ABSTRACT
SURVIVIN is an essential chromosomal passenger complex (CPC) subunit and participates in cell division. In this study, we used porcine oocyte as a model to investigate the roles of Survivin during porcine oocyte maturation. Survivin was highly expressed in germinal vesicle (GV) and germinal vesicle breakdown (GVBD) stages oocytes, mainly localized in the GV at GV stage and on the chromosomes after GVBD. We have used RNA interference to specifically deplete Survivin in oocytes during in vitro maturation (IVM). Immunofluorescence assay showed that Survivin-depleted oocytes failed to produce polar body in meiosisⅠ (failed to complete cytokinesis), and they were arrested in metaphaseⅠwith misaligned chromosomes. The homologous chromosomes in Survivin-depleted oocytes could not be separated normally. Moreover, both the phosphorylation levels of Aurora B and the mRNA level of Mad2L1 related to spindle assembly checkpoint (SAC) was decreased in Survivin-depleted oocytes, which thus inhibited the degradation of Cyclin B1 (CCNB1) to complete meiosis. Taken together, we conclude that Survivin is an important mediator of centromere and midbody docking of Aurora-B as well as its activity and regulates SAC and MPF activity during meiosis in porcine oocytes.
Keywords: Survivin, oocyte, metaphase I, spindle assembly checkpoint, chromosomes, pig
Introduction
The protein SURVIVIN has become valuable as its extensive expression in various human cancers and its potential as a target in cancer therapy [1]. As the smallest member of the inhibitors of apoptosis protein (IAPs) family, Survivin has been identified as part of the chromosomal passenger complex (CPC). CPC contains three additional subunits: the Aurora-B kinase, the inner centromere protein (INCENP), and Borealin/Dasra-B (Borealin) [2,3]. As the functional subunit, Aurora-B interacts with Survivin, INCENP and Borealin during cell division [4], and they show a typical chromosomal passenger localization pattern during mitosis at the inner centromere in (pro)metaphase, the central spindle during anaphase and the midbody during cytokinesis [5]. In prophase, the CPC is localized on chromosome arms and centromeres and regulates the chromosome structure after histone H3 phosphorylation [6,7]. During pro-metaphase and metaphase, it localizes exclusively to centromeres which is important for correcting non-bipolar kinetochores attachment and it also contributes to spindle formation and stability [8–11]. After metaphase, CPC moves to the spindle midzone, and then is located to the equatorial cortex and finally to the midbody, these processes are essential for the execution of cytokinesis [12]. Previous results indicated that knockdown of Survivin, INCENP or Borealin respectively disrupted Aurora-B localization and function, which led to similar mitotic defects [11,13–18]. It has been found that Aurora B bound to the C-terminal domain of INCENP (the so-called IN-box) and then targeted to centromeres by interacting with Survivin [3,19–21]. Aurora-B is activated at the G2–M transition and sustains activating until mitotic exit [22]. Thus, it seems that the functions of Aurora-B are mainly determined by its dynamic localization pattern [7]. But the relationship between the activity of Aurora-B and its correct location to the central spindle remained elusive during porcine oocyte maturation.
During mitosis, in addition to bipolar chromosome attachment, proper chromosome segregation also requires the alignment of every single chromosome before anaphase is initiated. This checkpoint mechanism inhibits the onset of anaphase until all chromosomes are attached to the mitotic spindle and properly aligned on the metaphase plate [21]. Spindle assembly checkpoint (SAC) is a mechanism which guarantees precise chromosome segregation by inhibiting the activity of the anaphase promoting complex/cyclosome (APC/C) in mitosis. The SAC involves various checkpoint proteins including Bub1, BubR1, Bub3, Mad1, Mad2, Mps1 and others. SAC prevents the transition of anaphase by inhibiting the anaphase promoting complex/cyclosome (APC/C) that regulates the proteolysis of various mitotic target protein, whose degradation is required for the beginning of anaphase [23,24]. Ablation of CPC proteins Aurora B and Survivin results in SAC silencing, which implied that the CPC might be part of the SAC [8,9,25,26]. Moreover, Aurora-B also could activate the SAC indirectly by generating unattached kinetochores during the correction process [27]. However, it is not clear whether Aurora B and the CPC are directly or indirectly involved in SAC activity.
SAC is activated by both unoccupied kinetochores and kinetochores with incorrect microtubule attachment tension. Aurora-B is required for phosphorylation of histone H3 on serine 10 [28,29] and for targeting of condensing to chromosomes [30], and these subunits are essential for chromosomes to achieve a stable metaphase orientation [29,31], which is possibly done by regulating the interaction of kinetochores with microtubules [31–33]. Ipl-1p (Aurora kinase in the budding yeast) is essential for the tension-sensitive arm of the spindle assembly checkpoint [34]. Studies using dominant-negative mutants and antibody injection have built a relationship between mammalian Survivin and Aurora-B in the spindle assembly checkpoint [35–37]. Survivin may contribute to the regulation of microtubule dynamics [38] and has been reported to be involved in spindle assembly [39,40]. In C. Elegans, Aurora-B/AIR-2 is also required for separation of homologous chromosomes in meiosis I [41]. To confirm the role of the CPC in SAC activity, it has been found that the kinetochore localization of Bub1 and BubR1 was dependent on Aurora B [15,25,26,37,42]. Interestingly, Aurora-B becomes essential for a nocodazole induced mitotic delay in the absence of Bub1, which implied that Bub1 and Aurora-B might collaborate to maintain the SAC [43,44].In the end, Survivin and Aurora-B are required for the execution of cytokinesis [38,45,46]. Above all, these results suggest that the CPC might play a direct role in SAC function, but the contribution of the individual CPC subunit protein for proper SAC mechanism remains unclear. In this study, we have used siRNA to deplete Survivin expression in porcine oocytes to study the roles of Survivin during porcine oocyte maturation.
