Abstract
Despite improvements in survival in metastatic melanoma with combined BRAF and mitogen-activated protein kinase/extracellular signal-regulated kinase inhibitor treatment, the overwhelming majority of patients eventually acquire resistance to both agents. Consequently, new targets for therapy in resistant tumors are currently being evaluated. Previous studies have identified p90 subfamily of ribosomal S6 kinase (p90RSK) family kinases as key factors for growth and proliferation, as well as protein synthesis via assembly of the 7-methyl-guanosine triphosphate cap-dependent translation complex. We sought to evaluate inhibitors of p90RSK family members: BI-D1870 and BRD7389, for their ability to inhibit both proliferation and protein synthesis in patient-derived melanoma cell lines with acquired resistance to combined treatment with the BRAF inhibitor vemurafenib and the mitogen-activated protein kinase/extracellular signal-regulated kinase inhibitor selumetinib. We found that the RSK inhibitors blocked cell proliferation and protein synthesis in multiple dual-resistant melanoma lines. In addition, single agent RSK inhibitor treatment was effective in drug-naïve lines, two of which are innately vemurafenib resistant. We also used Reverse Phase Protein Array screening to identify differential protein expression that correlates with BI-D1870 sensitivity, and identified prognostic biomarkers for survival in human melanoma patients. These findings establish p90RSK inhibition as a therapeutic strategy in treatmentresistant melanoma and provide insight into the mechanism of action.
INTRODUCTION
Melanoma currently represents the eighth leading cause of cancer-related death in the United States, causing an estimated 9,710 deaths in 2014, and with an increasing incidence of approximately 3% per year (Yamada et al., 2005). BRAF activating mutations occur in roughly half of melanoma cases, leading to constitutive activation of the mitogenactivated protein kinase (MAPK) pathway (Davies et al., 2002; Garnett and Marais, 2004). In 2011, the FDA approved vemurafenib, the first selective inhibitor of mutant BRAF, leading to a progression-free survival benefit of approximately 5e7 months, with a median overall survival of 15.9 months (Long et al., 2013; Sosman et al., 2012). Although this represented a significant improvement over historical overall survival of 6e10 months for patients on dacarbazine-based chemotherapy (Paraiso et al., 2010; Silva et al., 2014), the vast majority of patients treated with BRAF inhibitors develop progressive disease between 2 and 18 months (Baudy, 2012). The addition of clinical MAPK/ERK kinase (MEK) inhibitors such as trametinib or cobimetinib further extends progression-free survival by approximately 3e4 months, but the vast majority of tumors eventually acquire resistance to the combination therapy as well (Halaban et al., 2010; Held et al., 2013). Although extracellular signalregulated kinase (ERK), a downstream node of the MAPK pathway, has frequently been proposed as a third potential target for inhibition (Carlino et al., 2013; Rizos et al., 2014), investigation of targets downstream of ERK has been less extensive.
One of the targets of active ERK is the p90 subfamily of ribosomal S6 kinase (p90RSK) enzymes. These enzymes are directly phosphorylated by ERK (Boussemart et al., 2014), and are known to play a role in cell survival, cell cycle control, and regulation of protein synthesis (Romeo and Roux, 2011; Silva et al., 2014; Sulzmaier and Ramos, 2013). Specifically, p90RSK family members promote mammalian target of rapamycin complex 1 activity, enhancing mRNA translation through activation of ribosomal protein S6 (RPS6) and elongation initiation factor 4e (eIF4e), as well as inactivation of translational repressor 4E-binding protein 1, allowing for cap-dependent translation initiation (Gwin et al., 2011; Romeo et al., 2013; She et al., 2010). In addition to providing an additional layer of control over cell cycling and protein metabolism, posttranscriptional control of cap-dependent mRNA translation is a direct method of cellular regulation of energy homeostasis (Chen et al., 1996; Diehl et al., 1998). Glycolysis, a core metabolic pathway whose activity is greatly upregulated in cancer (Dell’ Antone, 2012; Koppenol et al., 2011), is also known to be partially regulated by p90RSK (Anjum et al., 2005). Although the importance of p90RSK as a regulator of protein translation in cancer has previously been demonstrated (Fujita et al., 2003; Romeo et al., 2013; Sutherland et al., 1993), the possible utility of inhibitors of RSK as therapies for dual BRAF and MEK-inhibitor-resistant tumors has yet to be explored. In this study, we investigate the use of two inhibitors of p90RSK family enzymes: BI-D1870 and BRD7389, in several BRAFi and MEKi dual-resistant melanoma cell lines. Additionally, we use reverse phase protein array (RPPA) data predicting sensitivity to RSK inhibition to identify a number of potential prognostic biomarkers for survival in late-stage melanoma.
RESULTS
Dual BRAF and MEK inhibitor treatment resistant cell lines are able to maintain new protein synthesis in the presence of both drugs
We first selected two BRAF-mutant drug-naïve patient-derived melanoma cell lines established by the Yale SPORE in Skin Cancer that have been previously shown to be sensitive to growth inhibition by both vemurafenib and selumetinib as single agents (Held et al., 2013). We independently verified the sensitivity of these lines: YUMAC and YURIF by plating cells in 96-well format and incubating them for 72 hours in the presence of varying concentrations of both drugs (Supplementary Table S1 online). Each cell line was then serially passaged in increasing concentrations of both vemurafenib and selumetinib until proliferative capacity was maintained at moderate levels of each inhibitor. At this time, lines were redesignated YUMACdr and YURIFdr (“dual resistant”) and again screened for drug response with BRAF and MEK inhibition. Growth inhibition with 3 μM vemurafenib and 150 nM selumetinib was minimal compared with growth inhibition achieved with parental lines at lower drug concentrations (Supplementary Figure S1 online).
