Abstract
Background:
Unbiased screening studies have repeatedly identified actin-related proteins as one of the families of proteins most influenced by neurotrauma. Nevertheless, the status quo model of cytoskeletal reorganization after neurotrauma excludes actin and incorporates only changes in microtubules and intermediate filaments. Actin is excluded in part because it is difficult to image with conventional techniques. However, recent innovations in fluorescent microscopy provide an opportunity to image the actin cytoskeleton at super-resolution resolution in living cells. This study applied these innovations to an in vitro model of neurotrauma.
New Method:
New methods are introduced for traumatizing neurons before imaging them with high speed structured illumination microscopy or lattice light sheet microscopy. Also, methods for analyzing structured illumination microscopy images to quantify post-traumatic neurite dystrophy are presented.
Results:
Human induced pluripotent stem cell-derived neurons exhibited actin organization typical of immature neurons. Neurite dystrophy increased after trauma but was not influenced by jasplakinolide treatment. The F-actin content of dystrophies varied greatly from one dystrophy to another.
Comparison with Existing Methods:
In contrast to fixation dependent methods, these methods capture the evolution of the actin cytoskeleton over time in a living cell. In contrast to prior methods based on counting dystrophies, this quantification scheme parameterizes the severity of a given dystrophy as it evolves from a local swelling to an almost-perfect spheroid that threatens to transect the neurite.
Conclusions:
These methods can be used to investigate genetic factors and therapeutic interventions that modulate the course of neurite dystrophy after trauma.
Keywords: Structured illumination microscopy, lattice light sheet microscopy, dystrophy, traumatic brain injury, human induced pluripotent stem cell derived neurons
Introduction:
Every year in the United States, traumatic brain injury (TBI) leads to about 56,000 deaths, 282,000 hospitalizations and 2.5 million emergency room visits[1]. Some consequences of TBI, such as skull fracture, are visible to the naked eye while others, such as contusion, hemorrhage and edema can be readily detected in computed tomography or magnetic resonance imaging. However, conventional imaging sequences cannot detect injury to neurites, which manifests in post-mortem histopathology as dystrophy i.e. the normally cylindrical neurites become misshapen, often assuming a ‘string of beads’ morphology [2]. Neurites can be extended up to 70% in vitro without snapping [3] and primary axotomy (i.e. instantaneous transection of a neurite by mechanical overload) is unlikely in closed head injury [4]. Nevertheless, hyperextension causes cellular dysfunction and/or death in the hours following injury. Swellings loaded with intracellular cargo vesicles emerge and intervening regions of the neurite narrow, creating the characteristic ‘string of beads’ morphology. In some cases, the neurite narrows down to nothing at the edge of a bead and pinches off, effectively transecting the neurite long after the initial mechanical insult. This process is known as secondary axotomy [4].
Cytoskeletal reorganization plays an important role in post-traumatic neurite dystrophy. After trauma, intermediate filaments compact [5] and microtubules become disrupted [6]. Local swellings or beads form near sites of microtubule disruption. These observations have motivated a model in which beads form because axonal transport fails and creates accumulations of intracellular cargo vesicles at sites of microtubule disruption [7]. This status quo model of post-traumatic neurite dystrophy does not include actin. Nevertheless, the actin cytoskeleton has the potential to play an important role in neurite dystrophy. Actin disruption generated a ‘string of beads’ morphology in non-neuronal cells with long processes [8]. Also, actin associated proteins have been identified in unbiased screening experiments as being particularly sensitive to trauma [9, 10]. Therefore, the role of the actin reorganization in neurite dystrophy after trauma merits investigation. There are convenient agents that modulate actin organization. For example, cytochalasin D and latrunculin disrupt actin organization while jasplakinolide, which is applied in this study, stabilizes actin organization. Nevertheless, investigations of actin reorganization after trauma have been impossible until recently due to the absence of appropriate imaging tools.
Prior studies of cytoskeletal changes in the neurite after trauma have relied on electron microscopy (EM) [11–13]. This technique excels at imaging microtubules and intermediate filaments but it struggles to capture images of the actin cytoskeleton because most EM fixation protocols disrupt actin [14]. Fortunately, super-resolution fluorescent microscopy tools have recently emerged that can image the actin cytoskeleton of the neurite in living neurons without fixation [15–17]. These tools have revealed that actin is organized in thick fibers running longitudinally throughout the neurite in immature neurons [18]. As neurons mature, a periodic membrane skeleton (PMS) replaces these fibers. The periodic membrane skeleton is a series of alternating, circumferential hoops of actin and spectrin supporting the plasma membrane [19]. It emerges first in the axons and then later in the dendrites [20]. Retraction of injured neurites requires disruption of the PMS [21], suggesting that the actin cytoskeleton can play an important role in cell morphology under stress.
A neurite transection trauma model that has previously been used to investigate the effects of neurotrauma on actin in the neurite was adopted for this experiment [22]. High speed structured illumination microscopy (SIM) was used to quantify the effect of trauma on actin in the neurite. A novel image analysis algorithm was used to quantify neurite dystrophy in these images. The trends noted in these images were further investigated by two channel imaging with a lattice light sheet microscope (LLSM). Experiments were conducted to test the hypothesis that transection would render the neurite dystrophic. Jasplakinolide was applied in some cases to determine if stabilizing the actin cytoskeleton would prevent neurites from becoming dystrophic after trauma.