Materials and methods
Porcine oocytes collection and culture
Porcine ovaries were obtained from a standard slaughterhouse, then transported to the laboratory in an insulated containers with saline at 38.5 ℃ in 1–2 h. Follicular fluid from 3–6 mm antral follicles was aspirated with an 10 ml injection syringe. COCs were selected and culture as reported [47]. The oocytes obtained from COCs were used for subsequent experiments.
Microinjection
For Survivin knockdown or overexpression experiment, 5–10 pL of Survivin siRNA (50 μM) or 1.0 µg/µL Survivin mRNA and control siRNA or ultrapure water was microinjected into the cytoplasm of GV oocytes with granulosa cell layers. A 1 μL drop of siRNA was placed in the dish to fill the micropipette, and injections were performed using an Ti-S inverted microscope (Ti-S, Nikon, Japan) equipped with micromanipulation equipment (Nikon, Japan) with 38.5℃. COCs were injected by femtojet 4i ® (Eppendorf, Germany) and injection pipette which had an inner diameter about 0.5 μm. After injection, oocytes were kept at room temperature (RT) for 10 min and then moved into the incubator for following experiment. The COCs with Survivin depletion were cultured in the medium supplied with 2 mM dbcAMP for 20 h [48] and then matured in vitro. Furthermore, the COCs with Survivin overexpression were cultured with 2 mM dbcAMP for 2 h and then matured in vitro.
For CCNB1 expression, we mixed 0.5 mg/mL CCNB1-eGFP mRNA and 50 µM survivin siRNA, and then injected the mixture into the cytoplasm of GV oocytes; in the control group, 0.5 mg/mL CCNB1-eGFP mRNA was injected into the cytoplasm. The microinjection operation was finished within 30 min. After injection, COCs were cultured in the medium supplied with 2 mM dbcAMP for 20 h [48] and then matured in vitro.
Antibodies, immunofluorescence and confocal microscopy
Antibodies used in the experiments were purchased from the following companies: rabbit monoclonal anti-Survivin (Cell Signaling Technology, Beverly, MA, USA); mouse monoclonal anti-α-tubulin (Sigma), anti-GAPDH (Invitrogen, Carlsbad, CA, USA); Secondary antibodies were purchased from ZhongShan Golden Bridge Biotechnology Co., Ltd (Beijing, China). Oocytes for immunofluorescent staining were fixed in 4% paraformaldehyde in PBS for 60 min at RT. Then, they were transferred to membrane permeabilization solution (0.5% Triton X-100) for 20 min and blocking buffer (1% BSA-supplemented PBS) for 1 h. At last, oocytes were incubated overnight at 4℃ with antibodies described above in appropriate dilutions. Then, the oocytes were mounted on glass slides and examined with a laser scanning confocal microscope (Zeiss LSM 510, Oberkochen, Germany).
cDNA cloning, mRNA synthesis and microinjection
Total RNA of each sample was isolated from 50 porcine oocytes using an Arcturus Pico Pure kit (Life Technologies, Grand Island, NY). Enhanced GFP (eGFP) cRNA was transcribed in vitro from pIVT-eGFP [49]. And 1 ng was added to each sample prior to RNA isolation as an internal control. The cDNA of each sample was obtained using a Fast Quant RT kit (Tiangen). Quantitative RT-PCR was performed according to the instruction in Super Real PreMix Plus kit (Tiangen), using cDNA of each sample. Relative gene expressions were calculated using the ΔCq method [50] with eGFP expression for normalization. The primers were listed as follows: GFP (F: GAACGGCATCAAGGTGAACT; R: TGCTCAGGTAGTGGTTGTCG); Survivin (F: CCTGGCAGCTCTACCTCAAG; R: GAAAGCACAACCGGATGAAT); Mad2l1 (F: CCAAGATGAAATCCGGTCA; R: TCAGAATTGGTTATGAACTGTGG); Mad2l2 (F: CAGACGAGCAGGATGTCCAC; R: GCTCGCTCTTCCACGTACAG); Cdc20 (F: TGTGCTCCATTCTCTGGTCTC; R: GGATGCCTTGGTGGATGA); Cdk1 (F: GGGTCAGCTCGCTACTCAAC; R: AGTTTTTGACGTGGGATGC); Ndc (F: CCTCTCCATGCAGGAGTTAAGA; R: GGTCTCGGGTCCTTGATTTTCT).
Vector construction and transcription in vitro
The cDNA of porcine oocytes, as the templates of Survivin and CCNB1 genes cloning, were obtained with the method of RNA isolation and reverse transcription as previously mentioned. Survivin and CCNB1 genes were cloned with DNA polymerase (Takara, Prime STAR Mix, DP214-02). The products were used for A-tailing and TA cloning with DNA A-Tailing kit (Takara, 6109), then transformed into competent Escherichia coli cells. Positive colonies were selected and plasmids were extracted with plasmid extraction kit (AxyGen, AP-MN-P-50). Survivin and CCNB1 were inserted into the pIVT vector and the pIVT-C-eGFP vector. After linearization, these vectors were used for in vitro transcription and tailing with mMESSAGE mMACHINE T7 kit (Thermo, AM1344). The mRNA was purified with RNAeasy kit (Tiangen) and stored at −80℃.