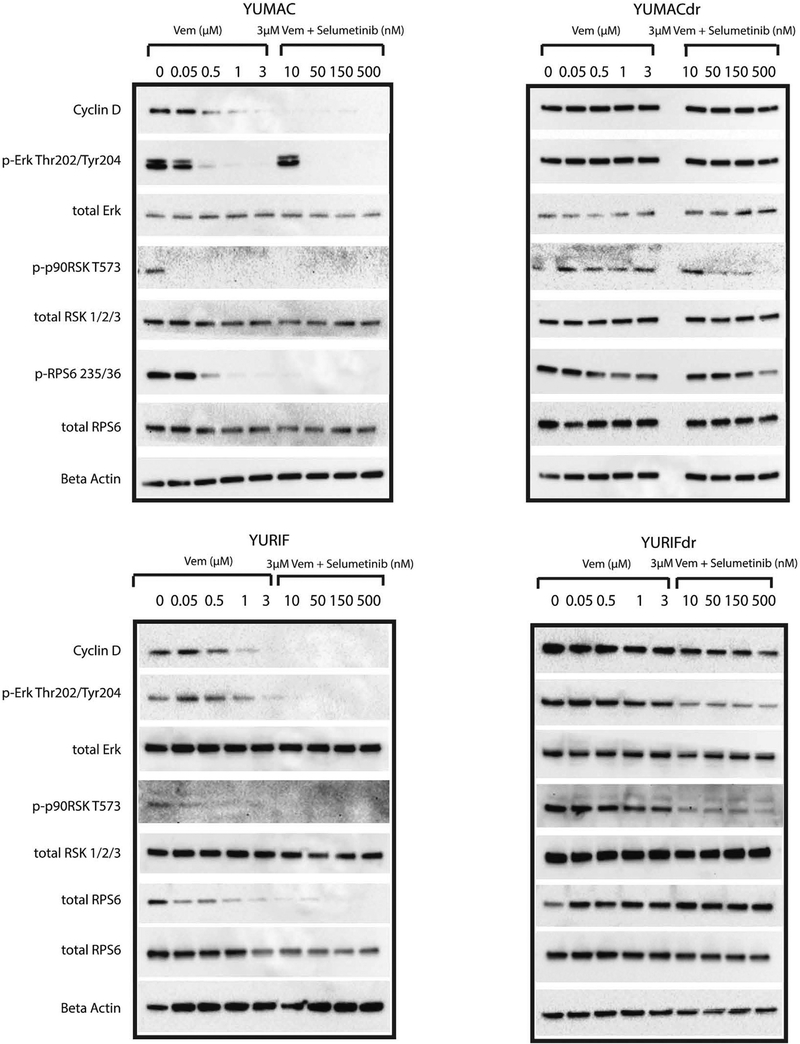
Previous studies of vemurafenib-resistant melanoma have explored the importance of de novo protein translation to continued survival and proliferation of cells exposed to treatment (Silva et al., 2014), focusing largely on activity of the phosphoinositide 3-kinase/Akt/mammalian target of rapamycin pathway. We evaluated whether signaling associated with protein translation capacity is maintained selectively in dual-resistant lines by culturing sensitive and acquired dual-resistant lines in the presence of vemurafenib with selumetinib (or vemurafenib alone) for 24 hours and beyond (Figure 1, Supplementary Figure S2 online). As predicted, although YUMACdr showed some decrease, YUMACdr and YURIFdr were both able to maintain the activity of key signaling nodes upstream of assembly of the capdependent translation initiation complex, such as ERK, p90RSK, and RPS6 (Romeo et al., 2013; Silva et al., 2014). Notably, YURIFdr showed virtually no decrease in phosphorylation of RPS6 even at high concentrations of both drugs. Conversely, parental YUMAC and YURIF showed significant loss of upstream signaling even at low doses of vemurafenib monotreatment. Importantly, these results also validated the importance of continued MAPK signaling to the maintenance of mammalian target of rapamycin pathway activity in BRAF-mutant melanoma, as has previously been established (Corcoran et al., 2013; Silva et al., 2014).
Figure 1. Dual vemurafenib and selumetinib treatment is insufficient to restrict protein translation in lines with dual BRAF and MEK inhibitor resistance.
Western blot of parental YUMAC and YURIF, as well as dual-resistant YUMACdr and YURIFdr, shows significant downregulation of new protein translation at low concentrations in YUMAC and YURIF but only at the highest concentrations in YUMACdr and YURIFdr. Erk, extracellular signal-regulated kinase; MEK, MAPK/ERK kinase; p90RSK, p90 subfamily of ribosomal S6 kinase; RPS6, ribosomal protein S6.
Inhibition of p90RSK in dual-resistant melanoma lines inhibits growth
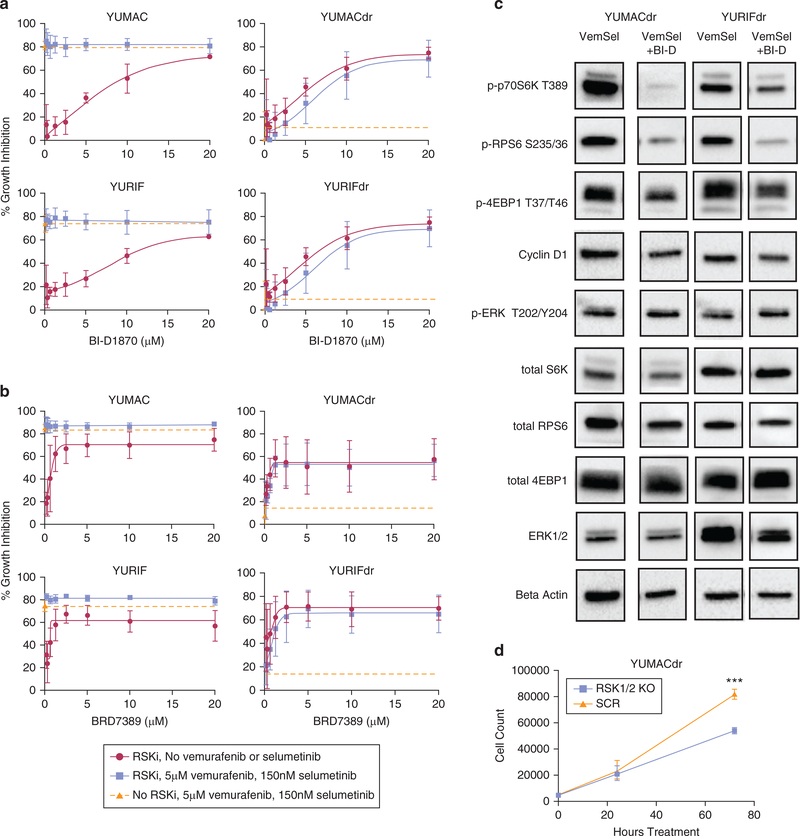
We tested the hypothesis that interfering with protein translation through inhibition of p90RSK is sufficient to significantly reduce proliferation. This hypothesis was supported by previous research that has demonstrated functional links between p90RSK family members and the phosphoinositide 3-kinase/Akt/mammalian target of rapamycin pathway by promoting assembly of mammalian target of rapamycin complex 1, which exerts control over assembly of the capdependent translation initiation complex (Roux et al., 2007; Shahbazian et al., 2006). We selected two established p90RSK inhibitors: BI-D1870 and BRD7389 (Fomina-Yadlin et al., 2010; Romeo et al., 2013) to evaluate in the presence of dual BRAF and MEK inhibition treatment. The first inhibitor, BI-D1870, showed significant growth inhibition when used as a single agent against the parental YUMAC and YURIF lines. BI-D1870 failed to add significant inhibition over and above combined vemurafenib and selumetinib inhibitor therapy, presumably owing to near-maximal inhibition achieved at the single combined vemurafenib and selumetinib dose evaluated at 72 hours (Figure 2a). As expected, vemurafenib and selumetinib treatment of the BRAF and MEK inhibitor-resistant YUMACdr and YURIFdr lines yielded only minimal inhibition of proliferation. In contrast, BI-D1870, both as a single agent and in combination with vemurafenib and selumetinib, significantly inhibited growth of both dual resistant lines, though there was no significant synergy of this agent with the BRAF and MEK inhibitors (Figure 2a). A similar pattern was observed with the second RSK inhibitor, BRD7389, which showed efficacy as a single agent in the parental lines and roughly equivalent efficacy in the dual resistant lines between the RSKi single agent and the RSKi, vemurafenib, and selumetinib combination conditions (Figure 2b).
