Abstract
The prognosis for patients with head and neck cancer (HNC) remains poor, owing to uncontrolled tumor invasion and metastasis. Epithelial-mesenchymal transition (EMT) serves an important role in this invasion and metastasis, and transient receptor potential polycystic 2 (TRPP2) enhances metastasis and invasion by regulating EMT in human laryngeal squamous cell carcinoma. The present study examined whether exosomes/TRPP2 small interfering RNA (siRNA) complexes were able to reduce EMT by suppressing TRPP2 expression in FaDu cells, a cell line of human pharyngeal squamous cell carcinoma. Using agarose gel electrophoresis, it was determined that exosome/TRPP2 siRNA complexes were stable in the presence of nucleases and serum. A fluorescence assay and western blotting analysis was performed, and it was reported that the FaDu cells took up exosomes, the exosomes effectively delivered TRPP2 siRNA into FaDu cells and that exosome/TRPP2 siRNA complexes significantly suppressed TRPP2 protein expression levels in FaDu cells. Furthermore, expression levels of E-cadherin were significantly increased, whereas expression levels of N-cadherin and vimentin were significantly decreased in FaDu cells transfected with TRPP2 siRNA. Thus, exosome/TRPP2 siRNA complexes markedly suppressed TRPP2 expression and EMT in FaDu cells. These results suggested that further development of exosome/TRPP2 siRNA complexes for use as an RNA-based gene therapy in the treatment of HNC is warranted.
Keywords: exosome, small interfering RNA, epithelial-mesenchymal transition, transient receptor potential polycystic 2, head and neck cancer
Introduction
Head and neck cancer (HNC) is one of the most common malignant neoplasms observed worldwide (1). Although early detection and therapeutic strategies for HNC, including radiotherapy, chemotherapy, immunotherapy and surgery, have substantially improved, the outcomes for patients with HNC remain poor, with an overall 5-year survival rate of only 50% (2,3). Therefore, it is important to find novel treatment strategies for HNC. Uncontrolled invasion and metastasis contribute to this poor prognosis, and recent study results have suggested that epithelial-mesenchymal transition (EMT) serves an essential role in cancer cell metastasis, invasion, radiotherapy resistance, drug resistance, immune evasion and the cancer stem-cell phenotype (4,5). Previous studies have demonstrated that a number of critical biomarkers, including E-cadherin, N-cadherin and vimentin, are involved in EMT (5,6). The present study reported that that the expression levels of transient receptor potential polycystic 2 (TRPP2, previously known as polycystin-2, PKD2 or PC2), a nonselective cation channel encoded by the PKD2 gene, are markedly increased in laryngeal squamous cell carcinoma. It was also previously determined that inhibition of TRPP2 protein expression via transfection with small interfering RNA (siRNA) markedly decreased the expression levels of vimentin and N-cadherin and increased E-cadherin expression levels in Hep2 cells (a cell line originating from human laryngeal squamous cell carcinoma) (5).
Targeted delivery using siRNA-based technology is a promising strategy for the treatment of a variety of diseases (7–10). However, certain characteristics of siRNA, including its polyanionic charge, poor stability against serum nuclease degradation, low permeability, immune response and toxicity, make it difficult to use in clinical practice (10,11). Exosomes, which are endogenous nano-sized vesicles that mediate cell-to-cell communication, have been demonstrated to carry RNA and freely enter cells (12–15). These characteristics provide an opportunity for the use of exosomes to deliver therapeutic siRNA to targeted cancer cells in cancer gene therapy.
In the present study, TRPP2 siRNA was delivered into FaDu cells (a cell line originating from human pharyngeal squamous cell carcinoma) using exosomes secreted from 293 cells. The packaging capacity of exosomes for TRPP2 siRNA, stability of the exosome/TRPP2 siRNA complex, and expression levels of EMT biomarkers were determined, and cell migration and invasion were assessed, in order to establish whether EMT is inhibited by exosome-delivery of TRPP2 siRNA, and whether this strategy warranted further development as a viable treatment option in HNC.
Materials and methods
Cell culture
The FaDu cells were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured in DMEM supplemented with 10% fetal bovine serum (FBS; both Thermo Fisher Scientific, Inc., Waltham, MA USA) depleted of exosomes, 100 U/ml penicillin and 0.1 mg/ml streptomycin. The cells were incubated in an atmosphere of 5% CO2 at 37°C.
Exosome purification
The exosomes were prepared from 293 cells purchased from the American Type Culture Collection. Briefly, 5 ml DMEM with 10% exosome-depleted FBS was added to 60 mm diameter culture dish containing 293 cells (2×106/ml). Following 48 h of incubation, the cells were harvested and the culture medium was centrifuged at 2,000 × g for 30 min at 4°C to eliminate cells and cell debris. The remaining supernatant was mixed with polyethylene glycol (PEG) (16,17). The exosomes were precipitated with an equal volume of PEG buffer (160 g/l PEG and 1 M NaCl) overnight at 4°C and centrifuged at 10,000 × g for 1 h at 4°C. The supernatant was removed, leaving the exosomes in the bottom of the tube. The exosomes were resuspended in 10 µl PBS, and a bicinchoninic acid protein assay kit (BestBio, Shanghai, China) was used to determine the protein concentration. The exosomes were stored at −80°C until use.
Transmission electron microscopy
The diameters of the exosome/siRNA (5′-AACCUGUUCUGUGUGGUCAGGUUAU-3′; Biomics Biotechnologies Co., Ltd., Jiangsu, China) nanoparticles in water were analyzed at 25°C. The exosome was fixed with 1 ml of 2.5% glutaraldehyde in 0.1 M sodium cacodylate solution (pH 7.0) for 1 h at 4°C. Samples were subsequently embedded in pure low viscosity embedding mixture using Spurr Low Viscosity Embedding kit (cat. no. 01916-1; Polysciences Inc. Warrington, PA, USA) using the embedding mold, according to the manufacturer's protocols, and baked for 24 h at 65°C. Transmission electron microscopy (HT7700; Hitachi, Ltd., Tokyo, Japan) was used to obtain images, operating at an acceleration voltage of 100 kV. The sample solution with TRPP2 siRNA (exosome/siRNA, 4:5 µg/nM) was placed onto a 300-mesh copper grid coated with carbon. Following deposition for 5 min, the surface water was removed with filter paper and air-dried. A 4 wt% aqueous uranyl acetate solution was used for positive staining at room temperature for 20 min.
