animals (5,6). By binding to the 3′-untranslated region (UTR) of specific genes to degrade mRNA or inhibit translation, miRNAs act as powerful post-transcriptional regulators of gene expression and have important functions in multicellular organisms (7). In various types of cancer, including OS, miRNAs function as tumor suppressors or oncogenes as identified in a number of studies (8–11). Among the non-coding RNAs, the Let7 miRNA family was the first known human miRNA, which consists of Let7a/b/c/d/e/f/g/i and miR-98 (12,13). Let7 is involved in embryonic development and participates in the proliferation and invasion of cancer cells and cancer stem cells (14,15).
However, the function of Let-7b in OS is not fully understood. Preliminary findings (Zhang et al, unpublished data) suggested that decreased expression of Let-7b was observed in OS cells; therefore, the aim of the present study was to investigate the potential functions and mechanisms of Let-7b in OS cells.
Materials and methods
Cell culture and transfection
The U2OS, MG63 and SAOS-2 human OS cell lines and the human normal osteoblastic hFOB 1.19 cell line were purchased from the American Type Culture Collection (Manassas, VA, USA). All cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 U/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.). All cells were incubated at 37°C in a humidified incubator containing 5% CO2. All transfections were performed using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol.
Tissue collection
A total of 60 primary OS tissues and their matched adjacent non-tumor tissues were collected by surgical resection from patients who presented at the Department of Orthopedics, Linyi Central Hospital (Linyi, China) between March 2010 and December 2012. The study was approved by the Ethics Committee of Linyi Central Hospital. All patients included in the present study had not received any other therapeutic treatment prior to surgery. Written informed consent was obtained from each patient. The tissues were immediately snap-frozen in liquid nitrogen following surgical resection and stored at −80°C in liquid nitrogen until use. The clinical characteristics of the patients are summarized in Table I. The Tumor-Node-Metastasis staging was performed according to Fukuma et al (16).
Table I.
Clinicopathological characteristics of the 60 patients with osteosarcoma.
| Let-7b expression | ||||
|---|---|---|---|---|
| Characteristic | n | Low (n=36) | High (n=24) | P-value |
| Sex | 0.830 | |||
| Male | 36 | 22 | 14 | |
| Female | 24 | 14 | 10 | |
| Age, years | 0.863 | |||
| <18 | 36 | 24 | 12 | |
| ≥18 | 34 | 22 | 12 | |
| Tumor size, cm | 0.045 | |||
| ≥5 | 32 | 23 | 9 | |
| <5 | 28 | 13 | 15 | |
| TNM stage | 0.001 | |||
| IIA | 29 | 11 | 18 | |
| IIB/III | 31 | 25 | 6 | |
| Lung metastasis | 0.020 | |||
| Yes | 31 | 23 | 8 | |
| No | 29 | 13 | 16 | |
TNM, Tumor-Node-Metastasis.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA from the tissues and cell lines were extracted using TRIzol® reagent (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. A total of 2 µg RNA was reverse-transcribed into cDNA using a PrimeScript First Strand cDNA Synthesis kit (Takara Bio, Inc., Otsu, Japan), according to the manufacturer's protocol. qPCR was performed on an ABI 7500 Real-time PCR detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The reaction conditions were 95°C for 10 min, and 35 cycles of 95°C for 30 sec and 60°C for 1 min. GADPH was used as the internal reference gene. To analyze the levels of miRNA expression, qPCR was performed using an All-in-One™ miRNA qRT-PCR kit (GeneCopoeia, Inc., Rockville, MD, USA), according to the manufacturer's protocol. The U6 gene was used as an internal control. The relative mRNA level was calculated using the 2−∆∆Cq method (17). The primers used were as follows: Let7b forward, 5′-TGAGGTAGTAGGTTGTGTGGTT-3′; and reverse, 5′-GCTGTCAACGATACGCTACCTA-3′; GAPDH forward, 5′-CCGTCTAGAAAAACCTGCC-3′; and reverse, 5′-GCCAAATTCGTTGTCATACC-3′; U6, forward 5′-CTCGCTTCGGCAGCACA-3′; and reverse 5′-AACGCTTCACGAATTTGCGT-3′ (all GeneCopoeia, Inc.).