Western blot
A total of 300 porcine oocytes per sample were mixed with SDS sample buffer and boiled for 5 min at 98℃ for SDS-PAGE. Western blotting was performed as described previously [48], using the antibody dilution anti-Survivin (1: 2000); anti-GAPDH (1: 2000); anti-β-actin (1: 5000); anti-Aurora B (1: 1000); anti – phosphorylated Aurora B (1: 1000); anti-Mad2 (1: 1000).
Statistical analysis
All experiments were repeated at least three times. Statistical analysis was performed using SPSS 22.0 statistical software. Data were expressed as mean ± S.E.M. P<0.05 was considered as statistically significant and P<0.01 was considered as great significant.
Result
Expression and distribution of survivin during porcine oocyte maturation in vitro
To investigate the role of Survivin in meiosis, we detected the expression of Survivin in the transcription and protein level, as well as the subcellular location of Survivin in porcine oocytes during maturation. Porcine cumulus oocyte complexes (COCs) were cultured for 0 h, 24 h, 28 h or 44 h, respectively, which were corresponded to GV, GVBD, MI and MII stage of meiosis. To determine the expression and distribution of Survivin, oocytes were denuded at each stage and harvest for quantificational real-time polymerase chain reaction (qRT-PCR), western blot and immunofluorescence staining. The qRT-PCR result showed that Survivin was highly expressed at GV and GVBD stage and decreased rapidly in MI stage (Figure 1(a)). The western blot results revealed that Survivin was maintained in GV stage and reduced in GVBD to MII (Figure 1(c,d)). Immunofluorescence results showed that Survivin was mainly co-localized with chromatins in GV and chromosomes in GVBD, MI and MII. Meanwhile there were small quantity of Survivin dispersed in the cytoplasm from GV to MI and disappeared in MII (Figure 1(b)), which suggested that survivin might take effects by affecting chromosome arrangement during porcine oocytes maturation.
Figure 1.

Expression and subcellular localization of Survivin during porcine oocytes meiotic maturation. (a) Relative expression of Survivin mRNA in GV, GVBD, MI and MII stages; (b) Subcellular localization of Survivin during porcine oocytes maturation. N-Ctr: negative control; Bar = 25 µm. (c) The protein level of Survivin in in GV, GVBD, MI and MII stages. Proteins derived from a total of 300 oocytes were loaded for each sample.
Survivin depletion disturbed the behavior of meiotic chromosomes
To investigate the role of Survivin in meiosis, we used RNA interference (RNAi) to deplete the expression of Survivin in porcine oocytes during maturation. It was shown that Survivin-depleted oocytes failed to release polar body in meios Ⅱ (failed to complete cytokinesis) (Figure 2(a,b)). qRT-PCR result showed that Survivin were fully diminished after Survivin RNAi (Figure 2(c)). Immunoblot analysis showed that Survivin was substantially repressed (Figure 2(d,e)). These results suggested that segregation of chromosomes had been disturbed in Survivin-depleted porcine oocytes. We also found that depletion of Survivin compromized the ability of oocyte to align metaphaseⅠchromosomes (Figure 3(a)), which was consistent with previous results. About 70% of anaphase and telophase cells displayed abnormalities in chromosome segregation (Figure 3(b)). We concluded that Survivin was apparently essential for accurate segregation of homologous chromosomes.
Figure 2.
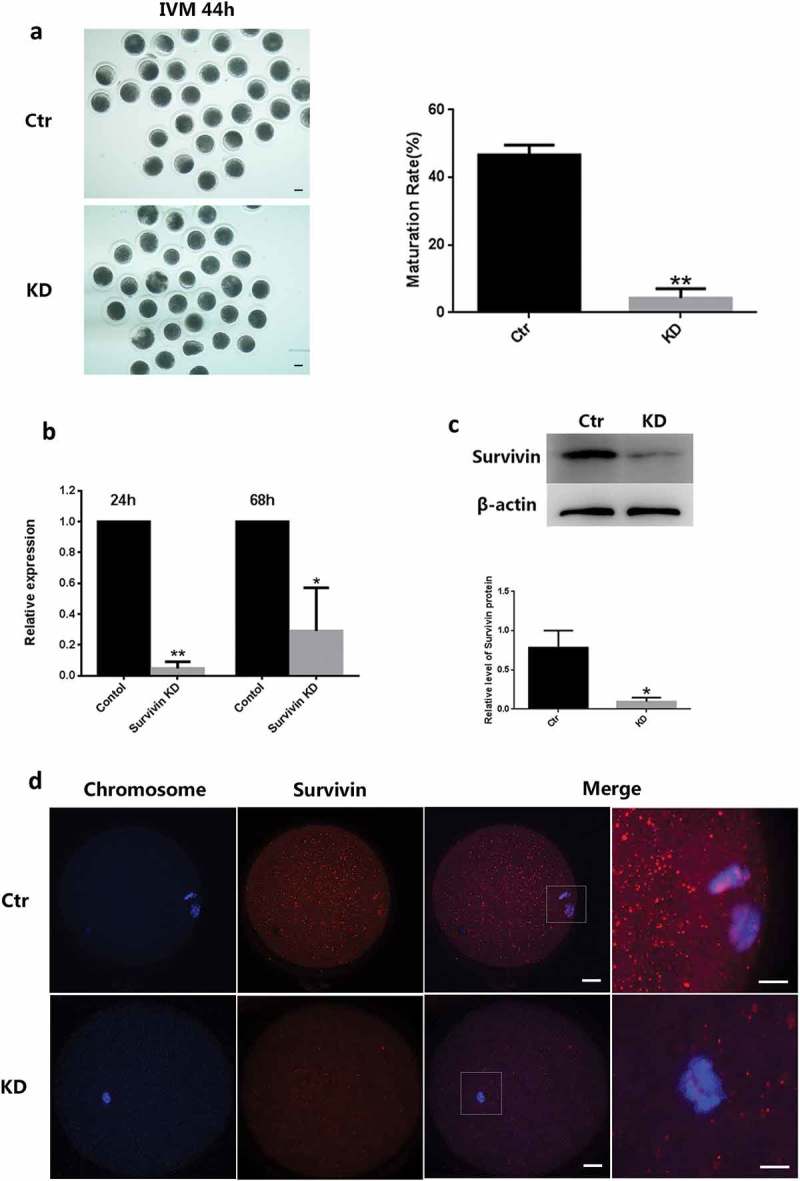
Survivin depletion led to oocytes arrested at the MI stage and impaired chromosome alignment. (a) Representative DIC images of oocytes and oocyte maturation percentage in the control and Survivin knockdown groups. Bar = 25 μm. ** P < 0.01 (b) Relative expression of Survivin mRNA in the control and Survivin knockdown groups after 24 h and 68 h siRNA injection. * P < 0.05, ** P < 0.01 (c) Expression of SURVIVIN protein in the siRNA-injected oocytes and relative levels of SURVIVIN protein between control and Survivin knockdown groups. Porcine COCs were injected with siRNA and incubated with dbcAMP for 20 h, followed by Western blotting. (d) Images of SURVIVIN (red) and chromosome (blue) in control and Survivin knockdown groups. Oocytes were stained with Survivin antibody to visualize the protein and co-stained with DAPI for chromosomes. Bar = 25 μm.
