Summary:
Understanding how cortical activity generates sensory perceptions requires a detailed dissection of the function of cortical layers. Despite our relatively extensive knowledge of their anatomy and wiring, we have a limited grasp for what each layer contributes to cortical computation. We need to develop a theory of cortical function that is rooted solidly in each layer’s component cell types and fine circuit architecture, and produces predictions that can be validated by specific perturbations. Here, we briefly review progress toward such a theory, and suggest an experimental roadmap toward this goal. We discuss new methods for the all-optical interrogation of cortical layers, for correlating in vivo function with precise identification of transcriptional cell type, and for mapping local and long range activity in vivo with synaptic resolution. The new technologies that can crack the function of cortical layers are finally on the immediate horizon.
In brief
In this perspective Adesnik and Naka outline the road map to overcome the existing conceptual and technical challenges for obtaining a detailed understanding for the role of cortical layers in sensory computation and perception.
Introduction
“At present we have no direct evidence on how the cortex transforms the incoming visual information. Ideally, one should determine the properties of a cortical cell, and then examine one by one the receptive fields of all the afferents projecting upon that cell.” – Hubel and Wiesel, 1962, Journal of Physiology
A primary goal of cortical physiology is to explain how the cortex transforms incoming information to generate perceptions. Despite more than half a century since the above statement was made, a detailed understanding of the mechanisms that mediate cortical transformations across the cortical layers remains remarkably incomplete. However, recent technological advances finally allow for the execution of the experiment that Hubel and Wiesel prescribed, as well as many other sophisticated assays that can overcome this conceptual challenge. First, we briefly review how existing data have motivated the available theories on the function of cortical layers, primarily with respect to sensory transformations. Next, we highlight the key data we lack that could confirm or invalidate these models or motivate new ones. Finally, we propose the new technologies and experiments that are needed to obtain the data that will allow us to arrive at a much more mechanistic, circuit-driven theory for the unique contributions of layer-specific circuits in sensory perception.
The cortical generation of sensory percepts can be thought of as a synthetic, hierarchical process or as one based largely in statistical inference. In the hierarchical model, neurons integrate their inputs to filter the sensory data, and transform it into an output spike train that encodes features of the stimulus. A simple feed-forward architecture composed of many neurons filtering their input in this manner should ultimately enable complex computations to mediate object identification and scene analysis (Hubel and Wiesel, 1962; Marr, 2010). The apparent feed-forward architecture of the primate visual system might help explain why object recognition is fast (Thorpe et al., 1996). In the framework of statistical inference, cortical circuits encode a generative model of the sensory environment, and recurrent interactions between cortical processing stages compare the expectations of the internally generated model with incoming data from the sensory apparatus (Bastos et al., 2012).
Two of the most compelling examples of the synthetic process are the encoding of edge orientation in the primary visual cortex (Hubel and Wiesel, 1959, 1962), and that of object or face selectivity in the inferotemporal cortex (Bruce et al., 1981; Gross et al., 1972). The emergence of orientation tuning stands as one of the few concrete examples of a de novo transformation that occurs in a layer of the primary visual cortex (V1), and can be well-explained by a simple feed-forward model involving the integration over a specific set of center-surround thalamic relay neurons (Hubel and Wiesel, 1962). Although the mechanistic details of an analogous feed-forward circuit for the generation of face selectivity are lacking, one can conceptualize a similar process where neurons exhibiting increasingly sophisticated feature tuning are built by summating over neurons with more elementary filtering properties (e.g., edges to contours, and contours to faces) (Chang and Tsao, 2017; Liu et al., 2016).
Based on this framework, one might expect that further de novo transformations would occur as sensory signals propagate through the layers of the cortex (e.g. from layer 4 to layer 2/3). Yet while ample data collected across cortical areas is consistent with the synthetic model, remarkably few, if any compelling examples of such transformations have been observed between cortical layers of a single sensory area, such as V1. Orientation tuning, direction selectivity, and ocular dominance are all observable within layer 4 neurons (Hubel and Wiesel, 1962; Sun et al., 2016). In other cortical areas, such as the somatosensory cortex, we have arguably even less insight into the synthesis of new response properties (Brecht, 2017). Receptive fields in the rodent barrel cortex and in the cat visual cortex tend to grow or change shape across layers (Brecht et al., 2003; Martinez et al., 2005), and some evidence supports the de novo generation of complex cells and contextual properties such as end-stopping between L4 and L2/3 in cats (Alonso and Martinez, 1998; Hubel and Wiesel, 1962; Martinez and Alonso, 2001). Yet the striking lack of concrete examples akin to orientation tuning, at least outside monkey V1, implies that the laminar circuity in a single cortical area is not set up to generate new types of feature selectivity. Furthermore, this hierarchical framework fails to account for a wide range of context-dependent phenomena observed in cortical activity, nor does it provide a compelling explanation for the profuse amount of feedback connections from higher cortical areas to lower ones(Felleman and Van Essen, 1991).
In contrast, the alternative framework that sees sensory processing as probabilistic inference can explain these ‘top down’ phenomena. In this scheme neurons in different cortical layers have unique roles to play in computing the conditional probabilities that a given pattern of afferent neural input represents a specific sensory stimulus (Bastos et al., 2012; Rao and Ballard, 1999). The core notion is that cortical neurons, moment to moment, compare afferent input from each preceding stage with an internal generative model of the sensory environment conveyed by top-down projections, a model based on both the recent past and accumulated experience. Predictions of this model are passed from higher to lower stages (both across layers in individual areas and between areas) through feedback connections, and neurons in earlier stages compare these predictions to errors indicated by deviation from the afferent, ‘bottom-up’ sensory data. In one version of this theory, principal cells in superficial cortical layers of primary sensory areas encode prediction errors, while those in deeper layers encode conditional expectations from which predictions are made (Bastos et al., 2012). Inhibitory interneurons within each cortical layer might be critical for canceling errors (i.e., incompatible predictions) when the predictive model matches the sensory data. This conceptual framework is attractive since one can assign specific functions to different layers and cell types that should be experimentally testable. However, at present, data supporting the inferential model is limited (for example, (Homann et al., 2017)). An intriguing variant on this theme is a ‘body model’ for somatosensory cortex in which the goal of S1 is to generate mental simulations of planned body actions. This model also assigns specific functions to each layer: body simulation to L4, sensory memory storage to L2/3, motor memory storage in L5, and relay of top-down drive from M1 through L6 and back to L4 (Brecht, 2017).
More recently, the canonical circuit has been conceptualized less in terms of layers and more in terms of cell-types which occupy specific layers and are connected by cell-type specific pathways (Harris and Shepherd, 2015). Although layer and cell-type are closely intertwined, this is an important distinction: a layer-centric view implicitly assumes that at least some basic cortical computations can be understood mechanistically by analyzing the activity of neurons in just one layer; the cell-type centered view assumes that we cannot achieve a satisfactory understanding of any computation without taking into account coordinated activity across multiple layers. While we retain layer as an organizing concept for the purposes of this perspective, we note that the experimental approaches we outline below apply equally well to cracking the function of cortical sublaminae or cortical cell-types. Nevertheless, we argue that the cortical literature on layers supports the notion that in a specific set of contexts, ensembles of neurons located in just one layer are sufficient to mediate key cortical computations, such as fast sensorimotor transformations. However, in most conditions, such as those involved in generating conscious sensory percepts, the basic unit of cortical computation is a neuronal ensemble spread across multiple layers, or spread across multiple layers and cortical areas.
Layer-specific features of 'canonical' cortical circuits
Our empirical knowledge of cortical layers from which we can build theories of their function comes from four types of exploration: 1) anatomy of single cells and their projections, 2) connectivity of pairs of cells according to their laminar location and cell type, 3) physiological responses to sensory stimuli, and 4) activation or suppression of neurons in discrete layers. Anatomy and connectivity are the most absolute in that they do not depend on brain state or type of sensory stimulation. They constrain the types of computations that layers can perform and the dynamics they can exhibit, but on their own provide limited insight into function. Conversely, physiological perturbations are much less absolute in that the resulting data will depend on the brain state and context in which they were obtained. However, they should provide the most direct insight into the different functions of cortical layers.
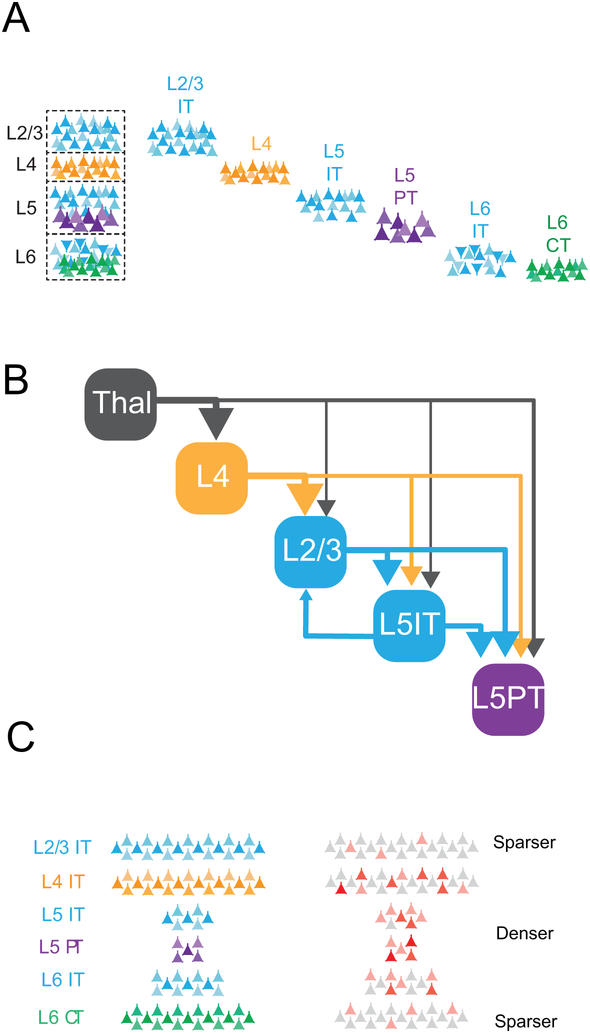
The often-repeated (although just as often maligned) notion of the ‘canonical cortical microcircuit’ is largely based on studies of anatomy and connectivity in rodents, primates and cats (Gilbert, 1983). These data have converged on a core model where thalamic input drives activity in a feed-forward and sequential fashion from L4, to L2/3, to L5 and out to other cortical and subcortical regions (Armstrong-James et al., 1992; Binzegger et al., 2004; Douglas and Martin, 1991; Lefort et al., 2009). While numerous examples of alternate connections exist (e.g., thalamus to other layers, L5 to L2/3, L4 to L5), the anatomy of these neurons (i.e., their intracortical axons and dendrites) matches well with paired electrophysiological recording and circuit mapping via optical approaches (see full citations below). As these and other pathways in the cortex have been extensively reviewed elsewhere (Callaway, 1998; Douglas and Martin, 2004; Feldmeyer, 2012; Gilbert, 1983; Harris and Shepherd, 2015; Thomson and Lamy, 2007) we will focus on data revealing the physiology and functional impact of the principal excitatory neurons in different cortical layers (Fig. 1). The long-range input/output logic of the canonical microcircuit is organized by layer. L4 neurons are thought to primarily target their local neighbors (Binzegger et al., 2004). The principal neurons of L2/3 are intratelencephalic (IT) cells, meaning that their long-range axons project only to targets within the telencephalon, such as other cortical areas and striatum. L5, sometimes called the primary cortical output layer, harbors IT cells as well as pyramidal tract (PT) cells, which send widely divergent projections to subcortical areas. L6 contains corticothalamic (CT) cells which provide a major feedback projection to thalamus, as well as IT cells.