Materials and Methods:
Cell Culture and Neurite Transection:
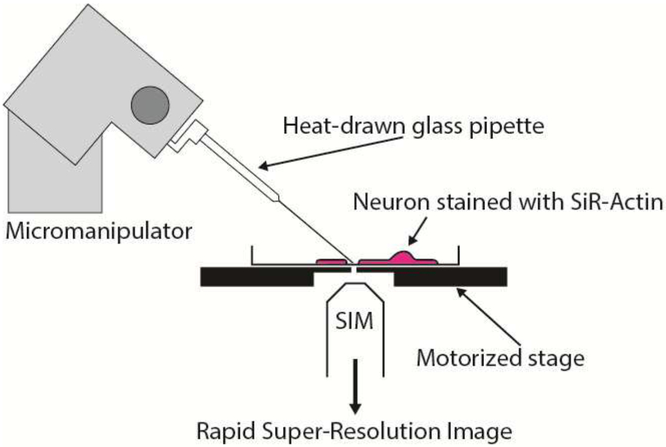
Human induced pluripotent stem cell-derived neurons (hiPSCNs) from a commercial supplier (Cellular Dynamics Inc., Madison, WI) were plated at 67,500 cells/cm2 in complete medium (iCell Neurons Maintenance Medium + iCell Neurons Medium Supplement + penicillin-streptomycin) on coverslips coated with 0.1 mg/mL poly-Lornithine (PLO). 50 μg/mL of laminin was added to the cell suspension at the time of plating. Experiments were performed on day in vitro (DIV) 3–6. The different geometries of the two microscopes necessitated different approaches to in vitro neurite transection. For SIM experiments, neurites were transected using a method based on that presented by Gallo [22]. The SIM employs a conventional, inverted microscope stand. For SIM experiments, neurites were transected using a glass capillary mounted on a micromanipulator (Leica). Either 1 μM jasplakinolide or vehicle (0.1% DMSO) was spiked into the culture media 20 minutes before the experiment. Borosilicate glass capillaries with an outer diameter of 1 mm and an inner diameter of 0.58 mm (Harvard Apparatus, 30–0019) were drawn to a narrow tip using a micropipette puller (Sutter Model P-97). To transect a neurite, the capillary needles were dipped in a 2 mg/ml solution of bovine serum albumin (BSA) in phosphate buffered saline (PBS) and then mounted in a micromanipulator. The lid was removed from the heated stage insert to allow access and the tip of the capillary was brought close to the surface of the cover glass. A suitable neurite was identified in a bright field image and the tip of the capillary needle was lowered into contact with the cover glass close to, but not on, this neurite. Then, the stage was translated rapidly so that the neurite moved across the tip of the needle and was transected (see Fig. 1). Transection was confirmed using the bright field image, the micromanipulator was withdrawn and the lid of the heated chamber was replaced.
Figure 1:
Schematic of transection apparatus for SIM experiments.
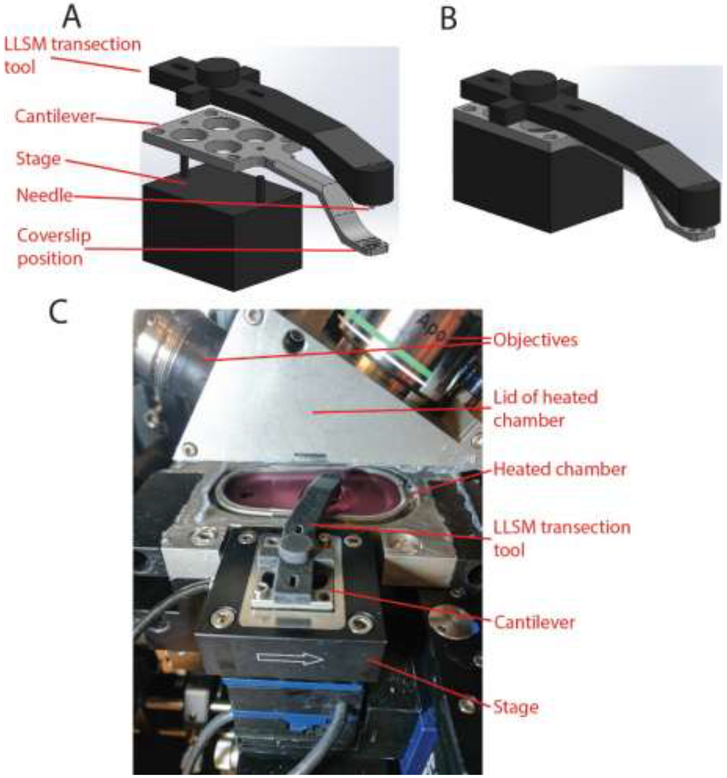
The LLSM does not have a conventional microscope stand or stage. The sample is surrounded by an inverted lens and a pair of upright objectives and is enclosed on all sides by a heated chamber during imaging. For LLSM experiments, the cover glass mounts on the end of a cantilever that sits on a positioning stage mounted outside the heated chamber (see Fig. 2). The cantilever passes through a small opening in the side of the heated chamber to position the coverslip appropriately with respect to the lenses. The cantilever engages with a pair of dowel pins on the stage that determine its position. A special tool was designed and fabricated to transect neurites in this setting. A structure was 3D printed with slots to engage the dowel pins on the stage and an arm that reached along the path of the cantilever. A hole was drilled in the end of this arm and a 0.9 mm diameter blunt ended needle was inserted in the hole and secured with two-part epoxy. The resulting tool is henceforth referred to as the LLSM transection tool. To transect neurites, the heated chamber surrounding the coverslip was opened and the LLSM transection tool was placed on top of the sample holder on the dowel pins. The needle in the LLSM transection tool was long enough to rest lightly on the cover glass when the tool was positioned in this manner. Sliding the tool along the slots engaging the dowel pins then transected neurites along a predictable, repeatable path on the coverslip. An .stl file of the tool geometry is provide in supplementary materials (see llsm_tool.stl).
Figure 2:
Transection apparatus for LLSM experiments. (A) Exploded view of the transection tool sitting on the cantilever which in turn sits on the stage. (B) Actual configuration of the transection tool, cantilever and stage for transection. (C) The transection tool in use. In this image, the lid of the heated chamber has been lifted to allow access to the sample. Note that the space around the sample is too small to accommodate a glass pipette mounted on a micromanipulator, even when the lid is open. The transection tool slides along the slots in the back end so that the needle protruding from the front end scratches the glass coverslip in the sample holder at a predictable location. Then, the transection tool is removed and the heated chamber is closed without moving the sample to begin imaging.
