Abstract
Midbrain dopamine (DA) neurons have diverse functions that can in part be explained by their heterogeneity. Although molecularly distinct subtypes of DA neurons have been identified by single-cell gene expression profiling, fundamental features such as their projection patterns have not been elucidated. Progress in this regard has been hindered by the lack of genetic tools to study DA neuron subtypes. Here, we develop intersectional genetic labeling strategies, based on combinatorial gene expression, to map the projections of molecularly defined DA neuron subtypes. We reveal distinct genetically-defined DAergic pathways arising from the substantia nigra pars compacta and from the ventral tegmental area that innervate specific regions of the caudate putamen, nucleus accumbens and amygdala. Together, the genetic toolbox and DA neuron subtype projections presented here constitute a resource that will accelerate the investigation of this clinically significant neurotransmitter system.
Introduction
How midbrain dopamine (DA) neurons underpin a spectrum of apparently unrelated behaviors and diseases is enigmatic1,2. One possibility is that the midbrain DAergic system is composed of functionally distinct neuron subtypes. Initially, DAergic neurons were classified into three anatomically-defined midbrain clusters located in the ventral tegmental area (VTA; A10), the substantia nigra pars compacta (SNc; A9), and retrorubral area (RR; A8)2. More recently, there has been increasing evidence of functional heterogeneity within these clusters, with subsets of DA neurons influencing specific behavioral outcomes (reviewed in3–7). Consistent with this, several putative subtypes of midbrain DA neurons have recently been defined on their distinct combinatorial gene expression profiles8–10. These initial single-cell profiling based studies provide a first-generation framework for disentangling the DAergic system into its constituent parts11,12. Key unresolved questions are whether these molecularly defined DA neuron subtypes have unique axonal projections, synaptic inputs, physiological properties, or functional roles.
The ability to genetically target DA neuron subtypes is critical for resolving these questions. Intersectional genetic paradigms exploit the expression of two or more genes with overlapping expression patterns to target a cell population with high selectivity13,14. Here, we developed three distinct intersectional genetic strategies to target DA neuron populations with fluorescent proteins and used these strategies to determine their broad axonal projections. Our results indicate that molecularly defined DA neuron subtypes have projection patterns that, despite some overlap, are unexpectedly distinctive even within defined target regions such as the caudate-putamen, nucleus accumbens, and amygdala.
Results
Generation and validation of the Th–2A–Flpo mouse line
To develop an intersectional platform for targeting DA neurons, we generated a Flp recombinase line driven from the tyrosine hydroxylase (Th) locus, a gene encoding the rate limiting enzyme in DA synthesis (Fig. 1A; see Methods). To determine the recombination potential of the Th–2A–Flpo driver, we crossed it with the Flp dependent reporters RC::FA (nlacZ, Fig. 1B) and RC::Frepe (mCherry, Fig. S1)13. Within midbrain DA clusters, we observed reporter staining within the SNc, VTA, interfascicular (IF), RR, and caudal linear (CLi) (Fig. 1C and S1). We found that most TH+ cells were recombined (βgal+TH+/total TH+; 99.9% ±0.01), indicating the Th-2A-Flpo line is highly efficient. We next quantified the percentage of βgal+ cell that co-express TH, and found high specificity of labeling (βgal+TH+/total βgal+; 93.0% ±0.1). The lower percentage of βgal+ cells that co-localize with TH, particularly observed in the IF, CLi and PAG/DR, is consistent with observations using Th-ires-Cre15,16, a fact that needs to be considered in the design and interpretation of experiments. At the level of the midbrain, we also observed labeled neurons in the region englobing the rostral linear and posterior hypothalamic (RLi/PH; FOXA2+, NURR1+, Ddc+, Dat–), as well as the interpeduncular nucleus (IPN; FOXA2–, NURR1–, Ddc–, Dat–; Fig. S2). Reporter was present in most brain regions with Th mRNA expression, including cortical interneurons, striatal interneurons, neurons in the medial forebrain, hypothalamic DA nuclei, subsets of Purkinje neurons, and hindbrain catecholaminergic cell groups (Fig. S3).
Figure 1. Generation and validation of Th–2A–Flpo.
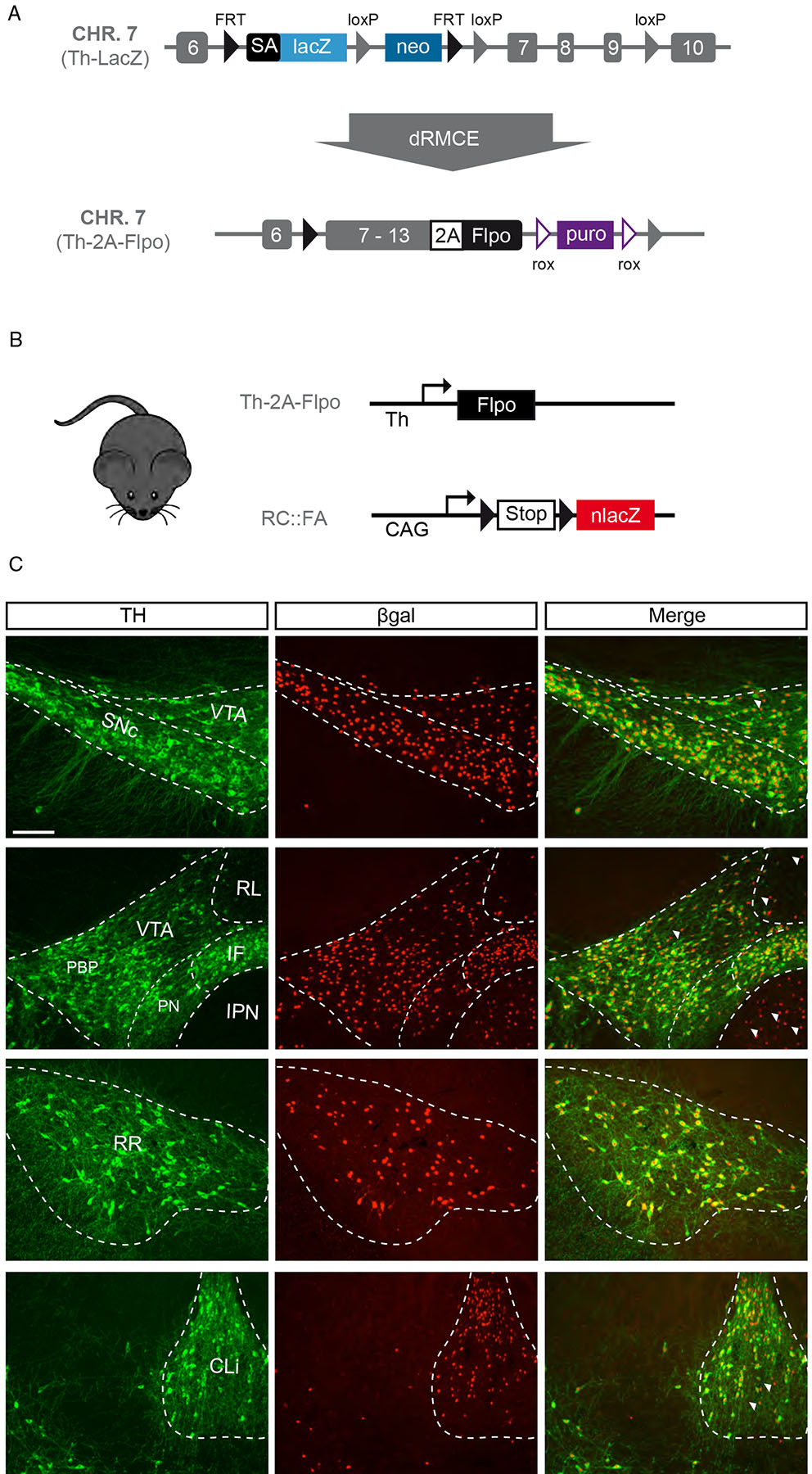
(A) We used dual Recombinase Mediated Cassette Exchange (dRMCE) to reengineer a previously generated Th–LacZ allele in which lacZ expression is driven by the tyrosine hydroxylase (Th) transcription by virtue of a splice acceptor (SA) located in the intron between the 6th and 7th exon. We converted this allele and replaced lacZ by exons 7 to 13 of the Th coding sequence, followed by the P2A peptide, and the recombinase Flpo.(B) Simplified schematic of Th–2A–Flpo crossed to reporter mouse RC::FA, designed to activate nlacZ expression in midbrain DA neuron clusters resulting in nuclear βgal expression (black triangles = FRT sites). (C) In Th–2A–Flpo; RC::FA midbrain, the large majority of TH+ neurons (green) of the substantia nigra pars compacta (SNc; 99.9% (±0.1)), the ventral tegmental area (VTA; 100.0%), interfascicular nucleus (IF; 100.0%), retrorubral area (RR; 100.0%), caudal linear nucleus (CLi; 100.0%), and periaqueductal gray/dorsal raphe regions (PAG/DR; 99.6% (±0.2); not shown) are βgal+ (red). We observed some βgal+ cells are TH– (examples are indicated with arrowhead), particularly in the rostral linear nucleus (RLi), interpeduncular nucIeus (IPN), and CLi. The fact that TH protein synthesis is required for Flpo production, because of effective P2A “autocatalytic” activity, suggests that these recombined cells produced TH protein, at least at some point during their developmental history. However, the majority of βgal+ cells also expressed TH in all DA neuron clusters: SNc (98.9% ±0.3), VTA (97.5% ±1.2), IF (85.8% ±5.8), CLi (88.1% ±2.4), RR (97.6% ±1.5) and the PAG/DR (68.4% ±5.1; not shown). n = 3.. PN = paranigral region, PBP = parabrachial pigmented area. Scale bar = 100 μm
Intersectional genetic strategies to map the projections of midbrain DA neuron subtypes
We next devised three distinct intersectional genetic strategies to target putative DA neuron subtypes. Strategy I uses a reporter mouse that labels with tdTomato only cells that express both Cre and Flpo recombinases (Ai65)17 (Fig. 2A). This approach is not limited by the diffusion of viral particles and therefore provides a more complete picture of the intersectional neuron population. However, one limitation of this approach is that several Cre lines may intersect with Th expression outside the midbrain, thereby labeling unwanted cells. Additionally for genes that have broader expression in DA neurons during development, this approach will label more cells than represented by the adult gene expression pattern. Strategy II exploits the INTRSECT viral system18, in which EYFP expression is activated uniquely in cells that express Cre and Flpo recombinases (Fig. 2B). By virtue of targeted viral delivery, this strategy has the advantage of avoiding most other TH+ populations in the brain, as well as labeling adult neurons based on their ongoing, rather than developmental, expression of Cre and Flpo. Strategy III uses a Dat-tTA strain (tetracycline transactivator driven by the Slc6a3 (Dat) locus19), in conjunction with the intersectional reporter mouse Ai8217, in which EGFP expression is driven by tetracycline response element (TRE) in a Cre dependent manner (Fig. 2C). The Dat locus has the advantage of being largely restricted to midbrain DA neurons16, barring few other neuronal populations (e.g. ventral premmamillary nucleus, PMv20). Together these complementary approaches form a powerful arsenal to target molecularly distinct DA neurons.
Figure 2. Three intersectional strategies to genetically target dopamine neuron subtypes.