Figure 3.
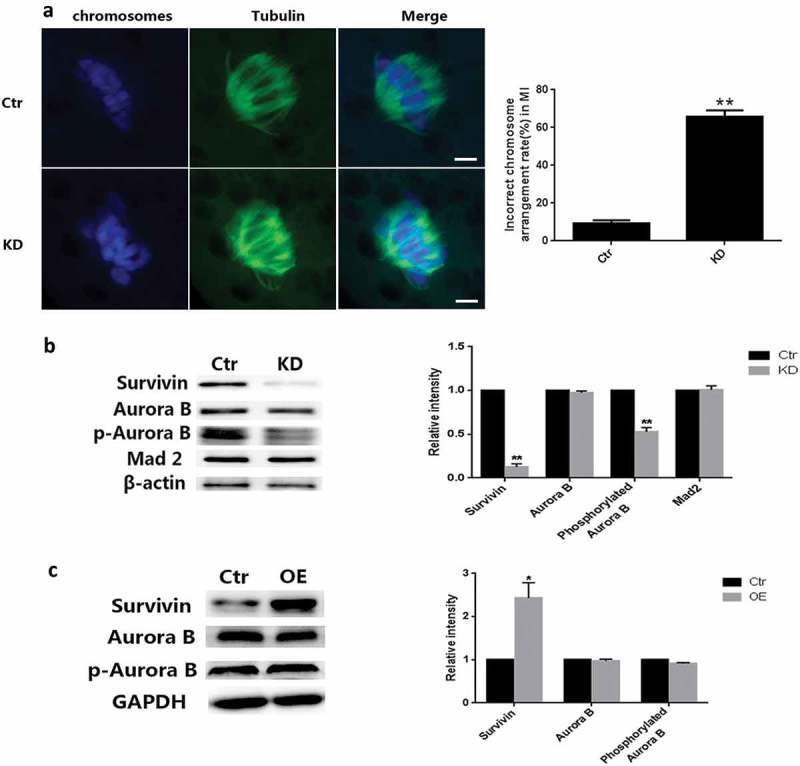
Survivin depletion led to chromosome misaligned and decrease of phosphorylated Auroa B. (a) Images of Spindle morphologies (green) and chromosome (blue) in control and Survivin knockdown groups. Oocytes were stained with α-tubulin-FITC antibody to visualize the spindles and co-stained with DAPI for chromosomes. Bar = 10 μm. (b) The percentage of spindle/chromosome defects in oocytes from the control and Survivin knockdown groups. (c) The protein level of Survivin, Aurora B, phosphorylated Aurora B and Mad2 examined by western blot in the control and Survivin knockdown groups in MI stage. And the relative level of each protein analysis was showed in (d). * P < 0.05, ** P < 0.01. (e) The protein level of Survivin, Aurora B, and phosphorylated Aurora B examined by western blot in the control and Survivin overexpression groups in MI stage. And the relative level of each protein analysis was showed in (f). * P < 0.05, ** P < 0.01.
Depletion of survivin decreased the phosphorylation of Aurora B
Our results indicates that chromosomes arrangement has anomalies in MI, and previous studies have demonstrated that Aurora B kinase activity was regulated by both Survivin binding and cell cycle-dependent phosphorylation [51]. Thus, we speculated that the activity of Aurora B kinase had been related to abnormal meiosis. To test this hypothesis, we detected the effect of Survivin depletion on Aurora B activity. We found that Survivin-depletion oocytes elicited a sustained decrease of phosphorylated Aurora B (Figure 3(c,e)), the key function subunit of CPC, which were associated with SAC activation. We next overexpressed Survivin by microinjecting synthesized Survivin mRNA to study its effect on the Aurora B kinase activity. Immunoblot analysis showed that Survivin level was increased, but there were no influence on phosphorylated Aurora B level (Figure 3(d,f)), which suggested that Aurora B was independent of the Survivin level.