Agarose gel electrophoresis
In the present study, it was assessed whether the exosome/TRPP2 siRNA complex enhanced the stability of TRPP2 siRNA in the presence of RNA nucleases or serum obtained from mice. Following incubation with nucleases for 0, 5, 15, 30, 60 and 120 min at 37°C, or incubation with serum for 120 min, heparin was added to release the TRPP2 siRNA [4:5 ratio of exosomes to TRPP2 siRNA (µg/nM)] in the exosome/TRPP2 siRNA complex groups. Agarose (0.9 g) was added to 100 ml Tris-acetate-EDTA buffer (40 mM Tris, 20 mM NaAc and 1 mM EDTA at pH 8.0) and dissolved by heating to 100°C. An ethidium bromide aqueous solution at a final concentration of 0.5 µg/ml was added to the dissolved gel. As the solution cooled to <50°C, the gel solidified. Once completely solidified, the gel was placed in a tank filled with electrophoresis solution. Electrophoresis was conducted at 30 V for 20 min, and the gel was placed under UV light to observe the siRNA bands. Exosomes (20 or 40 µl) containing proteins (4 µg/µl) were mixed with 200 nM TRPP2 siRNA, with or without serum obtained from mice (cat. no. HQ30078; Hongquan Biotechnology Co., Guangzhou, China).
Western blotting
Target proteins in FaDu cells were examined by western blotting, which was performed as previously described (18). The proteins were extracted from lysates of FaDu cells with a detergent extraction buffer that consisted of 150 mM NaCl, 50 mM Tris-HCl (pH 7.4), 1 mM disodium salt of EDTA, 0.1% SDS, 1% Triton X-100, and 1% sodium deoxycholate, sodium orthovanadate, sodium fluoride and leupeptin. A total of 30 µg protein per lane was separated via SDS-PAGE on a 10% gel and transferred to a polyvinylidene difluoride membrane. The membranes were blocked with Tris-buffered saline solution containing 10% nonfat milk and Tween-20 (0.1%) for 1 h at room temperature to block nonspecific binding sites. For immunoblotting, the membrane with the transferred proteins was incubated with specific primary antibodies overnight at 4°C (BIOSS, Beijing, China; anti-CD63, cat. no. bs-1523R) (Santa Cruz Biotechnology, Dallas, Texas, USA; anti-TRPP2 cat. no. sc-25749; anti-vimentin cat. no. sc-373717; anti-E-cadherin cat. no. sc-8426; and anti-N-cadherin cat. no. sc-393933), diluted 1:200. Subsequently, the membrane was washed with PBS in triplicate and incubated with the respective horseradish peroxidase-conjugated secondary antibody (dilution, 1:5,000; cat. no. E-AB-1003; Elabscience Biotechnology Co., Ltd, Wuhan City, China) at room temperature for 1 h. The resulting immunosignals were detected using an enhanced chemiluminescence detection system (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The optical densities of the protein bands were analyzed using ImageJ 1.50 software (National Institutes of Health, Bethesda, MD, USA). All protein bands were normalized to GAPDH located in the same lane and the results are expressed as the relative optical density.
Fluorescence assay
Purified exosomes were labeled with PKH26 red, a fluorescent dye, using a linker kit according to the manufacturer's instructions (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) (16). Briefly, 1 µl PKH26 was added to 250 µl diluent C for 5 min at room temperature. An equal volume of exosome-depleted serum was added to terminate the labeling reaction, and PEG precipitation was used to purify the exosomes. The PKH26-labeled exosomes were resuspended in 250 µl PBS. The PKH26-labeled exosomes (5, 10 and 20 µl) were added to FaDu cell culture medium in a 12-well plate with 1.8×106/ml density and incubated at 37°C for 12 h. For the control group, 250 µl dilution buffer was used in place of the exosomes. Following incubation, FaDu cells were washed with PBS. Uptake of the labeled exosomes by FaDu cells was determined using fluorescence microscopy (magnification, ×100).
To determine whether the exosomes were able to deliver TRPP2 siRNA into FaDu cells, 40 µl exosomes (4 µg/µl) was added to 50 ml Opti-MEM (Thermo Fisher Scientific, Inc.) for 5 min at room temperature. In addition, 10 µl fluorescein amidite (FAM)-labeled siRNA (1 µg/µl) was added to 50 ml Opti-MEM for 5 min at room temperature. Following 5 min incubation, the two solutions were mixed together and incubated for 20 min at room temperature, prior to adding to FaDu cells for transfection. For the control group, 40 µl dilution buffer replaced the exosomes. After 24 h from transfection, the fluorescence signal of the FAM-labeled TRPP2 siRNA in the FaDu cells was observed using fluorescence microscopy.
Wound healing assay
The FaDu cells were cultured in a 6-well plate until 100% confluent, washed with PBS and cultured overnight in low-serum (0.1%) DMEM. The sterile tips of 200 µl pipettes were used to create scratches or ‘wounds’ in the cell layer. The FaDu cells were rinsed with PBS to remove floating cells and were cultured in DMEM supplemented with 0.1% FBS. The lengths of the scratches in each well were recorded by capturing images using a fluorescence microscope (magnification, ×40; Nikon Corporation, Tokyo, Japan) at the same configuration 0, 24 and 48 h following the creation of the scratch. The relative distances that cells migrated through the scratched area were measured, and a percentage of ‘healing’ was calculated. The experiment was repeated four times.