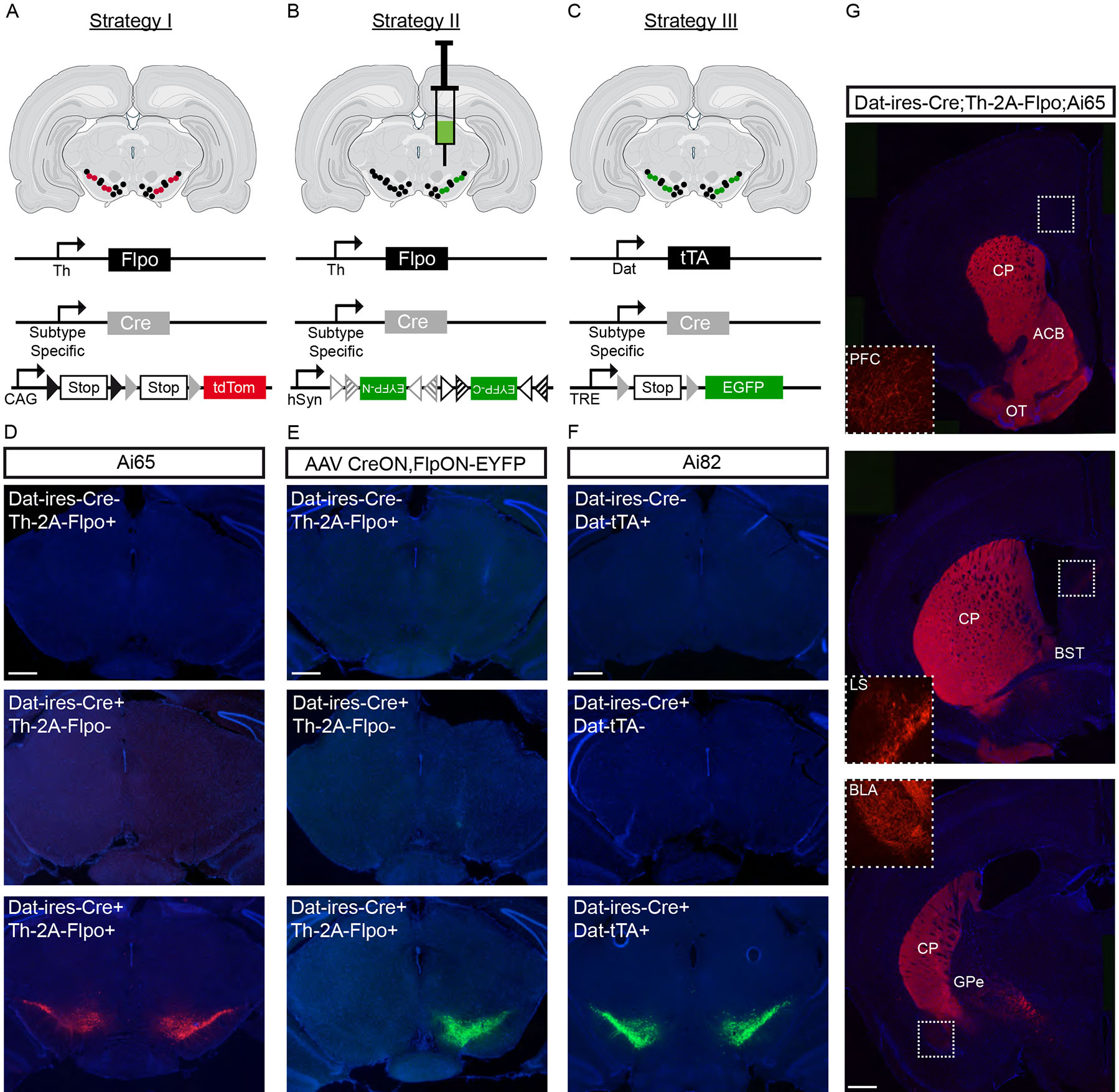
(A) A first strategy is to use subtype-specific Cre drivers in combination with Th–2A–Flpo and the intersectional reporter mouse Ai65 (or equivalent), which results in tdTomato expression only in cells expressing both Cre and Flpo recombinases. (B) A second strategy is to use subtype-specific Cre drivers, in combination with both Th–2AvFlpo and the INTRSECT viral system. This virus leads to EYFP expression in neurons expressing both Cre and Flpo recombinases. A simplified schematic of the INTRSECT virus is shown. (C) A third strategy is to use Cre drivers in conjunction with Dat–tTA and Ai82 intersectional reporter mice. In this reporter, EGFP expression is driven by Dat–tTA, following Cre recombination. (D) Validating Strategy I, we only detected tdTomato expression in midbrain DA neurons in mice with both Dat–ires–Cre and Th–2A–Flpo alleles, whereas no tdTomato was observed in the absence of Dat–ires–Cre or Th–2A–Flpo. (E) Validating strategy II, we observed strong EYFP fluorescence after viral injection of hSyn–CreON,FlpON–EYFP in mice with both Datvires–Cre and Th–2A–Flpo alleles, whereas no EYFP was observed in the absence of Dat–ires–Cre or Th–2A–Flpo. (F) For Strategy III, we observed EGFP labeling in Ai82+ mice having both Dat–ires–Cre and Dat–tTA alleles, whereas no fluorescence was observed in controls. (G) Using Dat–ires–Cre;Th–2A–Flpo;Ai65 mice to provide a comprehensive picture midbrain DA neuron projections, we observed tdTomato labeled fibers in the caudate putamen (CP), nucleus accumbens (ACB), olfactory tubercule (OT), prefrontal cortex (PFC), lateral septum (LS), bed nucleus of the stria terminalis (BST), basolateral amygdala (BLA), and external globus pallidus (GPe). We also observed very sparse tdTomato+ fibers in the lateral habenula, motor cortical areas, and rostral pontine regions (not shown). Grey triangles = loxP sites, grey triangle outline = lox2722, grey striped triangle = loxN, black triangles = FRT sites, black triangle outline = F3 sites, black striped triangle = F5. See Table S1 for number of animal replicates for each experiment. Scale bars = 400 μm
To validate Strategy I and to establish a pan-midbrain DA neuron projection map, we obtained Th–2A–Flpo;Dat–ires–Cre;Ai65 mice as well as control littermates. In mice with both Th–2A–Flpo and Dat–ires–Cre alleles, but not in controls, we observed strong fluorescence in midbrain DA neurons (Fig. 2D). We also observed tdTomato expressing neurons in the arcuate nucleus of the hypothalamus (ARH), but none in the PMv. Since ARH DA neurons do not contribute to ascending DAergic pathways2, these experiments revealed ascending DA pathways in their entirety, which was neither possible using the Th nor Dat promoter alone. As depicted in Figure 2G, tdTomato+ axonal fibers were observed in all known DA recipient regions. Thus, this intersectional genetic strategy provides a comprehensive picture of the projections of midbrain DA neurons. To establish the fidelity of Strategy II, we injected AAV–-CreON,FlpON–EYFP virus in the SNc of mice with only the Th–2A–Flpo or Dat–ires–Cre allele, as well as mice with both alleles. In mice with both Flpo and Cre alleles, but not either one alone, we observed strong fluorescence in DA neurons (Fig. 2E). Lastly, we validated Strategy III with Dat–ires–Cre;Dat-tTA;Ai82 mice, and observed robust fluorescence in DA cell bodies and axonal projections, but not in controls (Fig. 2F). These experiments demonstrate that our three complementary intersectional strategies display fidelity, and therefore allow analysis of genetically-defined DA neuron projections.
Projections of genetically defined DA neuron subtypes to caudate putamen
Guided by single-cell based classification studies and other literature, we elected to use five Cre lines (Ndnf, Sox6, Aldh1a1, Calb1 and Vglut2) in intersectional labeling experiments (Table S1), and elucidated three DA neuron subtypes in the SNc that displayed projections biased towards dorsolateral, ventromedial, and caudal CP.
Ndnf+/Sox6+ DA neuron projections cover most areas of the dorsal striatum
Ndnf is a gene encoding the neuron-derived neurotrophic factor which is expressed by at least two distinct SNc subtypes: the Aldh1a1+ subtype located in the ventral tier, and the Aldh1a1–subtype mainly located in the dorsal tier of the SNc but also in the dorsolateral VTA (a.k.a. parabrachial pigmented area; PBP)8,9. Using Strategy I (i.e. Ndnf–dgCre;Th–2A–Flpo;Ai65 mice; see methods for Cre activation), we observed tdTomato+ cells located in the SNc, dorsolateral VTA, and RR (Table S1). Co-labeling and quantification revealed that 98.5% (±0.5) expressed TH, 90.1% (±2.3) expressed SOX6, whereas only 1.3% (±0.7) expressed OTX2, consistent with previous single-cell gene expression studies8,9. Some tdtomato+ neurons were present outside the midbrain, precluding projection analysis. Using Strategy III (Ndnf–dgCre;Dat–tTA;Ai82 mice) resulted in numerous DA neurons expressing EGFP that were located in the SNc, dorsolateral VTA and RR (Fig. 3A,B). Similar to the results obtained with Strategy I, EGFP+ midbrain neurons were TH+ and the majority were SOX6+ (Fig. 3B). The lack of labeled neurons outside the midbrain made this strategy amenable for projection analysis (see Methods and Fig. S4). In these mice, TH+/EGFP+ axons follow the medial forebrain bundle (MFB), traverse the GPe, and innervate striatal targets. Within the dorsal striatum, fibers were observed at all rostrocaudal levels (Fig. 3F and S5), were particularly dense in the ventral portion of the intermediate (CPi) and caudal caudate putamen (CPc; Fig. 3H), but were significantly less dense in the tail of the caudate putamen (CPt; Fig. 3F and Fig. S5).
Figure 3. Dopamine neuron subtype projections to the caudate putamen.
