ABSTRACT
All animals have body burdens of polychlorinated biphenyls (PCBs) despite their ban decades ago. These and modern endocrine-disrupting chemicals (EDCs) such as the fungicide vinclozolin (VIN) perturb hormone signaling and lead to dysfunctions following prenatal exposures. Beyond direct exposures, transgenerational disease phenotypes can persist for multiple generations without subsequent exposure. The mechanisms of action of these EDCs differ: VIN is anti-androgenic while the PCB mixture Aroclor 1221 (A1221) is weakly estrogenic. Based on limited evidence for the inheritance of epimutations in germline, we measured DNA methylation in brain and sperm of rats. Pregnant dams were exposed from day 8–18 of gestation to low dosages of VIN, A1221, or the vehicle. To produce paternal lineages, exposed F1 males were bred with untreated females, creating the F2 and subsequently F3 generations. In adult F1 and F3 males, mature sperm was collected, and brain nuclei involved in anxiety and social behaviors (CA3 of the hippocampus; central amygdala) were selected for assays of epimutations in CpG islands using reduced representation bisulfite sequencing. In F1 sperm, VIN and PCBs induced differential methylation in 215 and 284 CpG islands, respectively, compared to vehicle. The majority of effects were associated with hypermethylation. Fewer epimutations were detected in the brain. A subset of differentially methylated regions were retained from the F1 to the F3 generation, suggesting a common mechanism of EDC and germline epigenome interaction. Thus, EDCs can cause heritable epimutations in the sperm that may embody the future phenotype of brain-behavior disorders caused by direct or transgenerational exposures.
KEYWORDS: Endocrine disrupting chemicals, vinclozolin, polychlorinated biphenyls, aroclor, DNA methylation, RRBS, transgenerational, inheritance, sperm, brain
Introduction
The developing fetus is highly sensitive to hormones, which are produced by its own endocrine glands or transferred to it from maternal and placental synthesis. A delicate internal hormonal milieu directs the developmental trajectory of hormone-sensitive organs, including reproductive tissues and the brain. Even small perturbations of this milieu during critical developmental periods can cause detrimental long-term effects on reproductive tissues, and alter sexual and affective behaviors in adulthood [1]. Endocrine-disrupting chemicals (EDCs), which change hormone signaling and actions, can alter developmental trajectories in directly exposed individuals and, in some instances, in subsequent generations [Reviewed in [2]]. More specifically, F3 descendants of animals given prenatal exposure to the anti-androgenic fungicide vinclozolin (VIN) during gonadal differentiation, but without any direct exposure to VIN, had a greater propensity for, and shorter latency to, the manifestation of disease phenotypes [3]. These F3 individuals also showed altered anxiety, social, and sexual behavior [4,5], and had changes in gene expression and metabolic activity in specific brain nuclei [5,6]. An endocrine-disrupting mixture of weakly estrogenic polychlorinated biphenyls (PCBs), Aroclor 1221 (A1221), also caused phenotypic changes through the F3 generation [7].
Fetal reprogramming of methylation by EDCs is thought to occur in primordial germ cells (PGCs), which are progenitors to sperm and ova [8]. During migration to the genital ridge, PGCs begin a proliferation process whereby active and passive demethylation cause a genome-wide erasure of the epigenome [9,10]. In parts of the body that are sexually differentiated, reestablishment of the epigenome occurs in a sex-specific manner that includes parental imprinting [11]. These periods of epigenome erasures and re-establishment are considered to be critical windows for natural hormone actions and for vulnerability to environmental insults. EDC exposure during this period can cause epimutations that become permanently embedded in the germline. If epimutations are the mechanism for transgenerational inherited diseased phenotypes, they must also escape erasure during PGC differentiation and be faithfully carried to future generations. Indeed, recent work on the epigenome of the male germline in F3 generation rats exposed to VIN demonstrated widespread alterations of DNA methylation and gene expression, and implicated the early (E13) germline [12]. However, other groups failed to replicate this work [13–15]. EDCs other than VIN have also been observed for their capacity to alter the epigenome. Atrazine, which interferes with estrogen signaling, caused epimutations in sperm of F1 – F3 males [16]. However, low-dose exposures to the xenoestrogen bisphenol-A during gestation did not alter DNA methylation in the hippocampus [17].
An unanswered question is whether and how epimutations in the germline due to EDC exposure may lead to changes in somatic gene expression in subsequent generations and the emergence of altered phenotypes. Presumably, the germline carries all the necessary information for proper establishment of tissue-specific epigenetic profiles that may be subject to dysregulation by epimutations. As for the brain, F3 individuals have altered behavioral phenotypes and gene expression [4–6,18], but the potential role of methylation has not been determined, nor related to the germline epigenetic state.
The current study fills this gap in knowledge and extends prior work by using environmentally relevant dosages of VIN and A1221, selected because they represent different classes of EDCs. VIN is primarily an anti-androgenic fungicide, and A1221 is a weakly estrogenic PCB mixture. These compounds have historical differences in usage, representing past (PCBs) and present (VIN) contaminants. Our study differs from previous work (e.g., [19]) by utilizing a breeding paradigm that focused exclusively on paternal transmission of epimutations. We analyzed DNA methylation of CpG islands in directly exposed (F1) or ancestrally exposed (F3) sperm. In the same animals, we selected two brain nuclei for epimutation analysis: the central amygdala, and CA3 of the hippocampus, that are a pivotal part of the neural network underlying the anxiety and social behavioral perturbations caused by transgenerational EDC inheritance. The F1 and F3 generations were selected as our focal animals because they are best-studied [3,12,19–22] and enabled us to put our results in the context of the published literature.
Results
Differentially methylated CpG islands in F1 and F3 sperm
Mature sperm retrieved from the cauda of the epididymis from F1 males (n = 5 per treatment group) and F3 males of paternal descent (n = 8 per treatment group) were analyzed for differentially methylated regions (DMRs) across CpG islands. CpG islands are regions of the genome with a higher than expected density of CG dinucleotides. Generally, a CpG island is greater than 200 base pairs in length, has a CG content of greater than 50%, and a CpG dinucleotide observed-to-expected ratio of greater than 0.6. CpG island annotations were retrieved from the University of California, Santa Cruz (UCSC) genome bioinformatics database. We used the entire CpG island as the unit of analysis whereby all methylated and non-methylated Cs in the CpG dinucleotide context were tested via a binomial logistic regression. A probe was considered hypomethylated if its average percent methylation was lower than the vehicle control, and hypermethylated if higher than the vehicle control. No absolute cutoff of percent methylation difference was used to filter these results. Here and elsewhere, statistical significance was set at p < 0.05 after applying the Benjamini and Hochberg false discovery rate (FDR) correction [23].
Sperm from VIN or A1221 males was compared to that of corresponding vehicle males in the same generation. Analysis of differentially methylated CpG islands in F1 individuals revealed that in comparison to the vehicle, there were more sites affected in the A1221 (284) lineage than the VIN (215) lineage. This relationship was reversed in the F3 descendants (142 and 207, respectively; Table 1 – Sperm). Of these, a similar proportion of DMRs (15% A1221) and 17% VIN) were located in intergenic regions and not directly associated with a known gene (Figure 1(a,b)). The majority of the identified DMRs were hypermethylated in all treatment groups (Table 1 – Sperm, Figure 2(a)). A table of all affected CpG islands and the gene they are associated with, if any, can be found in Supplemental Table 1.
Table 1.
Differentially methylated CpG islands.
| A1221 |
VIN |
|||||
|---|---|---|---|---|---|---|
| F1 | F3 | Both | F1 | F3 | Both | |
| Sperm | ||||||
| DMRs | 284 | 142 | 60 | 215 | 207 | 77 |
| Intergenic DMRs | 44 (15%) | 24 (17%) | 36 (17%) | 34 (16%) | ||
| Hypermethylated | 209 (74%) | 99 (70%) | 36 (60%) | 166 (77%) | 154 (74%) | 62 (81%) |
| Hypomethylated | 75 (26%) | 43 (30%) | 9 (15%) | 49 (23%) | 53 (26%) | 8 (10%) |
| Discordant | 15 (25%) | 7 (9%) | ||||
| CeA | ||||||
| DMRs | 101 | 143 | 20 | 124 | 106 | 25 |
| Intergenic DMRs | 12 (12%) | 31 (22%) | 17 (14%) | 18 (17%) | ||
| Hypermethylated | 69 (68%) | 107 (75%) | 11 (55%) | 101 (81%) | 81 (76%) | 19 (68%) |
| Hypomethylated | 32 (32%) | 36 (25%) | 2 (10%) | 23 (19%) | 25 (24%) | 0 (16%) |
| Discordant | 7 (35%) | 6 (16%) | ||||
| CA3 | ||||||
| DMRs | 91 | 136 | 26 | 74 | 89 | 19 |
| Intergenic DMRs | 11 (12%) | 30 (22%) | 11 (15%) | 15 (17%) | ||
| Hypermethylated | 33 (36%) | 122 (90%) | 11 (42%) | 37 (50%) | 42 (47%) | 13 (68%) |
| Hypomethylated | 58 (64%) | 14 (10%) | 0 (0%) | 37 (50%) | 47 (53%) | 3 (16%) |
| Discordant | 15 (58%) | 3 (16%) | ||||
Figure 1.
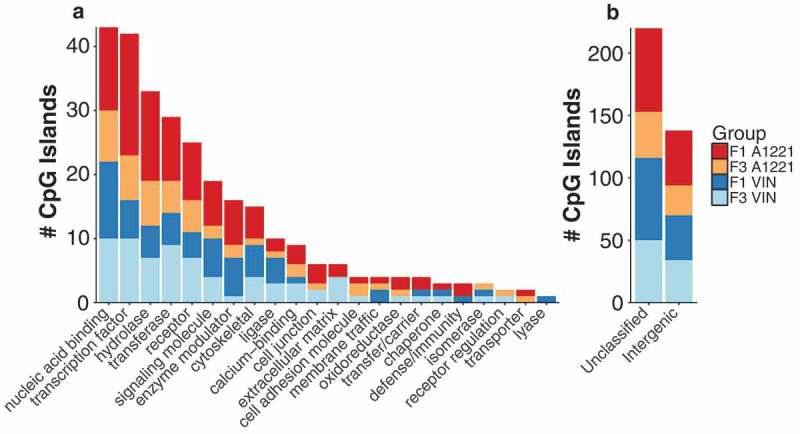
a) The functional category of CpG islands differentially methylated by treatment relative to vehicle control in F1 and F3 sperm are shown. No individual categories were significantly enriched. b) CpG islands that were either not classified or located in an intergenic region of the genome and not associated with a particular gene are shown.
Figure 2.