Western blot analysis
Proteins were extracted by lysing cells using radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) 72 h after transfection. The protein concentration was quantified using a bicinchoninic acid assay kit (Pierce; Thermo Fisher Scientific, Inc.). Equal amounts of proteins (30 µg) were subjected to SDS-PAGE (10% gel) and transferred onto polyvinylidene difluoride membranes (Thermo Fisher Scientific, Inc.). The membranes were blocked with 5% non-fat milk powder in PBS for 1 h at room temperature and was subsequently incubated with primary rabbit polyclonal anti-IGF1R antibody (1:1,000 dilution; cat. no. ab39675; Abcam, Cambridge, UK) and mouse monoclonal anti-human GADPH antibody (1:1,000 dilution; cat. no. sc-59540; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) at 4°C overnight. Following washing with Tris-buffered saline containing Tween-20 (0.5%) five times for 5 min each at room temperature, the membranes were incubated with the corresponding horseradish peroxidase-conjugated secondary antibodies goat anti-mouse IgG (cat. no. sc-2005) and anti-rabbit IgG (cat. no. sc-2004) (both 1:3,000; Santa Cruz Biotechnology, Inc.) at room temperature for 1 h. The signals detected were visualized using an enhanced chemiluminescence kit (Pierce; Thermo Fisher Scientific, Inc.). Densitometric evaluation of the proteins was performed using ImageJ (version 1.38; National Institutes of Health, Bethesda, MD, USA).
Flow cytometry assay
U2OS or SAOS-2 cells were seeded at 3×105 per well in 6-well plates and cultured to 85% confluence to analyze the cell cycle. Each group of cells was washed five times with PBS at room temperature, then collected by centrifugation at 1,000 × g for 5 min. All cells were resuspended in 500 µl PBS, and 5 µl Annexin V-FITC and 10 µl PI (both Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) were added at room temperature for 15 min. The flow cytometry assay was performed and analyzed by the FACSAria cell sorter within 1 h of incubation using the FACSuite software version 1.0 (both BD Biosciences, San Jose, CA, USA).
Cell proliferation assay
MTT assays (Sigma-Aldrich; Merck KGaA) were performed to determine the proliferation of cells, according to the manufacturer's protocol. To assess the proliferation rates, 1×106 cells in each group were seeded in 24-well plates and transfected with Let-7b miRNA mimic alongside a negative control (NC) (Let-7b mimic sequence, 5′-UGAGGUAGUAGGUUGUGUGGUU-3′; NC sequence 5′-UUCUCCGAACGUGUCACGUTT-3′) at a final concentration of 20 nM. After 24 h of transfection at 37°C, the cells were collected and 3,000 cells/well were seeded in 96-well plates and cultured for 0, 24, 48, 72 or 96 h. A total of 20 µl MTT solution (5 mg/ml) was added to each well and the cells were incubated for a further 4 h at 37°C. Next, the supernatant was removed and 200 µl dimethylsulfoxide was added to each well for 10 min at 37°C to dissolve the formazan product. The absorbance was determined using a Model 680 microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA) at a wavelength of 570 nm.
Invasion assay
Transwell assays were used to evaluate the invasive ability of OS cells. The Transwell chambers (8 µm pore size; Costar; Corning Incorporated, Corning, NY, USA) were coated with 60 µl Matrigel (BD Biosciences, San Jose, CA, USA), according to the manufacturer's protocol. The cells were collected 48 h after transfection and 2.5×105 cells per well were resuspended in 200 µl DMEM without FBS and then added into the upper chambers. The lower chambers were filled with 800 µl DMEM supplemented with 20% FBS. Following incubation for 24 h at 37°C, cells on the upper surface of the membrane were carefully removed with cotton swabs. The invading cells on the bottom of the membrane were fixed with 100% methanol for 10 min and stained with 0.5% crystal violet for 20 min at room temperature. The cells in five randomly selected fields were enumerated using an inverted microscope (CKX41; Olympus Corporation, Tokyo, Japan). All experiments were performed at least three times.
Bioinformatics analysis
Targetscan software (version 5.1; www.targetscan.org) was used to predicate the putative targets of Let-7b, according to the manufacturer's protocol.
Dual-luciferase reporter assay
Luciferase activities were determined using a Dual-Luciferase Reporter assay system (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocol. PGL3-IGF1R-3′UTR wild-type (WT) and PGL3-IGF1R-3′UTR mutant (Mut) were obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China). U2OS or Saos-2 cells were seeded in 96-well plates, co-transfected with the WT-IGF1R-3′UTR or Mut-IGF1R-3′UTR luciferase reporter plasmid and transfected with the Let-7b mimic or NC mimic respectively. After 48 h, luciferase activity was determined, and firefly luciferase activity was normalized to Renilla luciferase activity. Luciferase assays were performed in three independent experiments.