Cell migration and invasion assay
The 8-µm pore polycarbonate membranes (cat no. 3422; Corning Inc., Corning, NY, USA) of 24-well Transwell inserts were coated with Matrigel® (cat no. BD354277; BD Biosciences, San Jose, CA, USA). In each well, 30 µl of Matrigel® was added to the upper part of the insert and dried at 37°C to form a thin gel layer for 20 min. Serum-free medium (DMEM; 0.2 ml) was added to the upper chamber containing FaDu cells and medium (0.6 ml) supplemented with 20% FBS was added to the lower chamber. FaDu cells were seeded at a density of 1×105 in the upper part of the chamber. Following 48 h incubation in a 5% CO2 incubator at 37°C, the Transwell inserts were removed from the plates and the cells that had not migrated from the top of the membrane were wiped away using a cotton swab. Cells passing through the membrane pores were fixed with 4% paraformaldehyde was infiltrated for 5 min at room temperature and stained with DAPI for 3 min at room temperature. A fluorescence microscope (magnification, ×40; Nikon Corporation, Tokyo, Japan) was used to observe and count the number of cells that had successfully migrated through the membrane in four randomly selected areas.
Statistical analysis
SigmaPlot 13.0 software (Systat Software Inc., San Jose, California, USA) was used to analyze the collected data. All results are presented as the mean ± standard error of the mean. Two-tailed Student's t-tests were used to compare the results of different groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Characterizing the exosome/TRPP2 siRNA complex
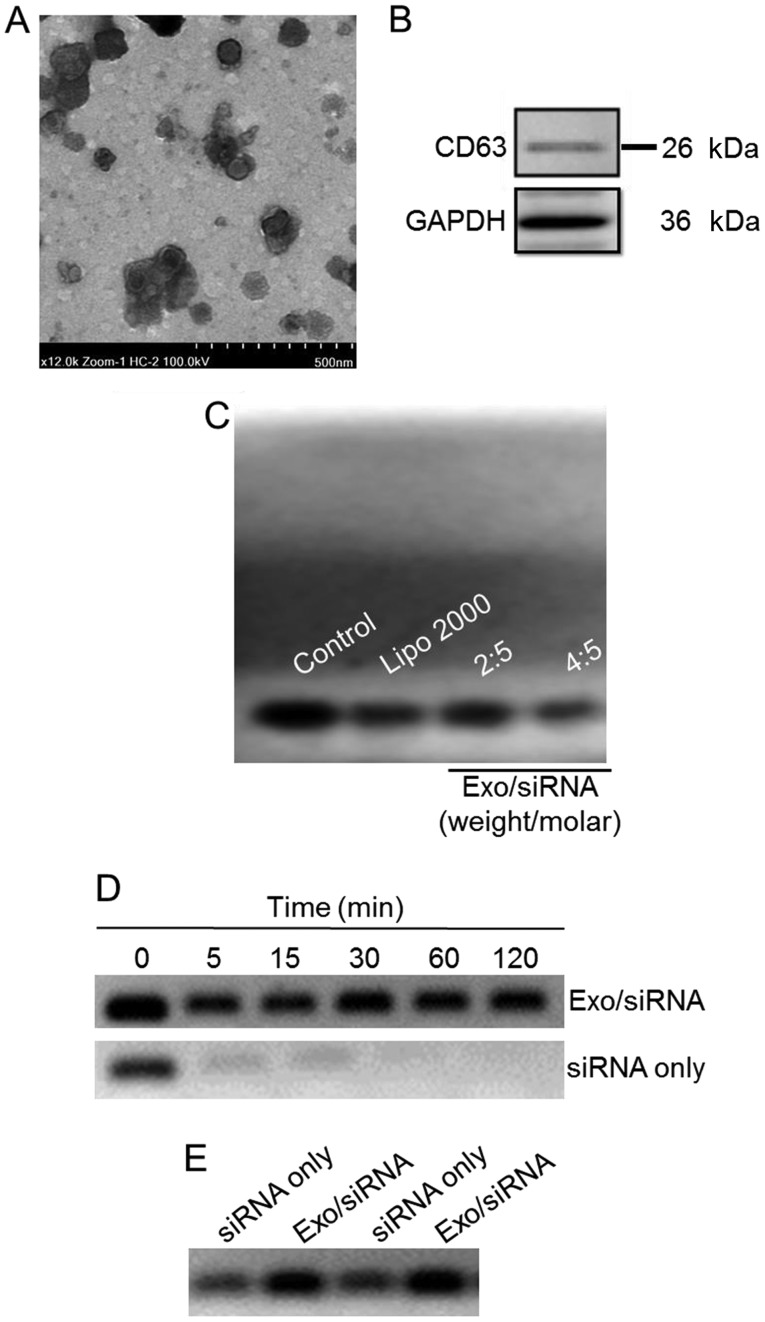
In order to view the morphology of exosome/TRPP2 siRNA, transmission electron microscopy was used to reveal spherical nanoparticles with diameters of 50–100 nm (Fig. 1A). Membranes of exosomes are enriched with numerous types of proteins, including cluster of differentiation (CD)63, CD9, CD37, CD53 and CD81 (19–22); CD63 and CD9 are considered to be specific markers for exosomes (20). In the present study, CD63 was probed in order to determine whether exosomes has been successfully extracted from 293 cells (Fig. 1B) and a gel retardation assay was used in order to determine the TRPP2 siRNA packaging capacity of the exosomes. As presented in Fig. 1C, exosomes encapsulated TRPP2 siRNA in a concentration-dependent manner. As the exosome to TRPP2 siRNA weight/molar (µg/nM) ratio increased, enhanced retardation of the TRPP2 siRNA band was observed, with the retardation of the TRPP2 siRNA band apparent at a 4:5 ratio of exosomes to TRPP2 siRNA (µg/nM) (Fig. 1C). These results indicated that exosomes were capable of encapsulating TRPP2 siRNA.
Figure 1.
Characterization of the exosome/TRPP2 siRNA complexes. (A) Representative transmission electron microscopy image displaying exosome/TRPP2 siRNA particles counterstained with 4% uranyl acetate. Scale bar, 500 nm. (B) Representative image presenting the protein expression of CD63, an exosomal marker. (C) Exosomes encapsulated TRPP2 siRNA (Exo/siRNA) in a concentration-dependent manner. The TRPP2 siRNA packaging capacity of exosomes was assessed. An agarose gel retardation assay was performed at different weight/molar ratios of exosomes to TRPP2 siRNA (µg/nM; Exo/siRNA). (D) Stability of TRPP2 siRNA only and TRPP2 siRNA encapsulated within exosomes against enzymatic degradation following incubation with RNA nucleases for 0, 5, 15, 30, 60 and 120 min or (E) in serum for 120 min in a 4:5 ratio of exosomes to TRPP2 siRNA (µg/nM) or naked siRNA (siRNA only). In the Exo/siRNA group, heparin was added to the exosome/TRPP2 complexes to release TRPP2 siRNA prior to conducting the gel retardation assay. Exo, exosome; Lipo, Lipofectamine®; CD, cluster of differentiation; siRNA, small interfering RNA; TRPP2, transient receptor potential polycystic 2; Exo, exosome.