Figure 1: Architectural principles of the cortical layers.
A) Diagram of the major excitatory cell types of the cortical layers, including L4 neurons that project locally, L2/3 and L5 IT (intratelencephalic) neurons that project intracortically, and L5 ‘pyramidal tract’ PT and L6 ‘corticothalamic’ CT neurons that project subcortically. B) Schematic of the ‘feed-forward’ architecture of the neocortex, emphasizing the increase in the specificity of translaminar targets across the layers. L6 circuitry has been excluded for clarity. C) Schematic of the change in the population size of principal cells in each layer (left) and the corresponding sparsity of their population responses to sensory stimuli (right).
Corticocortical pathways (i.e, inter-areal) are often conceptualized as being either feedforward, lateral, or feedback pathways (Felleman and Van Essen, 1991; Gamanut et al., 2018). Layer plays a key organizing role in this inter-areal hierarchical scheme (D'Souza and Burkhalter, 2017). Interestingly, as one moves along the ‘canonical’ pathway, translaminar connectivity becomes increasingly specific (Fig. 1). Primary sensory thalamus, constituting the input stage of the hierarchy, provides highly divergent output impinging on cell in all cortical layers (Cruikshank et al., 2010; Petreanu et al., 2009). L4 neurons exhibit strong recurrent intralaminar connectivity (Binzegger et al., 2004) and broadcast their output to all other layers but do not provide feedback to thalamus. Similarly, L2/3 has minimal feedback connectivity to L4, but connects densely to both types of pyramidal neurons in L5 (Adesnik and Scanziani, 2010; Lefort et al., 2009). In turn, L5 IT neurons exhibit dense, asymmetric connectivity onto PT neurons, and finally PT neurons appear to connect primarily only to other PT neurons, providing minimal feedback to any of the earlier layers (Yamawaki and Shepherd, 2015). An exception to this pattern of increasing selectivity is an ascending connection from L5 IT cells to L2/3 (Binzegger et al., 2004). As for L6, earlier models of the canonical microcircuit based primarily on data from monkeys and cats proposed that L6 receives major input from L5 and from superficial layers, and then ‘closes the loop’ by projecting back to L4 (Binzegger et al., 2004; Briggs and Callaway, 2001; Douglas and Martin, 2004; Gilbert, 1983). In rodents, L6 corticothalamic neurons project to L5a and to a lesser extent L4 (Kim et al., 2014) and receive strong long-range inputs (Kinnischtzke et al., 2016). There are putative discrepancies in the basic cortical circuitry between different mammalian species, implying that a unifying ‘canonical cortical circuit’ might not exist across mammals. However, despite their heterogeneity, existing data still indicate that cortical circuits across brain areas and species share some common functional principles that are key central to understanding their function.
Layer-specific perturbations: insights and challenges
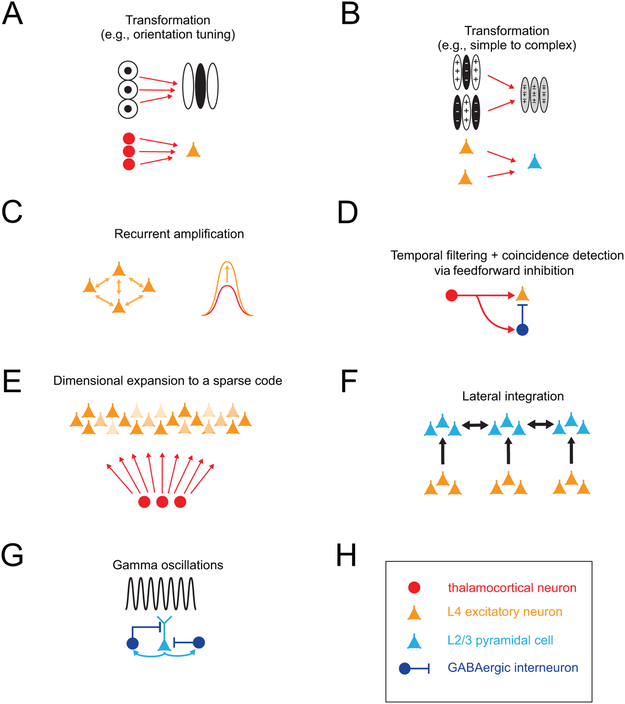
Understanding any neural circuit requires perturbing it and observing changes in the computations it performs. Examples of putative computations that cortical circuits implement are schematized in Fig. 2. These include summation that give rises to oriented edge detectors (Fig. 2A,B), signal amplification through recurrent excitation (Fig. 2C), coincidence detection (Fig. 2D), generation of a sparse code (Fig. 2E), lateral integration that might facilitate contour or boundary detection (Fig. 2F), and coding through synchronization (Fig. 2G). Although the anatomy and physiology of the neurons in any layer can help us build theories and propose hypotheses for how layers contribute to each of these computations, only manipulating the activity of specific layers or subsets of layers can test and validate these theoretical hypotheses. Prior to the advent of cell-type specific perturbations (via opto- or chemo-genetics), the primary tools for perturbation were chemical lesions (reversible or irreversible), cortical cooling, and electrical microstimulation. A common weakness of all these tools is that precisely calibrating the spatial extent of the perturbation is extremely challenging, and they cannot be absolutely layer specific since cortical neurons’ dendrites and axons often stretch across laminar boundaries. This last fact even muddies the concept of what a layer is in the cortex, since some deep layer pyramidal neurons derive much of their synaptic input from their dendrites that occupy different layers that the one in which their cell body resides (Larkum et al., 2018).
Figure 2: Examples of circuit motifs for basic sensory transformations and computations in L4 and L2/3.
A) Diagram of the thalamocortical circuit that generates orientation selectivity between the visual thalamus and primary visual cortex. Top: structure of receptive fields of the corresponding neurons. B) Diagram of a circuit between L4 and L2/3 that could generate complex cells from simple cells in V1. Top: structure of receptive fields of the corresponding neurons. C) Recurrent excitatory circuitry in L4 that linearly amplifies thalamocortical input. D) A simple feed-forward inhibitory circuit that enforces coincidence detection between thalamus and L4. E) Dimensional expansion of the sensory code between thalamus and cortical L4. F) Diagram of horizontal circuits in L2/3 that might contribute to contextual modulation. G) Schematic of the minimal recurrent excitatory/inhibitory circuit for generating gamma frequency oscillations in L2/3. H) Legend.
Layer-specific optogenetic manipulation overcomes this last problem, yet the results of both chemical and optogenetic perturbations of cortical layers have often challenged the canonical model of information flow across the cortical layers. For instance, reversibly blocking L4 activity (through chemically silencing specific layers of the visual thalamus) does not block much of the sensory evoked responses in L2/3 of the visual cortex of anesthetized cats (Malpeli, 1983)(but see (Martinez and Alonso, 2001)), but simultaneously suppressing higher visual cortex areas does (Mignard and Malpeli, 1991). In a similar vein, direct application of the action potential blocker lidocaine to the superficial layers of somatosensory cortex in has essentially no effect on whisker-evoked activity in L5 PCs in sedated and paralyzed rats (Constantinople and Bruno, 2013) but see (Wright and Fox, 2010). Direct optogenetic suppression of L4 in awake, locomoting mice leads to a modest reduction in sensory evoked activity in L2/3 of V1 or S1. However, it simultaneously leads to a potent disinhibition of activity in L5, due to a disynaptic translaminar inhibitory circuit between L4 and L5 (Pluta et al., 2015). Optogenetic suppression of L6 in awake mice also has a largely disinhibitory effect across most layers of cortex, an effect attributed to the deactivation of a broadly inhibiting translaminar inhibitory neuron (Bortone et al., 2014; Olsen et al., 2012). In one study optogenetic activation of L4, L2/3, or L5 in brain slices from mouse primary visual cortex revealed that activation of each of these layers suppressed activity within their own layer by recruiting potent recurrent inhibition, but could facilitate activity in downstream layers by generating a more favorable excitation/inhibition ratio (Adesnik, 2018), a feature previously only identified in L2/3 of the barrel cortex (Adesnik and Scanziani, 2010).
The outcomes of these experiments argue that the traditional, feed-forward, layercentric model of cortical activity can account for some but not all aspects of cortical processing. However, a major problem persists with both chemical and layer-specific optogenetic manipulations: owing to the highly inter-connected nature of neurons across layers and cortical areas, perturbation to cells in one layer will almost necessarily impact activity in other layers and areas. Therefore, it is difficult to attribute any measurable physiological or behavioral effect to the action of the pharmacologically or optogenetically targeted layer. While this is a criticism that can be more generally leveled at any sort of brain perturbation (Otchy et al., 2015), it stands as one of the key challenges in interpreting data gained from perturbation studies.
Layers acting alone
Perhaps the most fundamental, first-order question concerning cortical layers is whether the lamination is functionally relevant. Species such as birds exhibit high cognitive abilities but have no such layering in their pallium, and instead computation seems to be organized around cells clustered into ‘nuclei’ (Calabrese and Woolley, 2015). Strikingly, a mutant mouse with completely disorganized cortical layers has no immediately apparent sensory physiological deficits, suggesting that layers, per se, may not be critical for many aspects of cortical function (Guy and Staiger, 2017). This raises the alternative hypothesis that the layered structure of the neocortex is largely a consequence of early cortical development, and layers have no intrinsic function in the adult brain. Instead, they just happen to harbor unique cell types that, through their translaminar circuits, constitute the functional substrate of cortical computation. However, lamination is a conserved feature of brain circuits from insects to fish to mammals, particularly evident in structures such as the retina, cerebellum, tectum and the hippocampus (Striedter, 2005). In these structures, lamination organizes many aspects of synaptic connectivity and function, such as the segregation of ON and OFF pathways in the retina. Thus one is tempted to conclude that many circuits evolved lamination to achieve specific computational goals, even if it is not strictly necessary for computation in general.
We suspect that neural ensembles composed of co-active neurons distributed across layers are the major substrate of cortical computation. However, the selective impacts of the perturbation experiments described above imply that in certain circumstances, layers can have specific and identifiable effects on their local cortical circuit and even on behavioral output. These studies raise the possibility that for particular computations or behaviors only one cortical layer, or even a specific subtype within a single cortical layer, might be sufficient to execute the task based on its afferent input and its long-range output. This is surely possible from an anatomical perspective: principal neurons across all layers receive bottom-up thalamic inputs, most receive top-down cortical input from higher cortical areas, and many project a long range axon out of the cortex. If any of their long-range input pathways are sufficiently strong (or strong when provided in combination) many cortical neurons could operate independently of their local circuits. There is evidence to support this notion. A recent study identified a compact long-range circuit from the whisker to the barrel cortex in which a subset of L5 PCs projects directly back to pre-motor spinal trigeminal neurons that control whisker retraction (Matyas et al., 2010). Since L5 neurons can be driven directly by thalamus (Constantinople and Bruno, 2013), or through the conjunction of bottom-up and top-down motor cortical input (Manita et al., 2015), under specific conditions motor control of certain features of whisking (e.g., touch induced pumps (Deutsch et al., 2012)) might only require activation of these specific L5 PCs. If so, it would represent a complete sensorimotor behavior that could be cortically dependent, but does not require activity of superficial layers at all. In a similar vein, since other projection subtypes of L5 PCs target various sub-cortical nuclei, we suspect there might exist a suite of reflex-like sensorimotor behaviors that require minimal local processing within the primary sensory cortex, and instead rely exclusively on the integration of various long-range pathways to specific subtypes of L5 PCs. Conversely, since L2/3 PCs across species receive direct thalamic input and project to downstream cortical areas (Fitzpatrick et al., 1983; Meyer et al., 2010; Petreanu et al., 2009), various types of sensory computations in L2/3 could also be executed with limited involvement of L4-6. Taken together, this suggests that there may be specific behaviors and cortical computations that can depend on a single layer or even a single cell type within one layer.