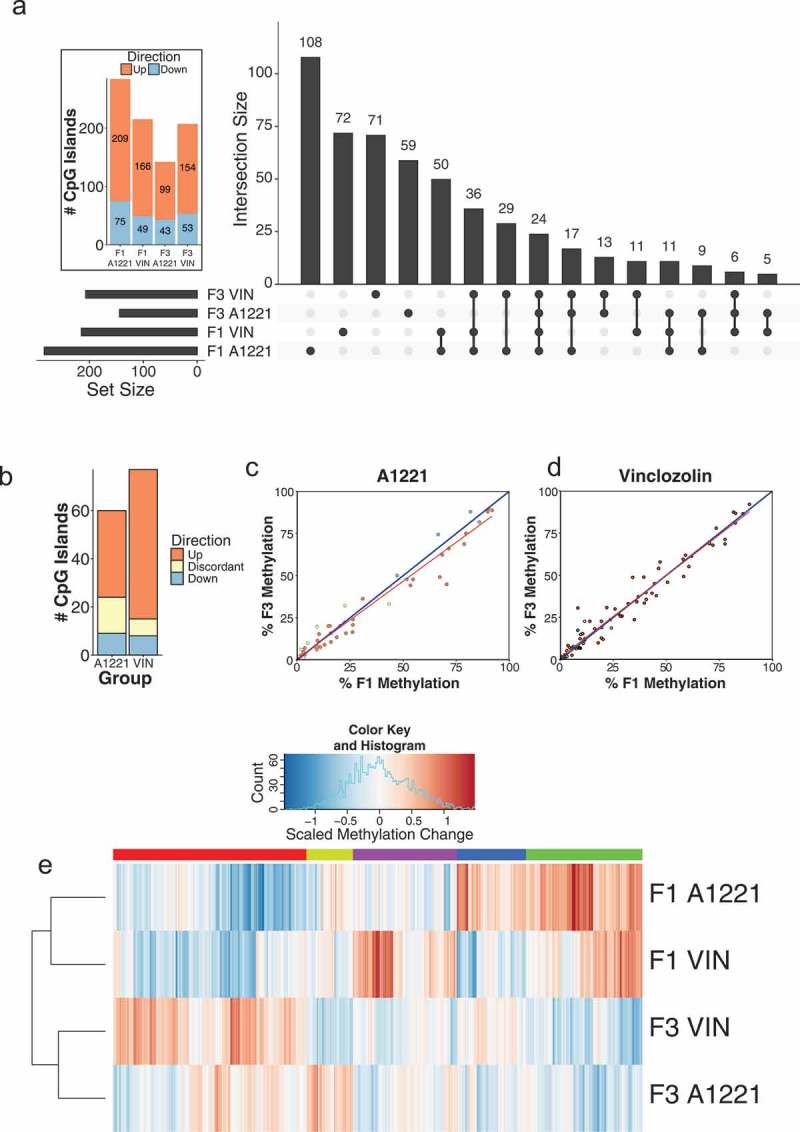
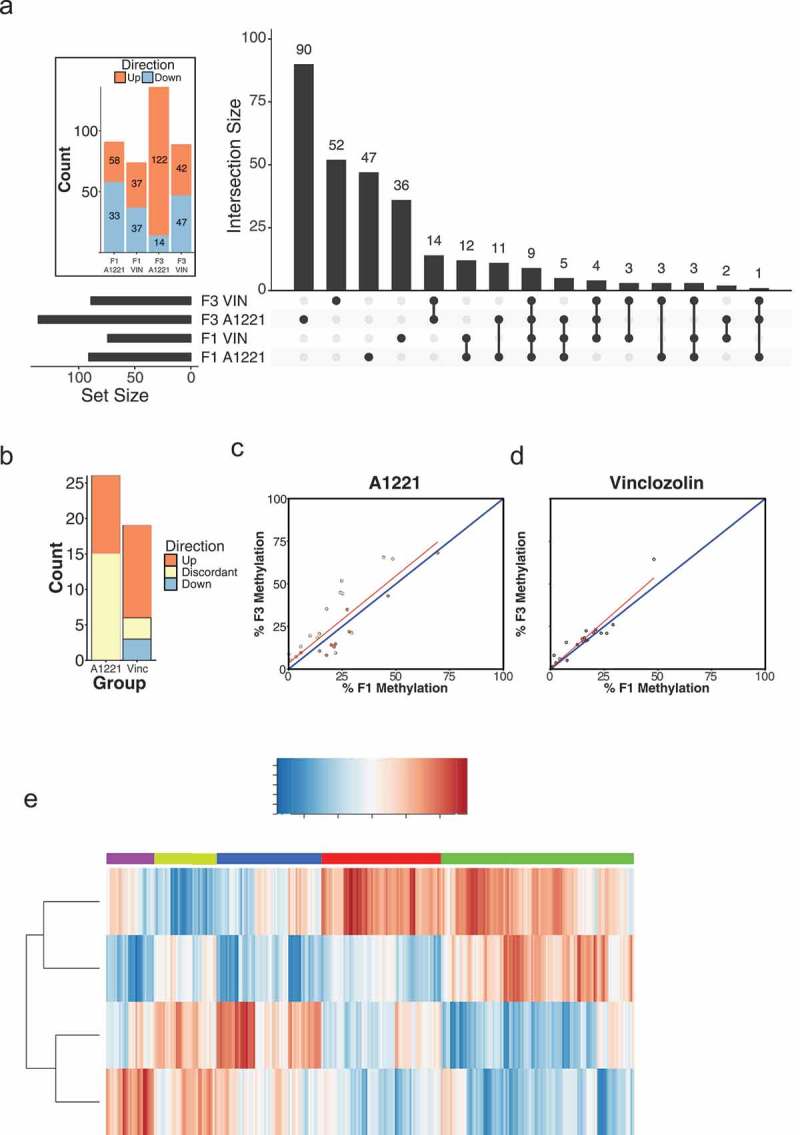
Differentially methylated (versus vehicle, p < 0.05 after FDR) CpG islands relative to vehicle control in F1 and F3 sperm are shown by generation and treatment. a) The intersection, the number of significantly altered CpG islands identified that are either unique or common between groups, is shown for all possible group combinations. Most identified CpG islands were unique to each generation and treatment, followed by within generation, then within treatment. The total number of altered CpG islands per treatment lineage and generation is shown in the figure inset; orange indicates CpG islands with increased methylation compared to vehicle, and blue indicates decreased methylation. b) The number and directionality of change of DMRs is shown between the F1 and F3 generations in relationship to vehicle. Orange indicates hypermethylation compared to vehicle in both the F1 and F3 generation, blue indicates hypomethylation in both generations, and yellow indicates that a CpG island is significantly affected in both generations but in opposite directions. VIN had more transgenerational DMRs and less discordance between generations than A1221. c & d) The % DNA methylation of probes affected in both the F1 and F3 generation were correlated within A1221 and VIN sperm, respectively. Data points below the 1:1 blue diagonal line have higher methylation in the F1 generation while those above have higher methylation in the F3 generation. e) Normalized % methylation relative to vehicle of all probes (CpG islands) affected across treatment groups is shown. Red indicates increased methylation compared to vehicle and blue indicates decreased methylation compared to vehicle at a given CpG island. Generation was a significant clustering factor (both AU/BP = 100/100%).
Gene ontology and gene category enrichment were performed using the Panther Classification System (version 13.1). In some instances, multiple CpG islands associated with the same gene were found to be differentially methylated due to treatment. These instances were treated as a single observation of a gene. Gene category enrichment did not identify any significantly enriched categories (Figure 1(a)). Gene ontology analysis identified significantly enriched biological categories, using Fisher’s Exact test with the Benjamini and Hochberg FDR [23] correction for multiple comparisons, for all groups except F3 A1221 males. Genes associated with nervous system development were enriched in all other groups compared to their corresponding vehicle (F1 A1221, p < 0.001, F1 VIN, p = 0.02, and F3 VIN p = 0.02). Cell differentiation and cellular component movement were also enriched in F1 A1221 sperm (p < 0.001 and p = 0.038, respectively). In F1 VIN sperm, neuron-neuron synaptic transmission (p = 0.046), gamete generation (p = 0.022), and cell differentiation (p = 0.031) were enriched.
We next identified the DMRs that were unique or common between treatment groups and across generations. DMRs unique to treatment and generation were the most populated categories (Figure 2(a)). There was substantial overlap of DMRs in the F1 generation; 121 DMRs were shared between A1221 (total of 284) and VIN (total of 215). Conversely, there was half as much overlap between treatments in the F3 generation. Within treatment and across generation (F1 and F3), there were 9 DMRs unique to A1221 and 11 DMRs unique to VIN (Figure 2(a)). We then analyzed the consistency of each DMR in the F1 and F3 generation within treatment group (Figure 2(b-d)). Of the 284 DMRs identified in F1 A1221 males, 60 (21%) were also identified in the F3 generation. To determine if the identified DMRs were consistent between generations, we compared the direction of change within each generation to the vehicle group of that generation. For example, if a DMR was hypermethylated compared to vehicle in the F1 generation and hypomethylated compared to vehicle in the F3 generation, we considered this effect discordant between generations. Discordance was slightly higher in individuals exposed to A1221 (25%, Table 1 – Sperm, Figure 2(b)). Of the 215 DMRs identified in F1 VIN males, 77 (36%) were also identified in the F3 generation. There were fewer DMRs in VIN than A1221 sperm found to be discordant (9%, Table 1 – Sperm, Figure 2(b)). A linear regression model was used to determine if the probes that were significantly altered by treatment in both the F1 and F3 generation deviated between directly (F1) and ancestrally exposed (F3) males. The model revealed that generation identity accounted for a similar proportion of variance in VIN (adjusted R2 = 0.954) and A1221 (adjusted R2 = 0.961) treated individuals (Figure 2(c,d)). Finally, cluster analysis was performed on all CpG islands altered by treatment or across generation. Generation was the best clustering variable of CpG island methylation. A1221 and VIN significantly occupied the same cluster in both the F1 and F3 generation (Both AU/BP = 100/100%, Figure 2(e) – AU is approximately unbiased p-value and BP is bootstrap probability value derived from hierarchical clustering and considered significant when > 95%).
Intergenic CpG islands and repetitive sequences
Of the CpG islands that we identified as altered by treatment in sperm samples, ~ 16% were not associated with a known gene and therefore categorized as intergenic. Analysis of the methylation status of all identified intergenic differentially methylated CpG islands revealed an average of 19.2% methylation with a broad range at individual loci (min < 1%, max = 97.6%). Of these, ~ 61% (84/138) were hypermethylated by EDC exposure. We then referenced our identified intergenic differentially methylated CpG islands to a database of approximately 400,000 known repetitive sequences, including transposable elements. A small subset (24/138) partially overlapped with known repeated sequences, exclusively on their 5’ end. These CpG islands had higher average DNA methylation (36.4%) than generic intergenic CpG islands (19.2%), with an equally broad range (min < 1%, max = 92.2%). This was reversed in CpG islands associated with known repetitive elements (65.5% hypomethylated, 15/24). All of the identified repetitive elements belonged exclusively to the long terminal repeats (17/24) or long interspersed elements (7/24) of retrotransposons.
Differentially methylated CpG islands in F1 and F3 brain
Central amygdala
The CeA was analyzed for differentially methylated CpG islands as described for sperm. Within F1 individuals, slightly more CpG islands were differentially methylated in VIN (124) compared to A1221 (101) males, a trend that was reversed in the F3 generation (143 for A1221, 106 for VIN; Table 1 – CeA) and mirrored by the proportion of DMRs located in intergenic regions (Figure 3(a,b)). Similar to sperm, the majority of affected CpG islands in CeA were hypermethylated by both EDCs compared to vehicle (Table 1 – CeA, Figure 4(a)). Gene ontology and classification enrichment analysis did not identify any significantly different categories (Figure 3(a)).
Figure 3.