Figure 2. p90RSK inhibition is an effective therapeutic strategy in dual-resistant melanoma lines.
RSK inhibitor treatment with (a) BI-D1870 or (b) BRD 3789 results in significant growth inhibition in both drug-naïve and acquired BRAF/MEK-inhibitor-resistant lines over 72 hours alone or in combination with vemurafenib and selumetinib. Growth inhibition with vemurafenib and selumetinib alone (no RSK inhibitor) plotted for comparison in green with error bars at 0 μM RSK inhibitor concentration. Mean ± SEM, N = 3. (c) Western blotting for signaling nodes involved in translation with 24-hour 5 μM BI-D1870, 5 μM vemurafenib, and 150 nM selumetinib triple therapy shows significant downregulation over dual Vem + Sel therapy alone. (d). shRNA-mediated knockdown of RSK1 and RSK2 leads to significant growth inhibition of YUMACdr treated with 5 μM vemurafenib and 150 nM selumetinib over 72 hours, P < 0.0001, mean ± SEM, N = 3. MEK, MAPK/ERK kinase; p90RSK, p90 subfamily of ribosomal S6 kinase; RPS6, ribosomal protein S6; SEM, standard error of the mean.
We next evaluated whether inhibition of p90RSK would significantly downregulate signaling nodes controlling new protein synthesis. These include RPS6, which is involved in ribosome assembly, p70S6 kinase (p70S6K), which plays a role in activating RPS6, and eukaryotic translation initiation factor 4E-binding protein 1, which inhibits translation initiation when hypophosphorylated (Roux et al., 2007). Triple treatment using vemurafenib and selumetinib along with the p90RSK inhibitor BI-D1870 significantly downregulated these key promoters of translational activity relative to dual treatment, including p-RPS6 S235/236, a known direct target of the RSK isoforms (Roux et al., 2007) (Figure 2c). These changes were observed at 24 hours of treatment: a time point before significant induction of apoptosis (Supplementary Figure S3 online), with continuation of the effect out to 48 hours (Supplementary Figure S4 online). This suggests that loss of protein translation preceded rather than resulted from cell death. Inhibition of p90RSK inhibited expression of Cyclin D1, a key cell cycle regulator that has been previously identified as a downstream target of p90RSK family kinases working through glycogen synthase kinase-3β (Chen et al., 1996; Diehl et al., 1998).
To directly determine whether loss of RSKs restricts growth in BRAF-driven melanoma, we used lentivirally infected shRNA to simultaneously knock down RSK1 and RSK2. A 72-hour proliferation assay showed significant inhibition in the RSK1/2 knockdown versus control, with minimal growth beyond plating density in the knockdown line (Figure 2d, Supplementary Figure S5 online). The two drugs have significant structural differences, and major differences in specificity, as BI-D1870 inhibits RSK1–4 and BRD7389 inhibits RSK1–3 (Fomina-Yadlin et al., 2010; Neise etal., 2013). With efficacy of shRNA RSK knockdown in growth inhibition, we conclude that on-target RSK inhibition by BI-D1870 and BRD7389 is important for the antiproliferative effects of both drugs.
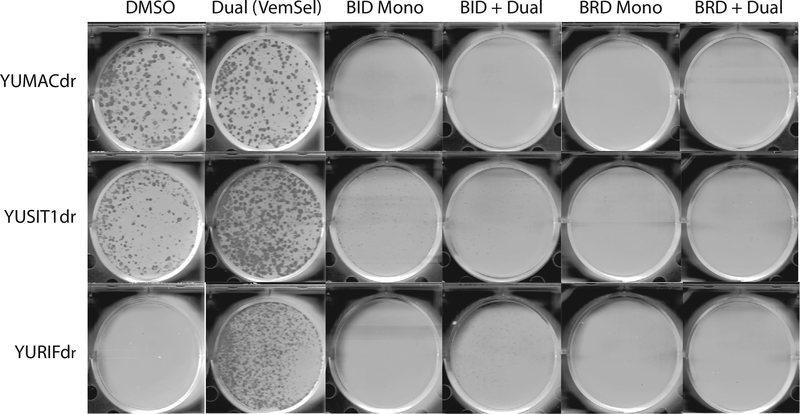
To verify the growth inhibitory effects of the p90RSK inhibitors in long-term culture, we performed colony formation assays over the course of 2 weeks with YUMACdr, YUSIT1dr, and YURIFdr. Treatment of all three cell lines with either of BI-D1870 and BRD7389 alone or in combination with vemurafinib and selumetinib nearly completely abrogated clonogenic growth of all three cell lines (Figure 3). FMK, a lower-potency RSK inhibitor selective for RSK1 and RSK2, but neither RSK3 nor RSK4 and SL0101, a low-potency pan-isoform inhibitor with an approximate EC50 of 50 μM in intact cells, were less effective but showed some inhibitory activity in combination with vemurafenib and selumetinib (Romeo et al., 2012, 2013) (Supplementary Figure S6 online). BI-D1870 and BRD7389 were also shown to show reduction in colony formation in a dose-dependent fashion (Supplementary Figure S7 online).