Protection of the exosome/TRPP2 siRNA complexes against nucleases and other enzymes in the bloodstream is critical for their use in siRNA-based therapies. Agarose gel electrophoresis was conducted to determine the stability of the exosome/TRPP2 siRNA complex. The agarose gel electrophoresis results indicated that naked (non-complexed; siRNA only) TRPP2 siRNA degraded within 5 min, whereas the TRPP2 siRNA encapsulated within exosomes remained intact, as no decrease in the density of the TRPP2 siRNA band was observed at any time examined (Fig. 1D and E). Hence, exosomes effectively protected TRPP2 siRNA against nuclease degradation and substantially enhanced TRPP2 siRNA stability.
Exosomes deliver TRPP2 siRNA into FaDu cells
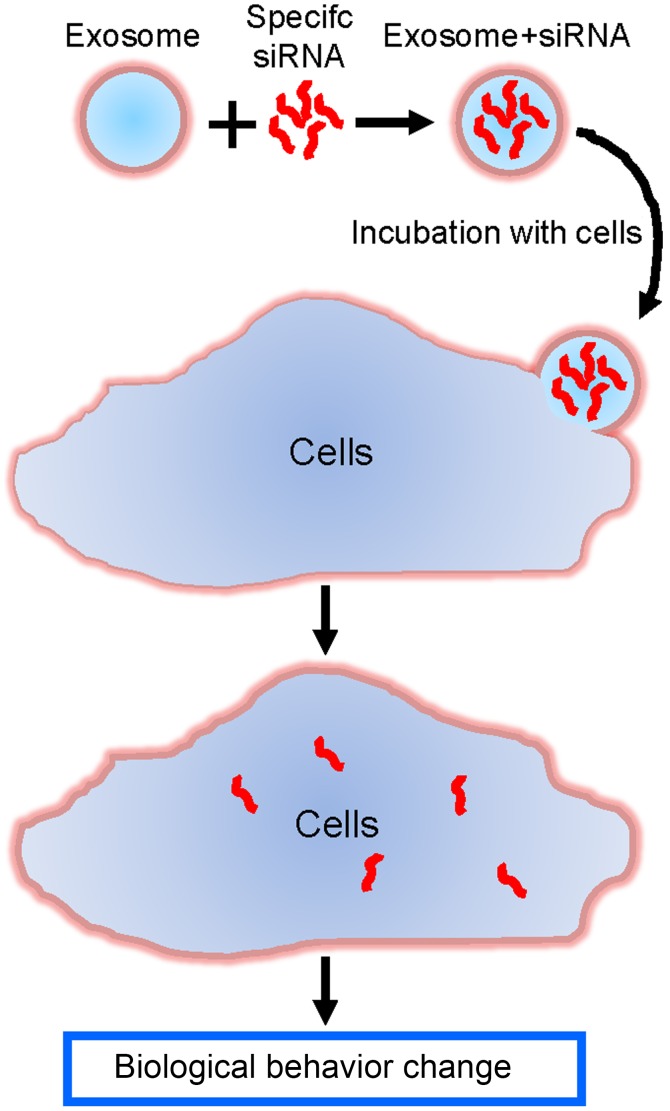
Exosomes/TRPP2 siRNA complexes were used as a tool to deliver TRPP2 siRNA into FaDu cells in order to regulate cellular biological behavior, and eventually develop a therapeutic approach to treat HNC (Fig. 2).
Figure 2.
A schematic representation of the formation of the exosome/TRPP2 siRNA complexes, which deliver siRNA into cells to regulate cellular biological behavior. siRNA, small interfering RNA.
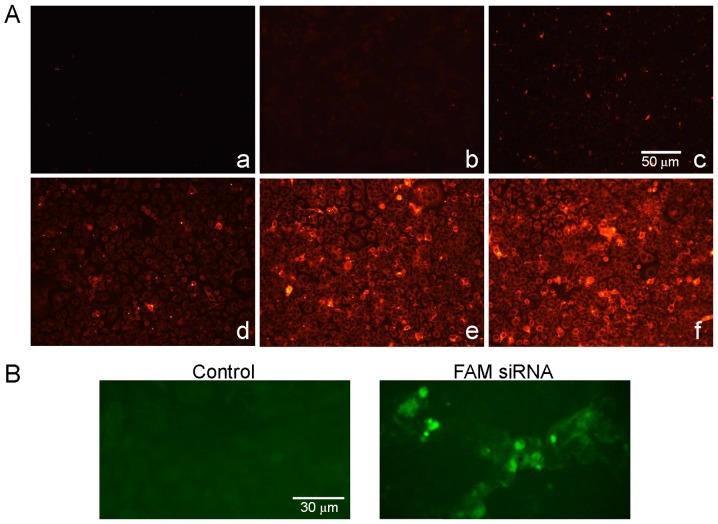
A fluorescence assay was used in order to determine whether FaDu cells were able to effectively uptake exosomes and whether exosomes were able to deliver TRPP2 siRNA into the FaDu cells. As presented in the fluorescence microscopy images in Fig. 3A, red fluorescence signals from the PKH26-labeled exosomes were observed in FaDu cells following transfection (Fig. 3Ad-f), whereas no red fluorescence signals were observed in the control group (Fig. 3Aa-c). Green fluorescence signals from FAM-labeled exosome/TRPP2 siRNA complexes, were observed in FaDu cells and not from controls (Fig. 3B). Together, these results demonstrated that FaDu cells uptake exosomes and that exosomes may have the ability to encapsulate and deliver TRPP2 siRNA into FaDu cells.