Structured Illumination Microscopy (SIM):
The structured illumination microscope used in these experiments is housed in the Advanced Imaging Center (AIC) at the Howard Hughes Medical Institute’s Janelia Research Campus and is optimized for speed [23]. 3D-SIM imaging was performed as previously described [24]. Briefly, excitation patterns were produced using a phase-only spatial light modulator (Bolder Vision Optik, BVO AHWP3). A mask system was used to select specifically the 0 and ±1 diffraction orders, which were then focused onto the back focal plane of a Zeiss Plan-Apochromat 100× 1.46NA objective. For optimal interference contrast in the sample, the polarity of the light was rotated to match the angle of the pattern using a liquid crystal variable retarder (LC, Meadowlark, SWIFT) and wave plates. Emissions were collected using interference filters and imaged onto a pair of sCMOS cameras (Hamamatsu, Orca Flash 4.0 v2 sCMOS). Samples were maintained at 37 °C and 5% CO2 using a stage-top incubator (H301, Okolabs, Naples, Italy). Raw image sets were reconstructed as described previously [25]. Cultures were stained overnight with 100 nM SiR-Actin in complete medium. One hour before the transection experiment, the cultures were moved into neurobasal medium containing 50 nM Sir-Actin, 15 mM HEPES and 2 μM verapamil and maintained in this medium throughout imaging. 1 μM jasplakinolide (or 0.1% DMSO vehicle) was spiked into the culture medium 20 minutes before the transection experiment. Images were acquired proximal to the site of transection. In this study, a complete 3D SIM image stack was acquired in 7.6–7.7 seconds, depending on how the top and bottom levels of the stack were set. Acquisition time reflects the time required to collect multiple images per plane at different phase angles and repeat this process for every plane in the stack. Therefore, it depends on the exposure time for each image, which was 20 ms for this study. Acquisition time could in theory be shortened by reducing the exposure time but this would also reduce the intensity of the image. Therefore, the minimum practical exposure time varies between studies based on the intensity of the fluorescent signal. A time series of 5 SIM images at 10 minute intervals was acquired from each cell. The spatial resolution of 3D SIM is 130 nm in the lateral direction and 400 nm in the axial direction for red light [23].
Lattice Light Sheet Microscopy (LLSM):
The lattice light sheet microscope (LLSM) used in these experiments is housed in the Advanced Imaging Center (AIC) at the Howard Hughes Medical Institute’s Janelia Research Campus. The system is configured and operated as previously described [26]. Briefly, cells were grown on 5 mm round glass coverslips (Warner Instruments, Catalog # CS-5R). Samples were illuminated by lattice light-sheet using 560 nm or 642 nm diode lasers (MPB Communications) at 30 % acousto-optic tunable filter (AOTF) transmittance and 50 mW initial box power through an excitation objective (Special Optics, 0.65 NA, 3.74-mm WD). Fluorescent emission was collected by detection objective (Nikon, CFI Apo LWD 25XW, 1.1 NA), and detected by a sCMOS camera (Hamamatsu Orca Flash 4.0 v2). Acquired data were deskewed as previously described [26] and deconvolved using an iterative Richardson-Lucy algorithm. Point-spread functions for deconvolution were experimentally measured using 200nm tetraspeck beads adhered to 5 mm glass coverslips (Invitrogen, Catalog # T7280) for each excitation wavelength. The night before the experiment, cultures were stained with 5 μM DiI in complete media for 20 min, then washed 3 times with complete media. 25 nM SiR-Actin in complete media was applied to the cells overnight. This staining solution was replaced with an identical, freshly prepared solution on the morning of the experiment. Cultures were imaged in 15 mM HEPES, 25 nM SiR-Actin, and 2μM Verapamil in neurobasal medium with 1× B27 supplement and 1× penicillin-streptomycin. The manufacturers of SiR-Actin recommend supplementing the media with verapamil to prevent the cells from pumping out the stain, thereby diminishing the fluorescent signal. Images were acquired proximal to the site of transection. A 30 ms exposure time was used so that it took 62.9 ms to acquire a two channel image at each plane of the stack (this value exceeds 60 ms because a small amount of time is required to read data from the camera). The total acquisition time for a 3D image stack depended on the size of the stack. 134 planes leading to an 8.4 second image stack acquisition time was a typical value. Acquisition intervals varied and are presented in the captions for the Movies 1, 3 and 4. In LLSM, spatial resolution in the imaging plane is determined by the standard Abbe formula familiar from conventional microscopy (Resolution = Wavelength / (2 × Numerical Aperture)). In plane resolution is 230 nm for light emitted by green fluorescent protein [26]. This value increases by 12% and 32% for DiI and SiR-Actin respectively because they emit at longer wavelengths. The resolution normal to the imaging plane can be as low as 370 nm [26] although this is a function of how the illumination light sheet is configured. In this study, the light sheet was configured to provide resolution of 500 nm normal to the imaging plane. Z step sizes during data collection were chosen to satisfy the Nyquist criterion; in most experiments Z stepping was oversampled to maximize the axial resolution.
Feature Quantification:
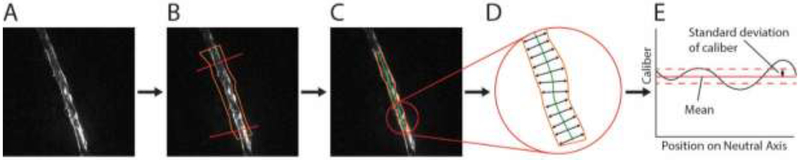
The degree of dystrophy in a neurite was quantified using SIM images. The 3D-SIM image stack was first converted into a maximum intensity projection along the z axis using the open source image processing software, Fiji. Then, a semi-automated process was used to segment a region of interest in MATLAB (MathWorks) (see Fig. 3). An operator blinded to experimental group defined the upper and lower limit of the region of interest (ROI) and then manually selected the corners of a polygon approximating the edge of the neurite between these limits. This polygonal shape was evolved using active contours toward the true segmentation of the neurite. Once the ROI was segmented, the neutral axis of the segmented area was obtained using a distance transform. Normals from the neutral axis were projected from every point along its length. The mean normal distance between the neutral axis and the edge of the segmentation was defined as the local caliber at every pixel on the neutral axis. The standard deviation of the local caliber along the neutral axis was used as a measure of dystrophy in a given image (see Fig. 3).
Figure 3:
Process for quantifying dystrophy. (A) The 3D-SIM image is converted into a maximum intensity projection. (B) A region of interest and an approximation of the segmentation are defined manually on the projected image. (C) Active contours are used to evolve the approximate segmentation towards the true segmentation. (D) The neutral axis is defined using distance transforms. The distance between the neutral axis and the edge along a line perpendicular to the neutral axis is determined and defined as the local caliber. (E) The standard deviation of the local caliber along the neutral axis is calculated to quantify the degree of dystrophy in the region of interest.