Depletion of survivin induced the decreased expression of SAC related genes
It was proved that CCNB1 regulated MPF activity and the activation of SAC. We found that CCNB1, which affected the activity of MPF critical for metaphase-anaphase transition, elicited a sustained existence in Survivin-depletion oocyte (Figure 4(a,b)). We concluded that the chromosomes arrangement anomalies caused by survivin depletion had characteristics of the activation of a phosphorylated Aurora B-dependent response. SAC and anaphase promoting complex/cyclosome (APC/C) are very important in meiosis. So we determined the expression of SAC and APC/C associated genes. The result showed that the expression of Mad2L1 and Cdk1 were decreased in Survivin-depletion oocytes. Meanwhile the other genes (Cdc25b, Cdc20 and Mad2L2) expression had no significant changes (Figure 4(c)).
Figure 4.
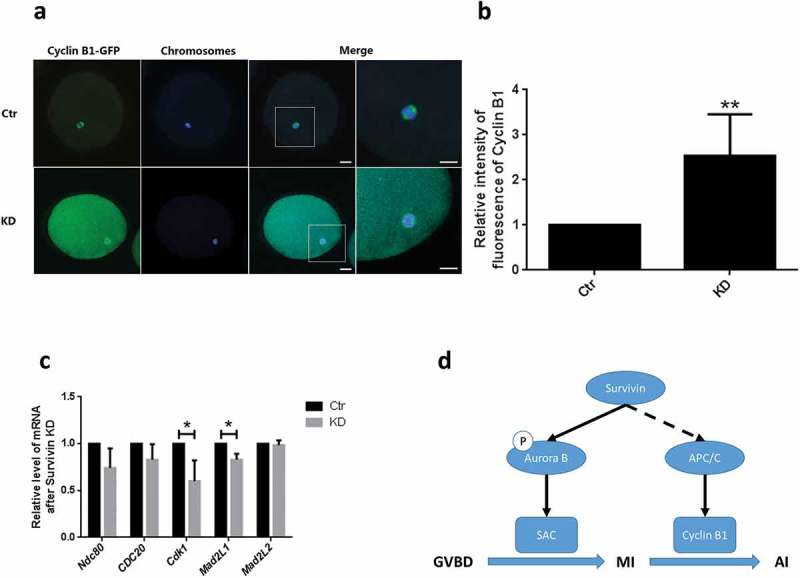
Survivin depletion inhibited Cyclin B1 degradation in oocytes at MI-AI. (a) Images of cyclin B1-GFP (green) in the control and Survivin knockdown groups. Cyclin B1 was immunolabled with GFP and examined by confocal microscopy. Bar = 25 μm (b) The average fluorescence intensity of Cyclin B1-GFP in the control and Survivin knockdown groups. (c) Relative expression of SAC or APC/C associated genes which were Cdc25b, Cdc20, Cdk1, Mad2L1 and Mad2L2. * P < 0.05. (d) Schematic illustrating function of Survivin during porcine oocytes maturation.
Discussion
In this study, we used porcine oocytes as a model to investigate the roles of SURVIVIN during porcine oocyte maturation. Our results demonstrated that Survivin regulated the phosphorylation levels of Aurora B and affected MPF activity to regulate SAC, which ensured that all chromosomes were accurately arrangement during meiosis in porcine oocytes (Figure 4(d)). These findings were consistent with the studies that Survivin-depleted HeLa cells and U2OS cells entirely lacked sister-chromatid segregation in mitosis [15,52]. Our observation was important to indicate the mechanism of chromosomes separation during porcine meiotic maturation.
The Aurora-B kinase was reported to promote bipolar kinetochore attachments by destabilizing kinetochore attachments [32,53]. Thus we propose that Survivin ensures the bipolar attachment of kinetochores by being essential for assembly of the CPC at centromeres in meiosis, and Survivin depletion leads to Aurora-B failing to assemble at centromeres in pro-metaphase in Survivin-depletion cells. Although INCENP owns a highly conserved centromere targeting domain in its N-terminus [54], the mechanism by which domain mediates centromeric localization remains controversial. Because INCENP interacts with Survivin by means of this N-terminal domain [3], and it is proved that Survivin mediates meiotic localization of the CPC [3]. And Aurora B enzymatic activity relies on accurate localization, so we suggested that Survivin regulated Aurora B activity by its location. In our experiments, Survivin-depletion oocytes elicited a sustained decrease of phosphorylated Aurora B; overexpressed Survivin did not increase the phosphorylated Aurora B, which implied that Survivin is associated with activated Aurora B by recruiting it to kinetochores.
Multiple correction process at kinetochores might be required until the bipolar attachment is finally achieved. Bipolar attachment generates tension by sister kinetochores and separates the CPC/Aurora B from the kinetochore targets spatially, which leads to the stabilization of correct attachments. Inhibition [55], depletion [3] or, as we have shown here, displacement of the CPC via Survivin-depletion from centromeres is associated with the generation of attached kinetochores, which silencesAurora B and induces chromosome misalignment. Cyclin B1 is a subunit in MPF and its degradation by active APC/C once oocytes enter into AI stage [56]. Meiosis will arrest at MI when Cyclin B1 accumulates, which were rely relied on MPF and APC activity. Survivin-depletion induced overriding of SAC may be associated with a mis-localization of the CPC from centromeres onto chromosome arms. This activity of mislocalization appears to be not related to DNA damage, but rather to its ability to MPF activity. So we assessed exogenous Cyclin B1-GFP degradation in survivin-depletion oocytes in MI oocytes. Survivin-depletion induced the sustaining of exogenous Cyclin B1 in MI oocytes which may extend AI stage.