Figure 3.
Exosomes deliver TRPP2 siRNA into FaDu cells. (A) Fluorescence images displaying the uptake of exosomes. The presence of red fluorescence signals indicates that exosomes were not observed in the control groups (a-c, 250 µl phosphate buffer saline), and were observed in the experimental groups (d-f, PKH26-labeled exosomes 5, 10 and 20 µl). (B) Delivery of TRPP2 siRNA into FaDu cells. The presence of the green fluorescent signals indicates that FAM-labeled TRPP2 siRNA was observed in the experimental group and not the control group. siRNA, small interfering RNA; FAM, fluorescein amidite; TRPP2, transient receptor potential polycystic 2.
Exosome/TRPP2 siRNA complexes suppress TRPP2 expression in FaDu cells
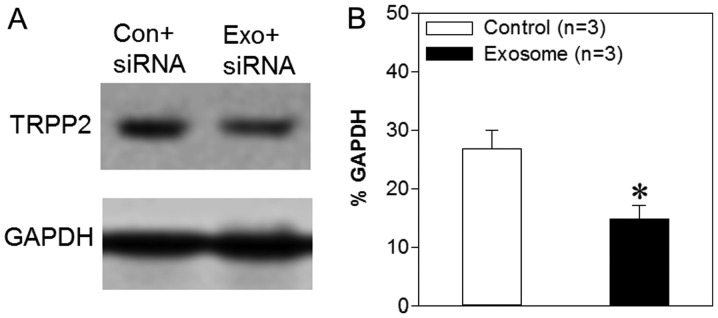
TRPP2, a nonselective cation channel encoded by the PKD2 gene, serves an important role in a number of cellular processes (23). A previous study demonstrated that TRPP2 expression levels are increased significantly in human laryngeal squamous cell carcinoma and that TRPP2 enhances metastasis in these cells by regulating EMT (5). The results presented above indicated that exosomes may be capable of delivering TRPP2 siRNA into FaDu cells in vitro; however, it was unknown as to whether TRPP2 siRNA achieved TRPP2-specific gene silencing. Thus, western blot analyses were used in order to assess the expression levels of TRPP2 in FaDu cells following transfection. The results demonstrated that TRPP2 expression levels were significantly decreased following the transfection of TRPP2 siRNA into FaDu cells compared with the control (FaDu cells transfected with scrambled siRNA) (Fig. 4). Thus, these findings indicated that exosomes may successfully deliver TRPP2 siRNA into FaDu cells and that TRPP2 siRNA may significantly suppress TRPP2 expression in FaDu cells.
Figure 4.
Effects of exosome/TRPP2 siRNA complexes on TRPP2 expression levels in FaDu cells. (A) Western blotting images and (B) summary data displaying TRPP2 expression levels in FaDu cells transfected with scrambled (Con) or TRPP2 (Exo) siRNA. The optical density of each protein was normalized to GAPDH. Values are presented as the mean ± standard error of the mean (n=3). *P<0.05 vs. control. siRNA, small interfering RNA; TRPP2, transient receptor potential polycystic 2; Exo, exosome; Con, control.
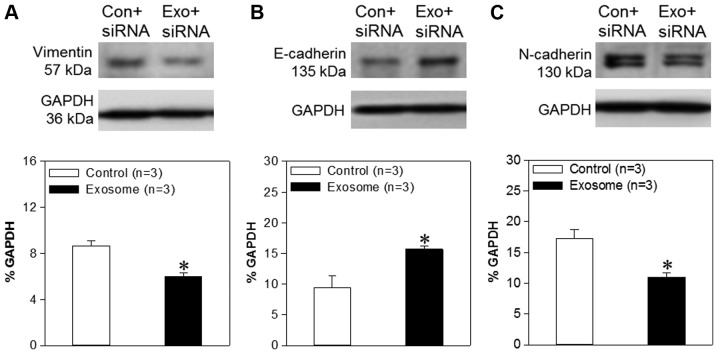
Exosome/TRPP2 siRNA complexes reduce EMT in FaDu cells
Previous studies have demonstrated that TRPP2 enhances metastasis and invasion by regulating EMT in human laryngeal squamous cell carcinoma (5). During EMT, E-cadherin expression levels, regarded as a prognostic biomarker for patients with numerous types of cancer, are significantly decreased, and cells produce more vimentin. In the present study, western blotting analyses were used in order to assess vimentin, E-cadherin and N-cadherin expression levels. The results demonstrated that the expression levels of N-cadherin and vimentin were decreased significantly in FaDu cells transfected with exosome/TRPP2 siRNA, compared with FaDu cells transfected with scrambled siRNA. Furthermore, E-cadherin levels were significantly increased in FaDu cells transfected with exosome/TRPP2 siRNA, compared with FaDu cells transfected with scrambled siRNA (Fig. 5), suggesting that exosome/TRPP2 siRNA complexes may reduce metastasis and invasion by inhibiting EMT. Taken together, these findings indicated that exosomes may be ideal vectors to deliver TRPP2 siRNA into FaDu cells, and that the exosome/TRPP2 siRNA complex is a potential siRNA-based therapy for HNC.
Figure 5.
Effects of TRPP2 siRNA on vimentin, E-cadherin and N-cadherin expression levels in FaDu cells. Western blot images and summary data displaying (A) vimentin, (B) E-cadherin and (C) N-cadherin protein expression levels in FaDu cells transfected with scrambled (Con) or TRPP2 (Exo) siRNA. The optical density of each protein was normalized to GAPDH. Values are presented as the mean ± standard error of the mean (n=3). *P<0.05 vs. control. siRNA, small interfering RNA; TRPP2, transient receptor potential polycystic 2; Exo, exosome; Con, control.