Results:
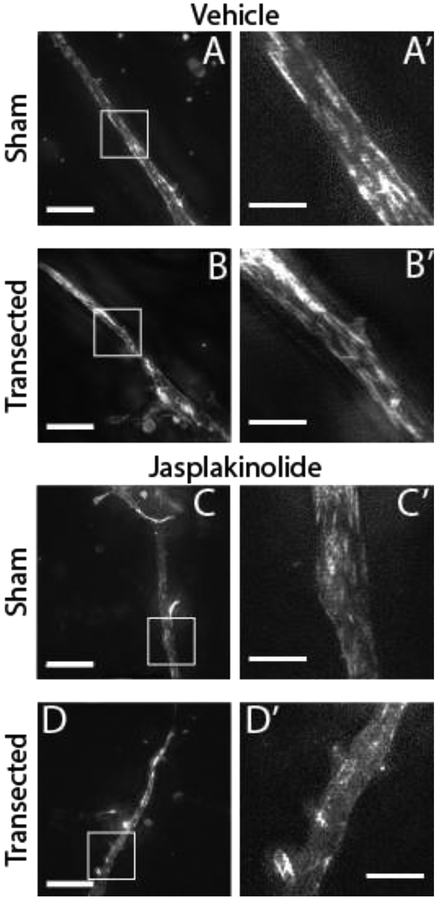
3D-SIM of SiR-Actin stained cultures revealed bundles of F-actin filaments in vehicle treated cells. These bundles were generally oriented more longitudinally than transversely with respect to the axis of the neurite. The neurite cortex stained brightly in some places but bundles were also observed in the interior of the neurite. Where cortical staining occurred, it was not periodic (see Fig. 4 and 5). There were fewer obvious actin filaments in jasplakinolide-treated neurites. However, the intensity of the SiR-Actin signal was also reduced in jasplakinolide-treated neurites, possibly because the SiR-Actin molecule is a derivative of jasplakinolide [16] and may compete with it for binding sites.
Figure 4:
Neurites contain longitudinal bundles of F-actin that are diminished by jasplakinolide treatment but not by transection. (A-D) Maximum intensity projections of 3D-SIM images of neurites at the first time point (mean 2.5 min post transection, SD 1.5 min) stained with SiR-Actin, with and without transection and/or jasplakinolide treatment (scale bars: Left = 10 μm, Right = 3 μm). Note that the dynamic range of the image was optimized independently for each neurite.
Figure 5:
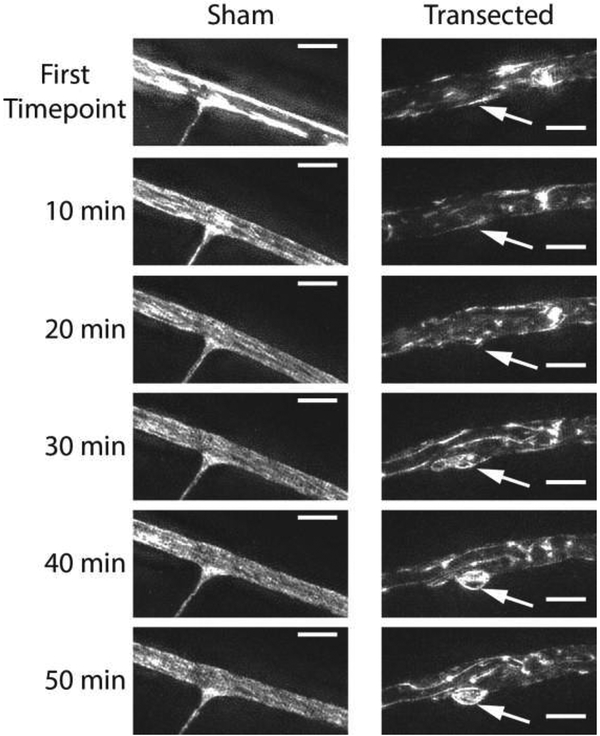
Time series of maximum intensity projections of 3D-SIM images of evolving dystrophic structures. (Left) Untransected neurite, no dystrophy forms. (Right) Region of transected neurite. The shape evolves continuously throughout the 50 minute time series due to the emergence of an F-actin positive dystrophic structure (arrow) (scale bars = 3 μm). Different dynamic ranges were used for the sham and transected time series but the dynamic ranges are consistent within each time series.
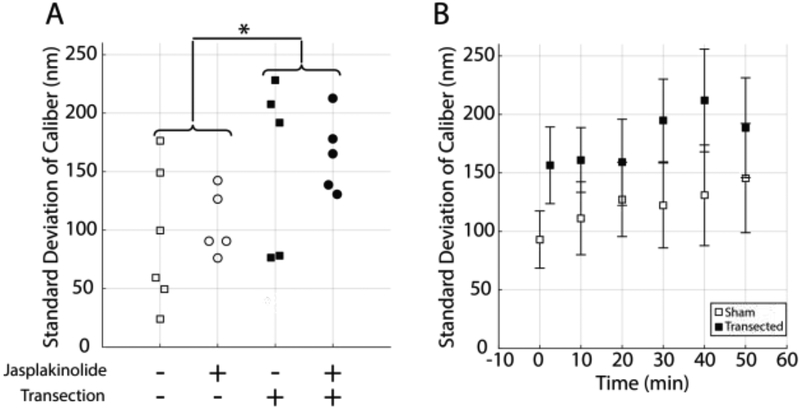
Transection caused a rapid change in the morphology of the neurite. The shapes of transected neurites evolved continuously over time, in contrast to the shapes of sham injured neurites, which remained constant (see Fig. 5). The standard deviation of the neurite caliber (the quantity used to measure neurite dystrophy) was higher in transected neurites than in naïve neurites at the first time point (ANOVA, p < 0.05), which was acquired an average of 2.5 minutes (S.D. 1.5 minutes) after transection (see Fig. 6A). There was no statistically significant effect of jasplakinolide treatment on neurite dystrophy at the first time point and no significant interaction between the effects of jasplakinolide and transection. Jasplakinolide treated neurites bleached more rapidly over time than their vehicle treated counterparts. Therefore, dystrophy was not quantified in jasplakinolide treated cultures after the first time point. In vehicle treated neurites, there was no statistically significant effect of time on neurite dystrophy and no interaction between the effect of time and the effect of transection (ANOVA, see Fig. 6B). Therefore, while the shapes of transected neurites changed over time, they did not on average become significantly more or less dystrophic.