In MI oocytes, sister kinetochores become attached to microtubules emanating from the spindle poles. If both kinetochores are attached in an end-on manner from opposite poles, the force balance leads to stable biorientation, which leads to all chromosomes uniformly arranged in the equatorial plane [57]. Once this process is finished, SAC is inactivated and oocytes start to enter into AI stage [58]. In our studies, we found the oocytes with Survivin depletion were arrested at MI stage and chromosomes were misaligned. Mad2 is the key submit of SAC, which is localized at kinetochores and combines with other functional groups to active SAC [59]. In addition, it can combine with Cdc20, and they together recruit other proteins to form meiosis checkpoint compounds (MCC) [60], which is the key factor in inhibition of the APC/C activity [58]. NDC80 is a component of kinetochore proteins which establish a connection between chromosomal centromeres and microtubes of spindle. NDC80 bridge is involved with the reductional division in meiosis I [61]. Related studies have reported that Aurora B kinase could interact with SAC [57], so we detected the expression of Mad2L1, Mad2L2, and Ndc80 in Survivin depleted oocytes. Our results showed that Survivin depletion decreased the expression of Mad2L1, which was consistent with the previous studies.
Taken together, our studies demonstrated that Survivin took critical roles in homologous chromosomes segregation during porcine oocyte meiotic maturation (Figure 4(d)). Both the phosphorylation levels of Aurora B and the expression of Mad2L1 related to spindle assembly checkpoint (SAC) were decreased in Survivin-depleted oocytes. Porcine oocytes could not complete the first meiotic division without SURVIVIN to release the first polar body. Our data provided important information that could potentially be used to culture porcine oocytes in vitro and provide more oocytes with high quality for research or embryo production.
Funding Statement
This research was supported by the Natural Science Foundation of Hubei Province (Grant# 2018CFA015); National Key Research and Development Program of China; Stem Cell and Translational Research (Grant# 2016YFA0100203); National Project for Breeding of Transgenic Pig (2016ZX08006-002).
Disclosure statement
The authors declare no competing financial interests.
Author contributions
C.L., T.L.Y, Z.W.N., T.W., Y.Y.G., S.Y.Y, L.J.H., and X.Z. conducted the experiments; L.C., T.L.Y and Y.L.M. analyzed the data, designed the experiments and wrote the manuscript. All authors reviewed the manuscript.
References
- [1].Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003. January 3;1:46–54. PubMed PMID: WOS:000180448400014; English. [DOI] [PubMed] [Google Scholar]
- [2].Jeyaprakash AA, Klein UR, Lindner D, et al. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell. 2007. October 19;131(2):271–285. PubMed PMID: WOS:000250507700018; English. [DOI] [PubMed] [Google Scholar]
- [3].Vader G, Kauw JJW, Medema RH, et al. Survivin mediates targeting of the chromosomal passenger complex to the centromere and midbody. EMBO Rep. 2006. January 7;(1):85–92. PubMed PMID: WOS:000236142200019; English DOI: 10.1038/sj.embor.7400562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Adams RR, Carmena M, Earnshaw WC. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 2001. February;11(2):49–54. PubMed PMID: 11166196. [DOI] [PubMed] [Google Scholar]
- [5].Vagnarelli P, Earnshaw WC. Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma. 2004. November;113(5):211–222. PubMed PMID: 15351889. [DOI] [PubMed] [Google Scholar]
- [6].Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nature Rev Mol Cell Bio. 2007. October;8(10):798–812. PubMed PMID: 17848966. [DOI] [PubMed] [Google Scholar]
- [7].Vader G, Medema RH, Lens SMA. The chromosomal passenger complex: guiding Aurora-B through mitosis. J Cell Biol. 2006. June 19;173(6):833–837. PubMed PMID: WOS:000238585700002; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ditchfield C, Johnson VL, Tighe A, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003. April 28;161(2):267–280. PubMed PMID: WOS:000182624200007; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hauf S, Cole RW, LaTerra S, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003. April 28;161(2):281–294. PubMed PMID: 12707311; PubMed Central PMCID: PMC2172906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gassmann R, Carvalho A, Henzing AJ, et al. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J Cell Biol. 2004. July 19;166(2):179–191. PubMed PMID: 15249581; PubMed Central PMCID: PMC2172304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Adams RR, Wheatley SP, Gouldsworthy AM, et al. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr Biol: CB. 2000. September 7;10(17):1075–1078. PubMed PMID: 10996078. [DOI] [PubMed] [Google Scholar]
- [12].Honda R, Korner R, Nigg EA. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol Biol Cell. 2003. August;14(8):3325–3341. PubMed PMID: WOS:000184877400021; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bishop JD, Schumacher JM. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B Kinase stimulates Aurora B kinase activity. J Biol Chem. 2002. August 2;277(31):27577–27580. PubMed PMID: 12048181; PubMed Central PMCID: PMC1855214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Andrews PD, Knatko E, Moore WJ, et al. Mitotic mechanics: the auroras come into view. Curr Opin Cell Biol. 2003. December;15(6):672–683. PubMed PMID: WOS:000187109200005; English. [DOI] [PubMed] [Google Scholar]