Figure 3. Long-term growth of dual resistant melanoma is greatly inhibited by triple therapy with BI-D1870 and BRD7389.
2D colony formation assays were performed with initial seeding of 5,000 cells per well. Cells were allowed to settle for 48 hours before being treated with 5 μM BI-D1870 or 5 μM BRD7389 ± 5 μM vemurafenib and 150 nM selumetinib for 14 days.Media and drug were replaced every 72 hours after commencement of drug treatment, N = 3.
Inhibition of proliferation in dual-resistant melanoma lines is due to a combination of G0/G1 arrest and induction of apoptosis
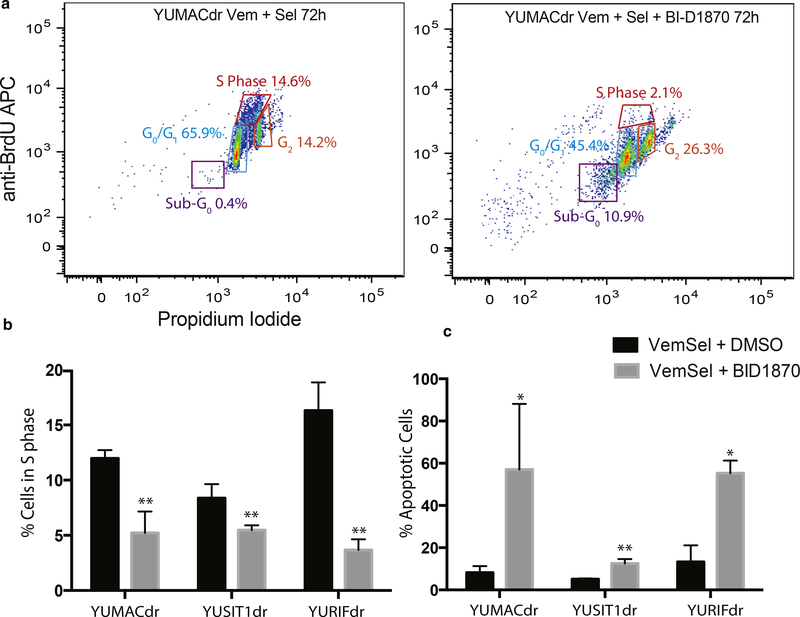
As both mitotic arrest and induction of apoptosis are potential mechanisms for reduced melanoma cell accumulation in the presence of p90RSK inhibitors, we next evaluated the primary mechanism of growth inhibition. RSK family proteins have been shown to regulate cell cycle progression via downstream effectors such as Cyclin D1 (Chen et al., 1996) and p27kip1 (Fujita et al., 2003), as well as members of the glycogen synthase kinase 3 family (Sutherland et al., 1993). We used BrdU and propidium iodide-based cell cycle analysis by flow cytometry to measure changes in cell cycle distribution with BI-D1870, vemurafenib, and selumetinib triple treatment versus dual vemurafenib and selumetinib treatment alone. A significant decrease in the number of cells in the S phase with the corresponding increase in G0/G1 fraction was observed with BI-D1870 triple treatment versus dual treatment alone starting at 12 hours and continuing out to 72 hours of treatment in all lines (Figure 4a, 4b, and Supplementary Figures S8 and S9 online).
Figure 4. p90RSK inhibition reduces proliferation in dual resistant melanoma through a mixture of apoptosis induction and growth arrest.
(a, b) Propidium iodide+BrdU cell cycle flow cytometry showed a significantly greater fold reduction in cells in the S phase after a 30-minute pulse followed by a 2-hour chase with 5 μM BI-D1870 triple therapy over dual 5 μM Vem + 150 nM Sel alone. *P < 0.05, **P < 0.01, mean ± SEM, N = 3. (c) Dual annexin V and propidium iodide flow cytometry showed a significantly larger increase in apoptosis induction by 5 μM BI-D1870 in 72-hour triple therapy over dual Vem + Sel alone. *P < 0.05, **P < 0.01, mean ± SEM, N = 3. APC, antigen-presenting cell; p90RSK, p90 subfamily of ribosomal S6 kinase; SEM, standard error of the mean.
RSK family kinases downregulate apoptosis by inhibiting both death-associated protein kinase (Anjum et al., 2005) and the apoptosis inducer Bad, a Bcl-2 family protein (Shimamura et al., 2000). In the setting of cell cycle analysis, apoptotic cells often appear as a DNA-fragmented sub-G0/G1 population when assayed (Chuang et al., 2005). We used dual propidium iodide and annexin V staining coupled with flow cytometry to assess changes in the fraction of sub-G0 cells after 72 hours of triple treatment with BI-D1870 in relation to dual treatment (Figure 4c). All three lines showed significant increases in the population of sub-G0/G1 cells with BI-D1870 triple treatment relative to dual BRAF and MEK inhibition.