Statistical analysis
All statistical analysis was performed using SPSS (version 19.0; IBM SPSS, Inc., Armonk, NY, USA). Results are expressed as the mean ± standard deviation of three independent experiments. Student's t-test was used to analyze the difference between two groups. One-way analysis of variance followed by Tukey's test was used to analyze the differences between multiple groups. Spearman's correlation analysis was used to investigate the correlation between IGF1R mRNA and Let-7b in T1-T2 stage osteosarcoma. P<0.05 was considered to indicate a statistically significant difference. The associations between the expression of Let-7b and the clinical characteristics of patients with OS were analyzed using a χ2 test.
Results
Let-7b is downregulated in OS cell lines and tissues
To reveal the function of Let-7b in OS progression, RT-qPCR analysis was used to explore the expression of Let-7b in the human OS cell lines U2OS, MG-63 and SAOS-2, and the human normal osteoblastic hFOB 1.19 cell line. As presented in Fig. 1A, the expression of Let-7b was significantly decreased in the OS cell lines. The clinical significance of Let-7b expression in OS was investigated in a total of 60 paired OS tissues and adjacent non-tumor tissues. It was observed that the Let-7b level was significantly lower in the OS tissues compared with the adjacent non-tumor tissues (Fig. 1B). On the basis of the mean value of Let-7b expression in patients with OS, tissues were divided into high and low Let-7b expression groups. As presented in Table I, low expression of Let-7b was significantly associated with increased tumor size, advanced Tumor-Node-Metastasis stage (16) and lung metastasis (P<0.05; Table I). However, there was no significant difference between Let-7b expression according to age or sex (Table I). These results indicated that Let-7b is downregulated in OS cells and tissues and therefore that Let-7b may serve a function in inhibiting the development of OS in humans.
Figure 1.
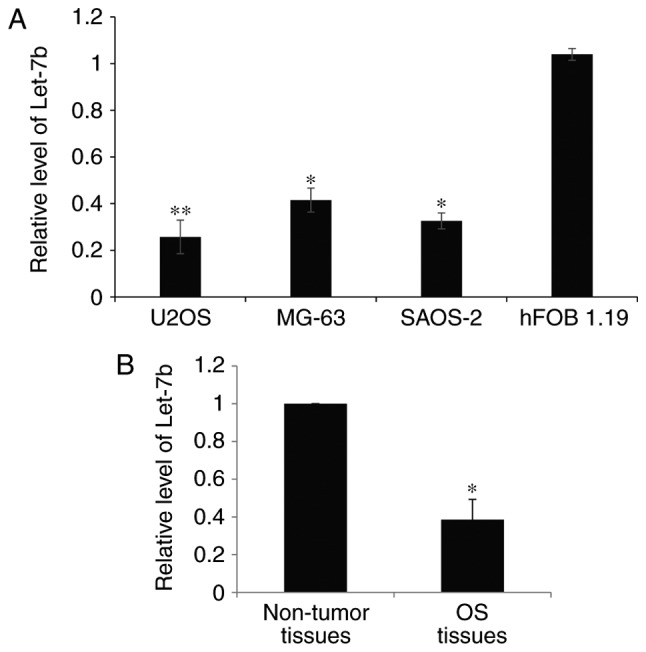
Let-7b is downregulated in osteosarcoma cell lines and osteosarcoma tissues. (A) RT-qPCR was used to determine the endogenous levels of Let-7b in human osteosarcoma cell lines U2OS, MG-63 and SAOS-2, and the normal human osteoblastic cell line hFOB 1.19. The expression levels of Let-7b were normalized to that of U6 RNA. *P<0.05, **P<0.01 vs. hFOB 1.19 cells. (B) The expression of Let-7b in a total of 60 paired osteosarcoma tissues and adjacent non-tumor tissues was determined by RT-qPCR. *P<0.05 vs. adjacent tissue. RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Let-7b inhibits the proliferation of OS cells