A holistic attack on cortical layer function
Why has the extensive physiological data collected over the last few decades provided few concrete examples of new transformations that occur across layers? This absence is problematic since it has precluded the generation and validation of hypotheses concerning the differential roles of layers in sensory computation. We can consider several explanations: first, higher order feature selectivity might not emerge across layers within a single area, and only across cortical areas; second, physiologists have, until recently, lacked the proper tools to observe these higher order features; third, most measurements have been made in too impoverished behavioral conditions; or fourth, data have been analyzed in the wrong conceptual framework (i.e., synthetic vs. inferential).
Cortical circuits in any given brain area can process sensory input from multiple modalities, encode behavior/movement related information, and directly contribute to motor control (Ibrahim et al., 2016; Iurilli et al., 2012; Matyas et al., 2010; Vinck et al., 2015). Accordingly, investigating the function of microcircuits in primary sensory cortex function by only focusing on sensory transformations is sure to lead to a partial understanding of cortical computation at best. Identifying ‘canonical computation’ will therefore require investigating how the ‘canonical circuit’ operates as but one functional part of a much larger architecture. While the corticocortical network is hierarchically organized, it is also massively recurrent and fundamentally sensorimotor (D'Souza and Burkhalter, 2017; Gamanut et al., 2018). It could be that certain computational principles of cortical microcircuits will be comprehensible only in the context of naturalistic, sensorimotor behaviors (Juavinett et al., 2018; Krakauer et al., 2017).
All of these issues enumerated above have likely hindered progress towards a fuller understanding of cortical layers and cortical computation in general. Based on the available data from primates, from the synthetic perspective it appears that major new transformations primarily emerge across cortical areas and not between layers of any individual area (Liu et al., 2016; Ziemba et al., 2016). Alternatively, rather than further refining encoding of sensory stimuli, different layers of sensory cortex might instead be specialized to perform different computations with respect to non-sensory factors such as the movement, motivation, or stored memories of an animal. Such computations might only become evident when the appropriate non-sensory factors are taken into account (Keller et al., 2012; Roelfsema et al., 1998; Saleem et al., 2013).
We propose that differentiating between these alternative hypotheses for the computational role of cortical layers requires a holistic approach to monitoring and manipulating activity along the cortical axis. Rarely does one have enough a priori knowledge for the specific path information will take under a specific condition such that focused, small scale recording will be sufficient. Furthermore, key types of transformations might only become apparent at the population level, and across neurons that are spatially intermingled with other ensembles and distributed across multiple cortical areas (Ma et al., 2006; Pouget et al., 2000; Wohrer et al., 2013). Thus, the appropriate holistic approach includes recording densely across all layers, identifying the cell types of the recorded cells within this volume, recording neural activity simultaneously in multiple connected brain regions (e.g., thalamus, secondary sensory areas, relevant motor cortices), and perturbing individual layers and cell types with high spatial and temporal precision. Just as importantly, behavioral data on motor actions, brain state, attentional focus, and potential goals should be acquired to inform all analysis. Where possible, experiments should target ethological behaviors in as natural context as the experimental design will permit. Applying this approach comparatively across different cortical areas and species will help reveal the circuit elements that are conserved in different cortical systems, which will be instrumental in identifying simplifying motifs. Whether one considers cortical computation from the synthetic or inferential vantage point, such an all-inclusive experimental approach can provide the comprehensive data needed to determine whether cortical circuits implement canonical computations, and if so, how specific layers and cell types contribute to specific computations or behaviors.
In particular, from such a data set one can execute several key critical tests. The first is an analysis of the neural activity across layers during a specific computation. One must determine which layers (and which cell types within each layer) show activity that can explain how the computation is mediated, and then track the transformation of the encoded signal across the layers, from cortical input to cortical output to motivate clear hypotheses for which layers are causally involved. A simple example of this process is the LGN to V1 transformation for the synthesis of oriented receptive fields, which was apparent even under conditions of sparse sampling since it probably dominates the first stage of V1 processing.
After observation, the second test requires a causal manipulation of the layer(s) and cell type(s) thought to be involved in a computational process based on the results of step one. To achieve this, first one would want to abolish the computation by eliminating the activity of the putative contributing layers. However, for the reasons mentioned above, loss-of-function or gain-of-function experiments cannot on their own entirely discriminate between various circuit models that could account for the same effect. To overcome this challenge, more sophisticated and more integrated approaches are required. Consider two examples: L6 activity has been proposed to control both the size of upper layer receptive fields and their gain by recruiting intracortical inhibition in cats and mice (Bolz and Gilbert, 1986; Olsen et al., 2012). Silencing L6 activity enhances upper layer responses and expands their receptive fields, but this could be due to direct intracortical effects, or it could operate indirectly through L6’s impact on the LGN. One study in awake mice, therefore, employed simultaneous recording from the LGN and V1 layers and demonstrated that the disinhibition of cortex during optogenetic stimulation of L6 preceded effects on the thalamus, arguing for a direct effect (Olsen et al., 2012).
Despite the elegance of this example, temporal offsets in the impacts of perturbations might not always be able to distinguish between competing models. Consider the possible explanations for the synthesis of complex receptive fields in L2/3. One model argues for a simple summation over phase-offset iso-oriented simple cells in L4 (Hubel and Wiesel, 1962; Martinez and Alonso, 2001) (Fig. 2G), while another argues that this computation depends on V2 feedback (Mignard and Malpeli, 1991), and a third proposes that appropriate summation over geniculocortical input directly to L2/3 is sufficient to account for it (Hoffman and Stone, 1971). One study showed that inactivating L4 suppresses L2/3 complex cell activity in anesthetized cats (Martinez and Alonso, 2001). This manipulation, however, only proves that L4 ascending input is required for L2/3 visually-evoked drive, but does not unequivocally demonstrate that complex cells emerge from the summation of L4 simple cells with spatially offset ON and OFF subfields. Instead, if one could photo-activate a small subset of iso-oriented L4 neurons with appropriately arranged subfields and recapitulate the non-phasic response of the L2/3 complex cell, one could distinguish between these models. Thus, a more general approach for revealing the mechanism of a transformation across layers with great certainty is to entirely reconstitute the computation by activating the appropriate ensemble of presynaptic neurons with extreme spatial and temporal precision. An alternative scheme is retrogradely and transynaptically labeling an ensemble of cortical neurons from one cortical starter cell with a GCaMP6-expressing rabies virus (Wertz et al., 2015). This permits measuring the functional responses of a large subset of putative presynaptic cells to a single cortical neurons. Employing a virus that drives a microbial opsin in addition to GCaMP6 could further enable photo-activation of the presynaptic ensemble for causal, functional tests.
While we have focused on how the above approach would apply to sensory transformations, a holistic attack on cortical layer function will also require a holistic perspective on how cortical layers and cell types contribute to behavior, including perceptual learning, decision-making, and motor control. In motor cortex, a key conceptual advance has been to investigate how cortical dynamics robustly control movements, instead of asking how cortical populations represent movement parameters (Churchland et al., 2012; Gallego et al., 2017; Michaels et al., 2016). By the same token, in the sensory cortex, it may be fruitful to shift the focus from asking how sensory information is represented and transformed per se and instead ask how sensory cortices and their component layers and cell types subserve the broader function of allowing an animal to flexibly adapt its behavior to changing environmental conditions in its environment.
Cracking layers new tools from the technical revolution in neuroscience
The holistic approach that understanding the cortex calls for is no longer beyond reach. Several emerging technologies will soon make combining large-scale recording with extremely precise perturbations of neural activity experimentally possible. We review several key advances and propose new experimental paradigms combining these advances that could address the function of cortical layers. The reader is referred to other reviews on this subject for more in-depth consideration of some of these approaches (Emiliani et al., 2015; Luo et al., 2018a).
Massively-parallel electrophysiology:
Unquestionably, measuring neural activity simultaneously across cortical layers and cortical input and output structures will be highly advantageous for gauging the computational role and impact of each cortical layer. For more than a decade, laminar multi-electrode arrays have represented the key tool in the cortical physiologist’s arsenal for addressing this challenge (Csicsvari et al., 2003), yet their relatively low channel count and large size have precluded achieving the needed scale. Over the same period, multiphoton calcium imaging has begun to provide an incredibly dense view of local neural computation, albeit in circumscribed regions and specific layers of the cortex (Stosiek et al., 2003). However, major advances in both silicon probe technology and large-scale calcium imaging place us at the threshold of probing neural activity at the scale and speed needed to address the problem at hand.
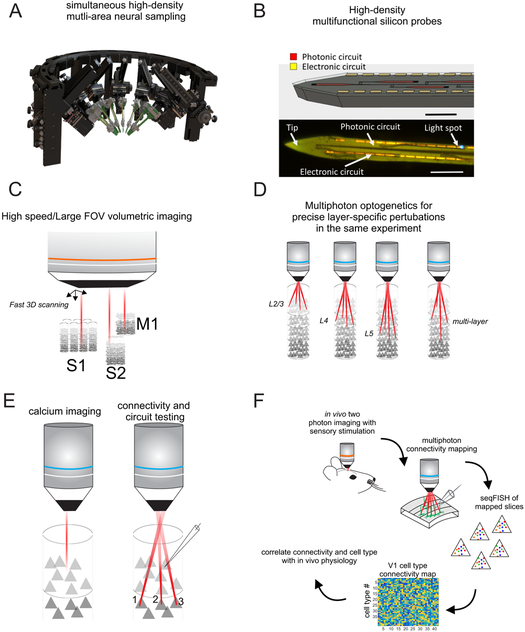
Leveraging new fabrication techniques, extremely compact multi-electrode arrays with up to 1,000 contacts now permit simultaneous physiological access to dozens of brain areas, and just as importantly, densely within and across layers (Jun et al., 2017; Shobe et al., 2015). Clever mechanical systems for inserting multiple such probes into the same head-fixed animal should allow tracking the activity across all layers of each area and across a series of connected areas in the cortical hierarchy simultaneously (Fig. 3A). This possibility is fundamental: it will allow investigators to track the evolution of neural activity, with the resolution of single neurons and single spikes, from massive populations of neurons representing the key nodes of computation. While these probes do net yet offer integrated micro-optics, structured illumination from a separate optical system could provide simultaneous high resolution optogenetic manipulation, at least for superficial cortical areas. Adding micro-LEDs (Scharf et al., 2016) or optical waveguides (Wu et al., 2013) directly onto these probes could provide optical control over their full extent so that investigators could optogenetically control specific layers within specific cortical areas (Fig. 3B). Furthermore, the integration of nano- or microfluidic ports into the same probes could additionally provide spatiotemporally resolved pharmacological manipulations that are needed for determining how various neuromodulators influence laminar interactions and computations (Canales et al., 2015).
Figure 3: New methods for dissection cortical layer function.