Inhibition of p90RSK significantly decreases 7-methyl-guanosine triphosphate (m7-GTP) cap-dependent protein translation in dual-resistant melanoma lines
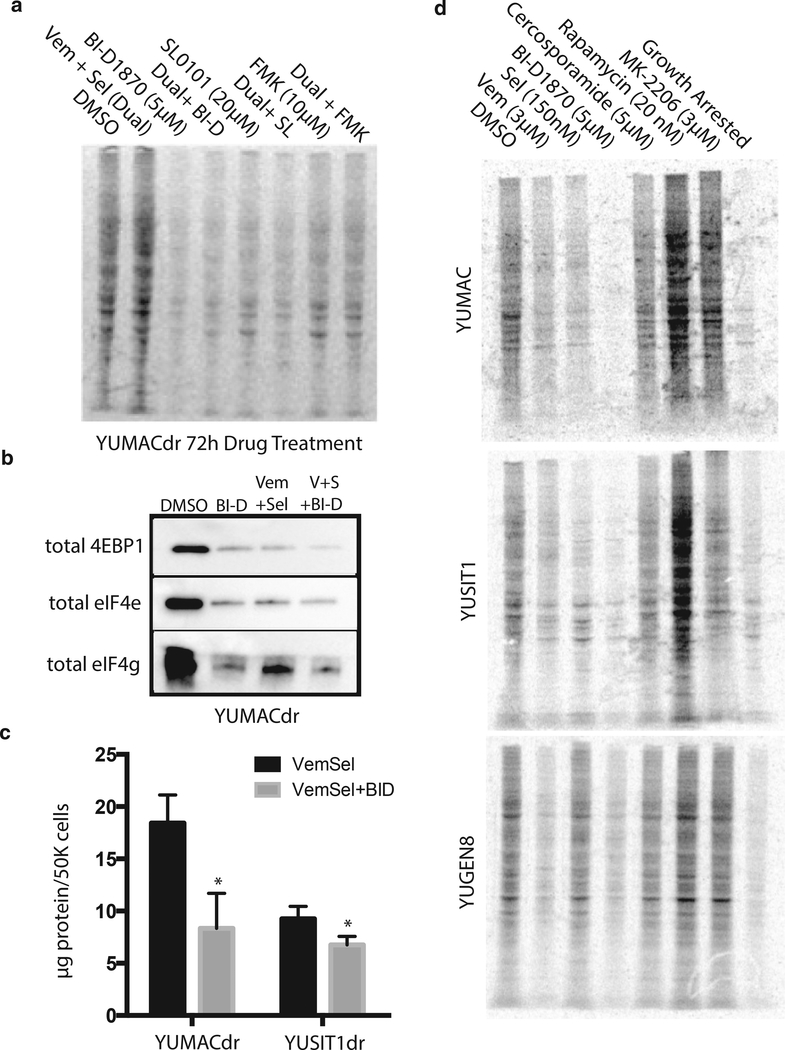
To verify that RSK inhibitors decreased new protein translation, we treated YUMACdr with BI-D1870, as well as the lower-potency RSK inhibitors FMK and SL0101 as either monotreatment or triple treatment with 3μM vemurafenib and 150 nM selumetinib. After drug incubation, we labeled cells with [35S]-methionine and [35S]-cysteine for 1 hour followed by SDS-PAGE and imaging of radiolabeled protein extracts. Global protein synthesis was markedly reduced after p90RSK inhibitor treatment (Figure 5a). Interestingly, BI-D1870 appeared to induce comparable decreases in translational activity with or without the presence of BRAF or MEK inhibitors and reduced new protein synthesis to a greater extent than the two lower-potency RSK inhibitors. We evaluated assembly of the cap-dependent translation initiation complex by performing m7-GTP- affinity chromatography of key complex components 4E-binding protein 1, eIF4e and eIF4g. Association of the protranslation eIF4 proteins with the affinity matrix was substantially reduced in the setting of both monotreatment and triple treatment (Figure 5b). Although the inhibitory complex member 4Ebinding protein 1 was somewhat reduced as well, the difference in magnitude was much smaller than eIF4e and eIF4g. Hence we conclude that this combination substantially inhibits assembly of the cap-dependent translation initiation complex. To determine if this reduction affects overall cellular protein content, we next measured average protein content per cell using the Bradford assay in conjunction with cell number normalization by the CyQUANT DNA content assay. Seventy-two hours of BID1870 triple combination significantly reduced protein content of both YUMACdr and YUSIT1dr in comparison with incubation with the dual combination (Figure 5c), suggesting that this inhibition of translational activity has a significant impact on the protein metabolic state of the cells. We evaluated these effects in additional drug-naïve lines incubated with a number of other agents that inhibit signaling through either MAPK or phosphoinositide 3-kinase/Akt/mammalian target of rapamycin. In all three lines tested, YUMAC, YUSIT1, and YUGEN8, 72-hour BI-D1870 monotreatment caused the greatest inhibition of protein synthesis among of all drugs tested as assessed by biosynthetic labeling with [35S]-methionine and [35S]-cysteine (Figure 5d).
Figure 5. Growth inhibition with p90RSK inhibitors reduces protein synthesis.
(a) Seventy-hour YUMACdr treatment with 5 μM BI-D1870, 20 μM SL0101, or 10 μM FMK alone or with 5 μM vemurafenib + 150 nM selumetinib inhibits protein synthesis as assessed by 1-hour 35S-labeled methionine/cysteine pulse. (b) m7-GTP-sepharose bead cap-dependent translation complex precipitation from YUMACdr lysate after 72 hours of BI-D1870 mono- or triple therapy. Normalized to cell number. (c) After 72 hours of BI-D1870+Vem+Sel triple therapy, approximately 50% reduction in protein content per cell was observed using the Bradford assay normalized to cell number. *P < 0.05, mean ± SEM, N = 3. (d) Screening of a panel of drugs on melanoma lines showed inhibition of protein synthesis with BID1870 equal to or greater than vemurafenib, selumetinib, PI3K/Akt/ mTOR, or MAPK inhibitors. eIF, elongation initiation factor; m7-GTP, 7-methyl-guanosine triphosphate; MAPK, mitogen-activated protein kinase; mTOR, mammalian target of rapamycin; PI3K, phosphoinositide 3-kinase; p90RSK, p90 subfamily of ribosomal S6 kinase; SEM, standard error of the mean.
Identification of potential biomarkers that correlate with RSK inhibitor sensitivity and melanoma overall survival
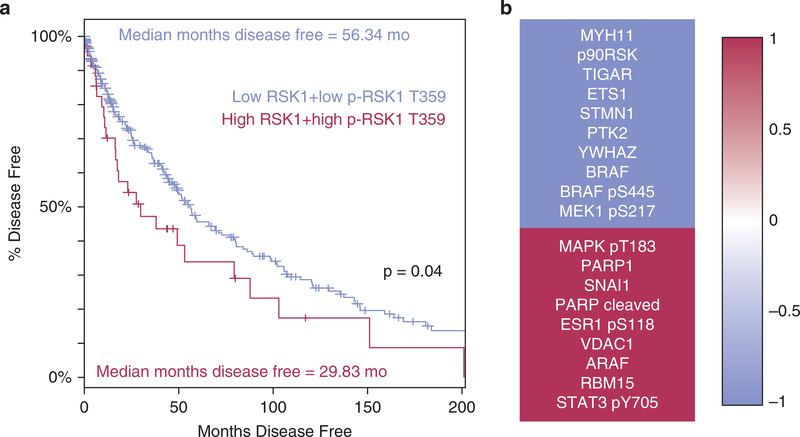
To confirm the clinical significance of p90RSK expression, we examined the correlation between RSK1 expression and phosphorylation and patient overall survival using data in The Cancer Genome Atlas (Cerami et al., 2012; Gao et al., 2013). We found that in a cohort of patients showing levels of both RSK1 and p-RSK1 T359 at least 1 standard deviation above zero-centered normalized mean, there was a median decrease in the overall survival of approximately 26.5 months (P = 0.04) (Figure 6a). This further confirmed the biological importance of correlates of p90RSK activity in human melanomas.