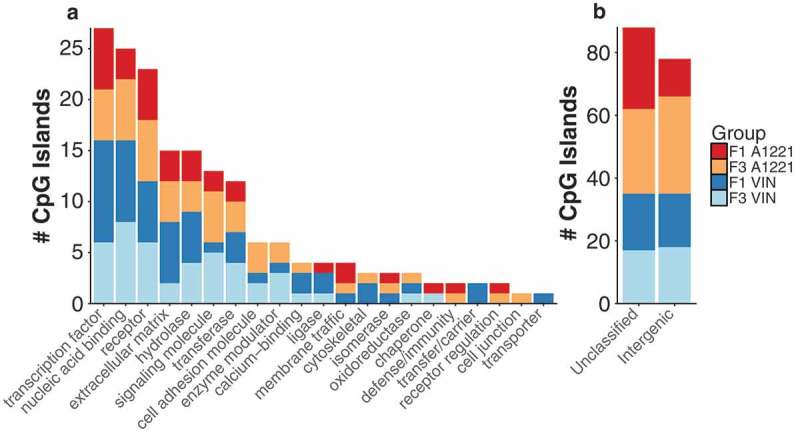
a) The functional category of CpG islands differentially methylated by treatment relative to vehicle control in F1 and F3 CeA are shown. No individual categories were significantly enriched. b) CpG islands that were either not classified or located in an intergenic region of the genome and not associated with a gene are shown.
Figure 4.
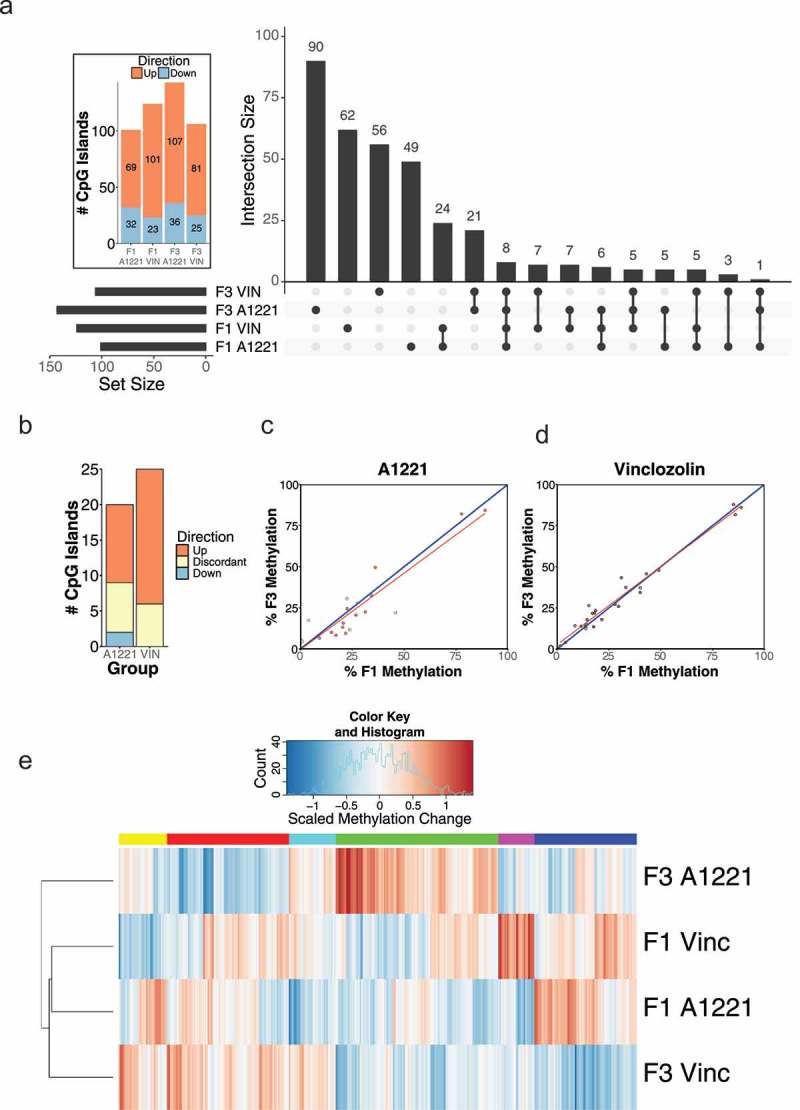
Differentially methylated (versus vehicle, p < 0.05 after FDR) CpG islands relative to vehicle control in F1 and F3 CeA are shown by generation and treatment. a) The intersection, the number of significantly altered CpG islands identified that are either unique or common between groups, is shown for all possible group combinations. Most identified CpG islands were unique to each generation and treatment, followed by within generation, then within treatment. The total number of altered CpG islands per treatment lineage and generation is shown in the figure inset; orange indicates CpG islands with increased methylation compared to vehicle, and blue indicates decreased methylation. b) The number and directionality of change of DMRs is shown between the F1 and F3 generations in relationship to vehicle. Orange indicates hypermethylation compared to vehicle in both the F1 and F3 generation, blue indicates hypomethylation in both generations, and yellow indicates that a CpG island is significantly affected in both generations but in opposite directions. VIN had more transgenerational DMRs and similar discordance between generations compared to A1221. c & d) The % DNA methylation of probes affected in both the F1 and F3 generation were correlated within A1221 and VIN CeA, respectively. Data points below the 1:1 blue diagonal line have higher methylation in the F1 generation while those above have higher methylation in the F3 generation. e) Normalized % methylation relative to vehicle of all probes (CpG islands) affected across treatment groups is shown. Red indicates increased methylation compared to vehicle and blue indicates decreased methylation compared to vehicle normalized within a given CpG island. F3 A1221 was found to be outside the cluster with all other groups (both AU/BP = 99/93%).
Analysis of differentially methylated CpG islands between treatments and across generations again showed that the majority were unique to treatment and generation (Figure 4(a)). Similarity by generation was the next most populated category; A1221 and VIN shared 43 DMRs in the F1 generation and 35 in the F3 generation. Few CpG islands were identified as exclusively altered within treatment group and between the F1 and F3 generation (A1221 = 9 and VIN = 11, Figure 4(a)). Analysis of the consistency of altered CpG islands across generation identified a similar proportion of affected sites between treatment, although there was a high proportion of discordance within the A1221 treatment (Table 1 – CeA, Figure 4(b-d)).
A linear regression model revealed that generation accounted for more variance in VIN individuals (adjusted R2 = 0.96) than in A1221 individuals (adjusted R2 = 0.86; Figure 4(c,d)). Finally, cluster analysis was performed on the methylation status of the differentially methylated CpG islands across all treatment groups relative to vehicle individuals. F3 A1221 was significantly outside the cluster containing all other treatment groups (AU/BP = 99/93% – Figure 4(e)). Both treatments in the F1 treatment were more related to each other than F3 VIN individuals, but this was not significant (AU/BP = 79/69%).
CA3 of the hippocampus
In the F1 generation, more differentially methylated CpG islands were identified due to A1221 (91) exposure than to VIN (74) exposure, an effect that was amplified in the F3 generation (136 and 89, respectively; Table 1 – CA3). A smaller proportion of affected CpG islands were in intergenic regions in directly exposed individuals compared to ancestrally exposed individuals (Table 1 – CA3, Figure 5(a,b)). Compared to sperm and CeA, differentially methylated CpG islands in CA3 had the highest proportion of hypomethylation (Figure 6(a)). This directionality was reversed in ancestrally treated A1221 individuals but persisted in VIN individuals (Table 1 – CA3, Figure 6(a)). Gene ontology and classification enrichment analysis did not identify any significantly enriched categories (Figure 5(a)).
Figure 5.
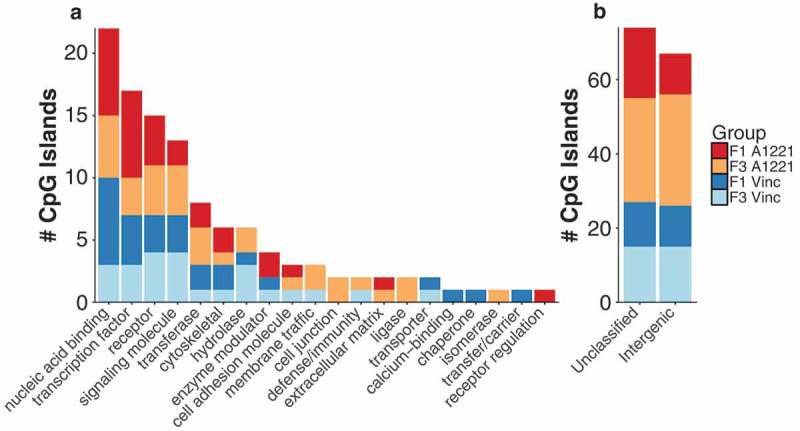
a) The functional category of CpG islands differentially methylated by treatment relative to vehicle control in F1 and F3 CA3 are shown. No individual categories were significantly enriched. Both F3 treatments had more altered unclassified and intergenic differentially methylated CpG islands. b) CpG islands that were either not classified or located in an intergenic region of the genome and not associated with a gene are shown.
Figure 6.
Differentially methylated (versus vehicle, p < 0.05 after FDR) CpG islands relative to vehicle control in F1 and F3 CA3 are shown by generation and treatment. a) The intersection, the number of significantly altered CpG islands identified that are either unique or common between groups, is shown for all possible group combinations. Most identified CpG islands were unique to each generation and treatment, followed by within generation, then within treatment. The total number of altered CpG islands per treatment lineage and generation is shown in the figure inset; orange indicates CpG islands with increased methylation compared to vehicle, and blue indicates decreased methylation. b) The number and directionality of change of DMRs is shown between the F1 and F3 generations in relationship to vehicle. Orange indicates hypermethylation compared to vehicle in both the F1 and F3 generation, blue indicates hypomethylation in both generations, and yellow indicates that a CpG island is significantly affected in both generations but in opposite directions. A1221 had more transgenerational DMRs but more discordance between generations compared to VIN. c & d) The % DNA methylation of probes affected in both the F1 and F3 generation were correlated within A1221 and VIN CA3, respectively. Data points below the 1:1 blue diagonal line have higher methylation in the F1 generation while those above have higher methylation in the F3 generation. e) Normalized % methylation relative to vehicle of all probes (CpG islands) affected across treatment groups is shown. Red indicates increased methylation compared to vehicle and blue indicates decreased methylation compared to vehicle normalized within a given CpG island. Generation was a significant clustering factor (both AU/BP = 100/100%).
Analysis of differentially methylated CpG islands again revealed that the majority of sites were unique to treatment and generation. Within treatment and between generation, there were more sites unique to A1221 than VIN exposed individuals (Figure 6(a)). Analysis of the consistency of altered CpG islands relative to vehicle showed a similar proportion of sites affected across generations and within A1221 or VIN; however, discordance was higher in A1221 than VIN rats (Table 1 – CA3, Figure 6(b)). A linear regression model again revealed that a larger proportion of variance in the model was accounted for by generation within the VIN treatment (adjusted R2 = 0.87, Figure 6(c)) than the A1221 treatment (adjusted R2 = 0.67, Figure 6(d)). Finally, cluster analysis was performed across treatment on the 343 identified differentially methylated CpG islands across all treatment groups relative to vehicle. Generation was the most salient effector of CpG island methylation. A1221 and VIN significantly occupied the same cluster in both the F1 and F3 generation (Both AU/BP = 100/100%, Figure 6(e)).