A) Schematic of a mechanical setup for the simultaneous insertion of multiple ultra-high density multielectrode arrays (courtesy of New Scale Technologies). B) Schematic of a multi-layer silicon probe integrating electrodes and optical waveguides for recoding and optogenetics (courtesy of V. Lanzio and S. Cabrini). C) Schematic of a very large field of view high speed volumetric two photon microscope for densely imaging across multiple layers and multiple connected cortical areas simultaneously. D) Schematic of using a holographic multiphoton microscope to selectively perturb individual layers in the same animal on different trials. E) Schematic of a paradigm for mapping the physiological responses of L2/3 and L4 cortical neurons with calcium imaging, and then optogenetically stimulating a precise ensemble of presynaptic L4 neurons while recording from a L2/3 neuron in attempt to recapitulate the sensory response properties of the L2/3 neuron. F) Sequence of proposed experiments for correlating the sensory physiology, synaptic connectivity, and transcriptionally defined cell types of densely imaged cortical neurons across layers.
Large-scale multiphoton imaging:
Major strides are also being made in scaling up the field of view, depth, and speed of multiphoton calcium imaging. New approaches for engineering the optical wavefront or point spread function of the ultrafast laser provides volumetric imaging of neural activity across depth, with limited compromise in the overall data acquisition speed (Lu et al., 2017; Prevedel et al., 2016; Theriault et al., 2014). Random access imaging via ultra-fast scanning or volumetric access with spatial light modulators represent alternative approaches (Katona et al., 2012; Reddy et al., 2008; Yang et al., 2016). These tools open up the possibility of capturing most, if not all of the neural activity of an entire layer in a single cortical area, or possibly even of two layers or more at a time. Further advances, such as splitting the scanning beam and multiplexing the volumetric imaging path, could, in theory, allow coverage of all neurons across all layers in a single area (Fig. 3C) (Cheng et al., 2011). Obtaining such comprehensive data sets on neurons imaged simultaneously across layers will potentially reveal types of transformations or forms of computations not readily apparent with sparser sampling techniques.
Just as sampling all cortical layers in a single cortical area extremely densely is surely advantageous, sampling a series of connected cortical areas simultaneously and at the same level of density could be even more revealing. For instance, dense sampling of L2/3 of V1 and multiple layers in V2 at the same time should unequivocally aid in better understanding how the output of L2/3 leads to new feature selectivity in V2. To achieve this, several groups have designed low magnification optical pathways that permit mesoscopic views of the cortex with single neuron resolution (Sofroniew et al., 2016; Stirman et al., 2016; Tsai et al., 2015). In the case of the mouse visual cortex, this places V1 and many of its associated V2-like downstream structures within the same accessible imaging field. For S1, one should be able to capture data in S1, S2, M1, and more frontal pre-motor areas all at the same time.
A major outstanding challenge for two-photon microscopy, however, is imaging the infragranular layers. Two photon imaging quality degrades with depth, and in species such as rats and primates that have thicker cortices than mice, L5 and L6 may be out of reach. Even in mice, imaging down to L6 is typically only possible when selectively labeling the deep layers, since out of focus multiphoton excitation of fluorophores in superficial layers leads to poor signal contrast. Approaches to overcome this challenge include implanting optics such as GRIN lenses and micro-prisms (Jung et al., 2004), the latter of which offers edge-on views of the cortical lamina (Andermann et al., 2013). These approaches, however, necessarily cause substantial tissue damage, a problem that may be avoided by employing three photon imaging, which can reach >1.2 mm into the rodent brain (Ouzounov et al., 2017). Taken together, some combination of these optical and technical advances will soon permit the measurement of neural activity at the requisite spatial and temporal scale to make major new discoveries in understanding the function of each layer in sensory coding.
Multiphoton optogenetics:
Although conventional optogenetic approaches readily provide potent and reversible control of genetically defined subtypes, one photon photostimulation with visible light offers little spatial precision due to limits on focusing light along the axial dimension and the scattering properties of cortical tissue. Multiphoton excitation, however, can provide tight spatial and temporal control of the photo-activated ensemble (Mardinly et al., 2018; Packer et al., 2012; Paluch-Siegler et al., 2015; Papagiakoumou et al., 2010; Pegard et al., 2017). In appropriate conditions, resolution can reach to the cellular level, permitting individual neurons to be photo-activated or photo-suppressed based not only on their genetic subtype, but also on their precise location and even based on their functional properties, such as their tuning to a specific stimulus feature (Packer et al., 2015; Rickgauer et al., 2014). With the introduction of computer-generated holography, the optical wavefront can be shaped in three dimensions, allowing for simultaneous targeting of neurons across layers. Over the past decade major strides have been made in engineering the multiphoton optical system and the opsin proteins themselves to offer potent control over large neural ensembles in the intact brain (Hernandez et al., 2016; Mardinly et al., 2018; Pegard et al., 2017; Prakash et al., 2012; Yang et al., 2018). Using this new technology makes it possible to execute a broad array of experiments targeting cortical layers, their cell types, or groups of cells across layers based on a common physiological property (Fig. 3D). For instance, it should now be possible to execute the experiment suggested by Hubel and Wiesel to address the mechanism of the synthesis of complex receptive fields in V1: one could map the receptive fields and orientation tuning of L4 neurons first with multiphoton calcium imaging, and then photo-stimulate an identified set of these neurons while monitoring activity in L2/3 neurons (Fig. 3E). This would allow investigators to literally execute the type of experiments called for by Hubel and Wiesel in their pioneering 1962 study.
More generally, multiphoton holographic optogenetics can be used to manipulate the activity of individual layers independently and all in the same animal. While prior studies have used one photon optogenetics and layer-specific Cre lines to activate or silence neurons specifically in a single layer, this does not allow the direct comparison of perturbations to each of the cortical layers in the same animal and under the same condition. It should also be noted that most layer-specific Cre lines that are available invariably label a subset of neurons in a given layer (Adesnik, 2018), leading to some ambiguity in how the data should be interpreted. With multiphoton optogenetics one can target the opsin to all cortical neurons in all layers, and use its superior spatial resolution to choose – moment to moment – which layer to perturb.
Multiphoton voltage imaging:
Although multiphoton calcium imaging has revolutionized population recording in the brain, it suffers several drawbacks, principally due to its reliance on calcium flux as an indirect measure of neural spiking. Calcium indicators, such as GCaMP6, provide only a highly temporally filtered measurement of spike rates changes (Chen et al., 2013), precluding precise assignment of spike times, and in many conditions may not report spike rates with a linear transfer function. One way to overcome these issues is to image voltage (and thus action potentials and potentially sub-threshold activity) directly. Achieving cellular resolution, millisecond precision voltage imaging of cortical layers would allow us to address major questions about cortical computation as it might be implemented by each layer. For example, correlated activity in cortical ensembles at fast time scales has been proposed to be a key means of encoding and computing aspects of sensory stimuli (Salinas and Sejnowski, 2001). Calcium imaging cannot properly report fine time scale correlations, and multi-electrode physiology cannot sample neuronal activity densely enough and does not as easily allow one to identify the genetic identity of the recorded cells (except via ‘opto-tagging’ which has various inherent drawbacks) (Lima et al., 2009). Multiphoton voltage imaging could, in principle, overcome these challenges and address the longstanding debate of the importance of spike timing in sensory coding in the cortex. Extending this type of analysis to data acquired simultaneously from neurons in multiple layers has the power to address whether and how spike timing, synchronization, and neuronal correlations contribute to the differential roles of cortical layers during sensory perception and behavior.
There are at least two major technical hurdles that must be overcome to achieve large scale, fast population voltage imaging: first, we require genetically targetable, fast voltage dyes with sufficient sensitivity to robustly report action potentials from many neurons simultaneously in vivo. Second, we require the optical system that can simultaneously sample the fluorescence of such a dye from large neuronal populations at a high enough rate to capture all emitted spikes. Recently, there has been dramatic progress in developing new, and ever more sensitive voltage dyes, including genetically encoded voltage indicators (GEVIs) or synthetic dyes that can be genetically targeted by ligation to a genetically expressed receptor. These have been reviewed elsewhere (Canepari et al., 2015; Luo et al., 2018b; Perron et al., 2009; Xu et al., 2017), so we focus on how existing voltage sensors meet the key attributes of the ideal voltage sensor for cracking cortical layers. The ideal sensor should be genetically targeted, show high voltage sensitivity, have fast kinetics, and be bright, photo-stable, and efficiently excited by multiphoton excitation. Of the available voltage dyes none meets all these requirements, but the rate of progress has been substantial. The ASAP and Arclight GEVIs show usable two photo cross-sections, but have comparatively less voltage sensitivity, slower kinetics, and faster photo-bleaching than the Archaerhodopsin (Arch) based GEVIs (Akemann et al., 2013; Chamberland et al., 2017; Xu et al., 2017). However, the latter show extremely low light sensitivity, precluding excitation in the multiphoton regime (Hochbaum et al., 2014). The Arch-based GEVIs can capture subthreshold responses with reasonable signal-to-noise (Piatkevich et al., 2018), and in restricted circumstances can be used in vivo with one photon illumination; however, their requirements for extremely high laser excitation powers have so far limited simultaneous in vivo imaging to less than a dozen neurons (Adam et al., 2018). Synthetic voltage dyes, which could be more rapidly customized to achieve the features of the ideal voltage sensor, represent a promising alternative path (Miller, 2016). They can be genetically targeted in a variety of ways, although the downside is that the dyes must be micro-injected into the brain, probably repeatedly for chronic imaging over several days. In light of these recent advances, we believe that a suitable voltage sensor for in vivo population voltage imaging is on the near horizon.
The second challenge – that of sampling the fluorescence of the voltage sensor across large populations of neurons at extremely high speeds, presents perhaps an even larger obstacle. Conventional raster scanning multiphoton imaging used with calcium dyes is 1-2 orders of magnitude too slow to sample voltage for capturing action potentials with millisecond precision. Random access multiphoton imaging could address this issue in part due to its much higher speed of beam steering (Bullen and Saggau, 1999), but in its current form might still not provide the sampling rates of the large populations ideally required, and will provide relatively low signal due to the very short dwell time of the laser on each neuron. A second alternative is to use temporally focused multiphoton holography – as described above for optogenetic stimulation – to illuminate a targeted ensemble of neurons and to capture their fluorescence emission on a high-speed camera (Bovetti et al., 2017; Tanese et al., 2017). The major advantage of such a scheme is that the target cells can be illuminated simultaneously, continuously, and across their entire somata, putatively providing large signals at a temporal rate only limited by the frame rate of the imaging sensor, which can reach into to 5-10 kHz range. The two challenges for such a system would be the limit of laser energy that can be delivered to the brain without causing heating or phototoxic damage, and computational challenges with de-mixing the scattered emission from the illuminated cells on the imaging sensor, particularly from densely labeled tissue where the somatic membranes of adjacent neurons can physically touch.
For any of these voltage imaging approaches, it seems likely that imaging a complete volume of tissue comparable to what can be achieved with two photon calcium imaging will be unachievable without truly dramatic improvements in voltage dyes. Instead, one can conceive of a hybrid approach, wherein cortical cells co-express a calcium indicator and a voltage dye. First, the response properties of large populations of cortical neurons can be probed via calcium flux, and then a restricted subset that show physiological features relevant to the experiment can be probed in a much more targeted fashion for voltage imaging. In the case of the sparse population codes in most cortical layers, such a hybrid approach should be particularly advantageous, as in many circumstances only a small fraction of the neurons in the imaging volume show significant responses to the stimulus set or behavioral task under study.