- [15].Carvalho A, Carmena M, Sambade C, et al. Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J Cell Sci. 2003. July 15;116(14):2987–2998. PubMed PMID: WOS:000184368100018; English. [DOI] [PubMed] [Google Scholar]
- [16].Sun SC, Wei L, Li M, et al. Perturbation of survivin expression affects chromosome alignment and spindle checkpoint in mouse oocyte meiotic maturation. Cell Cycle. 2009. October 15;8(20):3365–3372. PubMed PMID: 19806029. [DOI] [PubMed] [Google Scholar]
- [17].Sun SC, Liu HL, Sun QY. Survivin regulates Plk1 localization to kinetochore in mouse oocyte meiosis. Biochem Biophys Res Commun. 2012. May 18;421(4):797–800. PubMed PMID: 22554510. [DOI] [PubMed] [Google Scholar]
- [18].Jiang ZZ, Hu MW, Wang ZB, et al. Survivin is essential for fertile egg production and female fertility in mice. Cell Death Dis. 2014. March;27(5):e1154 . PubMed PMID: 24675472; PubMed Central PMCID: PMC3973204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Klein UR, Nigg EA, Gruneberg U. Centromere targeting of the chromosomal passenger complex requires a ternary subcomplex of Borealin, Survivin, and the N-terminal domain of INCENP. Mol Biol Cell. 2006. June;17(6):2547–2558. PubMed PMID: 16571674; PubMed Central PMCID: PMC1474794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lens SM, Rodriguez JA, Vader G, et al. Uncoupling the central spindle-associated function of the chromosomal passenger complex from its role at centromeres. Mol Biol Cell. 2006. April;17(4):1897–1909. PubMed PMID: 16436504; PubMed Central PMCID: PMC1415303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sessa F, Mapelli M, Ciferri C, et al. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol Cell. 2005. April 29;18(3):379–391. PubMed PMID: 15866179. [DOI] [PubMed] [Google Scholar]
- [22].Yasui Y, Urano T, Kawajiri A, et al. Autophosphorylation of a newly identified site of Aurora-B is indispensable for cytokinesis. J Biol Chem. 2004. March 26;279(13):12997–13003. PubMed PMID: 14722118. [DOI] [PubMed] [Google Scholar]
- [23].Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nature Rev Mol Cell Bio. 2007. May;8(5):379–393. PubMed PMID: 17426725. [DOI] [PubMed] [Google Scholar]
- [24].Bharadwaj R, Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004. March 15;23(11):2016–2027. PubMed PMID: 15021889. [DOI] [PubMed] [Google Scholar]
- [25].Vader G, Maia AF, Lens SM. The chromosomal passenger complex and the spindle assembly checkpoint: kinetochore-microtubule error correction and beyond. Cell Div. 2008. May 28;3:10 PubMed PMID: 18507820; PubMed Central PMCID: PMC2430558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lens SMA, Wolthuis RMF, Klompmaker R, et al. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. Embo J. 2003. June 16;22(12):2934–2947. PubMed PMID: WOS:000183724400007; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pinsky BA, Kung C, Shokat KM, et al. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nature Cell Biol. 2006. January;8(1):78–83. PubMed PMID: 16327780. [DOI] [PubMed] [Google Scholar]
- [28].Hsu JY, Sun ZW, Li X, et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell. 2000. August 4;102(3):279–291. PubMed PMID: 10975519. [DOI] [PubMed] [Google Scholar]
- [29].Adams RR, Maiato H, Earnshaw WC, et al. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J Cell Biol. 2001. May 14;153(4):865–880. PubMed PMID: 11352945; PubMed Central PMCID: PMC2192373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kaitna S, Pasierbek P, Jantsch M, et al. The aurora B kinase AIR-2 regulates kinetochores during mitosis and is required for separation of homologous Chromosomes during meiosis. Curr Biol. 2002. May 14;12(10):798–812. PubMed PMID: 12015116. [DOI] [PubMed] [Google Scholar]
- [31].Tanaka TU, Rachidi N, Janke C, et al. Evidence that the Ipl1-Sli15 (Aurora knase-INCENP) complex by altering promotes chromosome bi-orientation kinetochore-spindle pole connections. Cell. 2002. February 8;108(3):317–329. PubMed PMID: WOS:000173801600005; English. [DOI] [PubMed] [Google Scholar]
- [32].Kang J, Cheeseman IM, Kallstrom G, et al. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J Cell Biol. 2001. November 26;155(5):763–774. PubMed PMID: 11724818; PubMed Central PMCID: PMC2150868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001. December 1;15(23):3118–3129. PubMed PMID: 11731476; PubMed Central PMCID: PMC312839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Murata-Hori M, Wang YL. The kinase activity of aurora B is required for kinetochore-microtubule interactions during mitosis. Curr Biol. 2002. June 4;12(11):894–899. PubMed PMID: WOS:000176085500017; English. . [DOI] [PubMed] [Google Scholar]
- [35].Kallio MJ, Nieminen M, Eriksson JE. Human inhibitor of apoptosis protein (IAP) survivin participates in regulation of chromosome segregation and mitotic exit. FASEB J. 2001. December;15(14):2721–2723. PubMed PMID: 11687505. [DOI] [PubMed] [Google Scholar]
- [36].Kallio MJ, McCleland ML, Stukenberg PT, et al. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr Biol. 2002. June 4;12(11):900–905. PubMed PMID: 12062053. [DOI] [PubMed] [Google Scholar]
- [37].Oegema K, Desai A, Rybina S, et al. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol. 2001. June 11;153(6):1209–1226. PubMed PMID: 11402065; PubMed Central PMCID: PMC2192036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Li F, Ambrosini G, Chu EY, et al. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998. December 10;396(6711):580–584. PubMed PMID: 9859993. [DOI] [PubMed] [Google Scholar]
- [39].Giodini A, Kallio MJ, Wall NR, et al. Regulation of microtubule stability and mitotic progression by survivin. Cancer Res. 2002. May 1;62(9):2462–2467. PubMed PMID: WOS:000175265000003; English. [PubMed] [Google Scholar]