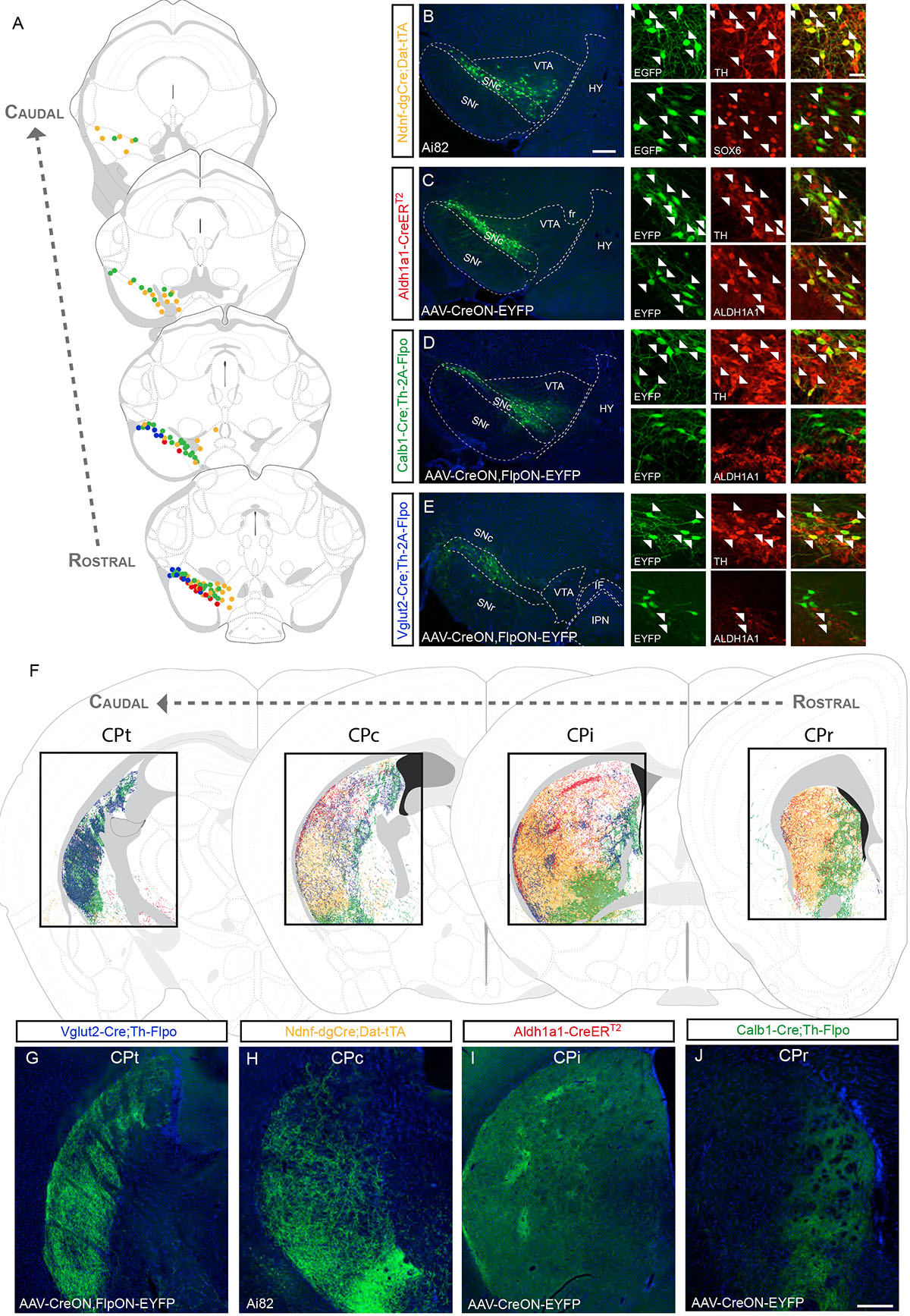
(A) Distribution of DA neuron subtypes used in the intersectional genetic experiments to trace projections in the caudate putamen (CP; each dot = 3 neurons; yellow = Ndnf experiment shown in B, red = Aldh1a1 experiment shown in C, green = Calb1 experiment shown in D, blue = Vglut2 experiment shown in E). (B) In Ndnf–dgCre;Dat–tTA;Ai82 mice, EGFP+ cell bodies are located throughout the SNc, the dorsolateral VTA and RR. The vast majority of these cells express TH and SOX6 (arrowheads). (C) Example of a Aldh1a1–CreERT2 brain injected in the SNc with AAV–CreON–EYFP virus. EYFP cells were positive for TH and ALDH1A1. (D) Injection of Calb1–Cre;Th–2A–Flpo mouse with a AAV–CreON,FlpON–EYFP virus in the SNc. Most EYFP+ cells did not express ALDH1A1. (E) Injection of a AAV–CreON,FlpON–EYFP virus in the SNc of Vglut2–Cre;Th–2A–Flpo mouse yields labeled neurons in the dorsolateral part of the SNc (corresponding in part to the pars lateralis). A small fraction of these neurons is ALDH1A1+ (arrowhead). (F) Distinctive areas of the dorsal striatum are innervated in the experiments described above, shown at different rostrocaudal levels (CPr = rostral, CPi = intermediate, CPc = caudal, CPt = tail). (G-J) Representative images from these experiments (See Table S1 for number of animal replicates for each experiment). Scale bars: B-E low mag. = 200 μm, high mag. = 40 μm; G-J =200 μm.
We considered the possibility that the less dense innervation of the CPt was due to incomplete recombination by the inducible Ndnf–dgCre. The transcription factor Sox6 has a similar expression pattern to Ndnf, being expressed in both Aldh1a1+ neurons in the ventral tier of the SNc, and in Aldh1a1–neurons in the dorsal tier of the SNc and the dorsolateral VTA8,9,21. To corroborate our findings using a second anchor gene, we generated a Sox6–FSF–Cre mouse line (Fig. S6). To validate the recombination specificity within the DAergic system, we crossed the Sox6-FSF-Cre with Th-2A-Flpo and the intersectional reporter RC::Frepe (Table S1). We found that 97.7% (±2.7) of EGFP+ neurons of the SNc were SOX6+, and that 82.7% (±1.9) of SOX6+/TH+ of the SNc were EGFP+. However, we could not use Strategy I for projection analysis since Th and Sox6 gene expression intersected in cell populations outside the midbrain.
To reveal the projections, we therefore used Strategy II, and injected AAV–CreON,FlpON–EYFP in the SNc of Sox6–FSF–Cre;Th–2A-Flpo mice. We observed EYFP+ cells mainly in the SNc, and few in the dorsolateral VTA (Fig. S6). The vast majority of labeled neurons co-expressed TH and SOX6 (Fig. S6), but only a subset of these neurons express ALDH1A1 (not shown). The overall projection pattern displayed similarities to Ndnf, with fibers observed only in the striatum and not in other known DAergic targets (Fig. S5; Table S1). Overall, Ndnf and Sox6 projections display important similarities (see Fig. S5 for discussion regarding the subtle differences). Interestingly, the CPt had significantly less dense innervation in both Sox6-FSF-Cre and Ndnf–dgCre experiments (Fig. 3F and S5). This suggests that the CPt receives a substantial innervation from a DA neuron subtype not defined by Ndnf and Sox6 expression.
Aldh1a1+ SNc DA neurons project principally to the dorsolateral caudate putamen
The gene Aldh1a1, which encodes an enzyme of the aldehyde dehydrogenase 1 family, is expressed by two distinct DA subtypes: one located in the ventral tier of the SNc, and the other located largely in the IF and ventromedial VTA (a.k.a. paranigral (PN))8,9. To genetically target Aldh1a1+ subtypes, we inserted the inducible recombinase CreERT2 at the start codon of the Aldh1a1 gene (Fig. S7). In tamoxifen (TAM) treated Aldh1a1–CreERT2, Ai9 mice, 93.9% (±1.0) recombined cells were ALDH1A1+ (Fig. S7). Since this experiment resulted in recombination non-DA neurons (i.e. cortex, hippocampus, hypothalamus and cerebellum), we employed intersectional Strategy I. In the midbrain of Aldh1a1–CreERT2;Th–2A–Flpo;Ai65 mice, ALDH1A1+ DA neurons were reliably labeled with tdTomato, but very few neurons were labeled outside the midbrain (mainly found in the ARH). We observed tdTomato+ fibers in the CP, the ACB, and the LS, but no fibers were observed in the PFC (Fig. S7). In the CP, co-staining with μ opioid receptor (MOR) antibody revealed that some MOR+ patches did indeed display higher density of ALDH1A1+ projections (Fig. S7), similar to what was previously reported by ALDH1A1 and MOR co-immunolabeling22.
To distinguish between the two Aldh1a1 expressing DA subtypes, we injected DIO–AAV–EYFP into either the SNc or VTA of Aldh1a1–CreERT2 mice (Fig. 3C; VTA injections will be discussed in the section on projections to the nucleus accumbens (ACB) and olfactory tubercle (OT)). In SNc injections, EYFP labeled cells were located in the ventral tier of the SNc, and co-expressed ALDH1A1, TH, SOX6, but not OTX2 (Fig. 3C, S7), consistent with previous DA neuron subtype classification8,9. In the representative injection displayed in Fig. 3C, we observed EYFP+ fibers along the MFB, GPe, the subcallosal streak and the CP. Within the CP, fibers were observed in most rostrocaudal levels, but were notably sparse in CPt (Fig. 3F and S5). Fibers were dense in the dorsolateral part of the CP and less dense ventrally, and were particularly concentrated in some striosomes in the CPi (Fig. 3F, I). Overall, projections from ALDH1A1+ neurons of the SNc appear to be a subset of the Sox6+/Ndnf+ projections.
Calbindin+ SNc DA neurons project to the ventromedial caudate putamen
Within the Sox6+/Ndnf+ population, Calb1 is enriched in Aldh1a1– neurons of the dorsal SNc8,9. Applying Strategy III (Calb1–Cre; Dat–tTA;Ai82), we observed EGFP+ neurons located in the dorsal SNc, which includes the dorsolateral-most region (pars lateralis in some anatomical schemes; SNpl; Fig. S8), as well as in the VTA. Within the SNc, 69.9% (±5.9) of EGFP+ neurons were SOX6+. Most of the SOX6– cells were found in the SNpl, suggesting that Calb1+ might label at least two distinct nigral DA neuron subtypes. WIth regard to projections, dense fibers were observed in medial, but not lateral regions of CPr, a region also innervated by Aldh1a1+ fibers (Fig. 3F). In the CPi and CPc, we observed projections that were mostly directed toward the ventromedial region with fewer fibers observed in the dorsolateral CP (Fig. S8). In contrast, we saw dense innervation throughout the CPt (Fig. S8).
To confirm the projection of nigral Calb1+ neurons, we used Strategy II and injected AAV–CreON,FlpON–EYFP virus in the SNc of Calb1–Cre;Th–2A–Flpo mice. Similar to the previous experiment (Fig. S8), the majority of EYFP+ neurons in the dorsal SNc expressed SOX6+, while cells labeled in the SNpl had low/absent SOX6 levels. Almost none of the EYFP+ cells co-expressed ALDH1A1 (Fig. 3D). In the CP, we observed the same pattern of projections as obtained with Strategy III (Fig. 3F,J and S5). Overall, two main points can be deduced from these data: first, the Sox6+/Ndnf+ DA neurons can be divided into at least two subtypes (Aldh1a1+ and Calb1+), the projections of each biased toward dorsolateral and ventromedial regions of the CP, respectively; and second, a Calb1+/Sox6–/Ndnf– population in the dorsolateral SNc (including the SNpl) could account for the innervation of the CPt.
Vglut2+ DA neurons of the lateral SNc project to the tail of the striatum
The gene encoding vesicular glutamate transporter (Slc17a6; Vglut2) has been reported mainly in DA neurons located in ventromedial VTA and IF5,23. However, the Allen Brain Atlas ISH database depicts the presence of Vglut2 mRNA in the dorsolateral region of the SNc. To demonstrate the existence of Vglut2+ DA neurons, we performed single-molecule RNA fluorescent in situ hybridization (smFISH) and observed several cells co-expressing Vglut2 and Th mRNA mainly in the lateral SNc (Fig. S9), consistent with a recent publication24. Vglut2 is known to be broadly expressed in DA neurons during development23, and unsurprisingly, when we applied Strategy I, we observed broad recombination in the SNc (Table S1). Thus, to determine the projections of Vglut2-expressing DA neurons in adult, we used Strategy II and injected AAV–CreON,FlpON–EYFP virus in the SNc of Vglut2–Cre;Th–2A–Flpo mice. We observed EYFP+ neurons in the dorsolateral part of the SNc (Fig. 3A, E), but less in the more medial SNc or the RR. These EYFP+ DA neurons expressed TH, but only 7.6% ± 0.6 co-expressed ALDH1A1 (Fig. 3E). The axonal projections were mostly absent from the CPr (Fig. 3F and S5), although some areas with concentrated axons were found in the CPi (Fig. 3F and S5). In the CPc, EYFP+ fibers were observed, partly overlapping with Aldh1a1 innervation in dorsolateral regions. In this experiment, the most densely innervated area was the CPt (Fig. 3G). Based on these results, we posit that the Vglut2+ DA neurons of the dorsolateral SNc largely correspond to the Calb1+/Sox6–population identified earlier, a subtype that has not been definitively demarcated by previous classification studies8,9. These neurons likely correspond to a population having a similar distribution in the SNc identified by retrograde virus tracing from the CPt25.