Discussion
The initial discovery that prenatal exposure to high dosages of VIN caused the transgenerational inheritance of disease phenotypes generations after initial exposure called attention to the potential of environmental contamination persisting long beyond the exposed individual’s lifetime [24,25]. Further work using this VIN model demonstrated somatic [3,26] and reproductive dysfunctions in males [24,25,27] and females [28], altered anxiety and social behaviors [4,5,18], disrupted metabolic activity [4,5], and changes in gene expression [4,5,18] in the brain. Since then, other EDCs, especially bisphenol A [22], tributyltin [29] and phthalates [30] have also been demonstrated to have adverse transgenerational phenotypes. Limited evidence from our lab also showed transgenerational effects of PCBs [7,31,32]. The current study extended prior work substantially by utilizing more realistic exposures to VIN (1 mg/kg in the current study, vs. 50–100 mg/kg in other studies), the inclusion of PCBs, and epigenetic endpoints in F1 and F3 sperm and brain.
The inheritance of disease phenotypes across generations is due, at least in part, to alterations in the germline [33,34]. For high-dose VIN, epimutations were observed in the male F3 germline [12] and sperm [19]. To our knowledge a single report evaluated genome wide epimutations in F1 and F3 sperm due to VIN exposure [21]. Using methylated DNA immunoprecipitation followed by hybridization to a promoter microarray (MeDIP-Chip), Beck et al. found that the F1 generation had fewer differentially methylated promoter regions compared to the ancestrally exposed F3 generation. They also reported negligible overlap between the generations, similar to our own finding as well as previous reports from other labs (e.g., [35,36]).
Here, we evaluated direct and transgenerational effects of low-dose VIN or PCBs on CpG island methylation across the genome in both sperm and brain. It is important to note several key differences between the methodology used in the current report and the ones that precede it. First, prior work utilized a breeding paradigm wherein F1 offspring from a VIN exposed litter were bred with a same-treatment partner to produce the F2 generation. The same paradigm (same-treatment matings among F2 males and F2 females) was used to yield the F3 generation [19]. By contrast, we used a breeding paradigm in which F1 treated males mated with untreated females to yield the F2 generation. This was repeated in the F2 generation, resulting in F3 males that derived from paternal transmission of the germline. Second, we injected a much lower and realistic dosage (1 mg/kg) of VIN exposure than previous reports (50–100 mg/kg, e.g., [21]). Third, we compared outcomes of VIN, an antiandrogenic fungicide, with those of A1221, a weakly estrogenic PCB mix, to determine whether different classes of EDCs have similar or different effects. Fourth, our lab has previously utilized a very short exposure window for A1221 (E16 and E18 only) to determine its effects during the beginning of the critical window of sexual differentiation [37,38]. Here, we extended the exposure window (E8-18) to encompass the period of germline erasure and re-establishment in fetal males (E8-14, e.g., [21]).
Sperm methylation is altered by A1221 and VIN
CpG islands are GC dense regions that typically contain low levels of DNA methylation in the germline [39] in part due to the propensity of methylated cytosine residues to spontaneously deaminate resulting in subsequent mutation [40]. About 70% of gene promoters include at least one CpG island [41] while ‘orphan’ CpG islands (not associated with a transcriptional start site) may initiate transcription of distant genes (reviewed in [42]). Notably, CpG islands in sperm have very low levels of methylation, presumably to protect against spontaneous deamination and subsequent mutation, and thereby maintaining the integrity of the germline [40]. McCarrey et al. proposed that the epimutations caused by VIN may also cause genomic instability in a subset of F3 individuals, termed ‘tertiary epimutations,’ that accelerate genetic mutation [43]. However, they found that C to T transitions were not statistically elevated due to treatment. In another study evaluating methylation status of DMRs in sperm due to ancestral high-dose VIN exposure, ~ 70% of affected DMRs were hypermethylated in F3 generation sperm [19,21]. In the current study, 74% of the differentially methylated CpG islands due to ancestral low-dose VIN exposure were hypermethylated. It is interesting that our rats with ancestral exposure to A1221 also had hypermethylation of a majority of the affected CpG islands (70%; Figure 2(a)). Organophosphate flame retardant exposure show a general effect of hypermethylation of imprinted genes in humans [44]. Other EDCs, however, show more equal distributions of hyper- and hypomethylation (DEHP [45]). Because hypermethylation of CpG islands in sperm causes genomic instability, an increased likelihood of spontaneous mutations may be passed to future generations. Further research is needed to confirm this possibility.
A subset of the differentially methylated CpG islands were categorized as intergenic. Previous work has hypothesized that DMRs in intergenic areas may be epigenetic control regions that alter tissue specific gene expression [3]. While CpG islands near promoters are generally hypomethylated in sperm [39], repetitive sequences and transposable elements located in intergenic space and outside of CpG islands are generally hypermethylated [46]. We identified a small subset of intergenic differentially methylated CpG islands that overlapped with repetitive elements from retrotransposons that were hypomethylated due to treatment. Considering that retrotransposons are functionally sensitive to methylation status [47], it is interesting to consider the possibility that EDC exposure may increase vulnerability to transposable elements.
The mechanism by which EDCs interact with and alter the epigenome is not currently known, although there is ample evidence to show epigenetic alterations in F1 individuals or in cell lines exposed to phthalates [48], parabens [49], diethylstilbestrol [50,51], and PCBs [52]. A particularly compelling theory [53] is that nuclear steroid hormone receptors, with which many EDCs interact either directly or indirectly, may be the point of convergence between EDCs and the epigenome. Interestingly, Casati et al., (2015) provided evidence that PCBs initiate downstream transcription of androgen sensitive genes via a histone demethylase, Jarid1b, that interacts with the androgen receptor [54]. Jarid1b catalyzes the removal of H3K4me2 and H3K4me3 histone methylation [55], important histone modifications that maintain a demethylated state within CpG islands [56] of genes associated with neural development and the germline [57]. VIN is typically considered anti-androgenic [58] but there is evidence that vinclozolin can also act as an agonist when there is a lack of competitive endogenous androgens [59]. PCBs are typically studied as estrogenic, anti-estrogenic, and anti-androgenic [60,61]. Furthermore, there is a prenatal testosterone surge in male rats late in gestation between E16 and E20, which peaks at E18 [62]. Thus, the EDCs in this study can affect testosterone signaling in a number of possible ways.
Methylation is altered by A1221 and VIN in the hippocampal CA3 and central amygdala
Prenatal gonadal steroid hormones are crucial to the proper development and sexual differentiation of the brain [63]. EDCs are well-established as interfering with this process in a timing, dose, and duration dependent manner [Reviewed in [64]]. Our previous work demonstrated that the brain is vulnerable to ancestral VIN exposure as evidenced by altered anxiety and social behaviors [4–6]. That steroid hormone dependent DNA methylation also contributes to proper sexual differentiation [65] suggests that EDCs may interfere with the brain methylome during early development. It is unclear if these alterations persist into adulthood or are consistently transferred between generations. The brain is fundamentally different from sperm. Alterations to the germline can be directly transferred to subsequent generations, and in the case of DNA methylation, presumably escape erasure and reestablishment of the epigenome. The brain is a somatic tissue, however, and whether there is a direct pathway to transfer information from germline to brain is unknown.
Here, we analyzed methylation in two discrete brain nuclei (CeA and CA3 of the hippocampus) previously identified as vulnerable to EDC exposure and involved in modulating behavioral phenotypes previously identified in the high-dose VIN model [4–6]. The CeA integrates sensory information and initiates physiological and behavioral output [66] while the CA3 of the hippocampus modulates fear and anxiety responses [67]. Prenatal exposure to both VIN and A1221 induced differential methylation of CpG islands in the brain, although these results should be interpreted cautiously in light of the small sample size (n = 4 per group). Interestingly, A1221 induced more changes (143 and 136) in the F3 generation (ancestral exposure) than due to direct exposure in the F1 generation (101 and 91) in CeA and CA3, respectively. The incidence of effects observed in intergenic regions not directly associated with a gene was approximately 2-fold higher in the F3 generation due to A1221 exposure than VIN. By contrast, VIN showed an equivalent number of DNA methylation alterations between generations. Moreover, fewer CpG islands were affected in the same direction between the F1 and F3 generation due to A1221 than VIN.
These results suggest two interesting conclusions. First, CpG islands in intergenic regions of the genome seem to be vulnerable to alteration by EDC exposure, particularly A1221. Intergenic, or orphan, CpG islands are often associated with non-coding RNAs or distant transcriptional start sites that are tissue specific and subjected to de novo methylation during development more so than CpG islands located in annotated promoters [68]. We identified 6 intergenic CpG islands with proximity to or overlapping non-coding RNAs called promoter-associated RNAs (pRNA; also referred to as nucleolar remodeling complex associated RNA; Table 2), which regulate ribosomal RNA transcription and interact with the epigenome by recruitment of de novo methyltransferase enzymes [69]. This class of non-coding RNAs is sensitive to the methylation status of nearby CpG islands; increased methylation results in decreased expression [70]. Interestingly, all 6 of these CpG islands were hypermethylated by both treatments, in both generations, and in both brain nuclei (Table 2). Moreover, non-coding RNAs are involved in the regulation of the epigenome [71]. We suggest that this class of non-coding RNA may be particularly vulnerable to EDC exposure and believe further investigation is warranted.
Table 2.
CpG islands affected in all treatment groups.