Figure 6. p90RSK is a potential clinical therapeutic target in BRAF-mutant melanoma.
(a) Patients indexed in The Cancer Genome Atlas were stratified into high and low RSK1 expressers, P = 3.9 × 10−7 patients with high p90RSK levels (n = 36) showed 26.5-month poorer disease-free survival versus patients who were low for both (n = 272), Kaplan-Meier estimate, P = 0.04. (b) RPPA screening of levels of 217 proteins in lysates collected from seven BRAF-mutant drug-naïve melanoma lines showed significant, P < 0.05, positive, and negative interaction between 20 proteins and BI-D1870 GI50. The top 10 genes with the strongest negative correlation of protein levels to drug potency are shown in red, whereas those with the strongest positive correlation between protein levels and drug potency are shown in blue. MAPK, mitogen-activated protein kinase; MEK, MAPK/ERK kinase; p90RSK, p90 subfamily of ribosomal S6 kinase; RPPA, reverse phase protein array; RPS6, ribosomal protein S6.
To identify potential biomarkers that might be used to predict RSK inhibitor sensitivity, we evaluated growth inhibition dose responses after BI-D1870 treatment in seven BRAF-mutant melanoma lines. We also used the RPPA analysis of 217 cancer-relevant proteins/epitopes (Langdon et al., 2015) and correlated the findings with BI-D1870 GI50 values. Many of the 20 most significantly correlated targets are located within or downstream of the MAPK pathway (Figure 6b, Supplementary Table S2 online). These included total RSK expression as one of the strongest correlates to BI-D1870 response (Pearson’s r = −0.88, P = 0.009), with BRAF expression (r = −0.71, P = 0.073) and the activating MEK phosphorylation site S217 (r = −0.70, P = 0.077) trending toward significance, all suggesting that melanomas with high levels of these proteins or phosphoepitopes are susceptible to p90RSK inhibition.
DISCUSSION
Despite major advances in melanoma therapeutics, the overwhelming majority of patients with stage IV melanoma still eventually acquire resistance to combined BRAF and MEK inhibitor treatment (Halaban et al., 2010; Held et al., 2013). Although estimates of the benefit vary, phase II and III trials have shown that the addition of an MEK inhibitor as combined treatment only increases progression-free survival on average by 2–16 weeks beyond BRAF inhibitor treatment alone (Flaherty et al., 2012; Larkin et al., 2014). p90RSK has been validated as a potential therapeutic target in breast (Gwin et al., 2011), prostate (Clark et al., 2005), and head and neck squamous cell cancer (Kang et al., 2010), among others. However, the evaluation of RSK inhibitors in the treatment of treatment-resistant melanoma has not been extensively explored.
Here we present evidence of the effectiveness of inhibitors of p90RSK in BRAF mutant melanoma cell lines that have acquired resistance to dual BRAF and MEK inhibitor treatment. We evaluated two structurally dissimilar RSK inhibitors with efficacy against RSK1–3, BI-D1870, and BRD7389, showing that treatment with these agents both as single agent therapy and as an adjunct to vemurafenib and selumetinib significantly inhibits melanoma proliferation. In addition to achieving this effect with two chemically dissimilar inhibitors, we have also demonstrated phenocopy of this effect through shRNA-mediated knockdown of RSK1 and RSK2, further supporting the importance of the on-target effects of these RSK inhibitors in slowing melanoma growth, as well as highlighting the importance of RSK biology to melanoma survival and proliferation in general. This appears to be mediated through significant downregulation of Cyclin D1 and GO/ G1 arrest in treated lines, as well as induction of apoptosis, suggesting that p90RSK plays an indispensable role in melanoma cell viability in addition to growth. RSK family members’ effects on these processes appear to be tied to regulation of protein translation derived from their ability to promote assembly of the cap-dependent translation initiation complex. These findings underscore the importance of further research on RSK inhibitors as a potential therapeutic modality in metastatic melanoma patients who experience disease progression on both BRAF and MEK inhibitors.
Our data in drug-naïve lines suggest that BI-D1870 is highly effective in melanoma lines that have high expression of p90RSK family members. This is especially important to note given that high-p90RSK melanomas show significantly decreased disease-free survival in analyzed patient data from The Cancer Genome Atlas. Consequently, p90RSK inhibitors may provide significantly improved survival in a subset of melanomas that had previously done very poorly on current first-line treatment. The efficacy of these drugs at low micromolar concentrations in melanoma lines previously demonstrated to be de novo resistant to BRAF and MEK inhibition (Held et al., 2013), 501Mel and YUKSI, also suggests that BI-D1870 and other p90RSK inhibitors might provide an increase in survival in patients who fail BRAF and MEK inhibitor treatment initially, as well as patients who are unable to tolerate these agents due to adverse events. As these agents have not yet been tested for in vivo efficacy in humans or mouse models, the further studies will also need to optimize route of administration and biological dosing, helping to translate these proof-of-principle data into new human-available therapies.
Overall, evidence from this study provides strong support for the further exploration of inhibitors of p90RSK as possible adjunct therapy in melanomas that show de novo and acquired resistance to BRAF and MEK inhibitors. It also provides evidence that growth inhibition correlates well with downregulation of new protein synthesis, leading to both induction of apoptosis and cell cycle arrest.