Projections of DA neuron subtypes to nucleus accumbens and olfactory tubercle
In this section, we will describe intersectional labeling experiments based on four Cre lines (Cck, Aldh1a1, Vglut2, and Sox6; see Table S1) and will discuss their projections to the ACB core, shell, and OT regions.
Cck+ DA neurons project to all regions of the ACB and OT
Applying Strategy I with Cck–Cre (Cck–Cre;Th–2A–Flpo;Ai65), we observed recombined cells across the VTA, the IF, and the CLi, as well as some neurons located in the dorsal SNc. In this experiment, we observed tdTomato+ fibers in all regions of the ACB and OT, although labeled neurons outside the midbrain precluded projection analysis (Table S1). Therefore, we used Strategy II and injected the intersectional virus into the VTA of Cck–Cre;Th–2A–Flpo mice (Fig. 4A–B and S10). In such experiments, the starter population had a similar distribution as in Strategy I (Fig. 4A–B). Some EYFP+ neurons were SOX6+, whereas other were OTX2+, indicating that at least two subtypes are labeled by this Cre line (Fig. 4B). Regarding projections, we observed fibers in the core and shell (medial, ventral and lateral) regions of the ACB, as well as throughout the OT (Fig. 4F,J and S10). Within the dorsal striatum, fibers were also observed in the medial part of the CPr and CPi (Fig. S4) in line with previous results demonstrating a VTA projection toward the medial CP26. Overall, Cck likely defines multiple populations demonstrating the necessity to employ more specific Cre drivers to label DA subtypes within the VTA.
Figure 4. Dopamine neuron subtype projections to the nucleus accumbens and olfactory tubercle.
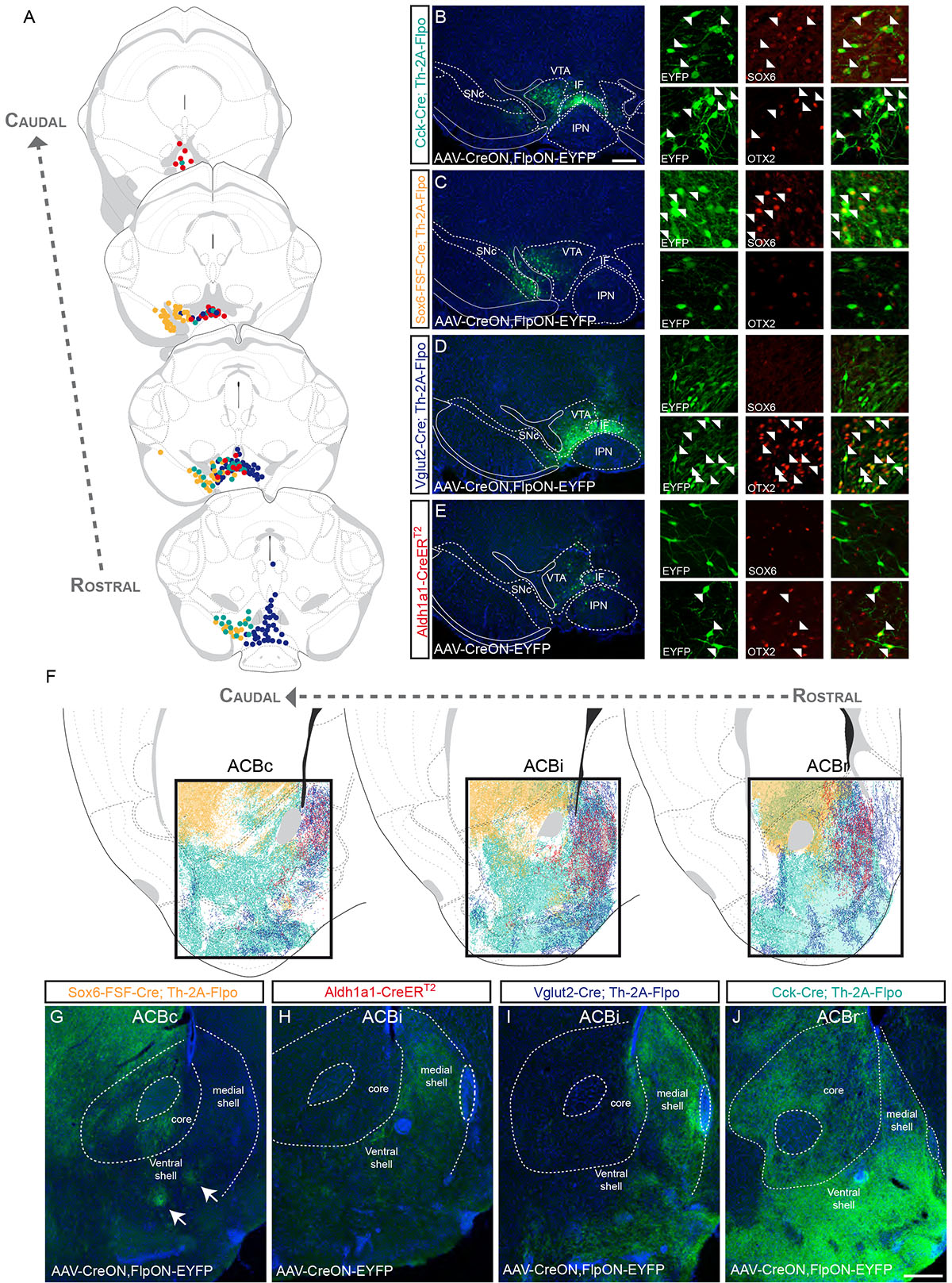
(A) Distribution of genetically labeled DA neurons with Vglut2–Cre (dark blue), Sox6–FSF–Cre (yellow), Cck–Cre (cyan) and Aldh1a1–CreERT2 (red) (each dot = 3 neurons). (B) Example of a Cck–Cre;Th–2A–Flpo mouse injected with AAV–CreON,FlpON–EYFP. EYFP+ cells either colocalized with SOX6 or OTX2 (arrowheads). (C) Example of a Sox6–FSF–Cre;Th–2AvFlpo mouse injected with AAV–CreON,FlpON–EYFP. In this mouse, EYFP+ cells colocalize only with SOX6 (white arrowhead) and not with OTX2. (D) Example of a Vglut2–Cre;Th–2A–Flpo mouse injected in the VTA with AAV–CreON,FlpON–EYFP. EYFP+ cells mostly express OTX2 (white arrowhead) and very few cells express SOX6. (E) An example of Aldh1a1–CreERT2 injected in the VTA with AAV–CreON–EYFP. EYFP+ cells are OTX2+ and SOX6–. (F) Projections of molecularly defined DA neurons to the nucleus accumbens at different rostral (ACBr), intermediate (ACBi), and caudal (ACBc) levels. Representative images are shown for Sox6–FSF–Cre (G), Aldh1a1–CreERT2 (H), Vglut2–Cre (I), and Cck–Cre (J) experiments. See Table S1 for number of animal replicates for each experiment. Scale bars: B-E low mag. = 200 μm, high mag. = 30 μm; G-J = 200 μm.
Sox6+ neurons in the dorsolateral VTA innervate the ACB core and lateral shell
To investigate the contribution of VTA Sox6+ neurons, we used Strategy II and injected AAV–CreON,FlpON–EYFP in the VTA of Sox6–FSF–Cre;Th–2A–Flpo mice. Recombined cells were mostly located in the dorsolateral portion of the VTA (PBP region), although some EYFP+ neurons were observed in the medial SNc as well (Fig. 4A, C). EYFP+ positive cells were SOX6+, but OTX2– (Fig. 4C). In the ACB, EYFP+ fibers were most dense in the ACB core and lateral shell (Fig. 4F and S10). Fewer fibers were found in the OT and the ACB medial shell (Fig. 4F–G), but interestingly, two small areas located in the ventral shell also receive Sox6+ innervation (Fig. 4G; arrows) that may correspond to the D2R-expression poor areas described by Gangarossa et al.27. Similar to SNc targeted injections in Sox6–Cre;Th–2A–Flpo mice, here we also observed projections to the medial CPr and CPi. No labeled fibers were found outside the striatum, suggesting that Sox6 might define DA neurons that predominantly innervate striatal targets.
Vglut2+ and Aldh1a1+ DA neurons of the VTA projects to ACB medial shell and OT
To investigate the projections of Vglut2-expressing DA neurons of the ventromedial VTA5,23, we employed Strategy II. Injections into the VTA resulted in EYFP+/TH+ cells largely in the PN region, IF and CLi, but also often resulted in EYFP+/TH– cells in the RLi (Fig. 4A, D). Immunolabeling revealed that few, if any, EYFP+/TH+ neurons were SOX6+, but the majority were OTX2+ (Fig. 4D). A subset of EYFP+/OTX2+ cells were ALDH1A1+ (not shown). Within the striatum, we found EYFP+ fibers most densely in the ACB medial shell, in a somewhat complementary manner to Sox6 projections (Fig. 4F, I and S10). In the OT, we found EYFP+ fibers in discrete, dense column-like structures (Fig. 4I and S10), in contrast to the Cck labeling experiments in which widespread OT labeling was observed. Similar results were obtained with Adcyap1–Cre (Table S1).
We next assessed the projections to the ACB from Aldh1a1+ DA neurons of the VTA, applying Strategy I (Aldh1a1–CreERT2;Th–2A–Flpo;Ai65). Focusing on the ACB, we observed a strong innervation of the ACB medial shell and OT, and significantly less fibers innervating the core and lateral shell (Fig. S7). To exclude the contribution of Aldh1a1 SNc neurons, we next injected a small volume of virus into the VTA of Aldh1a1–CreERT2 mice resulting in EYFP+ cells in the VTA, IF, and CLI regions (Fig. 4A, E). Of the EYFP+ cells 95.7% (±2.3) were OTX2+, and only 4.2% (±1.9) expressed SOX6 (Fig. 4E and S7). EYFP+ fibers were observed in the ACB medial shell and in the OT (Fig. 4F,H and S10). Taken together with Vglut2–Cre experiments, ALDH1A1+ neurons in the VTA are a subset of the Vglut2+ population, and accordingly, ALDH1A1+ projections in the medial shell are a subset of the Vglut2 projections.
Projections of genetically defined DA neurons to the amygdala
In Dat–ires–Cre;Th–2AvFlpo;Ai65 mouse brains, labeled fibers were observed in the central (CEA), intercalated nuclei (IA), the basolateral (BLA), and the posterior nuclei (Fig. 5A). Sparse tdTomato+ fibers were also observed in the lateral nucleus (LA), medial nucleus (MEA) and amygdalar cortical areas. To investigate if these amygdalar nuclei received a differential innervation for DA neuron subtypes, we used three Cre drivers (Vip, Cck, Vglut2; Table S1).
Figure 5. Dopamine neuron subtype projections to the amygdala.