Transsynaptic viral tracing:
Despite the power of the aforementioned approaches, none provide direct access to populations of cortical neurons that are synaptically coupled. Enriched synaptic connectivity is arguably a crucial hallmark of the neural ensembles – within or across layers - that represent the fundamental units of cortical computation. Genetic labeling strategies that can drive fluorescent proteins, activity indicators or actuators in synaptically coupled ensembles thus represent a key tool for dissecting the laminar basis of cortical computation. Despite existing knowledge of interlaminar connectivity, transsynaptic labeling strategies can become a key tool in the cortical physiologists’ arsenal for correlating the structure and function of cortical layers. The primary strategy to achieve this is retrograde transsynaptic labeling with deletion mutant rabies viruses that can only jump one synapse (Wickersham et al., 2007). This approach has gained wide adoption for mapping brain inputs to specific cell types, and, when the tracing is initiated from just one neuron, permits direct imaging of the spatial distribution and even response properties of large numbers of cortical neurons that converge onto single cells (Marshel et al., 2010; Rancz et al., 2011; Wertz et al., 2015; Wickersham et al., 2007). More recent studies have described anterograde transsynaptic viral tracers that can complement the retrograde rabies strategy (Lo and Anderson, 2011; Zeng et al., 2017; Zingg et al., 2017). These approaches have at least two caveats that have largely remained unresolved: to what extent are the labeled neurons truly synaptically connected, and what bias exists in the labeling, as clearly not all presynaptic cells are labeled; in other words, what is the false positive and false negative rate of the tracing. This issue was original addressed by ground truth paired whole cell recordings in organotypic slices (Wickersham et al., 2007); however, cultured slices harbor highly aberrant wiring patterns. Similar validation in brain slices or intact brain tissue, although perhaps vastly more difficult to achieve, is essentially lacking, implying that those relying on this tool are doing so somewhat blindly. These are challenging questions to address, but would be worthwhile to answer so that studies employing transsynaptic tracers could interpreted in the proper light. As an approach, transsynaptic tracing has the potential to directly reveal and permit the perturbation of the neural ensembles at the core of cortical function with relatively simple tools. Long-range projection patterns can also be probed with a combination of single-cell labeling and whole brain fluorescence imaging and ‘MAPseq’, which identifies projections by sequencing axontargeted DNA barcodes (Han et al., 2018).
Genetically labeling correlated neuronal ensembles:
An attractive alternative strategy for labeling neurons that exhibit correlated activity in vivo is through genetic approaches that drive reporter genes under the control of activity-dependent promoters, principally the c-fos locus (Barth et al., 2004). This general strategy allows one to express a fluorescent protein, activity dye or actuator in a large, brain wide ensemble of neurons that exhibit c-fos promoter activity in some restricted time frame. This tool could be particularly useful for labeling trans-laminar ensembles united by a common in vivo function for further functional imaging or pertrubation. Originally this strategy was done with GFP (Barth et al., 2004), but was then extended to driver systems including the tetracycline transactivator (tTa), Cre recombinase, inducible Cre, and, most recently, the avian receptor for pseudotyped viruses (Guenthner et al., 2013; Reijmers et al., 2007; Sakurai et al., 2016). It has been used to great effect to label neuronal ensembles in various brain nuclei that might encode memories because they were co-active during memory encoding, retrieval, or consolidation (Liu et al., 2012; Reijmers et al., 2007). The main caveats to this approach are that the c-fos promoter shows variable basal levels of activity according to cell type and cortical layer, and the relationships between neuronal activity, c-fos promoter activity, and downstream indicator/actuator expression remain largely undefined and may depend on cell type and other contingencies. Therefore, like viral transsynaptic labeling, identifying neural ensembles with activity-dependent transcription represents a particularly powerful tool in the neuroscientists’ toolbox that requires more thorough validation. Once further validated or suitably refined, however, it could likewise lead to transformative advances in our understanding of cortical layer function.
An alternative to c-fos-based labeling is turning on reporter transgenes with light. Such strategies include photo-activatable recombinases (including Cre and Cas9) that could allow expression of genetically encoded indicators or actuators with exquisite spatial precision when combined with multiphoton activation of the recombinase (Schindler et al., 2015). This could permit a range of new types of experiments not possible with existing approaches. For example, an experimenter can identify a functionally correlated ensemble of neurons with two photon imaging and then photo-label these neurons to drive expression of ChR2. Subsequently, these neurons can be stimulated with simple, fiber-optic based one photon photo-stimulation in the freely behaving condition. One could selectively photo-label neurons in just one cortical layers and compare the effects of perturbing their activity with a similar ensemble composed of neurons distributed across several layers. Although the pattern of optical stimulation would not have the specificity of multiphoton optogenetics, the target population would be just as specific and this approach would obviate the need for head-fixation or the complex hardware required for holographic optogenetics.
Cell typing by in situ RNA profiling:
All cortical layers are composed of multiple cell types that have been differentiated by their molecular markers, morphology, intrinsic physiology, and local and long-range connectivity. To understand how layers, by virtue of containing a discrete set of identifiable circuit elements, differentially contribute to cortical computation we need to determine how each cell type in each layer differentially encodes sensory stimuli or correlates with behavior. Indeed, abundant evidence indicates that distinct cell classes exhibit strikingly different responses to varying stimulus features, brain states, or behavioral choices. Furthermore, we would also like to determine the complete synaptic wiring diagram between all of these cell types in each layer, which is necessary for both conceptual and quantitative models of cortical circuit dynamics. While an absolute parsing of the number and distinction of cell types is a matter of ongoing and fruitful debate, the more important question is how one can identify the myriad cell types of neurons in the same tissue that can be probed in vivo. The most popular approach so far has been to genetically label a specific cardinal type using transgenic lines and identify them in vivo with two photon imaging or electrophysiologically with ‘opto-tagging’. The downside is that the number of cell types that can be identified in the same animal is limited by the number of independent reporter transgenes (with two or three being a typical practical limit). An alternative approach is to register in vivo imaged tissue with post-hoc immunostaining for multiple cell markers (typically up to three). This has been used to correlate sensory responses with three of the major markers for cortical GABAergic neurons (Kerlin et al., 2010). However, immunostaining is limited by both the number of available antibodies and the number of separable color channels. In situ RNA profiling is an exciting new technology for overcoming this limitation. This emergent technology (termed ‘seqFISH’ or ‘MerFISH’) uses in situ temporal multiplexed hybridization of a user-defined set of DNA barcodes to quantitatively identify the set of mRNAs each neuron in a tissue section expresses (Chen et al., 2015; Lubeck et al., 2014). Importantly, the appropriate set of barcodes to differentiate cortical neurons can be drawn from recent single cell RNA sequencing data sets of cortical neurons (Tasic et al., 2016). This approach was recently validated in hippocampal brain slices (Shah et al., 2016).
Based on this technology, we can propose two types of experiments to obtain powerful new datasets on the in vivo physiology and synaptic connectivity of all of the transcriptionally defined sub-classes that compose each cortical layer. First, one could sequentially image the activity of a volume of cortex in vivo, and then assign each imaged neuron to a specific transcriptionally defined cell class by post-hoc seqFISH and careful image registration (Fig. 3F). In one fell swoop, one could thus begin to define the physiological responses of >40 cell classes for highly quantitative, within tissue comparison. Second, one could map the monosynaptic connectivity among all the cortical cell types composing each layer by combining singe cell resolution multiphoton optogenetic mapping with patch clamp electrophysiology, ideally in vivo to avoid cutting any projections. In this latter scheme, one labels all cortical neurons with a soma-targeted opsin and then systematically maps presynaptic neurons to a single target patched neuron (Baker et al., 2016). Subsequently, one probes the mapped tissue with seqFISH. This then provides the transcriptional identity of both the target neurons and all of its positively identified presynaptic partners. Perhaps a particularly exciting possibility would be to combine all of these steps into a single pipeline: imaging a volume of cortical tissue in vivo to obtain physiological responses across layers, then mapping the monosynaptic inputs to a small number of these neurons with patch clamp and multiphoton stimulation, and finally assigning cell types with seqFISH. Such a scheme would allow physiologists to begin to directly correlate synaptic connectivity with the in vivo physiology of all the major subtypes of cortical neurons. It could provide, for example, a nearly complete picture for how complex cell receptive fields might emerge in L2/3 by identifying all of a complex cell’s presynaptic neurons and quantifying their synaptic strengths. More generally, it can take a type of transformation that is only observable across cortical layers and track it back to the presynaptic pool of neurons that should generate it, and provide much of the needed data to build a highly realistic model of the computation.
Neural data science:
Continued advances in data analysis, experimental design, modeling and theory are critical for harnessing and fully exploiting the new experimental techniques available for observing and perturbing cortical circuits. These topics have been addressed elsewhere (Paninski and Cunningham, 2018), so we will summarize the most relevant points. As the number of neurons that can be simultaneously recorded continues to grow exponentially, neuroscientists must adapt their analytical techniques. In addition to the data-engineering challenges associated with collecting large datasets, neural data typically requires extensive preprocessing: multi-electrode array data requires spike-sorting, and calcium imaging datasets require denoising/motion correction, demixing to isolate signals from individual neurons, and temporal deconvolution to recover estimates of spiking activity. These processes have historically been expensive in terms of time and labor as they have typically involved some level of manual involvement of the experimenter for quality control. An issue that must soon be addressed is that the pre-processing steps used by different groups varies considerably; as data sets grow, broadly accepted standards will become increasingly important for rigorously comparing the outcomes of different studies (Harris et al., 2016). One potential solution is the increasing development of fully automated pre-processing (such as spike sorting and calcium source separation) that requires minimal human input (Chung et al., 2017; Dhawale et al., 2017; Pnevmatikakis et al., 2016). Not only is such automation necessary for handling the greatly expanding data sets, they could ensure that different groups would extract the same neural signals given the same data, promoting reproducibility. The challenge is that these tools, just like the experimental approaches listed above, must be rigorously and extensively validated with technically challenging ground truth experiments involving simultaneous single cell electrophysiology (Harris et al., 2000). Thankfully, the development and adoption of new analysis tools has increasingly automated these steps, which eases a key bottleneck for data acquisition and should improve the quality and reproducibility of preprocessing. The scalability of these methods to ever larger populations and to longer timescales is also a priority.
Closed-loop experimental design:
As increasingly sophisticated manipulations become available to experimenters, the question of how to optimally design these manipulations so as to definitively test key hypotheses becomes increasingly important and challenging. For instance, the high spatial and temporal resolution of multiphoton optogenetics, where different ensembles of neurons can be photo-activated every few milliseconds within a large volume of thousands of neurons, presents a combinatorial explosion of different possible patterns with which to perturb the system. How the experimenter chooses among this nearly limitless repertoire of stimulation patterns to gain insight into the circuit is far from trivial. One useful direction is generating stimulus patterns that mimic endogenous dynamics, and then parametrically altering key parameters of these patterns such as the distribution of firing rates, spike times, and neuronal correlations.
A companion problem, however, is that perturbations of highly recurrent networks, such as cortical circuits, are just as difficult to interpret. Indeed, the canonical model has largely failed to predict the results of even simple, relatively non-specific manipulations of individual layers. Theoretically, highly recurrent networks can be dissected via simultaneous manipulations of multiple components. Yet even in very simple cases this is extremely challenging due to the combinatorial explosion of perturbations that might need to be performed (Kumar et al., 2013). Tools like multiphoton optogenetics provide a potential solution to this problem, but arguably an experimenter, no matter how knowledgeable, cannot predict by intuition alone which perturbations to implement to overcome these challenges.