MATERIALS AND METHODS
Single and triple agent screening
Cells were plated in optical-bottom 96-well plates (Thermo Scientific, Waltham, MA) at a density of 5 × 103 cells per well. Cells were grown in basal media for 48 hours before 150 ml of fresh media was added. Media with maximal drug concentration was used to perform serial dilution across wells using a multichannel pipette (Eppendorf, Hamburg, Germany). After 72 hours of treatment, cell numbers were counted by CyQUANT assay. IC50 values were calculated using Graphpad Prism 6 (GraphPad Software, La Jolla, CA). For knockdown experiments, cells were infected with MISSION anti-RSK shRNA (Sigma Aldrich, St. Louis, MO) or scramble using the Thermo Fisher Scientific ViraPower Lentiviral Expression system. Viruses were produced in 293T cells according to the manufacturer’s recommendations, and viral supernatant was added to melanoma cells with 8 μg/ml polybrene. Selection for cells expressing construct or scramble was performed using neomycin and knockdown was verified by western blot >96 hours after infection.
m7-GTP-sepharose cap-binding precipitation
After 72-hour incubation with either DMSO vehicle, 5 μM BI-D1870, dual 3 μM and 150 μM selumetinib, or triple treatment with all three drugs, cells were trypsinized and counted using the CyQUANT cell proliferation assay and lysed with at a normalized concentration based on cell number. Cells were then incubated overnight at 4 °C with m7-GTP-immobilized γ-aminophenyl-m7-GTP sepharose beads (Jena Bioscience, Jena, Germany) to purify cap-dependent translation initiation complex members. Beads were washed according to the manufacturer’s specifications and samples were boiled with Laemmli buffer supplemented with BME and analyzed by western blot.
Radiographic studies
Cysteine and methionine-free DMEM (Life Technologies, Carlsbad, CA) was used to equilibrate cells for 1 hour before label pulse. This was followed by 1-hour incubation with the same media supplemented with EasyTag EXPRESS35S Protein Labeling Mix using the manufacturer’s protocol (PerkinElmer, Waltham, MA). Collected lysates were separated by SDS-PAGE and read using a Storm 860 phosphorimager (GMI, Ramsey, MN).
Reverse phase protein arrays
Cells were lysed according to the instructions provided by the RPPA Core Facility at the MD Anderson Cancer Center (Houston, TX). Array hybridization was performed and initially analyzed at MD Anderson. RPPA data were analyzed in R v.3.1.2 (R Foundation for Statistical Computing, Vienna, Austria). A correlation matrix was constructed using the HMisc package for R (Harrell, 2015). The correlation method used was the Pearson product-moment correlation coefficient. A correlogram was created using the corrgram package for R (Wright, 2015). Gene set enrichment analysis was conducted using The Broad Institute’s Molecular Signature Database (Subramanian et al., 2005) with a false discovery rate of q = 0.01.
The cancer genome atlas analysis
RPPA protein expression and mRNA gene expression values for patients with melanoma were obtained from the SKCM (Skin Cutaneous Melanoma) database of The Cancer Genome Atlas through CBioPortal (Cerami et al., 2012; Gao et al., 2013). Disease-free survival of patients with melanoma was stratified by the expression level of RPS6KA1. A subgroup of 36 patients with a high protein level of RPS6KA1 and high RPS6KA1 phospho T359 was compared with the rest of the SKCM cohort using the log-rank test and Student’s t-test, for comparing survival and protein expression, respectively. High protein/phosphoprotein level was defined as 1 standard deviation above zero center normalized mean. Statistical significance was defined as α < 0.05.
Supplementary Material
ACKNOWLEDGMENTS
We would like to acknowledge Ruth Halaban and Antonella Bacchiocchi for providing melanoma cell lines from the Yale melanoma SPORE tissue bank. These studies were supported in part by grants from the NIH Yale SPORE in Skin Cancer, NCI P50 CA121974, Ruth L. Kirschstein, NRSA 1F30CA18059101, funded by the NCI, the American Skin Association, the Yale Cancer Center, P01 CA128814, and the Hervey Family Foundation. RPPA was supported by pilot funding from the Yale Cancer Center, NCI P30 CA16359. CGL was supported by training grant T32GM007324.
Abbreviations:
- eIF
elongation initiation factor
- ERK
extracellular signal-regulated kinase
- MAPK
mitogen-activated protein kinase
- MEK
MAPK/ERK kinase
- m7-GTP
7-methyl-guanosine triphosphate
- p90RSK
p90 subfamily of ribosomal S6 kinase
- RPPA
reverse phase protein array
- RPS6
ribosomal protein S6
Footnotes
CONFLICT OF INTEREST
The authors state no conflict of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at http://d.doi.org/10.1016/j.jid.2016.12.033.