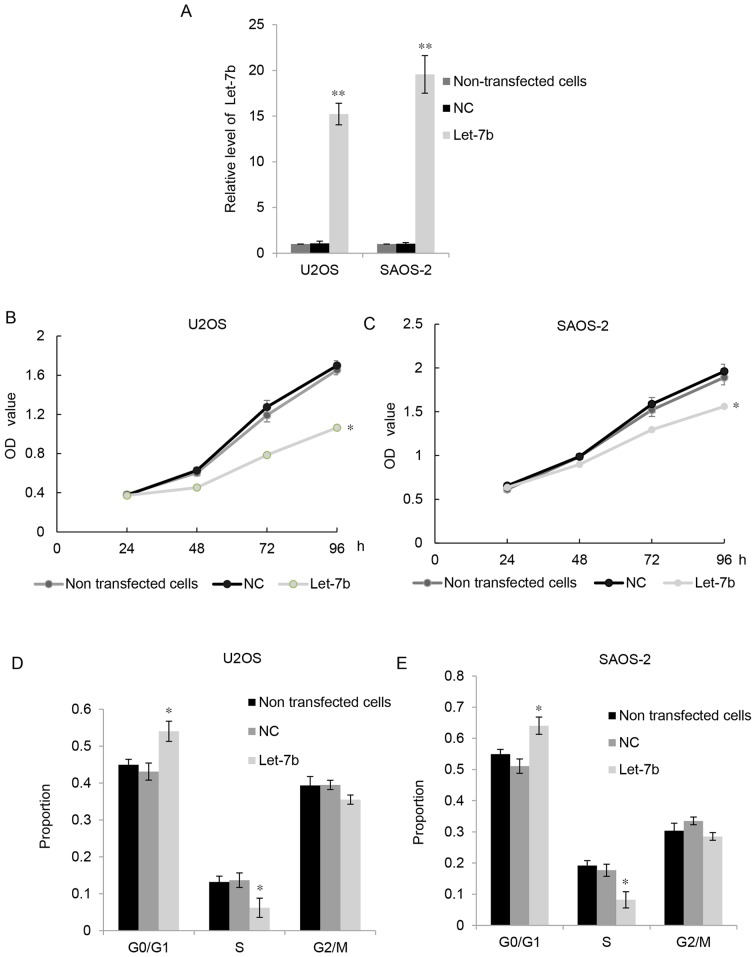
As Let-7b was downregulated in OS cell lines, U2OS and SAOS-2 cells were transfected with Let-7b. RT-qPCR indicated that, compared with the control groups, transfection with plasmids expressing Let-7b significantly upregulated Let-7b levels in U2OS and MG63 cells (Fig. 2A). MTT assays were performed to examine the proliferation of OS cells. As presented in Fig. 2B and C, the overexpression of Let-7b significantly suppressed the proliferation of U2OS and SAOS-2 cells compared with that of the control groups. Furthermore, flow cytometry was used to examine the cell cycle distribution of the U2OS and SAOS-2 cells. The results indicated that overexpression of Let-7b led to markedly high levels of cell cycle arrest at G1 stage (Fig. 2D and E). These results suggested that Let-7b exhibited suppressive effects on the proliferation of OS cells.
Figure 2.
Let-7b inhibits the proliferation of osteosarcoma cells. (A) The reverse transcription-quantitative polymerase chain reaction was used to determine Let-7b expression following transfection with NC or Let-7b mimic in U2OS and SAOS-2 cells. Non-transfected cells were used as a control. MTT assays were performed to determine the proliferation of (B) U2OS and (C) SAOS-2 cells following transfection with NC or Let-7b mimic. Non-transfected cells were used as control. Flow cytometry was used to determine the cell cycle distribution of (D) U2OS and (E) SAOS-2 following transfection with NC or Let-7b mimic. Non-transfected cells were used as a control. *P<0.05, **P<0.01 vs. control. Each sample was analyzed in triplicate. Results are presented as the mean ± standard error of the mean. NC, negative control; OD, optical density.
Let-7b suppresses the invasion of OS cells in vitro
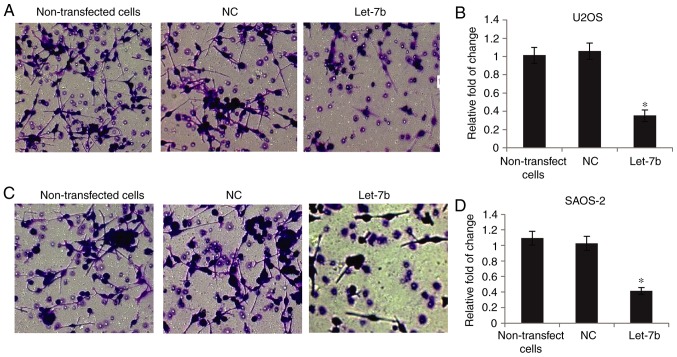
The invasive capability of OS cells was determined using Transwell assays. As presented in Fig. 3A and B, overexpression of Let-7b markedly suppressed U2OS invasion, compared with the non-transfected cells, whereas transfection with NC did not affect their invasion. Furthermore, SAOS-2 cell invasion was markedly decreased following overexpression of Let-7b (Fig. 3C and D). Together, these results suggested that Let-7b significantly inhibited the invasive ability of OS cells.
Figure 3.