Figure 6:
Transection rapidly increased neurite dystrophy. (A) At the first time point (mean 2.5 min post transection, SD 1.5 min), transection increased the standard deviation of caliber in 3D-SIM images (ANOVA, p < 0.05, n = 5–6). There was no significant effect of jasplakinolide treatment and no interaction between the effects of transection and jasplakinolide treatment. (B) In vehicle-treated cultures, transection increased the standard deviation of caliber across the time series but there was no statistically significant effect of time and no interaction between the effects of transection and time (ANOVA, p < 0.05, error bars = 1 standard error, n=5–6).
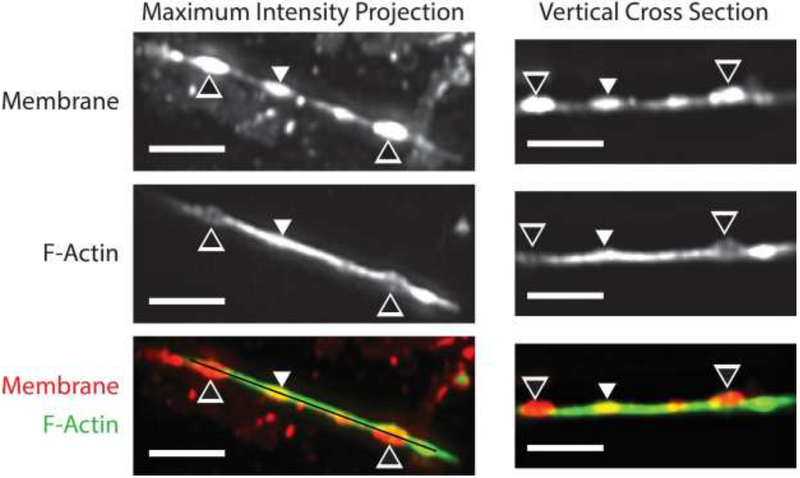
While F-actin positive dystrophic structures were observed in 3D-SIM images of transected neurites, these single channel images could not readily determine if F-actin negative dystrophic structures were also present. Three-dimensional, time series LLSM imaging of cultures stained with DiI and SiR-Actin revealed that neurite dystrophies can in fact be associated with either local loss or local accumulation of F-actin. DiI is a relatively phototoxic stain (this characteristic prohibited its use in 3D-SIM experiments). Nevertheless, cultures stained with both DiI and SiR-Actin tolerated time series LLSM imaging well due to the low phototoxic burden imposed by this instrument. In control experiments on sham injured cells, more than one hundred and fifty 2-channel, 3D images were acquired before phototoxicity-related changes in morphology were observed (see Movie 1). Fig. 7 shows an injured neurite at the first time point acquired after transection (approximately 5 minutes post transection). Neurite dystrophies are visible as bright ellipsoidal structures in the DiI channel (see Fig. 7 and Movie 2). In some of these structures, SiR-Actin signal is depressed relative to the rest of the neurite, implying local loss of F-actin. However, in another instance, SiR-Actin signal is elevated relative to the rest of the neurite and the neurite appears thicker in both the horizontal and vertical planes in the SiR-Actin channel, implying that the F-actin cortex is intact and bulges outwards along with the plasma membrane at this point.
Figure 7:
LLSM image of transected neurite. The black and white arrowheads point to dystrophies with low SiR-Actin signal relative to the rest of the neurite. The white arrowhead points to a dystrophy with high SiR-Actin signal relative to the rest of the neurite. The black line in the bottom left panel indicates the vertical plane of the cross-sectional images in the right column (scale bars = 5 μm).
As in 3D-SIM experiments, post-transection dystrophies formed quickly in LLSM experiments and were frequently already visible in the first image of the time series, acquired 3–5 minutes after transection. Some later dystrophy formation events were captured however. An example is shown in Movie 3 in which a dystrophy emerges over the course of approximately 100 seconds. Post-transection dystrophies were typically persistent in these experiments but fusion of two adjacent dystrophies into a single large dystrophy was also observed. These fusion events occurred rapidly (in 13 – 27 seconds) and unpredictably after varying periods of relative geometric stability post-transection (see Movie 4). Post-transection dystrophies ‘flickered’ (i.e. fluctuated under the action of thermal forces) both before and after fusion, implying that they have relatively low bending stiffness [27].
Discussion:
This study demonstrated the capacity of high speed SIM to quantify rapid change in neurite morphology. It also demonstrated that two channel LLSM with SiR-Actin and DiI staining can visualize changes in the actin cytoskeleton and compare them to changes in the overall shape of the neurite with high spatiotemporal resolution. Prior neuropathological studies of neurite dystrophy after trauma have noted the variety of shapes among dystrophic structures. In some cases, mild local swellings occur while in others, a true string-of-beads morphology emerges consisting of almost perfectly round spheres connected by narrow necks. Ovoid and even bilobulated structures have also been reported [4]. Over time, these swellings may resolve or evolve towards secondary axotomy. Therefore, a dynamic imaging tool that can meaningfully quantify the evolution of neurite dystrophy is urgently needed. Prior studies have counted dystrophies in fixed cultures [28, 29]. Counting captures the number of dystrophies but not their evolving severity. The current method exploits the high spatial resolution of SIM to measure the variation of neurite caliber along the neurite. This approach parameterizes the severity of each dystrophy in a living cell. In this study, dystrophy increased within 2.5 minutes of trauma but did not continue to increase over the following 47.5 minutes relative to shams (see Fig. 6). The effect of trauma at the 2.5 minute time point was not modulated by treatment with jasplakinolide, an agent that stabilizes actin. The fact that actin stabilization did not influence acute post-traumatic dystrophy suggests the observed increase in dystrophy arose from a loss of mechanical tension in the proximal portion of the neurite when it was transected rather than an active actin remodeling process. Jasplakinolide may be more influential at later times. However, this possibility could not be addressed with the current data set because the SiR-Actin signal declined in jasplakinolide treated samples after the first image to the point where it was considered inadequate for accurate segmentation of the neurite. This decline is presumed to be caused by competition between binding of jasplakinolide and SiR-Actin, which have similar chemical structures. Nevertheless, these results confirm that changes in neurite dystrophy can be detected and quantified in living neurites by applying the presented image analysis technique to SIM images of SiR-Actin stained neurites. While active actin remodeling did not determine the overall degree of dystrophy after trauma, there was clearly active, local remodeling of actin in traumatized neurites (see Fig. 5). By contrast, the actin cytoskeleton was stable in sham injured neurites and was not perturbed by repeated imaging (see Fig. 5). These data demonstrate the capacity of high speed SIM to capture post-traumatic remodeling of the actin cytoskeleton in living neurites.