REFERENCES
- Anjum R, Roux PP, Ballif BA, Gygi SP, Blenis J. The tumor suppressor DAP kinase is a target of RSK-mediated survival signaling. Curr Biol 2005;15: 1762–7. [DOI] [PubMed] [Google Scholar]
- Baudy AR, Dogan T, Flores-Mercado JE, Hoeflich KP, Su F, van Bruggen N, et al. FDG-PET is a good biomarker of both early response and acquired resistance in BRAFV600 mutant melanomas treated with vemurafenib and the MEK inhibitor GDC-0973. EJNMMI Res 2012;2:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussemart L, Malka-Mahieu H, Girault I, Allard D, Hemmingsson O, Tomasic G, et al. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature 2014;513:105–9. [DOI] [PubMed] [Google Scholar]
- Carlino MS, Saunders CA, Haydu LE, Menzies AM, Martin Curtis C Jr, Lebowitz PF, et al. (18)F-labelled fluorodeoxyglucose-positron emission tomography (FDG-PET) heterogeneity of response is prognostic in dabrafenib treated BRAF mutant metastatic melanoma. Eur J Cancer 2013;49: 395–402. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Juo PC, Curran T, Blenis J. Phosphorylation of c-Fos at the C-terminus enhances its transforming activity. Oncogene 1996;12:1493–502. [PubMed] [Google Scholar]
- Chuang JY, Tsai YY, Chen SC, Hsieh TJ, Chung JG. Induction of G0/G1 arrest and apoptosis by 3-hydroxycinnamic acid in human cervix epithelial carcinoma (HeLa) cells. In Vivo 2005;19:683–8. [PubMed] [Google Scholar]
- Clark DE, Errington TM, Smith JA, Frierson HF Jr, Weber MJ, Lannigan DA. The serine/threonine protein kinase, p90 ribosomal S6 kinase, is an important regulator of prostate cancer cell proliferation. Cancer Res 2005;65:3108–16. [DOI] [PubMed] [Google Scholar]
- Corcoran RB, Rothenberg SM, Hata AN, Faber AC, Piris A, Nazarian RM, et al. TORC1 suppression predicts responsiveness to RAF and MEK inhibition in BRAF-mutant melanoma. Sci Transl Med 2013;5:196ra98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949–54. [DOI] [PubMed] [Google Scholar]
- Dell’ Antone P Energy metabolism in cancer cells: How to explain the Warburg and Crabtree effects? Med Hypotheses 2012;79:388–92. [DOI] [PubMed] [Google Scholar]
- Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev 1998;12:3499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 2012;367:107–14. [DOI] [PubMed] [Google Scholar]
- Fomina-Yadlin D, Kubicek S, Walpita D, Dancik V, Hecksher-Sorensen J, Bittker JA, et al. Small-molecule inducers of insulin expression in pancreatic alpha-cells. Proc Natl Acad Sci USA 2010;107:15099–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Sato S, Tsuruo T. Phosphorylation of p27Kip1 at threonine 198 by p90 ribosomal protein S6 kinases promotes its binding to 14–3-3 and cytoplasmic localization. J Biol Chem 2003;278:49254–60. [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Marais R. Guilty as charged: B-RAF is a human oncogene. Cancer Cell 2004;6:313–9. [DOI] [PubMed] [Google Scholar]
- Gwin J, Drews N, Ali S, Stamschror J, Sorenson M, Rajah TT. Effect of genistein on p90RSK phosphorylation and cell proliferation in T47D breast cancer cells. Anticancer Res 2011;31:209–14. [PubMed] [Google Scholar]
- Halaban R, Zhang W, Bacchiocchi A, Cheng E, Parisi F, Ariyan S, et al. PLX4032, a selective BRAF(V600E) kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAF melanoma cells. Pigment Cell Melanoma Res 2010;23:190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell FE, Jr., Hmisc: Harrell miscellaneous. R package version 3.15–0, 2015. [Google Scholar]
- Held MA, Langdon CG, Platt JT, Graham-Steed T, Liu Z, Chakraborty A, et al. Genotype-selective combination therapies for melanoma identified by high-throughput drug screening. Cancer Discov 2013;3: 52–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Elf S, Lythgoe K, Hitosugi T, Taunton J, Zhou W, et al. p90 ribosomal S6 kinase 2 promotes invasion and metastasis of human head and neck squamous cell carcinoma cells. J Clin Invest 2010;120: 1165–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer 2011;11: 325–37. [DOI] [PubMed] [Google Scholar]
- Langdon CG, Held MA, Platt JT, Meeth K, Iyidogan P, Mamillapalli R, et al. The broad-spectrum receptor tyrosine kinase inhibitor dovitinib suppresses growth of BRAF-mutant melanoma cells in combination with other signaling pathway inhibitors. Pigment Cell Melanoma Res 2015;28: 417–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med 2014;371:1867–76. [DOI] [PubMed] [Google Scholar]
- Long GV, Wilmott JS, Haydu LE, Tembe V, Sharma R, Rizos H, et al. Effects of BRAF inhibitors on human melanoma tissue before treatment, early during treatment, and on progression. Pigment Cell Melanoma Res 2013;26: 499–508. [DOI] [PubMed] [Google Scholar]
- Neise D, Sohn D, Stefanski A, Goto H, Inagaki M, Wesselborg S, et al. The p90 ribosomal S6 kinase (RSK) inhibitor BI-D1870 prevents gamma irradiation-induced apoptosis and mediates senescence via RSKand p53-independent accumulation of p21WAF1/CIP1. Cell Death Dis 2013;4:e859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraiso KH, Fedorenko IV, Cantini LP, Munko AC, Hall M, Sondak VK, et al. Recovery of phospho-ERK activity allows melanoma cells to escape from BRAF inhibitor therapy. Br J Cancer 2010;102:1724–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizos H, Menzies AM, Pupo GM, Carlino MS, Fung C, Hyman J, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin Cancer Res 2014;20:1965–77. [DOI] [PubMed] [Google Scholar]
- Romeo Y, Moreau J, Zindy PJ, Saba-El-Leil M, Lavoie G, Dandachi F, et al. RSK regulates activated BRAF signalling to mTORC1 and promotes melanoma growth. Oncogene 2013;32:2917–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo Y, Roux PP. Paving the way for targeting RSK in cancer. Expert Opin Ther Targets 2011;15:5–9. [DOI] [PubMed] [Google Scholar]
- Romeo Y, Zhang X, Roux PP. Regulation and function of the RSK family of protein kinases. Biochem J 2012;441:553–69. [DOI] [PubMed] [Google Scholar]
- Roux PP, Shahbazian D, Vu H, Holz MK, Cohen MS, Taunton J, et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem 2007;282: 14056–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, et al. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J 2006;25:2781–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She QB, Halilovic E, Ye Q, Zhen W, Shirasawa S, Sasazuki T, et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell 2010;18:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura A, Ballif BA, Richards SA, Blenis J. Rsk1 mediates a MEK-MAP kinase cell survival signal. Curr Biol 2000;10:127–35. [DOI] [PubMed] [Google Scholar]
- Silva JM, Bulman C, McMahon M. BRAFV600E cooperates with PI3K signaling, independent of AKT, to regulate melanoma cell proliferation. Mol Cancer Res 2014;12:447–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med 2012;366:707–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulzmaier FJ, Ramos JW. RSK isoforms in cancer cell invasion and metastasis. Cancer Res 2013;73:6099–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland C, Leighton IA, Cohen P. Inactivation of glycogen synthase kinase3 beta by phosphorylation: new kinase connections in insulin and growth-factor signalling. Biochem J 1993;296(Pt 1):15–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright K corrgram: Plot a Correlogram. R package version 1.7, 2015.
- Yamada K, Brink I, Bisse E, Epting T, Engelhardt R. Factors influencing [F-18] 2-fluoro-2-deoxy-D-glucose (F-18 FDG) uptake in melanoma cells: the role of proliferation rate, viability, glucose transporter expression and hexokinase activity. J Dermatol 2005;32:316–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.