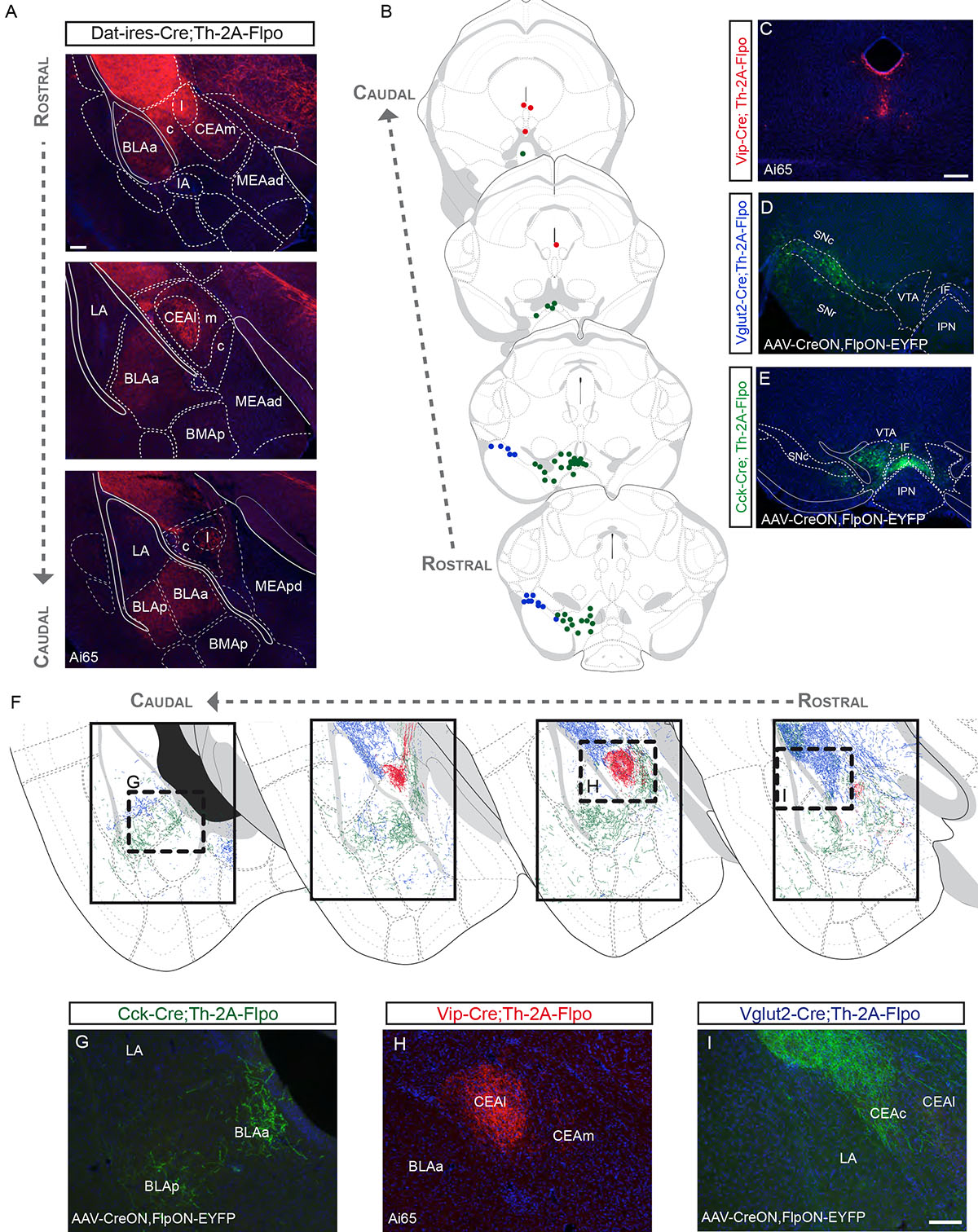
(A) Projections to the amygdala, as observed in Dat–ires–Cre;Th–2A–Flpo;Ai65 brains. (B) Distribution of DA neuron subtypes found to project to the amygdala labeled by Vip–Cre (red), Vglut2–Cre (blue), and Cck–Cre (green). (C) Labeled neurons in the Vip–Cre;Th–2A–Flpo;Ai65. (D) Labeled neurons by intersectional viral injections in the SNc of the Vglut2–Cre;Th–2A–Flpo. (E) EYFP+ labeled neurons by an injection in the VTA of a Cck–Cre;Th–2A–Flpo mouse. (F) Projections of molecularly defined DA neurons labeled with Vip–Cre (red), Vglut2–Cre (blue), and Cck–Cre (green). (G-I) Representative images from the experiments described above (See Table S1 for number of animal replicates for each experiment). Scale bars: A = 100 μm; C-E = 200 μm; G-I = 100 μm.
Vip+ DA neurons project to the lateral part of the central amygdala
In Vip–Cre;Th–2A–Flpo;Ai65 brain sections, we found tdTomato+ neurons in the CLi and PAG/DR (Fig. 5B–C), that co-localized with TH, but not with SOX6 or OTX2 (Fig. S11). Similar results were observed using Strategy III (Vip–Cre;Dat–tTA;Ai82; Fig. S11), although slightly fewer neurons were labeled. 67.0% (± 9.0) of EGFP+ neurons expressed TH. With both strategies, labeled fibers were found in the lateral part of the CEA, although some fibers were also observed in the medial part of the CEA (Fig. 5F,H; Fig. S11). Strikingly, no fibers were present in the BLA or other nuclei of the amygdala.
Vglut2+ DA neurons of the SNc innervate the capsular part of the central amygdala
The Vglut2+ DA neurons in the lateral SNc also contribute to the innervation of the amygdala (Fig. 5B, D), in addition to the CPt innervation already described. These cells send projections to the lateral part of the capsular division of the CEA (CEAcl), and are mostly excluded from other divisions of the CEA (Fig. 5F, I). We also observed sparse fibers reaching the ventral most LA nucleus and the posterior nucleus of the amygdala (PA; Fig. 5F).
Cck+/Vglut2+ DA neurons of the VTA innervate the basolateral amygdala
Labeling of Cck+ DA neurons using Strategy I resulted in tdTomato+ fibers in the medial and lateral part of the CEA (CEAm and CEAl), in the anterior and posterior BLA, as well as in the intercalated nuclei located in the external capsule (not shown). Using Strategy II, injections in the VTA resulted in EYFP+ fibers in a similar fashion, except for the CEAl (Fig. 5B, E, F, G). This difference in labeling between Strategy I and II could be explained by limited dispersion of viral particles, as these may not have reached Cck+ DA neurons that project to the CEAl. The Cck+ neurons that project to the CEAl are likely to be located in the DR/PAG and to encompass the Vip+ neurons. Similar to Cck, labeling Vglut2+ VTA neurons using Strategy II (Fig. 4D) resulted in a more restricted labeling within the amygdala. In this experiment, we principally observed a sparse innervation of the BLA (Table S1). Overall, the amygdala receives a varied innervation from genetically defined DA neurons located in the DR/PAG, SNc, and VTA.
Projections of genetically defined DA neurons to other regions
DA neuron subtype innervation of the prefrontal and entorhinal cortices
Using the Dat–ires–Cre driver in Strategy I (Dat–ires–Cre;Thv2A–Flpo;Ai65), we observed several tdTomato+ fibers in the infralimbic and prelimbic cortical areas (Fig. 6A). We also observed labeled fibers in the ENT and these were more concentrated in certain regions (Fig. 6D), reminiscent of the DAergic islands observed in Mingote et al28. To investigate which DA subtype innervates the PFC and ENT, we examined all previous experiments for the presence of fibers in these cortical areas. We observed EYFP+ fibers in the PFC and ENT using Strategy II with viral injectons in the VTA, with Cck–Cre (Fig. 6E, H), and Vglut2–Cre (Fig. 6I, L). In the PFC, axons were mostly found in the deep layers of the infralimbic and prelimbic cortices, but were more sparse when compared to Dat–ires–Cre;Th–2A–Flpo;Ai65 samples. Taken together, these data suggest that the mesocortical DA projections are derived, at least in part, from Vglut2+/Cck+ DA neurons that are located in the ventromedial VTA.
Figure 6. Dopamine neuron subtype projections to other brain regions.
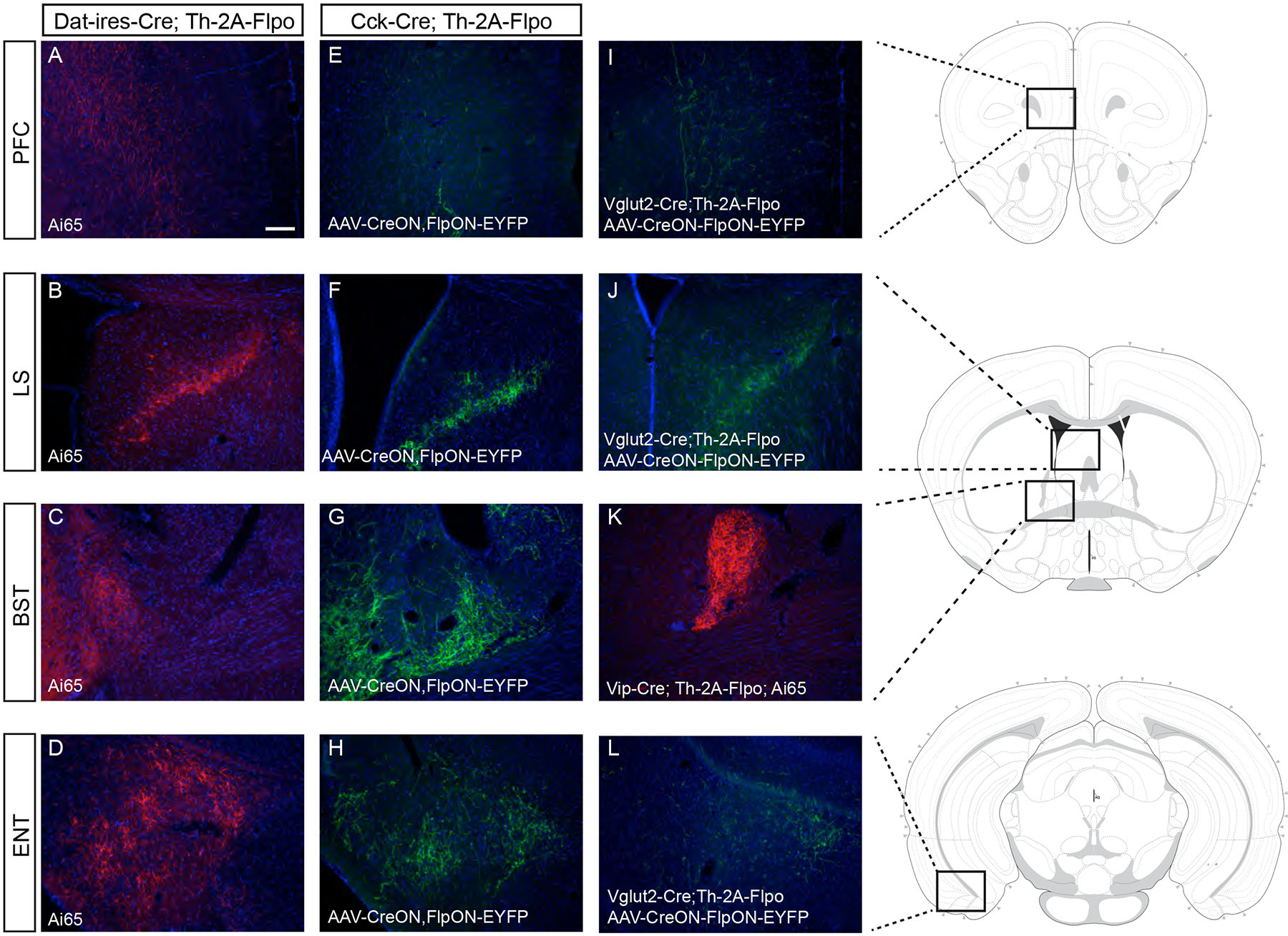
Intersectional genetic Strategy I with Dat–ires–Cre revealed DAergic projections to (A) the prefrontal cortex (PFC), (B) the lateral septum (LS), (C) the bed nucleus of the stria terminalis (BST) and (D) the entorhinal cortex (ENT). Using Strategy II in the VTA of Cck–Cre;Th–2A–Flpo mice we observed also an innervation of the PFC (E), LS (F), BST (G) and ENT (H). (I) EYFP+ fibers are observed in the PFC of Vglut2–Cre;Th–2A–Flpo mice after injection of AAV–CreON,FlpON–EYFP in the VTA. (J) EYFP+ fibers are observed in the LS of Vglut2–Cre;Th–2A–Flpo after injection of AAV–CreON,FlpON–EYFP in the VTA. (K) tdTomato+ fibers are observed in the BST of Vip–Cre;Th–2A–Flpo;Ai65 brains. (L) EYFP+ fibers are observed in the ENT of Vglut2–Cre;Th–2A–Flpo after injection of AAV–CreON,FlpON–EYFP in the VTA. See Table S1 for number of animal replicates for each experiment. Scale bar: A-L = 100 μm.