These are, to our knowledge, the first published images of the neuritic actin cytoskeleton in hiPSCNs. They revealed that F-actin was organized into thick, longitudinal fibers distributed throughout the neurite similar to previously reported actin trails [18]. This fibrous architecture is typical of immature neurons [14, 20]. There is no evidence of a PMS, even though the PMS has previously been imaged with SIM [30]. This actin architecture is consistent with other immature phenotypes in this cell type at this time point. They are unpolarized [31] and express a neonatal transcriptomic profile [32]. Little retraction was observed after transection. This response differs from that of mature chick axons, which retracted substantially when transected [22]. This difference could be a consequence of the relatively high laminin concentration used to coat coverslips in this study. Prior work with this cell type by our group has shown that they can retract their neurites [33] after trauma and that high laminin concentrations inhibit retraction [34].
The immaturity of hiPSCNs limits the range of questions that can be addressed with this model. However, it is worth noting that TBI is common in children aged 0–4 years [35]. The infant brain contains migrating, immature neurons until 18 months of age and these cells play an important role in development [36]. Also, techniques exist for accelerating maturation of hiPSCNs including modifying telomerases [37]. Rodent neurons mature rapidly in culture without such interventions, but hiPSCNs address important questions that cannot be addressed with rodent neurons. hiPSCNs retain the genetic identity of the donor patient from which they were generated. In addition, the genome can be edited to alter functionally significant genetic variants one by one [38]. Experiments with these cell populations isolate genetic factors influencing pathology from all the other factors that affect outcomes in the clinic. These methods have already been used to elucidate patient-specific mechanisms in amyotrophic lateral sclerosis[39], Alzheimer’s disease [40] and Parkinson’s disease [41]. There is an urgent need for similar experiments in neurotrauma. More than 30 major phase III clinical trials of TBI therapies have been conducted and all have failed [42], in part because genetic variation among patients obscures treatment effects. In vitro neurotrauma models employing hiPSCNs have the potential to identify genetic modulators of outcome, which would support clinical trial stratification and personalization of therapy.
An insult modeling neurotrauma is easier to achieve with high speed SIM than with the LLSM. High speed SIM employs a conventional microscope stand so the transection method developed by Gallo was applied without modification [22]. In the LLSM, the coverslip is less accessible because the microscope hardware occupies space both above it and below it. Therefore, a simple, novel tool was created to apply a repeatable transection insult in this setting (see Fig. 2). This tool can be fabricated quickly using 3D printing with a single, subsequent drilling step. It could be applied outside neurotrauma for conventional wound healing scratch assays on the LLSM. The high speed SIM is in theory compatible with large, stage mounted devices that simulate trauma by compressing [43] or shearing [44] hydrogels containing neural cells. However, these loading modes would be difficult to recreate on the LLSM. As imaging modalities, LLSM and high speed SIM had different strengths that complemented each other. Together, they offerreda more complete understanding of post-traumatic actin remodeling in the neurite than either could offer alone. The primary advantage of SIM relative to LLSM is its spatial resolution, which exceeds the diffraction limit. This spatial resolution allowed variations of actin organization within a neurite just a few microns wide to be observed (see Figs. 4 and 5). The primary advantage of LLSM relative to SIM is its low phototoxicity. This characteristic allowed two channel imaging with DiI alongside SiR-Actin so changes in actin organization could be compared to changes in the overall shape of the neurite. It also allowed more than a hundred 3D images to be acquired in a single time series (see Movie 1) so that rapid events occurring at unpredictable times could be captured (see Movies 3 and 4).
LLSM imaging placed the response of the actin cytoskeleton in the context of the overall shape change of the neurite. The low phototoxic load applied by the LLSM allowed times series 2 channel imaging to observe changes in the actin cytoskeleton and the overall neurite shape simultaneously (see Fig. 7 and Movie 2). Some post-traumatic swellings on neurites were SiR-Actin positive, consistent with a model in which tense F-actin fibers draw the neurite into a round shape. On the other hand, some post-traumatic swellings on neurites were SiR-actin negative, consistent with a model in which the actin cortex loses rigidity due to disassembly of actin fibers (see Fig. 7). This model of bead formation has previously been advanced for SVT2 cells [8]. It is worth noting that spheroidal shapes have a lower surface-to-volume ratio than cylinders so any process that tends to minimize the surface-to-volume ratio of the neurite forms spheroidal neurite dystrophies. Such dystrophies can be coherently explained by loss of cortical bending stiffness[8], increase in cortical tension[45], osmotic swelling [46] or transport failure[7]. Therefore, neurite dystrophy does not independently prove the existence of any one of these pathologies. The LLSM resolves the shapes of dystrophies sufficiently well to visualize fluctuations in the dystrophy surface over time (see Movies 3 and 4). These motions most likely arise from bending of the surface due to Brownian forces in the environment, a phenomenon known as ‘flickering’[27, 47]. LLSM time series also captured fusion events between neighboring dystrophies. These events occurred rapidly at unpredictable post-transection timepoints (see Movie 4). The capacity of the LLSM to image at high frequency for long periods is particularly useful in this context as it allows these unpredictable, rapid events to be captured.