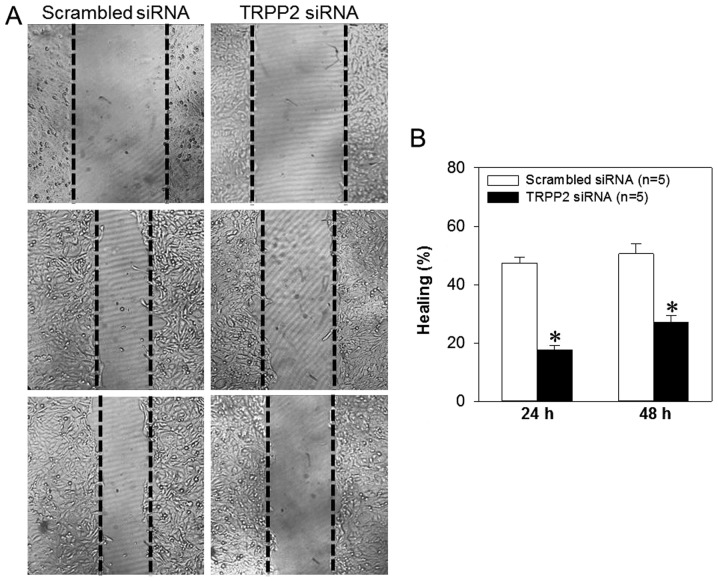
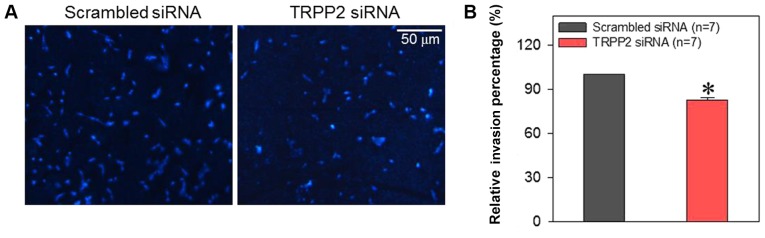
Exosome/TRPP2 siRNA complexes inhibit migration and invasion of FaDu cells
Cell migration and invasion are critically involved in the metastasis of laryngeal cancer. Therefore, FaDu cells were treated with exosome/TRPP2 siRNA complexes in order to determine whether this treatment slowed cell migration and invasion in vitro. When FaDu cells were transfected with exosome/TRPP2 siRNA complexes, wound healing data indicated that the migration speed of the FaDu cells was significantly decreased (Fig. 6). To investigate the effect of exosome/TRPP2 siRNA complexes on cell invasion, exosome/TRPP2 siRNA complex-transfected FaDu cells were cultured in Matrigel-coated Transwell inserts to simulate invasion through an extracellular matrix. The results demonstrated that compared with FaDu cells transfected with scrambled control siRNA, fewer of the cells transfected with exosome/TRPP2 siRNA complexes moved through the matrix (Fig. 7). These results indicated that exosome/TRPP2 siRNA complexes may inhibit FaDu cell migration and invasion in vitro.
Figure 6.
Effect of exosome/TRPP2 siRNA complexes on wound healing in FaDu cells. (A) Microscopy images (magnification, ×40) displaying the distance closed by FaDu cells transfected with scrambled siRNA or exosome/TRPP2 siRNA complexes at 0 h (top row), 24 h (middle row) and 48 h (bottom row) following the creation of the scratch. (B) Summary data presenting wound healing percentages at 24 and 48 h following scratching. Values are displayed as the mean ± standard error of the mean (n=5). *P<0.05 vs. respective scrambled siRNA group. siRNA, small interfering RNA; TRPP2, transient receptor potential polycystic 2.
Figure 7.
Effect of exosome/TRPP2 siRNA complexes on the invasiveness of FaDu cells. (A) Fluorescence images and (B) summary data displaying the invasiveness of FaDu cells transfected with scrambled siRNA or exosome/TRPP2 siRNA complexes. The cells that moved through the membrane were stained with DAPI (blue). Values are presented as the mean ± standard error of the mean. *P<0.05 vs. scrambled siRNA. siRNA, small interfering RNA; TRPP2, transient receptor potential polycystic 2.
Discussion
The present study investigated whether exosomes have the potential to be used for the delivery of siRNA into FaDu cells, and the role of the exosome/TRPP2 siRNA complex in the metastatic processes of these cells. The principal findings were as follows: i) Exosomes from 293 cells effectively encapsulated TRPP2 siRNA in a concentration-dependent manner, and the exosome/TRPP2 siRNA complex was stable in the presence of nucleases or serum obtained from mice; ii) FaDu cells were able to uptake exosome/TRPP2 siRNA complexes; iii) knockdown of the TRPP2 gene via exosome/TRPP2 siRNA transfection reduced TRPP2 expression in FaDu cells; iv) exosome/TRPP2 siRNA complex transfection significantly increased E-cadherin while significantly decreasing N-cadherin and vimentin protein expression levels; and v) exosome/TRPP2 siRNA complexes inhibited the ability of FaDu cells to migrate and invade. Taken together, these findings indicated that exosomes are able to deliver TRPP2 siRNA into FaDu cells. Furthermore, exosome/TRPP2 siRNA complexes may inhibit metastasis in HNC by regulating EMT. Therefore, these results provide evidence that further examination of the exosome/TRPP2 siRNA complex is required for the treatment of HNC.
HNC is a leading cause of cancer-associated illness and mortality worldwide, and >600,000 cases are diagnosed every year (24). Cetuximab, platinum and fluorouracil, the most common first-line drugs for the treatment of metastatic and recurrent HNC, are associated with a median overall survival of only 10 months (25,26). Although recent developments in immunotherapy, including checkpoint inhibitors, have been a breakthrough in the treatment of HNC, checkpoint inhibitors only benefit a minority of patients with relapsed and metastatic HNC (27,28). Therefore, effective therapies are still lacking in the treatment of HNC. Previous studies have demonstrated that TRPP2 knockdown significantly decreases ATP-induced Ca2+ release and vimentin and N-cadherin expression levels, yet enhances E-cadherin expression levels in Hep2 cells, suggesting that migration and invasion may be reduced through the inhibition of EMT (29). Those novel findings provided a potential therapeutic target in HNC. However, delivery of siRNA into cancer cells in vivo remained a major barrier due to the limiting characteristics of siRNA, including its polyanionic charge, low cell membrane permeability and low stability in the presence of serum nucleases (30,31). A number of siRNA delivery vehicles, including nonreplicating viruses, oncolytic virus platforms, adenovirus and proteins, have been widely investigated in order to address these challenges (32–35). Although a number of creative vectors have given promising results, their safety, biocompatibility and low transduction efficiency are notable concerns for the delivery of siRNA (30,31). Exosomes, produced endogenously from endosomes by numerous types of cells, are able to fuse with cancer cell membranes naturally, with promising safety, biocompatibility and transduction efficiency (30,36). The results of the present study strongly indicated that exosomes encapsulated TRPP2 siRNA in a concentration-dependent manner and effectively protected TRPP2 siRNA from nuclease degradation. Furthermore, it was demonstrated that FaDu cells took up the exosome/TRPP2 siRNA complexes. These results support the idea of exosomes being ideal carriers for the delivery of siRNA to specific cells. Although this particular exosome delivery system may not represent targeted delivery, the present study provides evidence to suggest that exosome delivery is a potential novel tool for the delivery of siRNA into the cells.