Let-7b suppresses the invasion of osteosarcoma cells in vitro. (A) Representative images and (B) quantitative data of the invasion of U2OS cells following transfection with NC or Let-7b mimic. Non-transfected cells were used as a control. (C) Representative images and (D) quantitative data of the invasion of SAOS-2 cells following transfection with NC or Let-7b mimic. Non-transfected cells were used as a control. *P<0.05 vs. control. Magnification, ×20. NC, negative control.
IGF1R is the direct target of Let-7b
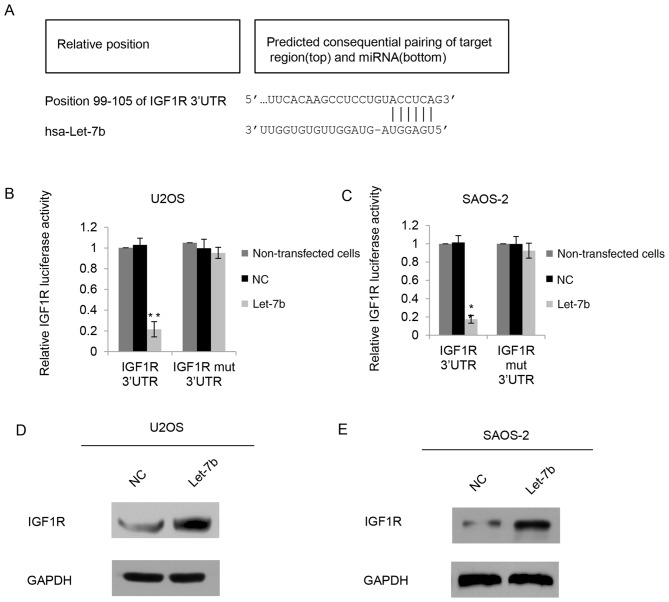
The function of miRNAs is to inhibit the expression of their target genes; therefore, the potential targets of Let-7b were investigated using Targetscan software. IGF1R was identified to be a putative target of Let-7b (Fig. 4A). To further identify the mechanisms of Let-7b affecting the biological behavior in OS, a luciferase activity assay was performed. A luciferase reporter vector containing the IGF1R 3′-UTR or a IGF1R mutant 3′-UTR was constructed, and the constructs were co-transfected with Let-7b or NC into U2OS cells. As presented in Fig. 4B, Let-7b significantly inhibited the luciferase activity of the IGF1R 3′-UTR reporter compared with that of the control groups, whereas the IGF1R mutant 3′-UTR eliminated the effect of Let-7b on luciferase activity, as there was no significant difference between the effects of the mutation and the control groups. In the SAOS-2 cells, a similar tendency was exhibited (Fig. 4C). Since miRNAs generally inhibit the expression of target genes at the post-transcriptional level, IGF1R protein expression was also determined in U2OS cells following Let-7b overexpression. The results from the western blot analysis identified that overexpression of Let-7b decreased the expression of endogenous IGF1R (Fig. 4D). Similarly, in SAOS-2 cells, IGF1R protein levels were decreased in the Let-7b transfection groups (Fig. 4E). Consistent with the luciferase reporter data, these results indicated that Let-7b bound directly to the 3-UTR of IGF1R, and therefore negatively regulated the expression of IGF1R.
Figure 4.
IGF1R is the direct target of Let-7b. (A) Predicted Let-7b-binding sites in the IGF1R 3′-UTR region identified using bioinformatic analyses. WT or mut Let-7b-binding sequence in the 3′UTR of IGF1R was subcloned into the pGL3 vector, and luciferase activity was determined in (B) U2OS or (C) SAOS-2 cells co-transfected with pGL3-WT-IGF1R-3′UTR or Mut-IGF1R-3′UTR luciferase reporter plasmids, and NC or Let-7b mimic, respectively. Non-transfected cells were used as a control. *P<0.05 and **P<0.01 vs. control. IGF1R protein expression was determined by western blot analysis in (D) U2OS or (E) SAOS-2 cells transfected with NC or Let-7b mimic. WT, wild-type; mut, mutant; NC, negative control; UTR, untranslated region; hsa, Homo sapiens.