Conclusions:
Reorganization of actin in the neurite after trauma is less well understood than reorganization of microtubules or intermediate filaments because actin is more difficult to image with EM. Recently developed imaging modalities were applied in this study to develop methods of observing reorganization of the actin cytoskeleton in living, traumatized neurons at high spatiotemporal resolution. Neurite dystrophy increased after trauma but was not influenced by jasplakinolide, an actin-stabilizing agent. This result suggests that in this case, dystrophy arose from loss of mechanical tension in the neurite rather than active reorganization of the actin cytoskeleton. These are the first super-resolution images in the literature of the actin cytoskeleton in the neurites of hiPSCNs. Actin was organized in a manner typical of immature neurons, consistent with other immature phenotypes reported for this cell type. High speed SIM provided sufficient spatial resolution to allow quantification of the variation of the axon caliber. This variation served as a measure of the degree of neurite dystrophy. LLSM provided long duration, high frequency, 3D images of traumatized neurons with spatial resolution approaching the diffraction limit. These images allow observation of unpredictable, rapid events after trauma and allow the remodeling of the actin cytoskeleton to be compared to overall shape change in the neurite.
Supplementary Material
Movie 1: A maximum intensity projection along the imaging axis of the LLSM (which is at a 31.8° angle to the coverslip) of an untransected neurite. No dystrophies were observed immediately or in the short term, but slowly evolving dystrophies apparently due to phototoxicity are observed beginning after approximately 150 frames (28 min 31 sec). Left greyscale and red channel = membrane. Center greyscale and green channel = F-actin. Interval 11.41 sec, playback at 30 frames per second. Scale bar 10 μm.
Movie 2: A rotating view of the neurite shown in Figure 7. Red-Green plane is parallel to the coverslip. Bounding box 8 μm × 8 μm × 32 μm.
Movie 3: A dystrophy emerging slowly after transection (on the right hand side of the field of view). Time step 13.37 seconds, 7 frames per second, bounding box 16 μm × 33 μm × 12 μm.
Movie 4: Dystrophies fusing after transection. Dystrophy fusion occurs within 13–27 seconds. Time step 13.37 seconds, 7 frames per second, bounding box 45 μm × 45 μm × 15 μm.
Highlights:
A novel means of transecting neurites mounted on a lattice light sheet microscope is presented.
A novel means of quantifying neurite dystrophy in a structured illumination microscopy image is presented.
The neurites of hiPSCNs have an immature actin cytoskeletal organization consisting of longitudinal filaments distributed throughout the neurite without periodic circumferential hoops in the neurite cortex.
Transection rapidly perturbs neurite shape but does not affect actin architecture.
2 channel, 3D lattice light sheet microscopy can detect brief, unpredictable events long after trauma and place change in the actin cortex in the context of overall change in neurite shape.
Acknowledgements:
The Advanced Imaging Center is jointly supported by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation. This work was funded in part by the National Institutes of Health (R21NS098129).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Taylor CA, et al. , Traumatic Brain Injury-Related Emergency Department Visits, Hospitalizations, and Deaths-United States, 2007 and 2013. MMWR Surveill Summ, 2017. 66(9): p. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson VE, Stewart W, and Smith DH, Axonal pathology in traumatic brain injury Exp Neurol, 2013. 246: p. 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith DH, et al. , High tolerance and delayed elastic response of cultured axons to dynamic stretch injury. J Neurosci, 1999. 19(11): p. 4263–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maxwell WL, Povlishock JT, and Graham DL, A mechanistic analysis of nondisruptive axonal injury: a review. J Neurotrauma, 1997. 14(7): p. 419–40. [DOI] [PubMed] [Google Scholar]
- 5.Pettus EH, et al. , Traumatically induced altered membrane permeability: its relationship to traumatically induced reactive axonal change. J Neurotrauma, 1994. 11(5): p. 507–22. [DOI] [PubMed] [Google Scholar]
- 6.Tang-Schomer MD, et al. , Mechanical breaking of microtubules in axons during dynamic stretch injury underlies delayed elasticity, microtubule disassembly, and axon degeneration. FASEB J, 2010. 24(5): p. 1401–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang-Schomer MD, et al. , Partial interruption of axonal transport due to microtubule breakage accounts for the formation of periodic varicosities after traumatic axonal injury. Exp Neurol, 2012. 233(1): p. 364–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bar-Ziv R, et al. , Pearling in cells: a clue to understanding cell shape. Proc Natl Acad Sci U S A, 1999. 96(18): p. 10140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loov C, et al. , Identification of injury specific proteins in a cell culture model of traumatic brain injury. PLoS One, 2013. 8(2): p. e55983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garland P, et al. , Soluble axoplasm enriched from injured CNS axons reveals the early modulation of the actin cytoskeleton. PLoS One, 2012. 7(10): p. e47552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yaghmai A and Povlishock J, Traumatically induced reactive change as visualized through the use of monoclonal antibodies targeted to neurofilament subunits. J Neuropathol Exp Neurol, 1992. 51(2): p. 158–76. [DOI] [PubMed] [Google Scholar]
- 12.Povlishock JT, Pathobiology of traumatically induced axonal injury in animals and man. Ann Emerg Med, 1993. 22(6): p. 980–6. [DOI] [PubMed] [Google Scholar]
- 13.Povlishock JT, et al. , Impact acceleration injury in the rat: evidence for focal axolemmal change and related neurofilament sidearm alteration. J Neuropathol Exp Neurol, 1997. 56(4): p. 347–59. [PubMed] [Google Scholar]
- 14.Zhong G, et al. , Developmental mechanism of the periodic membrane skeleton in axons. Elife, 2014. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang X, et al. , Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy. Nat Biotechnol, 2018. 36(5): p. 451–459. [DOI] [PubMed] [Google Scholar]
- 16.Lukinavicius G, et al. , Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat Methods, 2014. 11(7): p. 731–3. [DOI] [PubMed] [Google Scholar]
- 17.Maglione M and Sigrist SJ, Seeing the forest tree by tree: super-resolution light microscopy meets the neurosciences. Nat Neurosci, 2013. 16(7): p. 790–7. [DOI] [PubMed] [Google Scholar]