Aldh1a1+ DA neurons of the VTA project to the lateral septum
We observed strong innervation of the lateral septum (LS) in the Dat–ires–Cre;Th–2A–Flpo;Ai65 mouse (Fig. 6B). Analyzing all previous experiments, we observed labeled fibers in the LS using Strategy II only in Cck–Cre (Fig. 6F), Vglut2–Cre (Fig. 6J) and Aldh1a1–CreERT2 (Fig. S7) experiments. Altogether, our findings suggest that at least two types of Vglut2+/Cck+ DA neurons populate the VTA: Aldh1a1+ DA neurons that project to the ACB, OT and LS; and Aldh1a1– neurons that might contribute to these projections but also project to the PFC and ENT.
Vip+ and Cck+ DA neurons project to the bed nucleus of the stria terminalis
The BST receives an important innervation from midbrain DA neurons (Fig. 6C) and we observed consistent BST innervation with two Cre lines: Vip–Cre and Cck–Cre. In the case of Vip-Cre, using Strategy I, we observed dense tdTomato+ fibers in the oval and juxtacapsular nuclei of the BST, with sparse fibers also found in the anterolateral nucleus (Fig. 6K). In the case of Cck–Cre, using Strategy II, we observed EYFP+ fibers in the BST, mostly in the anterolateral nucleus, although sparse fibers were also observed in the oval and juxtacapsular nuclei as well (Fig. 6G). As observed in the CEA, the Cck+ VTA population labeled with Strategy II appears to project to the BST in a complementary manner to Vip+ PAG/DR DA neurons.
Discussion
As a step towards understanding the neuroanatomical correlates of DA neuron diversity, we developed intersectional genetic strategies to target DA neuron subtypes and demonstrated that they possess distinct but partly overlapping projection patterns, even within a given target region (Fig. 7; Table S1, Table S2). We demonstrated three distinct projection patterns arising from molecularly-defined DA neurons of the SNc and RR that segregate along the rostrocaudal, mediolateral and dorsoventral axes of the CP (Fig. 7). A first distinct projection pattern originates from Aldh1a1+/Sox6+/Ndnf+ neurons, which correspond to DA-1A8 or DA-SNC9. These neurons send fibers mainly to the CPr, CPi and CPc, which are particularly dense in the dorsolateral region and in some striosomes. Interestingly, this population projects predominantly to locomotor areas of the striatum29–31, and was shown to be particularly vulnerable in a mouse model of PD8. A second projection arises from the Calb1+/Sox6+/Aldh1a1– neurons of the SNc, which are located in the dorsal SNc as well as in the PBP (DA-1B8 or VTA19). These neurons densely innervate the medial CPr, partly overlapping with the Aldh1a1+ projection, as well as the ventromedial regions of the CPi and CPc in a somewhat complementary manner to Aldh1a1+. A key finding in this study is the genetic targeting of a Vglut2+/Calb1+/Sox6– DA neuron subtype located principally in the lateral SNc that sends dense projections to the CPt. SNc neurons projecting to the CPt have recently been identified as a separate class, based on unique inputs25. Imaging studies have shown CPt projecting DA neurons are strongly activated by novel cues and might encode general salience rather than error prediction, unlike other SNc DA neurons32. Taken together with our findings, this points to the possibility that this Vglut2+ DA neuron population is embedded in a unique circuit with distinct functional properties. Overall, we have demonstrated that SNc DA subtypes have topographically biased projections along the rostrocaudal, mediolateral and dorsoventral axes of the CP, possibly aligning with previously described functional subdivisions32–36. Consistent with this notion, recent studies have pointed to the fact that the striatum is also segregated in terms of its corticostriatal inputs29,30, as well as in terms of distributions of D1 and D2 expressing medium-spiny neurons37.
Figure 7. Remarkable specificity of genetically-defined dopamine neuron subtype projections within target regions.
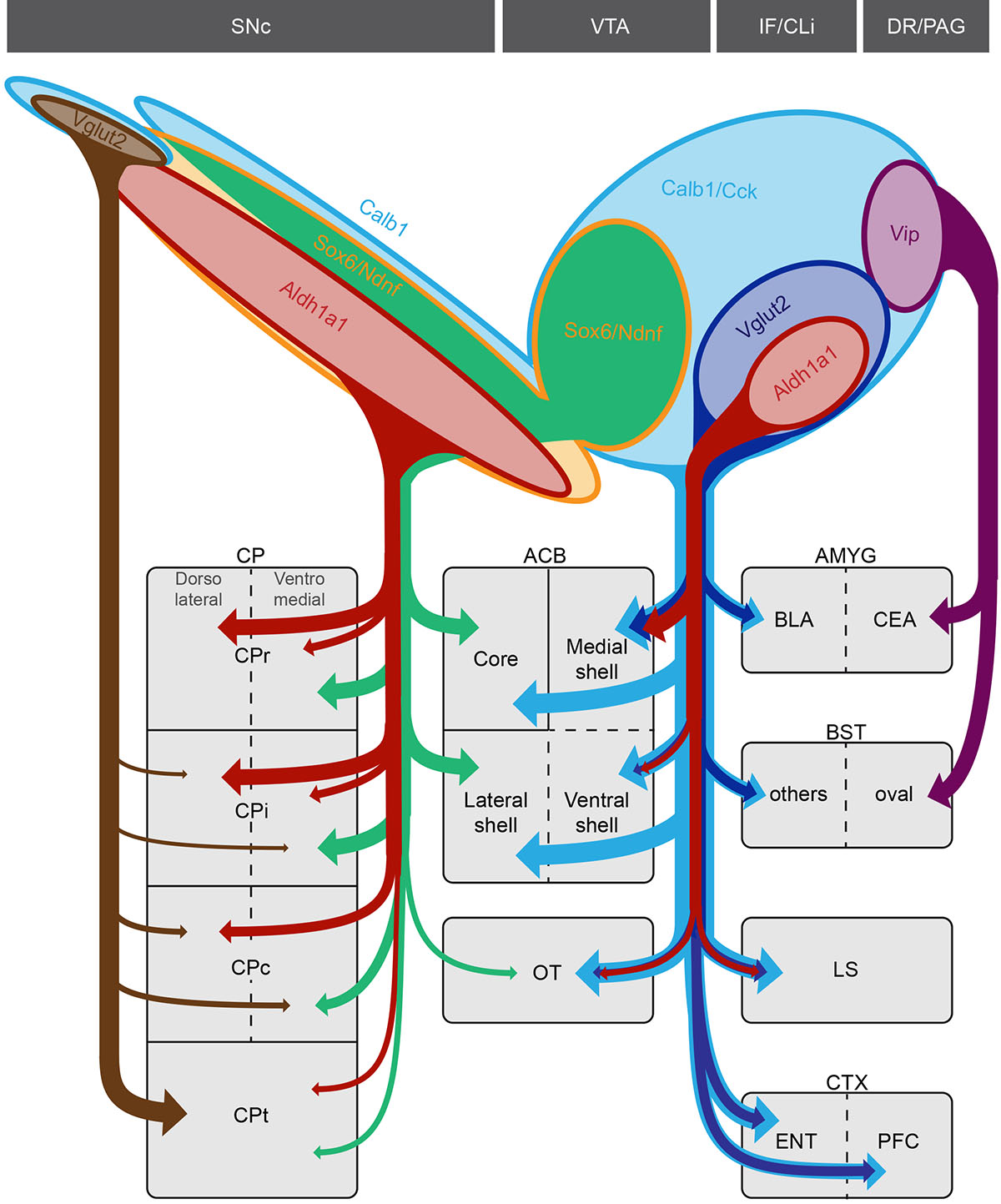
Top caption shows the approximate distribution of DA subtypes analyzed in this study. Vglut2+/Calb1+ DA neurons located in the SNc (brown) project most densely to the tail of the caudate putamen (CPt) extending ventrally to the capsular region of the amygdala (not depicted). Sox6+/Aldh1a1+ DA neurons of the SNc (red) project mainly to the rostral and intermediate caudate putamen (CPr and CPi respectively). Within the CP, Aldh1a1+ fibers are mainly observed in the dorsolateral region. The Calb1+/Sox6+ population of the SNc and dorsolateral VTA (green) send projections to the medial CPr and ventromedial CPi and CPc. This population also contributes projections to the lateral shell and core region of the nucleus accumbens. In the VTA, Aldh1a1+ DA neurons (red), which are also Cck+/Vglut2+, project to the medial shell of the ACB, olfactory tubercle (OT) and lateral septum (LS). Cck+/Vglut2+ (dark blue), but Aldh1a1-, contribute to the prefrontal (PFC) and entorhinal (ENT) cortices as well as the BLA. Finally, a Vip+ DA neuron population (purple) located in the dorsal raphe (DR) and periaqueductal gray (PAG) sends projections to the lateral part of the central amygdala (CEA) and the oval nucleus of the bed nucleus of the stria terminalis (BST). The Slc32a1+ subtype from Poulin et al. and La Manno et al. was not analyzed due to lack of unambiguous access.