IGF1R is involved in Let-7b-mediated proliferation and invasion of OS cells
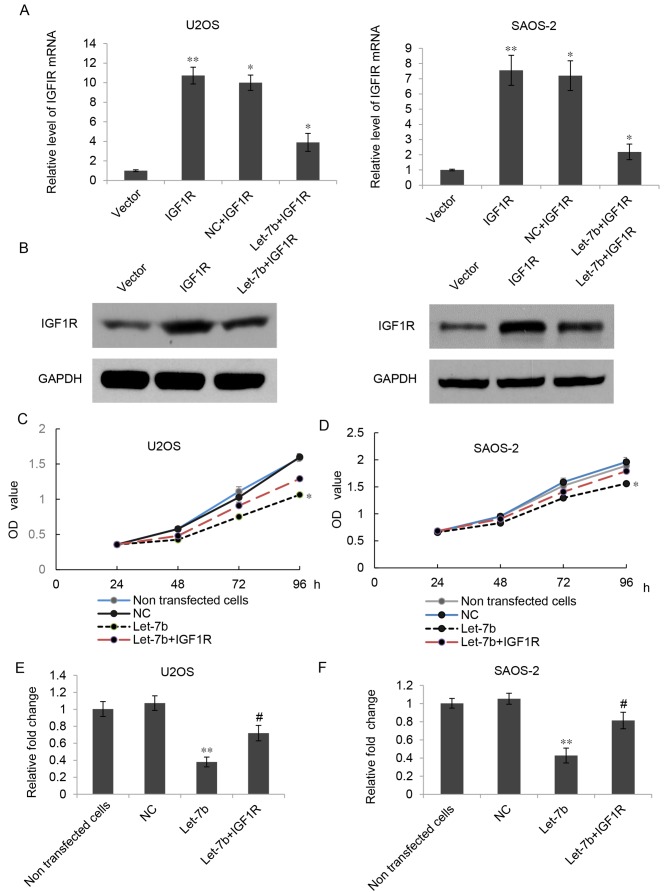
As IGF1R has been identified to be an oncogene in OS, and overexpression of Let-7b led to downregulation of IGF1R, it was hypothesized that Let-7b may suppress the proliferation and invasion of OS cells by regulating IGF1R. To verify this hypothesis, Let-7b-overexpressing U2OS and SAOS-2 cells were further transfected with a pcDNA3.1-IGF1R plasmid or a pcDNA3.1 vector. As presented in Fig. 5A and B, the mRNA and protein expression of IGF1R was markedly increased in the Let-7b+IGF1R group compared with that in the Let-7b group; however, transfection with the pcDNA3.1 vector did not affect the expression of IGF1R. The results of the MTT assay further indicated that the proliferation of U2OS and SAOS-2 cells was significantly increased in the Let-7b+IGF1R group when compared with that in the Let-7b group (Fig. 5C and D). A Transwell assay indicated that the overexpression of IGF1R could partially reverse the effects of Let-7b on the invasion of U2OS and SAOS-2 cells (Fig. 5E and F). Accordingly, these results suggested that IGF1R was indeed involved in the Let-7b-mediated suppressive effects in the proliferation and invasion of U2OS and SAOS-2 cells.
Figure 5.
IGF1R is involved in Let-7b-mediated proliferation and invasion of osteosarcoma cells. (A) Reverse transcription-quantitative polymerase chain reaction and (B) western blot analysis were used to determine the expression of IGF1R in U2OS and SAOS-2 cells transfected with Let-7b mimic or co-transfected with Let-7b mimic and IGF1R plasmid. MTT assays were used to determine the proliferation of (C) U2OS and (D) SAOS-2 cells transfected with Let-7b mimic or co-transfected with Let-7b mimic and IGF1R plasmid. Transwell assays were used to determine the invasion of (E) U2OS and (F) SAOS-2 cells. Results are expressed as the mean ± standard deviation. *P<0.05 and **P<0.01 vs. control; #P<0.05 vs. Let-7b. IGF1R, insulin-like growth factor 1 receptor; NC, negative control; OD, optical density.
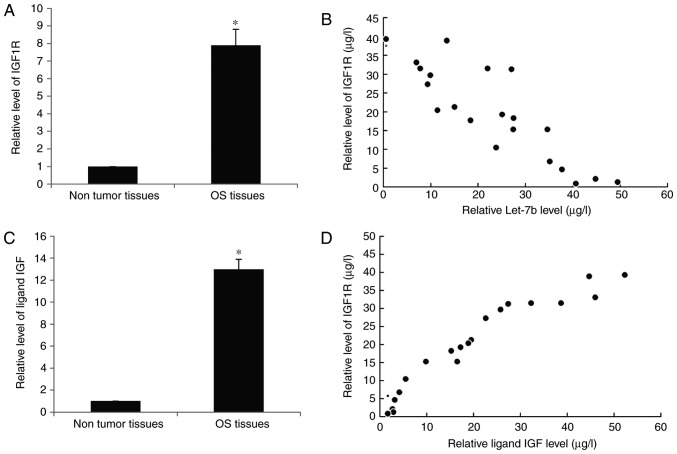
Increased IGF1R levels are inversely associated with Let-7b in OS tissues
Finally, the expression of IGF1R in human OS tissues was investigated. RT-qPCR results indicated that IGF1R was significantly increased in OS tissues compared with the adjacent non-tumorous tissues (Fig. 6A). Furthermore, a significant inverse correlation was identified between the expression of Let-7b and IGF1R in OS at T1-T2 stage (Fig. 6B), suggesting that the increased expression of IGF1R may be due to the downregulation of Let-7b. Furthermore, the expression of ligand insulin-like growth factor (IGF) in the serum of patients with OS was also upregulated (Fig. 6C) and exhibited a positive correlation with IGF1R (Fig. 6D), which represented an increase in IGF signal transduction.