Fortunately, new tools are making it increasingly possible to extract meaningful data in real time (Chung et al., 2017; Friedrich et al., 2017), enabling closed-loop experiments based on the online analysis of neural activity. This has two major implications: first, closed-loop design could address many of the challenges that arise as experiments become increasingly sophisticated. Closed-loop design will allow for the exploration of otherwise intractable spaces, such as mapping thousands of synaptic connections or estimating higher-order terms of nonlinear stimulus-response functions (Bolus et al., 2018; DiMattina and Zhang, 2013; Grosenick et al., 2015). Second, closed-loop design should enable whole new classes of experiments that could provide insight into cortical dynamics not obtainable with more conventional approaches. For instance, many recent studies of cortical computation have used dimensionality reduction and dynamical systems based approaches to identify latent structure in the activity of cortical populations. A particularly exciting direction for future studies will be to identify intrinsic manifolds in this space online and then perform optogenetic perturbations with respect to this structure (Jazayeri and Afraz, 2017) (e.g. compare effects of perturbations that allow the population to remain within the intrinsic manifold versus ones that push the population outside of it). More generally, while these analyses have begun to point to interesting differences between cortical areas (Russo et al., 2018; Seely et al., 2016), they have thus far been used in a manner that is largely agnostic to the layer and cell-type of the recorded neurons. An important goal for future work will be to identify how the structure of cortical population dynamics relates to the anatomical structure of the underlying microcircuits. Ultimately, we predict that closed loop design will allow the field to shift from the hammer toward the scalpel, forgoing manipulations that dramatically alter the activity of many neurons for ones that precisely perturb smaller ensembles while leaving the circuit as a whole within its normal regime of activity.
Missing tools in the cortical physiologists’ arsenal:
Despite the advent of new technical approaches for monitoring and manipulating neural activity, there is one key tool that is noticeably lacking in the neurophysiologist’s toolbox: the ability for synapse-specific silencing between user-defined pre- and post-synaptic neurons. Such a tool would be revolutionary for cracking the function of layers and their component cell types. Consider the challenge of understanding how L4 impacts other layers during sensory coding. Since L4 neurons target excitatory neurons in all other layers (including in L4 itself), the data obtained from simply suppressing L4 spiking does not allow one to disambiguate which of these synaptic connections was causally involved in the observed effect, whether it be on neural activity or behavior. If, instead, one could selectively silence just one of its output pathways (e.g., L4 to L2/3 or L4 to L5) one could obtain a more detailed understanding of its role in translaminar computation. Some investigators have employed synaptic terminal silencing with inhibitory opsins; the efficacy of this approach appears spotty and is difficult to confirm, and it is not useful for cracking cortical circuits in any case; intracortical circuits are too spatially intermingled. Instead, what is needed is an approach that is conceptually analogous to the ‘GRASP’ technology which allows one to target a split fluorescent reporter to two genetically defined classes of neurons (Feinberg et al., 2008); if they contact one another the fluorescent protein is reconstituted. Since splitting and reconstituting an integral membrane protein like an opsin seems implausible, alternative schemes must be developed. If this technical obstacle can be overcome, a whole new range of critical optogenetic experiments could be performed to dissect the function of each intracortical pathway.
Conclusion
With the advent of new technologies for large-scale recording and manipulation of neural activity across cortical layers, coupled with the possibility of identifying all of the cardinal cell types that compose each layer, we are on the threshold of obtaining the new data that could finally address how cortical layers differentially contribute to cortical computation. Mounting a holistic attack on cortical circuits with these new approaches, particularly in the appropriate behavioral paradigms, promises to provide fundamental new mechanistic insight into cortical circuit structure and function. Ultimately the proximal goal should be to distinguish between the competing conceptual frameworks that attempt to explain how cortical circuits generate perception. Probing the synthetic model requires us to reveal new types of transformations of sensory coding across and between the layers of each cortical area within the sensory hierarchy, if and when they exist. Ultimately, it is necessary to provide a complete picture for how complex feature selectivity (such as for faces) is synthesized across this hierarchy and how this depends on the activity and the connectivity among the cell types at each stage. Probing the inferential model requires the direct observation of neural activity that encodes predictions, prediction errors, and the sensory or motor memories that collectively lead to the moment-to-moment comparison between a generative model of the world and the sensory data that arrives via the sense organs.
Acknowledgements
The authors thank Dan Feldman, Elena Ryapolova-Webb and Benjamin Shababo for a critical reading of the manuscript, as well as New Scale Technologies for graphics, and Vittorino Lanzio and Stefano Cabrini for images of multifunctional electrodes. This work was funded by NINDS grant DP2NS087725-01, NEI grant R01EY023756-01, and the New York Stem Cell Foundation. H.A. is a New York Stem Cell Robertson Investigator.
Footnotes
Declaration of interests
H.A. has a patent related to this work. 3D Sparse Holographic Temporal focusing, 2016, L. Waller, N. Pegard, and H. Adesnik, Provisional Patent Application #62-429,017
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam Y, Kim JJ, Lou S, Zhao Y, Brinks D, Wu H, Mostajo-Radji MA, Kheifets S, Parot V, Chettih Sv et al. (2018). All-optical electrophysiology reveals brain-state dependent changes in hippocampal subthreshold dynamics and excitability. bioRxiv. [Google Scholar]
- Adesnik H (2018). Layer-specific excitation/inhibition balances during neuronal synchronization in the visual cortex. The Journal of physiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adesnik H, and Scanziani M (2010). Lateral competition for cortical space by layer-specific horizontal circuits. Nature 464, 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akemann W, Sasaki M, Mutoh H, Imamura T, Honkura N, and Knopfel T (2013). Two-photon voltage imaging using a genetically encoded voltage indicator. Sci Rep-Uk 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, and Martinez LM (1998). Functional connectivity between simple cells and complex cells in cat striate cortex. Nature neuroscience 1, 395–403. [DOI] [PubMed] [Google Scholar]
- Andermann ML, Gilfoy NB, Goldey GJ, Sachdev RNS, Wolfel M, McCormick DA, Reid RC, and Levene MJ (2013). Chronic Cellular Imaging of Entire Cortical Columns in Awake Mice Using Microprisms. Neuron 80, 900–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong-James M, Fox K, and Das-Gupta A (1992). Flow of excitation within rat barrel cortex on striking a single vibrissa. Journal of neurophysiology 68, 1345–1358. [DOI] [PubMed] [Google Scholar]
- Baker CA, Elyadat YM, Parra A, and Bolton MM (2016). Cellular resolution circuit mapping with temporal-focused excitation of soma targeted channelrhodopsin. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth AL, Gerkin RC, and Dean KL (2004). Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse. The Journal of neuroscience : the official journal of the Society for Neuroscience 24, 6466–6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos AM, Usrey WM, Adams RA, Mangun GR, Fries P, and Friston KJ (2012). Canonical Microcircuits for Predictive Coding. Neuron 76, 695–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binzegger T, Douglas RJ, and Martin KA (2004). A quantitative map of the circuit of cat primary visual cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience 24, 8441–8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolus MF, Willats AA, Whitmire CJ, Rozell CJ, and Stanley GB (2018). Design strategies for dynamic closed-loop optogenetic neurocontrol in vivo. J Neural Eng 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolz J, and Gilbert CD (1986). Generation of end-inhibition in the visual cortex via interlaminar connections. Nature 320, 362–365. [DOI] [PubMed] [Google Scholar]
- Bortone DS, Olsen SR, and Scanziani M (2014). Translaminar inhibitory cells recruited by layer 6 corticothalamic neurons suppress visual cortex. Neuron 82, 474–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovetti S, Moretti C, Zucca S, Dal Maschio M, Bonifazi P, and Fellin T (2017). Simultaneous high-speed imaging and optogenetic inhibition in the intact mouse brain (vol 7, 2017). Sci Rep-Uk 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecht M (2017). The Body Model Theory of Somatosensory Cortex. Neuron 94, 985–992. [DOI] [PubMed] [Google Scholar]
- Brecht M, Roth A, and Sakmann B (2003). Dynamic receptive fields of reconstructed pyramidal cells in layers 3 and 2 of rat somatosensory barrel cortex. J Physiol-London 553, 243–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs F, and Callaway EM (2001). Layer-specific input to distinct cell types in layer 6 of monkey primary visual cortex. Journal of Neuroscience 21, 3600–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce C, Desimone R, and Gross CG (1981). Visual Properties of Neurons in a Polysensory Area in Superior Temporal Sulcus of the Macaque. Journal of neurophysiology 46, 369–384. [DOI] [PubMed] [Google Scholar]
- Bullen A, and Saggau P (1999). High-speed, random-access fluorescence microscopy: II. Fast quantitative measurements with voltage-sensitive dyes. Biophysical journal 76, 2272–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese A, and Woolley SMN (2015). Coding principles of the canonical cortical microcircuit in the avian brain. Proceedings of the National Academy of Sciences of the United States of America 112, 3517–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway EM (1998). Local circuits in primary visual cortex of the macaque monkey. Annual review of neuroscience 21, 47–74. [DOI] [PubMed] [Google Scholar]
- Canales A, Jia XT, Froriep UP, Koppes RA, Tringides CM, Selvidge J, Lu C, Hou C, Wei L, Fink, Yv et al. (2015). Multifunctional fibers for simultaneous optical, electrical and chemical interrogation of neural circuits in vivo. Nat Biotechnol 33, 277–284. [DOI] [PubMed] [Google Scholar]
- Canepari M, Bernus O, and Zecevic D (2015). Membrane Potential Imaging in the Nervous System and Heart Preface. Membrane Potential Imaging in the Nervous System and Heart 859, V–Vi. [Google Scholar]
- Chamberland S, Yang HH, Pan MM, Evans SW, Guan SH, Chavarha M, Yang Y, Salesse C, Wu HD, Wu JC, et al. (2017). Fast two-photon imaging of subcellular voltage dynamics in neuronal tissue with genetically encoded indicators. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, and Tsao DY (2017). The Code for Facial Identity in the Primate Brain. Cell 169, 1013–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang SY, and Zhuang XW (2015). Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Goncalves JT, Golshani P, Arisaka K, and Portera-Cailliau C (2011). Simultaneous two-photon calcium imaging at different depths with spatiotemporal multiplexing. Nature methods 8, 139–U158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JE, Magland JF, Barnett AH, Tolosa VM, Tooker AC, Lee KY, Shah KG, Felix SH, Frank LM, and Greengard LF (2017). A Fully Automated Approach to Spike Sorting. Neuron 95, 1381–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchland MM, Cunningham JP, Kaufman MT, Foster JD, Nuyujukian P, Ryu SI, and Shenoy KV (2012). Neural population dynamics during reaching. Nature 487, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinople CM, and Bruno RM (2013). Deep cortical layers are activated directly by thalamus. Science 340, 1591–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruikshank SJ, Urabe H, Nurmikko AV, and Connors BW (2010). Pathway-Specific Feedforward Circuits between Thalamus and Neocortex Revealed by Selective Optical Stimulation of Axons. Neuron 65, 230–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csicsvari J, Henze DA, Jamieson B, Harris KD, Sirota A, Bartho P, Wise KD, and Buzsaki G (2003). Massively parallel recording of unit and local field potentials with silicon-based electrodes. Journal of neurophysiology 90, 1314–1323. [DOI] [PubMed] [Google Scholar]