EMT, in which cell adhesion is reduced and cell motility is enhanced, is a common phenomenon in cancer invasion and metastasis, and is associated with decreased E-cadherin expression levels and increased N-cadherin and vimentin expression levels. To inhibit EMT, TRPP2 siRNA was delivered into FaDu cells via exosomes isolated from 293 cells. The results demonstrated that TRPP2 expression levels were decreased significantly following TRPP2 siRNA transfection, and that decreased TRPP2 expression led to significantly decreased N-cadherin and vimentin expression levels and significantly increased E-cadherin expression levels. Together, these findings demonstrated that exosome/TRPP2 siRNA complexes may enter FaDu cells in order to reduce EMT, and may therefore inhibit the invasion and metastasis of FaDu cells. This RNA-based therapy provides a novel approach to the treatment of HNC, with improved biocompatibility and safety in addition to higher efficiency compared with radiotherapy, chemotherapy or immunotherapy.
In summary, it was demonstrated that exosomes isolated from 293 cells encapsulated TRPP2 siRNA in a concentration-dependent manner, and the exosome/TRPP2 siRNA complex remained stable in the presence of nucleases and serum. The FaDu cells effectively took up the exosome/TRPP2 siRNA complexes. Treatment with exosome/TRPP2 siRNA complexes markedly suppressed TRPP2 expression, EMT processes, and migration and invasion in FaDu cells. On the basis of these results, it may be hypothesized that the development of exosome/TRPP2 siRNA complexes as an RNA-based gene therapy in the treatment of HNC is warranted.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from National Natural Science Foundation of China (grant nos. 81371284, 81570403, 81600286 and U1732157); Anhui Provincial Natural Science Foundation (grant nos. 1408085MH157 and 1708085MH187); and the Science and Technology Research Project of Anhui Province (grant no. 1501041147).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
CW, YH and LC isolated and characterized the exosomes and wrote the manuscript. KL, TF, AJ and RZ performed the biological activity tests. XX, BS, JD and YL conceived and initiated the study. YL finalized the manuscript and supervised the entire study.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Liu W, Zhang B, Chen G, Wu W, Zhou L, Shi Y, Zeng Q, Li Y, Sun Y, Deng X, Wang F. Targeting miR-21 with sophocarpine inhibits tumor progression and reverses epithelial-mesenchymal transition in head and neck cancer. Mol Ther. 2017;25:2129–2139. doi: 10.1016/j.ymthe.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elzakra N, Cui L, Liu T, Li H, Huang J, Hu S. Mass spectrometric analysis of SOX11-binding proteins in head and neck cancer cells demonstrates the interaction of SOX11 and HSP90α. J Proteome Res. 2017;16:3961–3968. doi: 10.1021/acs.jproteome.7b00247. [DOI] [PubMed] [Google Scholar]
- 3.Xiang C, Lv Y, Wei Y, Wei J, Miao S, Mao X, Gu X, Song K, Jia S. Effect of EphA7 silencing on proliferation, invasion and apoptosis in human laryngeal cancer cell lines Hep-2 and AMC-HN-8. Cell Physiol Biochem. 2015;36:435–445. doi: 10.1159/000430110. [DOI] [PubMed] [Google Scholar]
- 4.Su Z, Li G, Liu C, Ren S, Deng T, Zhang S, Tian Y, Liu Y, Qiu Y. Autophagy inhibition impairs the epithelial-mesenchymal transition and enhances cisplatin sensitivity in nasopharyngeal carcinoma. Oncol Lett. 2017;13:4147–4154. doi: 10.3892/ol.2017.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu K, Shen B, Jiang F, Xia L, Fan T, Qin M, Yang L, Guo J, Li Y, Zhu M, et al. TRPP2 enhances metastasis by regulating epithelial-mesenchymal transition in laryngeal squamous cell carcinoma. Cell Physiol Biochem. 2016;39:2203–2215. doi: 10.1159/000447914. [DOI] [PubMed] [Google Scholar]
- 6.Natarajan J, Chandrashekar C, Radhakrishnan R. Critical biomarkers of epithelial-mesenchymal transition in the head and neck cancers. J Cancer Res Ther. 2014;10:512–518. doi: 10.4103/0973-1482.137926. [DOI] [PubMed] [Google Scholar]
- 7.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 8.Kurreck J. RNA interference: From basic research to therapeutic applications. Angew Chem Int Ed Engl. 2009;48:1378–1398. doi: 10.1002/anie.200802092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.López-Fraga M, Martinez T, Jiménez A. RNA interference technologies and therapeutics. From basic research to products BioDrugs. 2009;23:305–332. doi: 10.2165/11318190-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozcan G, Ozpolat B, Coleman RL, Sood AK, Lopez-Berestein G. Preclinical and clinical development of siRNA-based therapeutics. Adv Drug Deliv Rev. 2015;87:108–119. doi: 10.1016/j.addr.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Chen W, Xie H, Wei X, Yin S, Zhou L, Xu X, Zheng S. Biocompatible, chimeric peptide-condensed supramolecular nanoparticles for tumor cell-specific siRNA delivery and gene silencing. Chem Commun (Camb) 2014;50:7806–7809. doi: 10.1039/C4CC01061B. [DOI] [PubMed] [Google Scholar]