| F1 |
F3 |
|||||||
|---|---|---|---|---|---|---|---|---|
| Feature | Chromosome: Start – End | ID | VEH | A1221 | VIN | VEH | A1221 | VIN |
| Sperm | ||||||||
| 5_8S_rRNA | 1:11,903,554–11,916,838 | ENSRNOG00000051831 | 7.57 | 9.6 | 6.07 | 9.38 | 11.43 | 7.62 |
| pRNA | 1:11,961,296–11,975,108 | ENSRNOG00000061685 | 12.56 | 15.47 | 8.25 | 19.85 | 20.13 | 17.35 |
| NULL | 1:181,770,285–181,770,733 | NA | 3.89 | 0.43 | 2.27 | 1.73 | 0.81 | 0.46 |
| Basp1 | 2:77,586,518–77,587,151 | ENSRNOG00000046313 | 0.56 | 2.23 | 0.12 | 0.44 | 1.12 | 0.11 |
| AABR07012329.1 | 2:193,573,016–193,573,503 | ENSRNOG00000009602 | 2.12 | 3.67 | 2.65 | 2.34 | 3.11 | 3.05 |
| Ppp1r16b | 3:155,192,650–155,192,853 | ENSRNOG00000015614 | 3.38 | 10.97 | 3.63 | 3.8 | 7.48 | 8.31 |
| Sulf2 | 3:162,799,824–162,800,208 | ENSRNOG00000006052 | 2.67 | 9.36 | 8.91 | 3.79 | 6.21 | 6.71 |
| Apcdd1l | 3:171,884,884–171,885,204 | ENSRNOG00000028440 | 24.82 | 30.93 | 40.45 | 33.01 | 36.32 | 35.69 |
| Wnt4 | 5:155,667,522–155,667,924 | ENSRNOG00000013166 | 10.67 | 9.41 | 19.35 | 10.76 | 19.64 | 12.74 |
| Ptchd2 | 5:165,072,894–165,073,146 | ENSRNOG00000026447 | 13.87 | 26.23 | 34.87 | 14.46 | 20.75 | 23.67 |
| Cspg4 | 8:61,553,643–61,553,928 | ENSRNOG00000017208 | 13.49 | 26.29 | 19.36 | 17.44 | 24.03 | 22.66 |
| Spsb4 | 8:104,898,868–104,899,189 | ENSRNOG00000012862 | 50.23 | 67.23 | 58.98 | 46.7 | 49.15 | 61.84 |
| NULL | 9:38,201,817–38,202,041 | NA | 26.8 | 22.64 | 29.09 | 14.33 | 32.25 | 25.76 |
| Kcnj12 | 10:47,339,481–47,339,844 | ENSRNOG00000002303 | 34.49 | 70.53 | 47 | 34.21 | 44.88 | 57.83 |
| Evpl | 10:105,102,381–105,102,826 | ENSRNOG00000009343 | 82.57 | 66.44 | 77.61 | 77.41 | 74.4 | 71.28 |
| Orai1 | 12:38,982,126–38,982,802 | ENSRNOG00000001336 | 37.01 | 54.62 | 50.47 | 38.07 | 47.71 | 45.87 |
| Mn1 | 12:51,247,851–51,251,861 | ENSRNOG00000027489 | 2.09 | 3.03 | 3.05 | 3.19 | 3.94 | 5.22 |
| pRNA | 14:46,631,879–46,656,213 | ENSRNOG00000054197 | 19.57 | 22.48 | 24.06 | 20.07 | 22.26 | 18.66 |
| Rbp3 | 16:10,279,427–10,279,647 | ENSRNOG00000051911 | 12.74 | 14.83 | 16.23 | 11.95 | 20.04 | 18.24 |
| Spock1 | 17:7,797,172–7,797,430 | ENSRNOG00000012747 | 10.58 | 23.05 | 13.93 | 9.39 | 15.87 | 22.67 |
| Nedd4l | 18:60,580,983–60,581,707 | ENSRNOG00000017610 | 3.08 | 8.9 | 5.25 | 5.28 | 7.26 | 6.6 |
| NULL | 19:5,834,983–5,837,201 | NA | 8.27 | 5.08 | 7.36 | 7.69 | 9.91 | 7.31 |
| Zfhx3 | 19:42,764,737–42,765,779 | ENSRNOG00000014452 | 60.32 | 78.72 | 73.42 | 61.94 | 68.74 | 77.91 |
| Kcnk1 | 19:58,856,858–58,857,193 | ENSRNOG00000019937 | 46.57 | 53.39 | 58.08 | 38.46 | 44.23 | 49.25 |
| CeA | ||||||||
| pRNA | 1:11,961,296–11,975,108 | ENSRNOG00000061685 | 17.28 | 26.77 | 29.59 | 15.6 | 20.66 | 25.92 |
| pRNA | 5:91,124,273–91,141,016 | ENSRNOG00000053350 | 8.88 | 21.77 | 21.55 | 7.44 | 9.49 | 17.89 |
| pRNA | 14:46,565,300–46,566,607 | ENSRNOG00000053693 | 10 | 20.3 | 17.04 | 10.33 | 13.19 | 21.63 |
| pRNA | 14:46,579,886–46,580,733 | ENSRNOG00000052311 | 9.64 | 15.06 | 17.57 | 7.03 | 10.09 | 13.46 |
| pRNA | 14:46,631,879–46,656,213 | ENSRNOG00000054197 | 11.69 | 23.82 | 18.51 | 13.02 | 11.75 | 23.29 |
| pRNA | 14:46,678,413–46,678,837 | ENSRNOG00000054685 | 7.96 | 17.1 | 14.46 | 5.36 | 8.43 | 17.84 |
| NULL | 19:5,834,983–5,837,201 | NA | 13.65 | 20.67 | 18.02 | 11.53 | 15.72 | 21.78 |
| 5S_rRNA | 19:56,338,709–56,339,151 | ENSRNOG00000055067 | 27.76 | 22.47 | 27.91 | 24.72 | 30.51 | 27.18 |
| CA3 | ||||||||
| pRNA | 1:11,961,296–11,975,108 | ENSRNOG00000061685 | 20.36 | 28.19 | 29.15 | 14.13 | 21.98 | 26.03 |
| Kcnb1 | 3:163,935,121–163,937,341 | ENSRNOG00000046949 | 28.28 | 29.41 | 26.19 | 22.54 | 21.11 | 20.95 |
| pRNA | 5:91,124,273–91,141,016 | ENSRNOG00000053350 | 12.3 | 21.83 | 15.81 | 13.88 | 9.46 | 18.25 |
| Traf3 | 6:135,694,266–135,694,485 | ENSRNOG00000008145 | 78.82 | 44.29 | 48.16 | 32.7 | 65.58 | 64.38 |
| pRNA | 14:46,565,300–46,566,607 | ENSRNOG00000053693 | 9.03 | 19.83 | 16.58 | 10.47 | 14.21 | 22.43 |
| pRNA | 14:46,579,886–46,580,733 | ENSRNOG00000052311 | 10.3 | 14.42 | 17.28 | 7.03 | 10.67 | 17.09 |
| pRNA | 14:46,631,879–46,656,213 | ENSRNOG00000054197 | 13.11 | 21.21 | 20.02 | 10.85 | 13.09 | 21.47 |
| pRNA | 14:46,678,413–46,678,837 | ENSRNOG00000054685 | 8.81 | 17.66 | 14.75 | 5.1 | 8.12 | 17.69 |
| NULL | 19:5,834,983–5,837,201 | NA | 12.16 | 21.92 | 21.01 | 12.42 | 14.72 | 23.08 |
The second conclusion we draw from this work comes from the similarity in the proportion of CpG islands that were altered by EDC treatment in both sperm and brain tissue. Although the actual numbers were fewer in brain tissue and less consistent in direction, particularly due to A1221 treatment, this was an unexpected result. As previously mentioned, a mechanism by which changes in the epigenome of the brain are transferred to the germline, which in turn would impact the brain of subsequent generations, is not established. However, there is precedence for this connection from research on paternal stress, for which alterations to the germline and corresponding to alterations in neurobiological and endocrine outcomes were observed [72,73]. Specifically, paternal stress induced changes to the microRNA content of extracellular vesicles secreted in the epididymis that interact with mature sperm and subsequently alters stress reactivity in the following generation [73,74]. The hypothalamic-pituitary-adrenal control of stress involves the release of adrenal glucocorticoids. These steroid hormones influence development, and are involved in epigenetic modifications involved in developmental programming [75,76].
Comparison of F3 sperm and brain
Epigenetics stitches together what is passed genetically, and how the individual’s experiences modulates gene activity throughout life history. These edges are ragged, as some traits are more structural while others are ephemeral. Evolution directs this process, but the mechanism by which it is accomplished is development. The potential emergent properties of the components of a living individual are astounding. In mammals the possibility of truly ephemeral events such as experiences, usually deriving from significant change in the individual’s present environment as context, can have powerful effects on the epigenome. To understand epigenetics requires understanding development over both small and large timescales.
To visualize the data, we created gene clouds representing all differentially methylated CpG islands in F3 sperm, CeA and CA3 of A1221 or VIN compared to the vehicle counterparts 7(Figure 7; also represented tabularly in Supplemental Table 2). A1221 and VIN have different outcomes in the sperm and the brain. A Pearson’s Chi-Square revealed that the number of sites affected across the tissue sampled was dependent on treatment status (X2 (2) = 26.12, p < 0.001). While differentially methylated CpG islands are more numerous in sperm of VIN compared to A1221 males, DMRs in the brain are fewer in the VIN than A1221 lineage. Lastly, it is of note that the number of genes that overlap between sperm and brain is relatively small (A1221: Sperm – CA3 = 13; Sperm – CeA = 13; VIN: Sperm – CA3 = 16, Sperm – CeA = 15).
Figure 7.
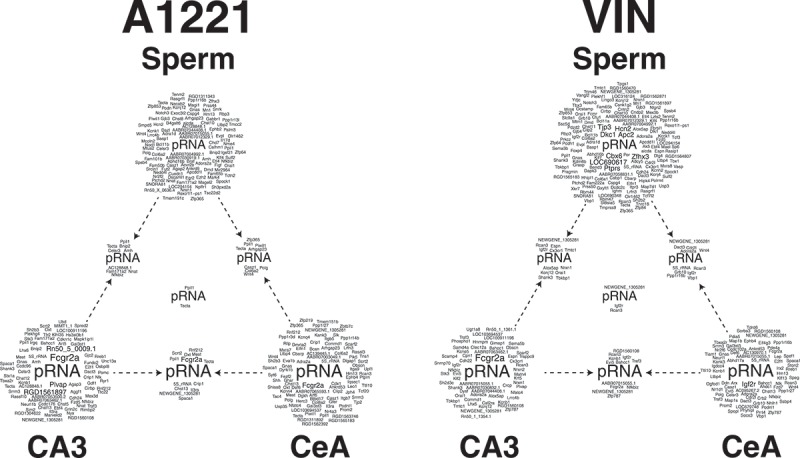
Topological view of gene clouds that contain the gene names of differentially methylated CpG islands associated with a known gene due to ancestral exposure to the weakly estrogenic PCB, Aroclor A1221 (A1221) or the anti-androgenic fungicide, Vinclozolin (VIN) relative to control of F3 male rats. Each vertex of a triangle represents a gene cloud that contains all affected genes due to treatment in F3 sperm, central amygdala (CeA), and CA3 of the hippocampus (clockwise). Each edge of a triangle represents differentially methylated CpG islands identified as significantly altered by treatment in both tissues from which an edge originates. The gene cloud at the center of a triangle represents known genes that are affected in all three tissues due to ancestral exposure. CpG islands that were not associated with a known gene (i.e., located in intergenic space) have been removed from this analysis. Within sperm, more sites are affected in VIN, but both brain regions have more affected sites due to A1221 treatment. The number of sites evident in both sperm and a brain area was low and unaffected by treatment.
In the process of developing into individuals, the organism becomes a vector for evolution. During this process, the organism is vulnerable to events that can alter normal trajectories. Some of these events can cause physiological and/or social harm outside of the reaction scope (i.e., factors and consequences that fall within normal variance). Studies of stress are approaching their centennial and we know many of the elements underlying the functional and structural plasticity in the brain in response to stress [77]. Recent evidence suggests an equally remarkable ability of the brain to recover from stress. This is not the case for prenatal exposure to EDCs.
Conclusions
Understanding the epigenetic transgenerational modifications that result from fetal exposure to ecologically relevant dosages of EDCs requires both molecular (reductionist) and organismal (emergent) levels of analysis. The brain, the organ of behavior, is a crux of this study, with the CeA and CA3 of the hippocampus playing important roles in anxiety, exploratory behavior, and learning and memory. The sperm, similar to ova, are unique from all other cells in that they encapsulate both the past (heritable modifications) as well as the present (life history experience). In other words, the gamete perpetuates what the organism has become. Females and males differ substantially in this regard: Females possess a limited number of ova at birth; whether experiences of the female as she ages is incorporated into epigenetic factors for evolution is not clear. Males, on the other hand, produce sperm throughout their adult life, thereby creating a potential of up-to-date exposures that can directly affect evolution.