- [40].Rogers E, Bishop JD, Waddle JA, et al. The aurora kinase AIR-2 functions in the release of chromosome cohesion in Caenorhabditis elegans meiosis. J Cell Bio. 2002. April 15;157(2):219–229. PubMed PMID: WOS:000176426700004; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kaestner P, Stolz A, Bastians H. Determinants for the efficiency of anticancer drugs targeting either Aurora-A or Aurora-B kinases in human colon carcinoma cells. Mol Cancer Ther. 2009. July;8(7):2046–2056. MCT-09-0323. PubMed PMID: 19584233. [DOI] [PubMed] [Google Scholar]
- [42].Morrow CJ, Tighe A, Johnson VL, et al. Bub1 and aurora B cooperate to maintain BubR1-mediated inhibition of APC/CCdc20. J Cell Sci. 2005. August 15;118(Pt 16):3639–3652. PubMed PMID: 16046481. [DOI] [PubMed] [Google Scholar]
- [43].Jiang ZZ, Hu MW, Wang ZB, et al. Survivin is essential for fertile egg production and female fertility in mice. Cell Death Dis. 2014. March 5:Artn E1154 PubMed PMID: WOS:000333754100058; English DOI: 10.1038/Cddis.2014.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Leverson JD, Huang HK, Forsburg SL, et al. The Schizosaccharomyces pombe aurora-related kinase Ark1 interacts with the inner centromere protein Pic1 and mediates chromosome segregation and cytokinesis. Mol Biol Cell. 2002. April;13(4):1132–1143. PubMed PMID: WOS:000175318400004; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Uren AG, Wong L, Pakusch M, et al. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol. 2000. November 2;10(21):1319–1328. PubMed PMID: 11084331. [DOI] [PubMed] [Google Scholar]
- [46].Gao YY, Chen L, Wang T, et al. Oocyte aging-induced Neuronatin (NNAT) hypermethylation affects oocyte quality by impairing glucose transport in porcine. Sci Rep. 2016. October;26(6):36008 . PubMed PMID: 27782163; PubMed Central PMCID: PMC5080544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nie ZW, Chen L, Jin QS, et al. Function and regulation mechanism of Chk1 during meiotic maturation in porcine oocytes. Cell Cycle. 2017;16(22):2220–2229. .PubMed PMID: 28933982; PubMed Central PMCID: PMC5736331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Igarashi H, Knott JG, Schultz RM, et al. Alterations of PLCbeta1 in mouse eggs change calcium oscillatory behavior following fertilization. Dev Biol. 2007. December 1;312(1):321–330. PubMed PMID: 17961538; PubMed Central PMCID: PMC2170533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pfaffl MW, Leverson JD, Huang H-K, et al. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001. May 1;29(9):e45 PubMed PMID: 11328886; PubMed Central PMCID: PMC55695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bolton MA, Lan WJ, Powers SE, et al. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and in a complex inase activity is phosphorylation. Mol Biol Cell. 2002. September;13(9):3064–3077. PubMed PMID: WOS:000178268300007; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Fortugno P, Wall NR, Giodini A, et al. Survivin exists in immunochemically distinct subcellular pools and is involved in spindle microtubule function. J Cell Sci. 2002. February 1;115(Pt 3):575–585. PubMed PMID: 11861764. [DOI] [PubMed] [Google Scholar]
- [52].Lampson MA, Renduchitala K, Khodjakov A, et al. Correcting improper chromosome-spindle attachments during cell division. Nature Cell Biol. 2004. March;6(3):232–237. PubMed PMID: 14767480. [DOI] [PubMed] [Google Scholar]
- [53].Ainsztein AM, Kandels-Lewis SE, Mackay AM, et al. INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J Cell Bio. 1998. December 28;143(7):1763–1774. PubMed PMID: 9864353; PubMed Central PMCID: PMC2175214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Becker M, Stolz A, Ertych N, et al. Centromere localization of INCENP-aurora B is sufficient to support spindle checkpoint function. Cell Cycle. 2010. April 1;9(7):1360–1372. PubMed PMID: WOS:000276369300032; English. [DOI] [PubMed] [Google Scholar]
- [55].Zeng X, King RW. An APC/C inhibitor stabilizes cyclin B1 by prematurely terminating ubiquitination. Nat Chem Biol. 2012. February 26;8(4):383–392. PubMed PMID: 22366722; PubMed Central PMCID: PMC3307893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kitajima TS, Ohsugi M, Ellenberg J. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell. 2011. August 19;146(4):568–581. PubMed PMID: 21854982. [DOI] [PubMed] [Google Scholar]
- [57].Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol. 2012. November 20;22(22):R966–80. PubMed PMID: 23174302. [DOI] [PubMed] [Google Scholar]
- [58].Tipton AR, Wang K, Link L, et al. BUBR1 and closed MAD2 (C-MAD2) interact directly to assemble a functional mitotic checkpoint complex. J Biol Chem. 2011. June 17;286(24):21173–21179. PubMed PMID: 21525009; PubMed Central PMCID: PMC3122179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Tipton AR, Tipton M, Yen T, et al. Closed MAD2 (C-MAD2) is selectively incorporated into the mitotic checkpoint complex (MCC). Cell Cycle. 2011. November 1;10(21):3740–3750. PubMed PMID: WOS:000296572100029; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Li XX, Dawe RK. Fused sister kinetochores initiate the reductional division in meiosis I. Nature Cell Biol. 2009. September;11(9):1103–U122. PubMed PMID: WOS:000269482500011; English. [DOI] [PubMed] [Google Scholar]
- [61].Shandilya J, Medler KF, Roberts SG. Regulation of AURORA B function by mitotic checkpoint protein MAD2. Cell Cycle. 2016. August 17;15(16):2196–2201. PubMed PMID: 27341405; PubMed Central PMCID: PMC4993431. [DOI] [PMC free article] [PubMed] [Google Scholar]