- 12.Lai RC, Yeo RW, Tan KH, Lim SK. Exosomes for drug delivery-a novel application for the mesenchymal stem cell. Biotechnol Adv. 2013;31:543–551. doi: 10.1016/j.biotechadv.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Tan A, Rajadas J, Seifalian AM. Exosomes as nano-theranostic delivery platforms for gene therapy. Adv Drug Deliv Rev. 2013;65:357–367. doi: 10.1016/j.addr.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 15.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Zhang L, Li Y, Chen L, Wang X, Guo W, Zhang X, Qin G, He SH, Zimmerman A, et al. Exosomes/microvesicles from induced pluripotent stem cells deliver cardioprotective miRNAs and prevent cardiomyocyte apoptosis in the ischemic myocardium. Int J Cardiol. 2015;192:61–69. doi: 10.1016/j.ijcard.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rider MA, Hurwitz SN, Meckes DG., Jr ExtraPEG: A polyethylene glycol-based method for enrichment of extracellular vesicles. Sci Rep. 2016;6:23978. doi: 10.1038/srep23978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao R, Zhou M, Li J, Wang X, Su K, Hu J, Ye Y, Zhu J, Zhang G, Wang K, et al. Increased TRPP2 expression in vascular smooth muscle cells from high-salt intake hypertensive rats: The crucial role in vascular dysfunction. Mol Nutr Food Res. 2015;59:365–372. doi: 10.1002/mnfr.201400465. [DOI] [PubMed] [Google Scholar]
- 19.Hurwitz SN, Nkosi D, Conlon MM, York SB, Liu X, Tremblay DC, Meckes DG., Jr CD63 regulates epstein-barr virus LMP1 exosomal packaging, enhancement of vesicle production, and noncanonical NF-κB signaling. J Virol. 2017;91(pii):e02251–16. doi: 10.1128/JVI.02251-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khushman M, Bhardwaj A, Patel GK, Laurini JA, Roveda K, Tan MC, Patton MC, Singh S, Taylor W, Singh AP. Exosomal markers (CD63 and CD9) expression pattern using immunohistochemistry in resected malignant and nonmalignant pancreatic specimens. Pancreas. 2017;46:782–788. doi: 10.1097/MPA.0000000000000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Escola JM, Kleijmeer MJ, Stoorvogel W, Griffith JM, Yoshie O, Geuze HJ. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J Biol Chem. 1998;273:20121–20127. doi: 10.1074/jbc.273.32.20121. [DOI] [PubMed] [Google Scholar]
- 22.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–D1244. doi: 10.1093/nar/gkr828. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsiokas L. Function and regulation of TRPP2 at the plasma membrane. Am J Physiol Renal Physiol. 2009;297:F1–F9. doi: 10.1152/ajprenal.90277.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 25.Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, Erfan J, Zabolotnyy D, Kienzer HR, Cupissol D, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359:1116–1127. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- 26.Argiris A, Harrington KJ, Tahara M, Schulten J, Chomette P, Ferreira Castro A, Licitra L. Evidence-based treatment options in recurrent and/or metastatic squamous cell carcinoma of the head and neck. Front Oncol. 2017;7:72. doi: 10.3389/fonc.2017.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferris RL, Blumenschein G, Jr, Fayette J, Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE, Even C, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016;375:1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chow LQ, Haddad R, Gupta S, Mahipal A, Mehra R, Tahara M, Berger R, Eder JP, Burtness B, Lee SH, et al. Antitumor activity of pembrolizumab in biomarker-unselected patients with recurrent and/or metastatic head and neck squamous cell carcinoma: Results from the phase Ib KEYNOTE-012 expansion cohort. J Clin Oncol. 2016;34:3838–3845. doi: 10.1200/JCO.2016.68.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J, Guo J, Yang Y, Jiang F, Chen S, Wu K, Shen B, Liu Y, Du J. Tumor necrosis factor alpha accelerates Hep-2 cells proliferation by suppressing TRPP2 expression. Sci China Life Sci. 2017;60:1251–1259. doi: 10.1007/s11427-016-9030-5. [DOI] [PubMed] [Google Scholar]
- 30.Lee K, Jang B, Lee YR, Suh EY, Yoo JS, Lee MJ, Lee JY, Lee H. The cutting-edge technologies of siRNA delivery and their application in clinical trials. Arch Pharm Res. 2018;41:867–874. doi: 10.1007/s12272-018-1069-4. [DOI] [PubMed] [Google Scholar]
- 31.Liu F, Wang C, Gao Y, Li X, Tian F, Zhang Y, Fu M, Li P, Wang Y, Wang F. Current transport systems and clinical applications for small interfering RNA (siRNA) drugs. Mol Diagn Ther. Jun 20; doi: 10.1007/s40291-018-0338-8. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 32.Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 33.Zhang YA, Nemunaitis J, Samuel SK, Chen P, Shen Y, Tong AW. Antitumor activity of an oncolytic adenovirus-delivered oncogene small interfering RNA. Cancer Res. 2006;66:9736–9743. doi: 10.1158/0008-5472.CAN-06-1617. [DOI] [PubMed] [Google Scholar]
- 34.DeGroot LJ, Zhang R. Viral mediated gene therapy for the management of metastatic thyroid carcinoma. Curr Drug Targets Immune Endocr Metabol Disord. 2004;4:235–244. doi: 10.2174/1568008043339875. [DOI] [PubMed] [Google Scholar]
- 35.Yao YD, Sun TM, Huang SY, Dou S, Lin L, Chen JN, Ruan JB, Mao CQ, Yu FY, Zeng MS, et al. Targeted delivery of PLK1-siRNA by ScFv suppresses Her2+ breast cancer growth and metastasis. Sci Transl Med. 2012;4:130ra148. doi: 10.1126/scitranslmed.3003601. [DOI] [PubMed] [Google Scholar]
- 36.Darband SG, Mirza-Aghazadeh-Attari M, Kaviani M, Mihanfar A, Sadighparvar S, Yousefi B, Majidinia M. Exosomes: Natural nanoparticles as bio shuttles for RNAi delivery. J Control Release. 2018;289:158–170. doi: 10.1016/j.jconrel.2018.10.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.