Collectively, our findings demonstrate that two common endocrine disruptors are capable of causing aberrant DNA methylation of CpG islands in sperm and the brain and that a few of these sites are faithfully transmitted for multiple generations. EDC exposure has been shown to be associated with altered DNA methylation in humans for dioxins [78], flame retardants like organophosphates [44] and polybrominated diphenyl ethers [79], phthalates [80], and PCBs [81]. To our knowledge, a similar study has not been performed for VIN in humans. The precise functional outcome of the effects presented here is not known but it is clear that humans are subject to similar outcomes. These data provide areas of interest for further investigation.
Materials and methods
Animal husbandry and EDC treatment
Adult male and female Sprague-Dawley rats were purchased from Harlan (Indianapolis, Indiana) and shipped to the University of Texas at Austin. All animal work was conducted using humane procedures that were approved by the Institutional Animal Care and Use Committee at the University of Texas at Austin (AUP-2016–00029) following NIH guidelines. After two weeks of habituation to the housing facility, daily vaginal smears were performed on females to determine cycle status. On proestrus, these rats were tested for receptivity by random pairing with a male rat. If a female was receptive (lordosis) to mounting by a male, the pair was left together overnight. If sperm were present the next morning in vaginal smears, the female was single-housed and classified as embryonic day (E)0. These animals are subsequently referred to as F0 dams. All animals, dams, and litters were maintained in a temperature controlled housing facility (~ 22 C) on a 10:14 light:dark cycle and were provided with ad libitum access to a low phytoestrogen rat chow (Envigo, #2019) and tap water.
The breeding scheme is shown in Figure 8. EDCs were administered by daily intraperitoneal injection to F0 pregnant dams on days E8 through E18, 1–2 hours before lights off. These were the animals injected with EDCs in the study; further treatment was not administered to F1 – F3 animals. The treatment groups, to which pregnant F0 dams were randomly selected, were either (1) the vehicle (6% dimethylsulfoxide (DMSO), Sigma D4540 in sesame oil), (2) 1 mg/kg A1221 (Accustandard, C-221N), or (3) 1 mg/kg VIN (Chem Service Inc., N-13,745). A1221 and VIN were diluted into the vehicle. Two F0 dams each were injected with vehicle, A1221, and VIN. Following the final injection on E18, cotton nesting material was added to each dam’s cage. The day of birth was called postnatal day (PND) 0. On PND 1 all litters were culled to 5 males and 5 females or as close as possible, and on PND 21 all litters were weaned 2–3 same-sex siblings per cage. To generate an F2 generation, F1 males (2 per litter) were mated on ~ PND 90 to untreated breeder females purchased and shipped from Harlan. This step was repeated with F2 generation males (2 per litter) to yield the F3 generation (Figure 8).
Figure 8.
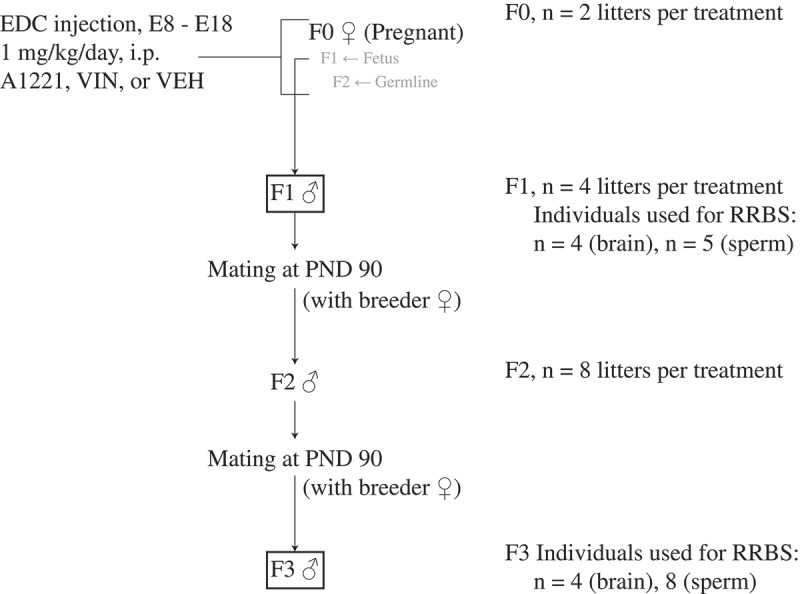
The breeding scheme that was used to produce F1 and F3 lineage rats for analysis is shown. Briefly, pregnant F0 dams were treated with a 1 mg/kg/day i.p. injection of either A1221, VIN, or vehicle (VEH: 6% DMSO in sesame oil) from embryonic (E) day 8–18. At the time of treatment, the F1 fetus and the F2 germline are directly exposed. F1 males were mated at PND 90 to colony acclimated breeder females purchased from Harlan. F2 males from the resulting litter were mated to untreated breeder females at PND 90 resulting in an F3 litter. Only F1 (directly exposed) and F3 (ancestrally exposed) males were used in these experiments.
The rationale for doses of VIN and A1221 is as follows. The VIN dose used here (1 mg/kg/day) is substantially lower than that used in similar experiments (50 or 100 mg/kg/day) [21,82]. The 1 mg/kg/day dose is also very close to the calculated lowest no observed adverse effect level (1.2 mg/kg/day [83]). Average daily intake of VIN due to consumption in humans has been estimated at 0.003 μg/kg/day in the United States in the early 1980s [84]. However, occupational exposures are higher as shown in Dutch farmers (0.1 mg/kg/day [85]). Human exposure to PCBs has fluctuated since their use was banned in 1979. Recent analysis of PCB concentrations in older individuals living along the upper Hudson River showed an average of 3.6 μg/kg (serum wet weight) [86]. Analysis of fetal exposure of PCBs in pregnant mothers from the Netherlands found that cord blood contained a sum of congeners of 0.45 μg/kg and a much higher concentration in breast milk 428 μg/kg [87]. As our exposure route is intraperitoneal injection to a pregnant rat dam, only a small portion of that dose is passed to the fetus. A 1 mg/kg PCB dose in a pregnant rat is estimated to result in a 2 μg/kg exposure to the fetus [88].
Sperm and brain tissue collection
F1 and F3 male rats were rapidly euthanized by decapitation at PND 120. Mature sperm was isolated by removing the cauda of the epididymis, dicing it into small pieces, placing it in 600 μl of warm phosphate-buffered saline (PBS), and mixed by rotating for 10 minutes. The PBS containing mature sperm was removed to a separate collection tube, replaced, and allowed to mix for an additional 10 minutes. The supernatant from both mixing steps were combined and centrifuged at 2500 g for 5 minutes. The PBS was decanted and the sperm pellet frozen at −80 C until use.
Within 2 minutes of decapitation, the rat’s brain was removed and slowly submerged in −40 C isopentane. Whole brains were stored at −80 C until use. Each brain was sliced coronally at 450 μm on a cryostat (CryoStar NX50, Thermo Fisher) and the central amygdala (CeA) and CA3 of the hippocampus were bilaterally punched from two sequential slices using a 1 mm Palkovits punch and stored at −80 C in RNA/DNAse-free microfuge tubes.
DNA extraction and purification
DNA was extracted from mature sperm using modifications of an established protocol to ensure minimal somatic cell contamination [89]. Briefly, the sperm were washed twice with PBS and subjected to two consecutive lysis steps. The first, containing 0.1% SDS and 0.5% Triton-X, lysed somatic cells contamination in the sperm pellet. The second, consisting of 100 mM tris(hydroxymethyl)aminomethane chloride (Tris-HCL – Sigma-Aldrich, T8168), 10 mM ethylenediaminetetraacetic acid (EDTA – NEB, 7011V), 1M dithiothreitol (DTT – Thermo Fisher, R0861), and 10% Proteinase-K, lysed the remaining mature sperm. Subsequent DNA isolation was performed using the Qiagen DNeasy Blood and Tissue kit according to the manufactures protocol (Qiagen, 69,504). For brain tissues, we used the AllPrep DNA/RNA mini kit according to the manufacture’s established protocols (Qiagen, 80204). This kit isolates first RNA then DNA; only the DNA fraction was used for this study.
Library preparation
Library preparation was performed by modification of previously established protocols [90] and the manufacturer’s protocols (NEB, E7535). DNA was quantified using the Quantifluor dsDNA system (Promega, E2670) and diluted to 20 ng/μl in low TE (1M Tris-HCl and 0.5M EDTA). DNA (sperm: 1 μg; brain 150 ng) was digested with MspI overnight at 37 C (NEB, R0106). Gap-filling and A-tailing were performed in a single step with 3’ -> 5’ exo- Klenow fragment (NEB, M0212) and an abundance of dATP (10 mM) in a dNTP (1 mM) mixture at 30 C for 20 minutes followed by 37 C for 20 minutes (NEB, N0446). The enzymes and dNTPs were removed and size selection was performed with a 2.0X concentration of Agencourt AMPure XP beads (Beckman Coulter, A63880) and eluted in 0.1X TE.
Adaptor ligation was performed using methylated adaptors (Index Primers Set 1 for sperm and 1–4 for brain, NEB, E7535) and TA/Blunt ligase master mix (NEB, M0367) for 15 minutes at room temperature. The USER enzyme (NEB, M5505) was then added to cleave the circular adaptors for 15 minutes at 37 C. Samples were again subjected to bead cleanup at 1.0X as described above. Adaptor ligated and size selected DNA was then subjected to bisulfite conversion according to the manufacture’s protocols (NEB, E3318) and subjected to bead cleanup at 4.0X concentration. Library enrichment was performed via PCR (sperm: 12 cycles, brain: 15 cycles) using a Hotstart Taq Polymerase and according to the manufacture’s protocol (NEB, M0490) in a 10 mM dNTP mix (each). Final libraries were then subjected to two consecutive bead clean up steps at 0.9X concentration as described above. The final libraries were again quantified and checked for appropriate size and quality using the High Sensitivity DNA Kit (Agilent, 5067–4626).
All libraries were sent to the Genomic Sequencing and Analysis Facility at the University of Texas at Austin for sequencing. Sperm libraries (F1: n = 5 per group, F3: n = 8 per group) were sequenced on an Illumina HiSeq 4000 at 150 paired-end base pairs and 10 million reads per sample. Sperm samples were equally distributed by treatment group and sample across 4 lanes in 3 different sequencing runs. Brain libraries (4 per group per generation) were sequenced in a single run using an Illumina NextSeq 500 at 75 paired-end base pairs and 7 million reads per sample. Individual samples were sequenced with unique barcodes and were not pooled. Final n’s are shown in Figure 8.
Bisulfite conversion quality control
The quality of data that is produced from a bisulfite-sequencing (BS-Seq) experiment depends in large part on the completeness of the bisulfite reaction during library preparation. If the bisulfite reaction is incomplete, unmethylated cytosine would be read and interpreted as methylated. In data processing, these erroneous methylation reads would be impossible to parse from accurate reads or filtered from the data set. We utilized two metrics to determine the completeness of the bisulfite reaction used in this study.