Figure 6.
Increased IGF1R levels are inversely correlated with Let-7b in OS tissues. (A) The mRNA expression of IGF1R in the OS tissues and their matched adjacent non-tumor tissues was assessed using RT-qPCR analysis. (B) IGF1R mRNA level was inversely correlated with Let-7b in osteosarcoma T1-T2 stage using Spearman's correlation analysis. (C) The mRNA expression of IGF in the OS tissues and their matched adjacent non-tumor tissues was assessed using RT-qPCR analysis. (D) The IGF mRNA level was positively correlated with IGF1R in OS T1-T2 stage using Spearman's correlation analysis. *P<0.05 vs. Non tumor tissues. IGF1R, insulin-like growth factor 1 receptor; IGF, insulin-like growth factor; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; OS, osteosarcoma; T stage, Tumor stage.
Discussion
It has been identified previously that a number of oncogenes and tumor suppressors serve key functions in the progression of OS (18). Therefore, understanding the underlying molecular mechanisms of OS may enable the development of effective therapeutic strategies (19).
IGF1R is a widely expressed cell membrane receptor and it is involved in different cancer types (20,21). As identified previously, increased expression of IGF1R in OS is associated with the development of tumor metastasis and indicates a poor prognosis, and such molecules usually represent an attractive therapeutic target (22,23). These results are consistent with the results of the present study that IGF1R is significantly increased in OS tissues compared with the adjacent non-tumorous tissues. By targeting cadherin 2 and IGF1R, miR-194 suppressed OS cell proliferation and metastasis in vitro and in vivo (24). miR-133a inhibited OS cellular proliferation and invasion via targeting IGF1R (25). miR-16 inhibited OS cell proliferation by targeting IGF1R (9). In the present study, it was identified that Let-7b was another regulator for IGF1R in OS. Let-7b bound directly to the 3-UTR of IGF1R, and therefore negatively regulated the expression of IGF1R.
A single miRNA is able to silence the expression of multiple genes simultaneously. It is reasonable to propose the hypothesis that, in addition to IGF1R, additional target genes are involved in suppression of OS by Let-7b. Using TargetScan, putative targets of Let-7b were identified, of which several were associated with tumor growth or metastasis. These predictions were the experimental basis for the present study of the regulatory mechanism of Let-7b.
In conclusion, the results of the present study indicated that Let-7b suppressed the proliferation and invasion of OS cells in vitro. Overexpression of Let-7b downregulated the protein level of IGF-IR, and there was a significant inverse correlation between the expression of Let-7b and IGF1R in OS at T1-T2 stage, which suggested the function of Let-7b as a tumor suppressor in OS progression by repression of the expression of IGF1R. In the future, investigating further putative target genes and understanding the molecular mechanisms of the contribution of Let-7b to OS are required. Let-7b may prove to be a promising gene therapeutic agent.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
KZ conceived and designed the study. WW and YL performed transwell and MTT assays. AG analyzed the data. DY performed tissue collection and wrote the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Linyi Central Hospital (Linyi, China). Written informed consent was obtained from each patient.