We uncovered distinct projection patterns arising from genetically defined DA neuron populations in the VTA and other medial clusters (IF, CLi, and DR/PAG). An Aldh1a1+ DA neuron subtype (DA-2B8 or DA-VTA29) sends axonal projections principally to the ACB medial shell, OT and LS. Aldh1a1+ cells in the VTA were found also to express Otx238, NeuroD69,39, Adcyap18,9, Lpl8,9, and Vglut28. Consistent with this latter finding, optogenetic stimulation of DA fibers in the ACB shell and OT elicits strong glutamatergic currents28,40, and VGLUT2+/TH+ fibers are found in the ACB shell and OT41. Interestingly, rats learn to self-administer amphetamine, cocaine or DA agonists directly into the medial shell and OT, but do not learn self-administration into the lateral shell or ACB core, pointing to the relevance of this genetically defined circuit for drug addiction6. A second deduced DAergic pathway, arising from Vglut2+/Aldh1a1– neurons of the VTA, also sends projections to the ENT and PFC. DA projections to the ENT and PFC have also been shown to co-release glutamate28,42, and do not arise from the same neurons that project to the ACB43. A third DA pathway, defined by Vip, sends projections to the CEA and BST. This population is likely part of the DAergic pathway arising from the DR/PAG, which has been shown to be involved in social behavior44 and wakefulness15. A fourth DAergic pathway arises from a Calb1+/Sox6+/Ndnf+ VTA population located in the PBP region and dorsal SNc, and is mainly directed toward the core and lateral shell of the ACB and the medial CP as discussed above, in a pattern complementary to VTA Vglut2+ projections. The Calb1+/Sox6+/Ndnf+ projections are consistent with previous analyses showing that lateral shell projecting DA neurons are distinct from those projecting to the medial shell or PFC43,45,46. These cells are distinct from the other VTA DA neurons as they do not express Otx2, Adcyap1 or Vglut2, and seem to correspond to the population DA-1B8 and DA-VTA19.
In summary, we have harnessed the power of intersectional genetic paradigms to access several molecularly defined DA neuron subtypes, and have provided an initial framework to dissect DA neuron diversity. Although this framework is likely to be refined in future, we envision that the tools generated here, in conjunction with an improved arsenal of intersectional reporter mice13,17 and viruses18, for neuron ablation47, excitation48 or inhibition49,50, will provide a starting point for functional interrogation of DA neuron subtypes in normal and diseased states.
Materials and Methods
Generation of transgenic mice
Th–2A–Flpo
To generate Th–2A–Flpo mice, we obtained KOMP targeted ES cell clone Th_C0951 from the KOMP repository Mutant Mouse Reginal Resource Center (MMRRC). This clone has been genetically modified such that the Engrailed splice acceptor (SA) followed by ires–lacZ cassettes have been inserted in the intron following exon 6 of the Th gene. We used dual Recombinase Mediated Cassette Exchange (dRMCE)52 to reengineer this LacZ allele into the Th–2A–Flpo allele. To achieve this we generated a targeting vector that included exons 7 to 13 of Th coding sequence, the autocalytic peptide P2A, Flpo, WPRE sequence, a bovine growth hormone polyadenylation signal (bGH pA) followed by puromycin selection cassette flanked by rox sites. Since the two coding sequences are separated by virtue of a ribosome skipping event that occurs at the glycyl-prolyl peptide bond at the C-terminus of the 2A peptide (effectively, an autocatalytic “cleavage”)53, by design, any cell that is translating TH protein will also synthesize Flpo. The targeting construct was electroporated in KOMP ES cells in combination with the plasmid pDIRE (Addgene: #26745), which induces high level of iCre and Flpo expression, and we selected for puromycin (the replacement selection cassette). We screened 62 clones by PCR using multitude combinations of primers, out of which 63% were positive for the presence of Flpo. Of these clones 26% had lost the lacZ and neomycin suggesting a correct integration at the Th locus and a successful RMCE. Locus targeting was confirmed by sequencing of PCR products spanning the entire region and the junctions. These reengineered ES clones were injected into blastocyst by Northwestern University Transgenic and Targeted Mutagenesis Laboratory (TTML). The puromycin selection cassette was later removed by crossing Th–2A–Flpo mice to CAG–Dre transgenic mice (MMRRC: 032246-UCD)54.
Aldh1a1–CreERT2
Since Aldh1a1 is express in DA neuron progenitors55, we opted to generate an inducible Cre. The short (2 Kb) and long arm (4 Kb) of the targeting vector were PCR cloned from RP23–129n10 BAC clone into pCRII–topo. A CreERT2 fragment from pCreERT2 56, a WPRE fragment (pLemir; Open Biosystems), bGH pA were subcloned into pBS-SK+. The short arm and long arm were cloned upstream and downstream respectively of this construct. Site mutagenesis was used to align CreERT2 open reading frame with the endogenous ATG locus of Aldh1a1 gene57. We then added a PGK–Neo cassette flanked with FRT sites (pK-11; gift from Dr. Meyers58) after the WPRE and bGH pA. In addition, we added a DTA selection cassette (pRosaPAm1) downstream of long homology arm. DNA was linearized with SwaI and both constructs were electroporated in PRXB6/N ES cells by Northwestern University TTML. Correctly targeted clones were identified with long range PCR and validated by sequencing.
Sox6–FSF–Cre
The 2 Kb short and 6 Kb long arms as well as a 200bp fragment containing the ATG (exon 3) were obtained by PCR from RP24–83n9 BAC clone. The 5637bp FRT–STOP–Puro–FRT cassette from FSF–TOPO (Addgene #22774) vector was cloned in between the short arm and the 200bp fragment, in the intronic region between Sox6 exon 2 and 3. A NLS–CrevpolyA cassette was cloned by aligning the ORF with the endogenous ATG and preceding 6Kb long arm. A DTA–polyA cassette was cloned at the 3’ end of the long arm for negative selection. The final construct (20,974bp) was linearized with PacI and electroporated in PRXB6/N ES cells by Northwestern University TTML. Correctly targeted clones were identified with long range PCR and sequenced.
Mice breeding and husbandry
Mice were maintained and euthanized according to the protocols approved by the Northwestern University Animal Care and Use Committee. Animals were provided food and water ad libitum and were maintained on a regular 14h:10h day/night cycle at no more than five adult animals per cage. Animals were maintained on the C57Bl/6 background by backcrossing to females (Charles River; Strain 574). Both males and females were used in this study, as there is no evidence in the literature of differential DAergic projection between gender. The mice used in this study were obtained in part from the Jackson Laboratory: Ai65 (stock #021875), Ai82 (stock #023532), Dat–ires–Cre (stock #006660), Cck–ires–Cre (stock #012706), Vip–ires–Cre (stock #010908), and Vglut2–ires–Cre (stock #016963). Others were generously provided by the Allen Institute for Brain Science (Calb1–ires–Cre and Ndnf–ires–dgCre59), Dr. Susan Dymecki (RC::FA and RC::Frepe60), and Dr. Xiaoxi Zhuang (Dat–tTA61). All experimental animals were heterozygous for the recombinase or tTA transgenes. For induction of Ndnf–dgCre 4–8 week old mice were injected by with Trimethoprim in a procedure modified from Sando et al.62 as described below. In short, 15mg of Trimethoprim lactate (Sigma; T0667) was dissolved in 500μL H2O, mixed with 500μL of 2X PBS. The mice were injected at a concentration of 0.170μg/g, for three injections 48h apart. To induce Cre recombination in Aldh1a1–CreERT2 mice, we performed five daily injections (1mg/40g) of tamoxifen (Sigma; T5648) diluted in corn oil (Sigma; C8264) to a final concentration of 10mg/mL, in 4–8 week old Aldh1a1–CreERT2 mice. We waited at least 3 weeks after the last tamoxifen or trimethoprim injection before proceeding with perfusion for adequate reporter expression.
Stereotaxic surgeries
Mice were anesthetized with 3% isoflurane and placed onto a stereotaxic apparatus, kept under anesthesia with 2% isoflurane. Mice were then given a subcutaneous injection of Buprenex (0.1mg/kg). The top of the head was shaved and scrubbed with Betadine followed by 70% Ethanol. An incision was made to reveal the skull. Using bregma as a reference point, the stereotaxic coordinates of the VTA (RC: −3.4mm, ML:−0.4mm; DV: −4.2mm) or the SNc (RC:−3.1mm; ML: −1.5mm: DV: −4.2) were used to obtain the location of where a small hole was drilled in the skull. The needle of a Hamilton Neuros Syringe filled with CreON,FlpON–EYFP INTRSECT viruses (AAVdj–hSyn–Con/Fon–EYFP, titer 1.1×1013vg/ml; AAVdj–nEF–Con/Fon–EYFP, titer 2.4×1012vg/ml; as previously generated18) was slowly lowered into the injection site and held for five minutes. Injection of 0.1 to 0.5μL of virus was made over five minutes. The needle was removed five minutes after the end of the injection. The skull was washed with sterile PBS and the wound was closed using staples. Mice were removed from the stereotaxic apparatus, and a subcutaneous injection of Rimadyl (5.0 mg/kg) was given. Mice were perfused 4 to 6 weeks after viral injections.
Immunofluorescence
Adults mice (more than 8 weeks of age) were perfused with cold 4% PFA in PBS, the brains were dissected and fixed overnight in the same solution, cryoprotected in 30% sucrose PBS solution, sectioned at 25μm using a freezing microtome, and stored in −20°C in cryoprotectant solution. For immunofluorescence on free-floating sections, we initially rinsed in PBS, blocked in 5% donkey serum, 0.3% Triton X-100 in PBS, and incubated overnight at 4°C with primary antibodies diluted in blocking solution: rabbit anti-ALDH1A1 (Cat# ab23375; Abcam; 1:200)8, goat anti-βgal (Cat# 4600–1409; Biogenesis; 1:1500)20, rabbit anti-βgal (Cat# 55976; Cappel; 1:1500)63, goat anti-FOXA2 (Cat# sc-6554; Santa Cruz; 1:50)64, chicken anti-GFP (Cat# ab13970; abcam; 1:1,500)20, rabbit anti-NURR1 (Cat# sc-990; Santa Cruz; 1:500)8, goat anti-OTX2 (Cat# GT15095; Neuromics; 1:200)8, rabbit anti-SOX6 (Cat# ab30455; Abcam; 1:500)8, rabbit anti-TH (Cat# P40101; Pel-Freez; 1:500)20, sheep anti-TH (Cat# P60101–0; Pel-Freez; 1:500)8, and rabbit anti-VIP (Cat# ab43841; Abcam; 1:500)8. Sections were rinsed in PBS+0.05% Tween20 solution and incubated with secondary antibodies generated in donkey and conjugated with appropriate fluorophores (Alexa488, Alexa555, Alexa647; Molecular Probes) diluted 1:250 in blocking solution and counterstained with DAPI (Sigma), rinsed in PBS+0.05% Tween20, and finally coverslipped with Gelvatol.
Image acquisition and processing
Epifluorescence images were acquired on a Leica DM5000 and Olympus Slide Scanner VS120, whereas confocal images were acquired on Nikon A1 confocal (Northwestern Cell Imaging Facility; NCI-CCSG-P30-CA060553). For the dorsal striatum, we imaged level 41 (CPr; +1.345mm from bregma), level 53 (CPi; +0.145mm from bregma), level 61 (CPc; −0.655mm from bregma), and finally level 69 (−1.455mm from bregma; aka the tail of the striatum CPt) of the Allen Reference Atlas (ARA) based on a previously published cortical-striatal projection analysis29. For the nucleus accumbens, we imaged level 41 (ACBr; +1.345mm from bregma), level 44 (ACBi; 1.045mm from bregma), and level 47 (ACBc; 0.745mm from bregma) of the ARA. For the amygdala, we imaged level 66 (−1.155mm from bregma), level 70 (−1.555mm from bregma), level 73 (−1.855mm from bregma), and level 77 (−2.255mm from bregma). To generate the axonal traces for figure depiction, the images were converted to single channel and inverted, and thresholded in Fiji. The images were then vectorized in Adobe Illustrator (CC2018) using the Image trace function and superposed to Allen Reference Atlas. Images brightness and contrast was adjusted in Fiji and figures were assembled in Adobe Illustrator (CC2018).