First, we processed a control library that contained PhiX genomic DNA, which has no DNA methylation, as a quality control agent and a metric of the efficiency of the bisulfite conversion. This library was prepared in tandem with other samples and treated identically. A bisulfite reaction should convert all cytosine residues that are not methylated to uracil and leave all methylated cytosine residues unaltered. Hence, a PhiX BS-Seq library should not result in any cytosine reads. Any that are found can be used as a proxy for how well the bisulfite conversion performed. We found that <0.5% of all methylation reads obtained from the PhiX library were read as cytosine indicating a > 99.5% bisulfite conversion rate.
Second, non-embryonic stem cells in mammals only have DNA methylation in the CpG context [91]. Other organisms contain DNA methylation in the CHG (H = any nucleotide) or the CHH context [e.g., [92]]. By parsing the number of methylation reads that were obtained in the CHG and CHH context (should not be present) from those were found in the CpG context (might be present), we were again able to calculate the efficiency of the bisulfite reaction. We found that < 0.1% of all methylation reads were obtained from the CHG and CHH context.
Last, we aligned the PhiX control library to the rat reference genome. Alignment produced effectively no alignment (<0.1%) between the PhiX library and the rat reference genome. When compared to the PhiX reference genome, 86.7% of the reads obtained from the PhiX control library found unique best hits indicating little or no cross-contamination during mass processing.
Bioinformatics pipeline and statistical analyses
Bioinformatics were performed at the Texas Advanced Computing Center (TACC) using the Stampede, Stampede2, and Lone Star 5 compute clusters. Reads from the Illumina HiSeq 4000 and NextSeq 500 were merged if necessary and analyzed for quality using FastQC. Adaptor and quality trimming were performed with trim galore and cutadapt and were again analyzed for quality with FastQC. Reads were aligned against the rat (Rnor_6.0) genome using Bowtie (version 2.3.2) and Bismark (version 0.14.5) to identified bisulfite mutated reads. Aligned reads were filtered for non-bisulfite conversion > 1% and methylation calls were extracted using Bismark (version 0.14.5). Aligned methylation calls were analyzed with SeqMonk (version 1.38.2, Babraham Bioinformatics).
Reduced representation bisulfite sequencing (RRBS) is a read-efficient protocol to achieve deep coverage of CpG islands, which were the target of this study. Although we could not perform loci-specific confirmation via alternate methods, RRBS consistently performs well compared against traditional methods and gold standards of DNA methylation analysis [93,94]. CpG island annotations were obtained from the University of California, Santa Cruz (UCSC) genome bioinformatics database, which included 20,989 CpG islands. A full database of CpG island annotations used for this study can be found in Supplemental Table 2. Annotations for nested repeated elements were also obtained from the University of California, Santa Cruz (UCSC) genome bioinformatics database, which includes 401,017 nested repeats. We further classified CpG islands by their relationship to known or predicted coding regions. Promoter CpG islands were classified as CpG islands upstream and within 1 kb of a known transcriptional start site. Intronic and exonic CpG islands were classified as those fully contained within the coding region of a gene and overlapping known introns or exons. 3-prime CpG islands were classified as those overlapping the last exon of a gene and extending downstream of the 3-prime exon. Finally, intergenic CpG islands were classified as those that were not within 2kb in either direction of the CpG island. The methylation status of CpG islands in VIN or A1221 samples was statistically compared to vehicle samples using a logistic regression analysis (SeqMonk, R). CpG islands were considered statistically different at <0.05 after a Benjamini & Hochberg FDR correction was applied [23]. An absolute percent cut off of percent methylation difference between comparison groups was not applied. Downstream analyses and figure creation was performed in R (version 3.3.2) with the following packages; dplyr, tidyr, ggplot2, heatmap.2, UpSetR, and pvclust.
Funding Statement
NIH 1RO1 ES023254; National Institute of Environmental Health Sciences [5RO1 ES023254].
Acknowledgments
We would like to thank the Genomic Sequencing and Analysis facility, Jessica Podnar and Dennis Wylie in particular, at The University of Texas at Austin for their wonderful work and advice on library preparation and analysis. The computational resources provided by the Texas Advanced Computing Center were essential for the analyses in this paper. We also thank Lindsay Thompson for her logistical support and assistance with animal treatment, and Dr. Krittika Krishan and Dr. Amanda Holley for their help maintaining the animal colony and tissue collection.
Author Contributions
Conceptualization: R.G., A.C.G., D.C.; Methodology: R.G.; Investigation: R.G., M.S., L.T.; Writing – Original Draft: R.G.; Writing – Review & Editing: R.G., A.C.G., D.C.; Funding Acquisition: A.C.G., D.C.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here
References
- 1.Wilson CA, Davies DC.. The control of sexual differentiation of the reproductive system and brain. Reproduction. 2007. February;133(2):331–359. [DOI] [PubMed] [Google Scholar]
- 2.Gore AC, Chappell VA, Fenton SE, et al. EDC-2: the endocrine society’s second scientific statement on Endocrine-Disrupting Chemicals. Endocr Rev. 2015. December;36(6):E1–E150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skinner MK, Manikkam M, Haque MM, et al. Epigenetic transgenerational inheritance of somatic transcriptomes and epigenetic control regions. Genome Biology BioMed Central. 2012. October 3;13(10):R91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crews D, Gillette R, Scarpino SV, et al. Epigenetic transgenerational inheritance of altered stress responses. Proc Natl Acad Sci. 2012. June 5;109(23):9143–9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gillette R, Miller-Crews I, Nilsson EE, et al. Sexually dimorphic effects of ancestral exposure to vinclozolin on stress reactivity in rats. Endocrinology. 2014. October;155(10):3853–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillette R, Miller-Crews I, Skinner MK, et al. Distinct actions of ancestral vinclozolin and juvenile stress on neural gene expression in the male rat. Front Genet. 2015;6:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mennigen JA, Thompson LM, Bell M, et al. Transgenerational effects of polychlorinated biphenyls: 1. Development and physiology across 3 generations of rats. Environ health. BioMed Cent. 2018. February 20;17(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gardner RL. The relationship between cell lineage and differentiation in the early mouse embryo. Results Probl Cell Differ. 1978;9:205–241. [DOI] [PubMed] [Google Scholar]
- 9.Ohno R, Nakayama M, Naruse C, et al. A replication-dependent passive mechanism modulates DNA demethylation in mouse primordial germ cells. Development. Oxford University Press for the Company of Biologists Limited. 2013. July;140(14):2892–2903. [DOI] [PubMed] [Google Scholar]
- 10.Kawasaki Y, Lee J, Matsuzawa A, et al. Active DNA demethylation is required for complete imprint erasure in primordial germ cells. Sci Rep Nature Publishing Group. 2014. January;13(4):3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seisenberger S, Andrews S, Krueger F, et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012. December 28;48(6):849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skinner MK, Haque CG-BM, Nilsson E, et al. Environmentally induced transgenerational epigenetic reprogramming of primordial germ cells and the subsequent germ line. PLoS ONE. 2013. July 15;8(7):e66318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider S, Kaufmann W, Buesen R, et al. Vinclozolin - The lack of a transgenerational effect after oral maternal exposure during organogenesis. Reprod Toxicol. 2008. [2008 April];25(3):352–360. [DOI] [PubMed] [Google Scholar]
- 14.Inawaka K, Kawabe M, Takahashi S, et al. Maternal exposure to anti-androgenic compounds, vinclozolin, flutamide and procymidone, has no effects on spermatogenesis and DNA methylation in male rats of subsequent generations. Toxicol Appl Pharmacol. 2009. June 1;237(2):178–187. [DOI] [PubMed] [Google Scholar]
- 15.Schneider S, Marxfeld H, Gröters S, et al. Vinclozolin–no transgenerational inheritance of anti-androgenic effects after maternal exposure during organogenesis via the intraperitoneal route. Reprod Toxicol. 2013. June;37:6–14. [DOI] [PubMed] [Google Scholar]
- 16.McBirney M, King SE, Pappalardo M, et al. Atrazine induced epigenetic transgenerational inheritance of disease, lean phenotype and sperm epimutation pathology biomarkers. PLoS ONE. 2017;12(9):e0184306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aiba T, Saito T, Hayashi A, et al. Does the prenatal bisphenol A exposure alter DNA methylation levels in the mouse hippocampus?: an analysis using a high-sensitivity methylome technique. Genes Environ BioMed Central. 2018;40(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skinner MK, Anway MD, Savenkova MI, et al. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior PLoS ONE. 2008. Nov 18;3(11):e3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerrero-Bosagna C, Settles M, Lucker B, et al. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS ONE. 2010. September 30;5(9):e13100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolstenholme JT, Goldsby JA, Rissman EF. Transgenerational effects of prenatal bisphenol A on social recognition. Horm Behav. 2013. November 1;64(5):833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beck D, Sadler-Riggleman I, Skinner MK. Generational comparisons (F1 versus F3) of vinclozolin induced epigenetic transgenerational inheritance of sperm differential DNA methylation regions (epimutations) using MeDIP-Seq. Environ Epigenet. 2017. July;3(3):1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drobna Z, Henriksen AD, Wolstenholme JT, et al. Transgenerational effects of bisphenol a on gene expression and dna methylation of imprinted genes in brain. Endocrinology. 2018. January;159(1):132–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc. 1995;57(1):289–300. [Google Scholar]
- 24.Anway MD, Cupp AS, Uzumcu M, et al. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005. June 3;308(5727):1466–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anway MD, Memon MA, Uzumcu M, et al. Transgenerational effect of the endocrine disruptor vinclozolin on male spermatogenesis. J Androl. 2006. July 12;27(6):868–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerrero-Bosagna C, Covert TR, Haque MM, et al. Epigenetic transgenerational inheritance of vinclozolin induced mouse adult onset disease and associated sperm epigenome biomarkers. Reprod Toxicol. 2012. December;34(4):694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skinner MK, Anway MD. Seminiferous cord formation and germ-cell programming: epigenetic transgenerational actions of endocrine disruptors. Ann N Y Acad Sci. 2005. December;1061:18–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilsson E, Larsen G, Manikkam M, et al. Environmentally induced epigenetic transgenerational inheritance of ovarian disease. PloS one. 2012. May 3;7(5):e36129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chamorro-García R, Diaz-Castillo C, Shoucri BM, et al. Ancestral perinatal obesogen exposure results in a transgenerational thrifty phenotype in mice. Nat Commun. 2017. December 8;8(1):2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brehm E, Rattan S, Gao L, et al. Prenatal exposure to di(2-ethylhexyl) phthalate causes long-term transgenerational effects on female reproduction in mice. Endocrinology. 2018. February 1;159(2):795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steinberg RM, Walker DM, Juenger TE, et al. Effects of perinatal polychlorinated biphenyls on adult female rat reproduction: development, reproductive physiology, and second generational effects. Biol Reprod. 2008. [2008 June];78(6):1091–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gore AC. Developmental programming and endocrine disruptor effects on reproductive neuroendocrine systems. Front Neuroendocrinol. 2008. June;29(3):358–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jirtle RL, Skinner MK. Environmental epigenomic and disease susceptibility. Nature Rev Genet. 2007;8:253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crews D. Epigenetic modifications of brain and behavior: theory and practice. Horm Behav. 2011;59:393–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skinner MK, Guerrero-Bosagna C. Role of CpG deserts in the epigenetic transgenerational inheritance of differential DNA methylation regions. BMC Genomics. 2014. August;20(15):692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skinner MK, Ben Maamar M, Sadler-Riggleman I, et al. Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease. Epigenetics Chromatin. 2018. February;11(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gillette R, Reilly MP, Topper VY, et al. Anxiety-like behaviors in adulthood are altered in male but not female rats exposed to low dosages of polychlorinated biphenyls in utero. Horm Behav. 2017. January;87:8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reilly MP, Weeks CD, Topper VY, et al. The effects of prenatal PCBs on adult social behavior in rats. Horm Behav. 2015. July 1;73(C):47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schilling E, Rehli M. Global, comparative analysis of tissue-specific promoter CpG methylation. Genomics. 2007. September;90(3):314–323. [DOI] [PubMed] [Google Scholar]