Patient consent for publication
Patients provided consent for the publication of the data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Valery PC, Laversanne M, Bray F. Bone cancer incidence by morphological subtype: A global assessment. Cancer Causes Control. 2015;26:1127–1139. doi: 10.1007/s10552-015-0607-3. [DOI] [PubMed] [Google Scholar]
- 2.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the surveillance, epidemiology, and end results program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broadhead ML, Clark JC, Myers DE, Dass CR, Choong PF. The molecular pathogenesis of osteosarcoma: A review. Sarcoma. 2011;2011:959248. doi: 10.1155/2011/959248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou AJ, Geller DS, Gorlick R. Therapy for osteosarcoma: Where do we go from here? Paediatric Drugs. 2008;10:315–327. doi: 10.2165/00148581-200810050-00005. [DOI] [PubMed] [Google Scholar]
- 5.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Developmental Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 8.Liang J, Liu C, Qiao A, Cui Y, Zhang H, Cui A, Zhang S, Yang Y, Xiao X, Chen Y, et al. MicroRNA-29a-c decrease fasting blood glucose levels by negatively regulating hepatic gluconeogenesis. J Hepatol. 2013;58:535–542. doi: 10.1016/j.jhep.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Wang Q, Wang GD, Wang HS, Huang Y, Liu XM, Cai XH. miR-16 inhibits cell proliferation by targeting IGF1R and the Raf1-MEK1/2-ERK1/2 pathway in osteosarcoma. FEBS Lett. 2013;587:1366–1372. doi: 10.1016/j.febslet.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Ji F, Zhang H, Wang Y, Li M, Xu W, Kang Y, Wang Z, Wang Z, Cheng P, Tong D, et al. MicroRNA-133a, downregulated in osteosarcoma, suppresses proliferation and promotes apoptosis by targeting Bcl-xL and Mcl-1. Bone. 2013;56:220–226. doi: 10.1016/j.bone.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 11.Kim HR, Shin CH, Lee H, Choi KH, Nam DH, Ohn T, Kim HH. MicroRNA-1908-5p contributes to the oncogenic function of the splicing factor SRSF3. Oncotarget. 2017;8:8342–8355. doi: 10.18632/oncotarget.14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 13.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 14.Lan FF, Wang H, Chen YC, Chan C, Ng SS, Li K, Xie D, He ML, Lin MC, Kung HF. Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-Myc and upregulation of p16(INK4A) Int J Cancer. 2011;128:319–331. doi: 10.1002/ijc.25336. [DOI] [PubMed] [Google Scholar]
- 15.Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, Shi Y. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Nati Acad Sci USA. 2010;107:1876–1881. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuma H, Beppu Y, Yokoyama R, Omiya K. TNM classification of bone and soft tissue sarcomas. Gan to kagaku Ryoho (Japanese) 1997;24:1035–1040. [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Leichter AL, Sullivan MJ, Eccles MR, Chatterjee A. MicroRNA expression patterns and signalling pathways in the development and progression of childhood solid tumours. Mol Cancer. 2017;16:15. doi: 10.1186/s12943-017-0584-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang N, Song WX, Luo J, Haydon RC, He TC. Osteosarcoma development and stem cell differentiation. Clin Orthop Relat Res. 2008;466:2114–2130. doi: 10.1007/s11999-008-0335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S, Ferber A, Miura M, Baserga R. Mitogenicity and transforming activity of the insulin-like growth factor-I receptor with mutations in the tyrosine kinase domain. J Biol Chem. 1994;269:32558–32564. [PubMed] [Google Scholar]
- 21.Kirschner LS. Inhibition of IGF-1R in adrenocortical carcinoma. Lancet Oncol. 2015;16:356–357. doi: 10.1016/S1470-2045(15)70130-0. [DOI] [PubMed] [Google Scholar]
- 22.Maniscalco L, Iussich S, Morello E, Martano M, Gattino F, Miretti S, Biolatti B, Accornero P, Martignani E, Sánchez-Céspedes R, et al. Increased expression of insulin-like growth factor-1 receptor is correlated with worse survival in canine appendicular osteosarcoma. Vet J. 2015;205:272–280. doi: 10.1016/j.tvjl.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Cao Y, Roth M, Piperdi S, Montoya K, Sowers R, Rao P, Geller D, Houghton P, Kolb EA, Gill J, Gorlick R. Insulin-like growth factor 1 receptor and response to anti-IGF1R antibody therapy in osteosarcoma. PLoS One. 2014;9:e106249. doi: 10.1371/journal.pone.0106249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han K, Zhao T, Chen X, Bian N, Yang T, Ma Q, Cai C, Fan Q, Zhou Y, Ma B. MicroRNA-194 suppresses osteosarcoma cell proliferation and metastasis in vitro and in vivo by targeting CDH2 and IGF1R. Int J Oncol. 2014;45:1437–1449. doi: 10.3892/ijo.2014.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen G, Fang T, Huang Z, Qi Y, Du S, Di T, Lei Z, Zhang X, Yan W. MicroRNA-133a inhibits osteosarcoma cells proliferation and invasion via targeting IGF-1R. Cell Physiol Biochem. 2016;38:598–608. doi: 10.1159/000438653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.