Quantitative analysis
To quantify starter populations expressing the fluorescent protein, and assess the colocalization with TH and subtype specific markers, we imaged 10X images on a Leica DM5000 microscope. For every Cre line quantified, we counted at least 6 images representative of the rostro-caudal distribution of DA neurons, in at least 3 different brains.
Statistic and reproducibility
The results are reported as average per brain (± standard error of the mean). No statistical analysis was performed in this study. No data point was excluded. No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those reported in previous publication8,20. The number of animal used in each experiment is listed in Table S1. For each experiment, mice were arbitrarily assigned for stereotaxic surgery, or were processed for imaging accordingly to the available genotype. Data collection and analysis were not performed blind to the conditions of the experiments.
Reporting Summary
Further information on experimental design and analysis is available in the Life Sciences Research Reporting Summary linked to this article.
Reagent and data availability
All newly generated mouse lines will be made available upon request. The original axonal tracing data that support the findings depicted in Figures 3 to 5 are available online (labs.feinberg.northwestern.edu/awatramani/data/poulin-et-al-2018.html). Additional data or reagents are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We apologize to the authors of the excellent work that could not be acknowledged due to space restrictions. The authors wish to thank Sangrag Ganguli, Manuel Jurado, and Idil Oksuz for their technical assistance, Dr. Simon Pieraut and Dr. Anton Maximov for help with trimethropin injection protocol, Dr. Xiaoxi Zhuang and Dr. Bradford Lowell for sharing mouse reagents. This work was supported by NIH grants R01NS06977 and R01NS047085 to CSC; R01MH110556–01A1 to DAD; R01MH110556–01A1, 1R21NS072703–01A1, R01NS096240–01, NARSAD and Paul Ruby Foundation to RA; and grants from MJFF and CIHR to JFP.
References
- 1.Berke JD What does dopamine mean? Nat Neurosci 1 (2018). doi: 10.1038/s41593-018-0152-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Björklund AA & Dunnett SB Dopamine neuron systems in the brain: an update. Trends Neurosci 30, 194–202 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Lammel S, Lim BK & Malenka RC Reward and aversion in a heterogeneous midbrain dopamine system. Neuropharmacology 76 Pt B, 351–359 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roeper J Dissecting the diversity of midbrain dopamine neurons. Trends Neurosci 36, 336–342 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Morales M & Margolis EB Ventral tegmental area: cellular heterogeneity, connectivity and behaviour. Nat Rev Neurosci 18, 73–85 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Ikemoto S Dopamine reward circuitry: two projection systems from the ventral midbrain to the nucleus accumbens-olfactory tubercle complex. Brain Res Rev 56, 27–78 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bromberg-Martin ES, Matsumoto M & Hikosaka O Dopamine in motivational control: rewarding, aversive, and alerting. Neuron 68, 815–834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poulin J-F et al. Defining midbrain dopaminergic neuron diversity by single-cell gene expression profiling. Cell Rep 9, 930–943 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.La Manno G et al. Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167, 566–580.e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hook PW et al. Single-Cell RNA-Seq of Mouse Dopaminergic Neurons Informs Candidate Gene Selection for Sporadic Parkinson Disease. Am. J. Hum. Genet 102, 427–446 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poulin J-F, Tasic B, Hjerling-Leffler J, Trimarchi JM & Awatramani RB Disentangling neural cell diversity using single-cell transcriptomics. Nat Neurosci 19, 1131–1141 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Brignani S & Pasterkamp RJ Neuronal Subset-Specific Migration and Axonal Wiring Mechanisms in the Developing Midbrain Dopamine System. Front. Neuroanat 11, 55 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dymecki SM, Ray RS & Kim JC Mapping cell fate and function using recombinase-based intersectional strategies. Meth Enzymol 477, 183–213 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Awatramani RB, Soriano P, Rodriguez C, Mai JJ & Dymecki SM Cryptic boundaries in roof plate and choroid plexus identified by intersectional gene activation. Nat Genet 35, 70–75 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Cho JR et al. Dorsal Raphe Dopamine Neurons Modulate Arousal and Promote Wakefulness by Salient Stimuli. Neuron 94, 1205–1219.e8 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Lammel S et al. Diversity of transgenic mouse models for selective targeting of midbrain dopamine neurons. Neuron 85, 429–438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madisen L et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fenno LE et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat Methods 11, 763–772 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Xie Z, Turkson S & Zhuang X A53T human α-synuclein overexpression in transgenic mice induces pervasive mitochondria macroautophagy defects preceding dopamine neuron degeneration. J. Neurosci 35, 890–905 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nouri N & Awatramani RB A novel floor plate boundary defined by adjacent En1 and Dbx1 microdomains distinguishes midbrain dopamine and hypothalamic neurons. Development 144, 916–927 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panman L et al. Sox6 and Otx2 control the specification of substantia nigra and ventral tegmental area dopamine neurons. Cell Rep 8, 1018–1025 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Sgobio C et al. Aldehyde dehydrogenase 1-positive nigrostriatal dopaminergic fibers exhibit distinct projection pattern and dopamine release dynamics at mouse dorsal striatum. Sci. Rep 7, 5283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trudeau L-E et al. The multilingual nature of dopamine neurons. Prog Brain Res 211, 141–164 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinkellner T et al. Role for VGLUT2 in selective vulnerability of midbrain dopamine neurons. J. Clin. Invest 128, 774–788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menegas W et al. Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. Elife 4, e10032 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerfen CR, Herkenham M & Thibault J The neostriatal mosaic: II. Patch- and matrix-directed mesostriatal dopaminergic and non-dopaminergic systems. The Journal of neuroscience 7, 3915–3934 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gangarossa G et al. Distribution and compartmental organization of GABAergic medium-sized spiny neurons in the mouse nucleus accumbens. Front Neural Circuits 7, 22–22 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mingote S et al. Functional Connectome Analysis of Dopamine Neuron Glutamatergic Connections in Forebrain Regions. J. Neurosci 35, 16259–16271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hintiryan H et al. The mouse cortico-striatal projectome. Nat Neurosci 19, 1100–1114 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh SW et al. A mesoscale connectome of the mouse brain. Nature 508, 207–214 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yetnikoff L, Lavezzi HN, Reichard RA & Zahm DS An update on the connections of the ventral mesencephalic dopaminergic complex. Neuroscience 282C, 23–48 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Menegas W, Babayan BM, Uchida N & Watabe-Uchida M Opposite initialization to novel cues in dopamine signaling in ventral and posterior striatum in mice. Elife 6, 988 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lerner TN et al. Intact-Brain Analyses Reveal Distinct Information Carried by SNc Dopamine Subcircuits. Cell 162, 635–647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howe MW & Dombeck DA Rapid signalling in distinct dopaminergic axons during locomotion and reward. Nature 535, 505–510 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin HH et al. Dynamic reorganization of striatal circuits during the acquisition and consolidation of a skill. Nat Neurosci 12, 333–341 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorn CAC, Atallah HH, Howe MM & Graybiel AMA Differential Dynamics of Activity Changes in Dorsolateral and Dorsomedial Striatal Loops during Learning. Neuron 66, 15.–15 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gangarossa G et al. Spatial distribution of D1R- and D2R-expressing medium-sized spiny neurons differs along the rostro-caudal axis of the mouse dorsal striatum. Front Neural Circuits 7, 124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Salvio M et al. Otx2 controls neuron subtype identity in ventral tegmental area and antagonizes vulnerability to MPTP. Nat Neurosci 13, 1481–1488 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Khan S et al. Survival of a Novel Subset of Midbrain Dopaminergic Neurons Projecting to the Lateral Septum Is Dependent on NeuroD Proteins. J. Neurosci 37, 2305–2316 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stuber GD, Hnasko TS, Britt JP, Edwards RH & Bonci A Dopaminergic terminals in the nucleus accumbens but not the dorsal striatum corelease glutamate. The Journal of neuroscience 30, 8229–8233 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hnasko TS, Hjelmstad GO, Fields HL & Edwards RH Ventral tegmental area glutamate neurons: electrophysiological properties and projections. The Journal of neuroscience 32, 15076–15085 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabanova A et al. Function and developmental origin of a mesocortical inhibitory circuit. Nat Neurosci 18, 872–882 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Lammel S et al. Unique properties of mesoprefrontal neurons within a dual mesocorticolimbic dopamine system. Neuron 57, 760–773 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Matthews GA et al. Dorsal Raphe Dopamine Neurons Represent the Experience of Social Isolation. Cell 164, 617–631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beier KT et al. Circuit Architecture of VTA Dopamine Neurons Revealed by Systematic Input-Output Mapping. Cell 162, 622–634 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baimel C, Lau BK, Qiao M & Borgland SL Projection-Target-Defined Effects of Orexin and Dynorphin on VTA Dopamine Neurons. Cell Rep 18, 1346–1355 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Duan B et al. Identification of spinal circuits transmitting and gating mechanical pain. Cell 159, 1417–1432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sciolino NR et al. Recombinase-Dependent Mouse Lines for Chemogenetic Activation of Genetically Defined Cell Types. Cell Rep 15, 2563–2573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ray RS et al. Impaired respiratory and body temperature control upon acute serotonergic neuron inhibition. Science 333, 637–642 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bourane S et al. Gate control of mechanical itch by a subpopulation of spinal cord interneurons. Science 350, 550–554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods reference list
- 51.Skarnes WC et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osterwalder M et al. Dual RMCE for efficient re-engineering of mouse mutant alleles. Nat Methods 7, 893–895 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Donnelly ML et al. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J. Gen. Virol 82, 1013–1025 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Anastassiadis K et al. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis Model Mech 2, 508–515 (2009). [DOI] [PubMed] [Google Scholar]
- 55.Wallén A et al. Fate of mesencephalic AHD2-expressing dopamine progenitor cells in NURR1 mutant mice. Exp. Cell Res. 253, 737–746 (1999). [DOI] [PubMed] [Google Scholar]
- 56.Feil R, Wagner J, Metzger D & Chambon P Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun 237, 752–757 (1997). [DOI] [PubMed] [Google Scholar]
- 57.Hsu LC, Chang WC, Hoffmann I & Duester G Molecular analysis of two closely related mouse aldehyde dehydrogenase genes: identification of a role for Aldh1, but not Aldh-pb, in the biosynthesis of retinoic acid. Biochem J 339 (Pt 2), 387–395 (1999). [PMC free article] [PubMed] [Google Scholar]
- 58.Meyers EN, Lewandoski M & Martin GR An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet 18, 136–141 (1998). [DOI] [PubMed] [Google Scholar]
- 59.Tasic B et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci 19, 335–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jensen P et al. Redefining the serotonergic system by genetic lineage. Nat Neurosci 11, 417–419 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen L et al. Unregulated cytosolic dopamine causes neurodegeneration associated with oxidative stress in mice. J Neurosci 28, 425–433 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sando R et al. Inducible control of gene expression with destabilized Cre. Nat Methods 10, 1085–1088 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joksimovic M et al. Spatiotemporally separable Shh domains in the midbrain define distinct dopaminergic progenitor pools. Proc. Natl. Acad. Sci. U.S.A. 106, 19185–19190 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anderegg A et al. An Lmx1b-miR135a2 regulatory circuit modulates Wnt1/Wnt signaling and determines the size of the midbrain dopaminergic progenitor pool. PLoS Genet. 9, e1003973 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