- 40.Razin A, Riggs AD. DNA methylation and gene function. Science. 1980. November 7;210(4470):604–610. [DOI] [PubMed] [Google Scholar]
- 41.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci. 2006. January 31;103(5):1412–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011. May 15;25(10):1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCarrey JR, Lehle JD, Raju SS, et al. Tertiary epimutations - A novel aspect of epigenetic transgenerational inheritance promoting genome instability. PLoS ONE. 2016;11(12):e0168038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soubry A, Hoyo C, Butt CM, et al. Human exposure to flame-retardants is associated with aberrant DNA methylation at imprinted genes in sperm. Environ Epigenet. 2017. January;3(1):dvx003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stenz L, Escoffier J, Rahban R, et al. Testicular dysgenesis syndrome and long-lasting epigenetic silencing of mouse sperm genes involved in the reproductive system after prenatal exposure to DEHP. PLoS ONE. 2017;12(1):e0170441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molaro A, Hodges E, Fang F, et al. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011. September 16;146(6):1029–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet. 1998. October;20(2):116–117. [DOI] [PubMed] [Google Scholar]
- 48.Kang SC, Lee BM. DNA methylation of estrogen receptor alpha gene by phthalates. J Toxicol Environ Health Part A. 2005. December 10;68(23–24):1995–2003. [DOI] [PubMed] [Google Scholar]
- 49.Park CJ, Nah WH, Lee JE, et al. Butyl paraben-induced changes in DNA methylation in rat epididymal spermatozoa. Andrologia. 2012. May;44(Suppl 1):187–193. [DOI] [PubMed] [Google Scholar]
- 50.Hilakivi-Clarke L. Maternal exposure to diethylstilbestrol during pregnancy and increased breast cancer risk in daughters. Breast Cancer Res. 2014;16(2):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sato K, Fukata H, Kogo Y, et al. Neonatal exposure to diethylstilbestrol alters expression of DNA methyltransferases and methylation of genomic DNA in the mouse uterus. Endocr J. 2009;56(1):131–139. [DOI] [PubMed] [Google Scholar]
- 52.Casati L, Sendra R, Colciago A, et al. Polychlorinated biphenyls affect histone modification pattern in early development of rats: a role for androgen receptor-dependent modulation? Epigenomics. 2012. February;4(1):101–112. [DOI] [PubMed] [Google Scholar]
- 53.Casati L, Sendra R, Sibilia V, et al. Endocrine disrupters: the new players able to affect the epigenome. Front Cell Dev Biol. 2015;3:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Casati L, Sendra R, Poletti A, et al. Androgen receptor activation by polychlorinated biphenyls: epigenetic effects mediated by the histone demethylase Jarid1b. Epigenetics. 2013. October;8(10):1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kristensen LH, Nielsen AL, Helgstrand C, et al. Studies of H3K4me3 demethylation by KDM5B/Jarid1B/PLU1 reveals strong substrate recognition in vitro and identifies 2,4-pyridine-dicarboxylic acid as an in vitro and in cell inhibitor. FEBS J. 2012. June;279(11):1905–1914. [DOI] [PubMed] [Google Scholar]
- 56.Yan H, Zhang D, Liu H, et al. Chromatin modifications and genomic contexts linked to dynamic DNA methylation patterns across human cell types. Sci Rep. 2015. February;12(5):8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmitz SU, Albert M, Malatesta M, et al. Jarid1b targets genes regulating development and is involved in neural differentiation. Embo J. 2011. November 16;30(22):4586–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kelce WR, Monosson E, Gamcsik MP, et al. Environmental hormone disruptors: evidence that vinclozolin developmental toxicity is mediated by antiandrogenic metabolites. Toxicol Appl Pharmacol. 1994. June;126(2):276–285. [DOI] [PubMed] [Google Scholar]
- 59.Wong C, Kelce WR, Sar M, et al. Androgen receptor antagonist versus agonist activities of the fungicide vinclozolin relative to hydroxyflutamide. J Biol Chem. 1995. August 25;270(34):19998–20003. [DOI] [PubMed] [Google Scholar]
- 60.Jansen HT, Cooke PS, Porcelli J, et al. Estrogenic and antiestrogenic actions of PCBs in the female rat: in vitro and in vivo studies. Reprod Toxicol. 1993;7:237–248. [DOI] [PubMed] [Google Scholar]
- 61.Sager DB. Effect of postnatal exposure to polychlorinated biphenyls on adult male reproductive function. Environ Res. 1983;31:76–94. [DOI] [PubMed] [Google Scholar]
- 62.Ward IL, Ward OB, Affuso JD, et al. Fetal testosterone surge: specific modulations induced in male rats by maternal stress and/or alcohol consumption. Horm Behav. 2003. May;43(5):531–539. [DOI] [PubMed] [Google Scholar]
- 63.McEwen BS. Sexual differentiation of the brain. Nature. 1981. June;291(5817):610. [DOI] [PubMed] [Google Scholar]
- 64.Walker DM, Gore AC. Epigenetic impacts of endocrine disruptors in the brain. Front Neuroendocrinol. 2017. January;44:1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nugent BM, Schwarz JM, McCarthy MM. Hormonally mediated epigenetic changes to steroid receptors in the developing brain: implications for sexual differentiation. Horm Behav. 2011. March;59(3):338–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gilpin NW, Herman MA, Roberto M. The central amygdala as an integrative hub for anxiety and alcohol use disorders. Biol Psychiatry. 2015. May 15;77(10):859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Engin E, Treit D. The role of hippocampus in anxiety: intracerebral infusion studies. Behav Pharmacol. 2007. September;18(5–6):365–374. [DOI] [PubMed] [Google Scholar]
- 68.Illingworth RS, Gruenewald-Schneider U, Webb S, et al. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010. September 23;6(9):e1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmitz KM, Mayer C, Postepska A, et al. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010. October 15;24(20):2264–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ghoshal K, Majumder S, Datta J, et al. Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J Biol Chem. 2004. February 20;279(8):6783–6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014. February 17;9(1):3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dias BG, Ressler KJ. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat Neurosci. 2014. January;17(1):89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodgers AB, Morgan CP, Bronson SL, et al. Paternal stress exposure alters sperm microRNA content and reprograms offspring HPA stress axis regulation. J Neurosci. 2013. May;33(21):9003–9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chan JC, Nugent BM, Morrison KE, et al. Epididymal glucocorticoid receptors promote intergenerational transmission of paternal stress. bioRxiv. 2018;Jan:1;:321976. [Google Scholar]
- 75.McEwen BS. Steroid hormones and brain development: some guidelines for understanding actions of pseudohormones and other toxic agents. Environ Health Perspect. 1987. October;74:177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thomassin H, Flavin M, Espinas ML, et al. Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J EMBO Press. 2001. April 17;20(8):1974–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McEwen BS. Stress-induced remodeling of hippocampal CA3 pyramidal neurons. Brain Res. 2016. August 15;1645:50–54. [DOI] [PubMed] [Google Scholar]
- 78.Pilsner JR, Shershebnev A, Medvedeva YA, et al. Peripubertal serum dioxin concentrations and subsequent sperm methylome profiles of young Russian adults. Reprod Toxicol. 2018. June;78:40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Suvorov A, Shershebnev A, Wu H, et al. Perinatal exposure to low dose 2,2“,4,4-”tetrabromodiphenyl ether (BDE-47) alters sperm DNA methylation in adult rats. Reprod Toxicol. 2018. January;75:136–143. [DOI] [PubMed] [Google Scholar]
- 80.Wu H, Estill MS, Shershebnev A, et al. Preconception urinary phthalate concentrations and sperm DNA methylation profiles among men undergoing IVF treatment: a cross-sectional study. Hum Reprod. 2017. November 1;32(11):2159–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kobayashi N, Miyauchi N, Tatsuta N, et al. Factors associated with aberrant imprint methylation and oligozoospermia. Sci Rep Nature Publishing Group. 2017. February;10(7):42336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stouder C, Paoloni-Giacobino A. Transgenerational effects of the endocrine disruptor vinclozolin on the methylation pattern of imprinted genes in the mouse sperm. Reproduction. 2009. [2010 January 25];139(2):373–379. [DOI] [PubMed] [Google Scholar]
- 83.Environmental Protection Agency Reregistration eligibility decision: vinclozolin (EPA publication no. 738-R-00-023). Rockville, MD: US Environmental Protection Agency; 2000October24 1–130. [Google Scholar]
- 84.Gartrell MJ, Craun JC, Podrebarac DS, et al. Pesticides, selected elements, and other chemicals in adult total diet samples, October 1980-March 1982. J Assoc Off Anal Chem. 1986. January;69(1):146–159. [PubMed] [Google Scholar]
- 85.Juhler RK, Larsen SB, Meyer O, et al. Human semen quality in relation to dietary pesticide exposure and organic diet. Arch Environ Contam Toxicol. 1999. October;37(3):415–423. [DOI] [PubMed] [Google Scholar]
- 86.Fitzgerald EF, Belanger EE, Gomez MI, et al. Polychlorinated biphenyl exposure and neuropsychological status among older residents of upper Hudson River communities. Environ Health Perspect. 2008. February;116(2):209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schantz SL, Widholm JJ, Rice DC. Effects of PCB exposure on neuropsychological function in children. Environ Health Perspect. 2003. March;111(3):357–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Takagi Y, Aburada S, Hashimoto K. Transfer and distribution of accumulated (14C) polychlorinated biphenyls from maternal to fetal and suckling rats. Arch Environ Contam Toxicol. 1986;15(6):709–715. [DOI] [PubMed] [Google Scholar]
- 89.Mao S, Goodrich RJ, Hauser R, et al. Evaluation of the effectiveness of semen storage and sperm purification methods for spermatozoa transcript profiling. Syst Biol Reprod Med. 2013. October;59(5):287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boyle P, Clement K, Gu H, et al. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 2012. October 3;13(10):R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gruenbaum Y, Stein R, Cedar H, et al. Methylation of CpG sequences in eukaryotic DNA. FEBS letters. 1981. February 9;124(1):67–71. [DOI] [PubMed] [Google Scholar]
- 92.Lyko F, Ramsahoye BH, Jaenisch R. DNA methylation in drosophila melanogaster. Nature. 2000. November 30;408(6812):538–540. [DOI] [PubMed] [Google Scholar]
- 93.Bock C, Tomazou EM, Brinkman AB, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010. October;28(10):1106–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chatterjee A, Macaulay EC, Ahn A, et al. Comparative assessment of DNA methylation patterns between reduced representation bisulfite sequencing and sequenom epityper methylation analysis. Epigenomics. 2017. June;9(6):823–832. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