- D'Souza RD, and Burkhalter A (2017). A Laminar Organization for Selective Cortico-Cortical Communication. Frontiers in neuroanatomy 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch D, Pietr M, Knutsen PM, Ahissar E, and Schneidman E (2012). Fast Feedback in Active Sensing: Touch-Induced Changes to Whisker-Object Interaction. PloS one 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawale AK, Poddar R, Wolff SBE, Normand VA, Kopelowitz E, and Olveczky BP (2017). Automated long-term recording and analysis of neural activity in behaving animals. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMattina C, and Zhang KC (2013). Adaptive stimulus optimization for sensory systems neuroscience. Frontiers in neural circuits 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas RJ, and Martin KA (1991). A functional microcircuit for cat visual cortex. The Journal of physiology 440, 735–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas RJ, and Martin KA (2004). Neuronal circuits of the neocortex. Annual review of neuroscience 27, 419–451. [DOI] [PubMed] [Google Scholar]
- Emiliani V, Cohen AE, Deisseroth K, and Hausser M (2015). All-Optical Interrogation of Neural Circuits. Journal of Neuroscience 35, 13917–13926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg EH, VanHoven MK, Bendesky A, Wang G, Fetter RD, Shen K, and Bargmannl CI (2008). GFP reconstitution across synaptic partners (GRASP) defines cell contacts and Synapses in living nervous systems. Neuron 57, 353–363. [DOI] [PubMed] [Google Scholar]
- Feldmeyer D (2012). Excitatory neuronal connectivity in the barrel cortex. Frontiers in neuroanatomy 6, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felleman DJ, and Van Essen DC (1991). Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex 1, 1–47. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick D, Itoh K, and Diamond IT (1983). The Laminar Organization of the Lateral Geniculate-Body and the Striate Cortex in the Squirrel-Monkey (Saimiri-Sciureus). Journal of Neuroscience 3, 673–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich J, Zhou PC, and Paninski L (2017). Fast online deconvolution of calcium imaging data. PLoS computational biology 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego JA, Perich MG, Miller LE, and Solla SA (2017). Neural Manifolds for the Control of Movement. Neuron 94, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamanut R, Kennedy H, Toroczkai Z, Ercsey-Ravasz M, Van Essen DC, Knoblauch K, and Burkhalter A (2018). The Mouse Cortical Connectome, Characterized by an Ultra-Dense Cortical Graph, Maintains Specificity by Distinct Connectivity Profiles. Neuron 97, 698–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert CD (1983). Microcircuitry of the Visual-Cortex. Annual review of neuroscience 6, 217–247. [DOI] [PubMed] [Google Scholar]
- Grosenick L, Marshel JH, and Deisseroth K (2015). Closed-Loop and Activity-Guided Optogenetic Control. Neuron 86, 106–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross CG, Rocha-Miranda CE, and Bender DB (1972). Visual properties of neurons in inferotemporal cortex of the Macaque. Journal of neurophysiology 35, 96–111. [DOI] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, and Luo L (2013). Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron 78, 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, and Staiger JF (2017). The Functioning of a Cortex without Layers. Frontiers in neuroanatomy 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YY, Kebschull JM, Campbell RAA, Cowan D, Imhof F, Zador AM, and Mrsic-Flogel TD (2018). The logic of single-cell projections from visual cortex. Nature 556, 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KD, Henze DA, Csicsvari J, Hirase H, and Buzsaki G (2000). Accuracy of tetrode spike separation as determined by simultaneous intracellular and extracellular measurements. Journal of neurophysiology 84, 401–414. [DOI] [PubMed] [Google Scholar]
- Harris KD, Quiroga RQ, Freeman J, and Smith SL (2016). Improving data quality in neuronal population recordings. Nature neuroscience 19, 1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KD, and Shepherd GM (2015). The neocortical circuit: themes and variations. Nature neuroscience 18, 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez O, Papagiakoumou E, Tanese D, Fidelin K, Wyart C, and Emiliani V (2016). Three-dimensional spatiotemporal focusing of holographic patterns (vol 7, pg 11928, 2016). Nature communications 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochbaum DR, Zhao Y, Farhi SL, Klapoetke N, Werley CA, Kapoor V, Zou P, Kralj JM, Maclaurin D, Smedemark-Margulies N, et al. (2014). All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nature methods 11, 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman KP, and Stone J (1971). Conduction velocity of afferents to cat visual cortex: a correlation with cortical receptive field properties. Brain research 32, 460–466. [DOI] [PubMed] [Google Scholar]
- Homann J, Koay SA, Glidden AM, Tank DW, and Berry MJ (2017). Predictive Coding of Novel versus Familiar Stimuli in the Primary Visual Cortex. bioRxiv. [Google Scholar]
- Hubel DH, and Wiesel TN (1959). Receptive fields of single neurones in the cat's striate cortex. The Journal of physiology 148, 574–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubel DH, and Wiesel TN (1962). Receptive fields, binocular interaction and functional architecture in the cat's visual cortex. The Journal of physiology 160, 106–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim LA, Mesik L, Ji YY, Fang Q, Li HF, Li YT, Zingg B, Zhang LI, and Tao HW (2016). Cross-Modality Sharpening of Visual Cortical Processing through Layer-1-Mediated Inhibition and Disinhibition. Neuron 89, 1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iurilli G, Ghezzi D, Olcese U, Lassi G, Nazzaro C, Tonini R, Tucci V, Benfenati F, and Medini P (2012). Sound-Driven Synaptic Inhibition in Primary Visual Cortex. Neuron 73, 814–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri M, and Afraz A (2017). Navigating the Neural Space in Search of the Neural Code. Neuron 93, 1003–1014. [DOI] [PubMed] [Google Scholar]
- Juavinett AL, Erlich JC, and Churchland AK (2018). Decision-making behaviors: weighing ethology, complexity, and sensorimotor compatibility. Current opinion in neurobiology 49, 42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JJ, Steinmetz NA, Siegle JH, Denman DJ, Bauza M, Barbarits B, Lee AK, Anastassiou CA, Andrei A, Aydin C, et al. (2017). Fully integrated silicon probes for high-density recording of neural activity. Nature 551, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JC, Mehta AD, Aksay E, Stepnoski R, and Schnitzer MJ (2004). In vivo mammalian brain Imaging using one- and two-photon fluorescence microendoscopy. Journal of neurophysiology 92, 3121–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katona G, Szalay G, Maak P, Kaszas A, Veress M, Hillier D, Chiovini B, Vizi ES, Roska B, and Rozsa B (2012). Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nature methods 9, 201–208. [DOI] [PubMed] [Google Scholar]
- Keller GB, Bonhoeffer T, and Hubener M (2012). Sensorimotor Mismatch Signals in Primary Visual Cortex of the Behaving Mouse. Neuron 74, 809–815. [DOI] [PubMed] [Google Scholar]
- Kerlin AM, Andermann ML, Berezovskii VK, and Reid RC (2010). Broadly Tuned Response Properties of Diverse Inhibitory Neuron Subtypes in Mouse Visual Cortex. Neuron 67, 858–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Matney CJ, Blankenship A, Hestrin S, and Brown SP (2014). Layer 6 Corticothalamic Neurons Activate a Cortical Output Layer, Layer 5a. Journal of Neuroscience 34, 9656–9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnischtzke AK, Fanselow EE, and Simons DJ (2016). Target-specific M1 inputs to infragranular S1 pyramidal neurons. Journal of neurophysiology 116, 1261–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer JW, Ghazanfar AA, Gomez-Marin A, MacIver MA, and Poeppel D (2017). Neuroscience Needs Behavior: Correcting a Reductionist Bias. Neuron 93, 480–490. [DOI] [PubMed] [Google Scholar]
- Kumar A, Vlachos I, Aertsen A, and Boucsein C (2013). Challenges of understanding brain function by selective modulation of neuronal subpopulations. Trends in neurosciences 36, 579–586. [DOI] [PubMed] [Google Scholar]
- Larkum ME, Petro LS, Sachdev RNS, and Muckli L (2018). A Perspective on Cortical Layering and Layer-Spanning Neuronal Elements. Frontiers in neuroanatomy 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort S, Tomm C, Floyd Sarria JC, and Petersen CC (2009). The excitatory neuronal network of the C2 barrel column in mouse primary somatosensory cortex. Neuron 61, 301–316. [DOI] [PubMed] [Google Scholar]
- Lima SQ, Hromadka T, Znamenskiy P, and Zador AM (2009). PINP: a new method of tagging neuronal populations for identification during in vivo electrophysiological recording. PloS one 4, e6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, She L, Chen M, Liu TY, Lu HDD, Dan Y, and Poo MM (2016). Spatial structure of neuronal receptive field in awake monkey secondary visual cortex (V2). Proceedings of the National Academy of Sciences of the United States of America 113, 1913–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, and Tonegawa S (2012). Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature 484, 381–U415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo L, and Anderson DJ (2011). A Cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron 72, 938–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu RW, Sun WZ, Liang YJ, Kerlin A, Bierfeld J, Seelig JD, Wilson DE, Scholl B, Mohar B, Tanimoto M, et al. (2017). Video-rate volumetric functional imaging of the brain at synaptic resolution. Nature neuroscience 20, 620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, and Cai L (2014). Single-cell in situ RNA profiling by sequential hybridization. Nature methods 11, 360–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo LQ, Callaway EM, and Svoboda K (2018a). Genetic Dissection of Neural Circuits: A Decade of Progress. Neuron 98, 256–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo LQ, Callaway EM, and Svoboda K (2018b). Genetic Dissection of Neural Circuits: A Decade of Progress (vol 98, pg 256, 2018). Neuron 98, 865–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma WJ, Beck JM, Latham PE, and Pouget A (2006). Bayesian inference with probabilistic population codes. Nature neuroscience 9, 1432–1438. [DOI] [PubMed] [Google Scholar]
- Malpeli JG (1983). Activity of cells in area 17 of the cat in absence of input from layer a of lateral geniculate nucleus. Journal of neurophysiology 49, 595–610. [DOI] [PubMed] [Google Scholar]
- Manita S, Suzuki T, Homma C, Matsumoto T, Odagawa M, Yamada K, Ota K, Matsubara B, Inutsuka A, Sato M, et al. (2015). A Top-Down Cortical Circuit for Accurate Sensory Perception. Neuron 86, 1304–1316. [DOI] [PubMed] [Google Scholar]
- Mardinly AR, Oldenburg IA, Pegard NC, Sridharan S, Lyall EH, Chesnov K, Brohawn S, Waller L, and Adesnik H (2018). Precise multimodal optical control of neural ensemble activity. Nature neuroscience. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marr D (2010). Vision : a computational investigation into the human representation and processing of visual information (Cambridge, Mass.: MIT Press; ). [Google Scholar]
- Marshel JH, Mori T, Nielsen KJ, and Callaway EM (2010). Targeting single neuronal networks for gene expression and cell labeling in vivo. Neuron 67, 562–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez LM, and Alonso JM (2001). Construction of complex receptive fields in cat primary visual cortex. Neuron 32, 515–525. [DOI] [PubMed] [Google Scholar]
- Martinez LM, Wang Q, Reid RC, Pillai C, Alonso JM, Sommer FT, and Hirsch JA (2005). Receptive field structure varies with layer in the primary visual cortex. Nature neuroscience 8, 372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyas F, Sreenivasan V, Marbach F, Wacongne C, Barsy B, Mateo C, Aronoff R, and Petersen CCH (2010). Motor Control by Sensory Cortex. Science 330, 1240–1243. [DOI] [PubMed] [Google Scholar]
- Meyer HS, Wimmer VC, Hemberger M, Bruno RM, de Kock CPJ, Frick A, Sakmann B, and Helmstaedter M (2010). Cell Type-Specific Thalamic Innervation in a Column of Rat Vibrissal Cortex. Cerebral Cortex 20, 2287–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels JA, Dann B, and Scherberger H (2016). Neural Population Dynamics during Reaching Are Better Explained by a Dynamical System than Representational Tuning. PLoS computational biology 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignard M, and Malpeli JG (1991). Paths of Information-Flow through Visual-Cortex. Science 251, 1249–1251. [DOI] [PubMed] [Google Scholar]