- 18.Ganguly A, et al. , A dynamic formin-dependent deep F-actin network in axons. J Cell Biol, 2015. 210(3): p. 401–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu K, Zhong G, and Zhuang X, Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science, 2013. 339(6118): p. 452–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’Este E, et al. , STED nanoscopy reveals the ubiquity of subcortical cytoskeleton periodicity in living neurons. Cell Rep, 2015. 10(8): p. 1246–51. [DOI] [PubMed] [Google Scholar]
- 21.Unsain N, et al. , Remodeling of the Actin/Spectrin Membrane-associated Periodic Skeleton, Growth Cone Collapse and F-Actin Decrease during Axonal Degeneration. Sci Rep, 2018. 8(1): p. 3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallo G, Myosin II activity is required for severing-induced axon retraction in vitro. Exp Neurol, 2004. 189(1): p. 112–21. [DOI] [PubMed] [Google Scholar]
- 23.Fiolka R, et al. , Time-lapse two-color 3D imaging of live cells with doubled resolution using structured illumination. Proc Natl Acad Sci U S A, 2012. 109(14): p. 5311–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shao L, et al. , Super-resolution 3D microscopy of live whole cells using structured illumination. Nat Methods, 2011. 8(12): p. 1044–6. [DOI] [PubMed] [Google Scholar]
- 25.Gustafsson MGL, et al. , Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophysical Journal, 2008. 94(12): p. 4957–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen BC, et al. , Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science, 2014. 346(6208): p. 1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brochard F and Lennon JF, FREQUENCY SPECTRUM OF FLICKER PHENOMENON IN ERYTHROCYTES. Journal De Physique, 1975. 36(11): p. 1035–1047. [Google Scholar]
- 28.Monnerie H, et al. , Dendritic alterations after dynamic axonal stretch injury in vitro. Exp Neurol, 2010. 224(2): p. 415–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hemphill MA, et al. , A possible role for integrin signaling in diffuse axonal injury. PLoS One, 2011. 6(7): p. e22899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qu Y, et al. , Periodic actin structures in neuronal axons are required to maintain microtubules. Mol Biol Cell, 2017. 28(2): p. 296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura H, et al. , Quantitative analysis of intraneuronal transport in human iPS neurons. J Pharmacol Sci, 2015. 128(4): p. 170–8. [DOI] [PubMed] [Google Scholar]
- 32.Dage JL, et al. , Pharmacological characterisation of ligand-and voltage-gated ion channels expressed in human iPSC-derived forebrain neurons. Psychopharmacology (Berl), 2014. 231(6): p. 1105–24. [DOI] [PubMed] [Google Scholar]
- 33.Sherman SA, et al. , Stretch Injury of Human Induced Pluripotent Stem Cell Derived Neurons in a 96 Well Format. Sci Rep, 2016. 6: p. 34097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips JK, et al. , Method for High Speed Stretch Injury of Human Induced Pluripotent Stem Cell-derived Neurons in a 96-well Format. J Vis Exp, 2018(134). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faul MXL, Wald MM, Coronado V, Traumatic brain injury in the United States: emergency department visits, hospitalizations, and deaths, 2002–2006. 2010, CDC, National Center for Injury Prevention and Control: Atlanta, GA. [Google Scholar]
- 36.Paredes MF, et al. , Extensive migration of young neurons into the infant human frontal lobe. Science, 2016. 354(6308): p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vera E, Bosco N, and Studer L, Generating Late-Onset Human iPSC-Based Disease Models by Inducing Neuronal Age-Related Phenotypes through Telomerase Manipulation. Cell Rep, 2016. 17(4): p. 1184–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim HS, et al. , Genomic editing tools to model human diseases with isogenic pluripotent stem cells. Stem Cells Dev, 2014. 23(22): p. 2673–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiskinis E, et al. , Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell, 2014. 14(6): p. 781–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Israel MA, et al. , Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature, 2012. 482(7384): p. 216–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beevers JE, Caffrey TM, and Wade-Martins R, Induced pluripotent stem cell (iPSC)-derived dopaminergic models of Parkinson’s disease. Biochem Soc Trans, 2013. 41(6): p. 1503–8. [DOI] [PubMed] [Google Scholar]
- 42.Kabadi SV and Faden AI, Neuroprotective strategies for traumatic brain injury: improving clinical translation. Int J Mol Sci, 2014. 15(1): p. 1216–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bar-Kochba E, et al. , A Fast Iterative Digital Volume Correlation Algorithm for Large Deformations. Experimental Mechanics, 2015. 55(1): p. 261–274. [Google Scholar]
- 44.Cullen DK and LaPlaca MC, Neuronal response to high rate shear deformation depends on heterogeneity of the local strain field. J Neurotrauma, 2006. 23(9): p. 1304–19. [DOI] [PubMed] [Google Scholar]
- 45.Markin VS, et al. , Biomechanics of stretch-induced beading. Biophys J, 1999. 76(5): p. 2852–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Budde MD and Frank JA, Neurite beading is sufficient to decrease the apparent diffusion coefficient after ischemic stroke. Proc Natl Acad Sci U S A, 2010. 107(32): p. 14472–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu FY, Haley SC, and Zidovska A, On the origin of shape fluctuations of the cell nucleus. Proceedings of the National Academy of Sciences of the United States of America, 2017. 114(39): p. 10338–10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1: A maximum intensity projection along the imaging axis of the LLSM (which is at a 31.8° angle to the coverslip) of an untransected neurite. No dystrophies were observed immediately or in the short term, but slowly evolving dystrophies apparently due to phototoxicity are observed beginning after approximately 150 frames (28 min 31 sec). Left greyscale and red channel = membrane. Center greyscale and green channel = F-actin. Interval 11.41 sec, playback at 30 frames per second. Scale bar 10 μm.
Movie 2: A rotating view of the neurite shown in Figure 7. Red-Green plane is parallel to the coverslip. Bounding box 8 μm × 8 μm × 32 μm.
Movie 3: A dystrophy emerging slowly after transection (on the right hand side of the field of view). Time step 13.37 seconds, 7 frames per second, bounding box 16 μm × 33 μm × 12 μm.
Movie 4: Dystrophies fusing after transection. Dystrophy fusion occurs within 13–27 seconds. Time step 13.37 seconds, 7 frames per second, bounding box 45 μm × 45 μm × 15 μm.