- Miller EW (2016). Small molecule fluorescent voltage indicators for studying membrane potential. Curr Opin Chem Biol 33, 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SR, Bortone DS, Adesnik H, and Scanziani M (2012). Gain control by layer six in cortical circuits of vision. Nature 483, 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otchy TM, Wolff SBE, Rhee JY, Pehlevan C, Kawai R, Kempf A, Gobes SMH, and Olveczky BP (2015). Acute off-target effects of neural circuit manipulations. Nature 528, 358–363. [DOI] [PubMed] [Google Scholar]
- Ouzounov DG, Wang TY, Wang MR, Feng DD, Horton NG, Cruz-Hernandez JC, Cheng YT, Reimer J, Tolias AS, Nishimura, Nv et al. (2017). In vivo three-photon imaging of activity of GCaMP6-labeled neurons deep in intact mouse brain. Nature methods 14, 388–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, Peterka DS, Hirtz JJ, Prakash R, Deisseroth K, and Yuste R (2012). Two-photon optogenetics of dendritic spines and neural circuits. Nature methods 9, 1202–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, Russell LE, Dalgleish HWP, and Hausser M (2015). Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nature methods 12, 140–U186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluch-Siegler S, Mayblum T, Dana H, Brosh I, Gefen I, and Shoham S (2015). All-optical bidirectional neural interfacing using hybrid multiphoton holographic optogenetic stimulation. Neurophotonics 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paninski L, and Cunningham JP (2018). Neural data science: accelerating the experiment- analysis-theory cycle in large-scale neuroscience. Current opinion in neurobiology 50, 232–241. [DOI] [PubMed] [Google Scholar]
- Papagiakoumou E, Anselmi F, Begue A, de Sars V, Gluckstad J, Isacoff EY, and Emiliani V (2010). Scanless two-photon excitation of channelrhodopsin-2. Nature methods 7, 848–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegard NC, Mardinly AR, Oldenburg IA, Sridharan S, Waller L, and Adesnik H (2017). Three-dimensional scanless holographic optogenetics with temporal focusing (3D-SHOT). Nature communications 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron A, Mutoh H, Akemann W, Gautam SG, Dimitrov D, Iwamoto Y, and Knopfel T (2009). Second and third generation voltage-sensitive fluorescent proteins for monitoring membrane potential. Front Mol Neurosci 2, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petreanu L, Mao T, Sternson SM, and Svoboda K (2009). The subcellular organization of neocortical excitatory connections. Nature 457, 1142–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatkevich KD, Jung EE, Straub C, Linghu CY, Park D, Suk HJ, Hochbaum DR, Goodwin D, Pnevmatikakis E, Pak N, et al. (2018). A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nat Chem Biol 14, 352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta S, Naka A, Veit J, Telian G, Yao L, Hakim R, Taylor D, and Adesnik H (2015). A direct translaminar inhibitory circuit tunes cortical output. Nature neuroscience 18, 1631–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pnevmatikakis EA, Soudry D, Gao Y, Machado TA, Merel J, Pfau D, Reardon T, Mu Y, Lacefield C, Yang W, et al. (2016). Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron 89, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouget A, Dayan P, and Zemel R (2000). Information processing with population codes. Nature Reviews Neuroscience 1, 125–132. [DOI] [PubMed] [Google Scholar]
- Prakash R, Yizhar O, Grewe B, Ramakrishnan C, Wang N, Goshen I, Packer AM, Peterka DS, Yuste R, Schnitzer MJ, et al. (2012). Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nature methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevedel R, Verhoef AJ, Pernia-Andrade AJ, Weisenburger S, Huang BS, Nobauer T, Fernandez A, Delcour JE, Golshani P, Baltuska A, et al. (2016). Fast volumetric calcium imaging across multiple cortical layers using sculpted light. Nature methods 13, 1021–U1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancz EA, Franks KM, Schwarz MK, Pichler B, Schaefer AT, and Margrie TW (2011). Transfection via whole-cell recording in vivo: bridging single-cell physiology, genetics and connectomics. Nature neuroscience 14, 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao RPN, and Ballard DH (1999). Predictive coding in the visual cortex: a functional interpretation of some extra-classical receptive-field effects. Nature neuroscience 2, 79–87. [DOI] [PubMed] [Google Scholar]
- Reddy GD, Kelleher K, Fink R, and Saggau P (2008). Three-dimensional random access multiphoton microscopy for functional imaging of neuronal activity. Nature neuroscience 11, 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijmers LG, Perkins BL, Matsuo N, and Mayford M (2007). Localization of a stable neural correlate of associative memory. Science 317, 1230–1233. [DOI] [PubMed] [Google Scholar]
- Rickgauer JP, Deisseroth K, and Tank DW (2014). Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields. Nature neuroscience 17, 1816–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelfsema PR, Lamme VAF, and Spekreijse H (1998). Object-based attention in the primary visual cortex of the macaque monkey. Nature 395, 376–381. [DOI] [PubMed] [Google Scholar]
- Russo AA, Bittner SR, Perkins SM, Seely JS, London BM, Lara AH, Miri A, Marshall NJ, Kohn A, Jessell TM, et al. (2018). Motor Cortex Embeds Muscle-like Commands in an Untangled Population Response. Neuron 97, 953–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai K, Zhao S, Takatoh J, Rodriguez E, Lu J, Leavitt AD, Fu M, Han BX, and Wang F (2016). Capturing and Manipulating Activated Neuronal Ensembles with CANE Delineates a Hypothalamic Social-Fear Circuit. Neuron 92, 739–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem AB, Ayaz A, Jeffery KJ, Harris KD, and Carandini M (2013). Integration of visual motion and locomotion in mouse visual cortex. Nature neuroscience 16, 1864–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas E, and Sejnowski TJ (2001). Correlated neuronal activity and the flow of neural information. Nature Reviews Neuroscience 2, 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf R, Tsunematsu T, McAlinden N, Dawson MD, Sakata S, and Mathieson K (2016). Depth-specific optogenetic control in vivo with a scalable, high-density muLED neural probe. Sci Rep 6, 28381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler SE, McCall JG, Yan P, Hyrc KL, Li MJ, Tucker CL, Lee JM, Bruchas MR, and Diamond MI (2015). Photo-activatable Cre recombinase regulates gene expression in vivo. Sci Rep-Uk 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seely JS, Kaufman MT, Ryu SI, Shenoy KV, Cunningham JP, and Churchland MM (2016). Tensor Analysis Reveals Distinct Population Structure that Parallels the Different Computational Roles of Areas M1 and V1. PLoS computational biology 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Lubeck E, Zhou W, and Cai L (2016). In Situ Transcription Profiling of Single Cells Reveals Spatial Organization of Cells in the Mouse Hippocampus. Neuron 92, 342–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shobe JL, Claar LD, Parhami S, Bakhurin KI, and Masmanidis SC (2015). Brain activity mapping at multiple scales with silicon microprobes containing 1,024 electrodes. Journal of neurophysiology 114, 2043–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew NJ, Flickinger D, King J, and Svoboda K (2016). A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirman JN, Smith IT, Kudenov MW, and Smith SL (2016). Wide field-of-view, multiregion, two-photon imaging of neuronal activity in the mammalian brain. Nat Biotechnol 34, 857–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stosiek C, Garaschuk O, Holthoff K, and Konnerth A (2003). In vivo two-photon calcium imaging of neuronal networks. Proceedings of the National Academy of Sciences of the United States of America 100, 7319–7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striedter GF (2005). Principles of brain evolution (Sunderland, Mass: Sinauer Associates; ). [Google Scholar]
- Sun WZ, Tan ZC, Mensh BD, and Ji N (2016). Thalamus provides layer 4 of primary visual cortex with orientation- and direction-tuned inputs. Nature neuroscience 19, 308–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanese D, Weng JY, Zampini V, De Sars V, Canepari M, Rozsa B, Emiliani V, and Zecevic B (2017). Imaging membrane potential changes from dendritic spines using computergenerated holography. Neurophotonics 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao ZZ, Levi B, Gray LT, Sorensen SA, Dolbeare Tv et al. (2016). Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nature neuroscience 19, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriault G, Cottet M, Castonguay A, McCarthy N, and De Koninck Y (2014). Extended two-photon microscopy in live samples with Bessel beams: steadier focus, faster volume scans, and simpler stereoscopic imaging. Front Cell Neurosci 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson AM, and Lamy C (2007). Functional maps of neocortical local circuitry. Frontiers in neuroscience 1, 19–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe S, Fize D, and Marlot C (1996). Speed of processing in the human visual system. Nature 381, 520–522. [DOI] [PubMed] [Google Scholar]
- Tsai PS, Mateo C, Field JJ, Schaffer CB, Anderson ME, and Kleinfeld D (2015). Ultra-large field-of-view two-photon microscopy. Optics express 23, 13833–13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinck M, Batista-Brito R, Knoblich U, and Cardin JA (2015). Arousal and Locomotion Make Distinct Contributions to Cortical Activity Patterns and Visual Encoding. Neuron 86, 740–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz A, Trenholm S, Yonehara K, Hillier D, Raics Z, Leinweber M, Szalay G, Ghanem A, Keller G, Rozsa B, et al. (2015). PRESYNAPTIC NETWORKS. Single-cell-initiated monosynaptic tracing reveals layer-specific cortical network modules. Science 349, 70–74. [DOI] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann KK, and Callaway EM (2007). Retrograde neuronal tracing with a deletion-mutant rabies virus. Nature methods 4, 47–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohrer A, Humphries MD, and Machens CK (2013). Population-wide distributions of neural activity during perceptual decision-making. Prog Neurobiol 103, 156–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright N, and Fox K (2010). Origins of Cortical Layer V Surround Receptive Fields in the Rat Barrel Cortex. Journal of neurophysiology 103, 709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Stark E, Im M, Cho IJ, Yoon ES, Buzsaki G, Wise KD, and Yoon E (2013). An implantable neural probe with monolithically integrated dielectric waveguide and recording electrodes for optogenetics applications. J Neural Eng 10, 056012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YX, Zou P, and Cohen AE (2017). Voltage imaging with genetically encoded indicators. Curr Opin Chem Biol 39, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki N, and Shepherd GMG (2015). Synaptic Circuit Organization of Motor Corticothalamic Neurons. Journal of Neuroscience 35, 2293–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WJ, Carrillo-Reid L, Bando Y, Peterka DS, and Yuste R (2018). Simultaneous two- photon imaging and two-photon optogenetics of cortical circuits in three dimensions. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WJ, Miller JEK, Carrillo-Reid L, Pnevmatikakis E, Paninski L, Yuste R, and Peterka DS (2016). Simultaneous Multi-plane Imaging of Neural Circuits. Neuron 89, 269–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng WB, Jiang HF, Gang YD, Song YG, Shen ZZ, Yang H, Dong X, Tian YL, Ni RJ, Liu Y, et al. (2017). Anterograde monosynaptic transneuronal tracers derived from herpes simplex virus 1 strain H129. Mol Neurodegener 12, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemba CM, Freeman J, Movshon JA, and Simoncelli EP (2016). Selectivity and tolerance for visual texture in macaque V2. Proceedings of the National Academy of Sciences of the United States of America 113, E3140–E3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingg B, Chou XL, Zhang ZG, Mesik L, Liang F, Tao HW, and Zhang LI (2017). AAV-Mediated Anterograde Transsynaptic Tagging: Mapping Corticocollicular Input-Defined Neural Pathways for Defense Behaviors. Neuron 93, 33–47. [DOI] [PMC free article] [PubMed] [Google Scholar]