Abstract
Premise of the Study
Access to improved crop cultivars is the foundation for successful agriculture. New cultivars must have improved yields that are determined by quantitative and qualitative traits. Genotype‐by‐environment interactions (GEI) occur for quantitative traits such as reproductive fitness, longevity, height, weight, yield, and disease resistance. The stability of genotypes across a range of environments can be analyzed using GEI analysis. GEI analysis includes univariate and multivariate analyses with both parametric and non‐parametric models.
Methods and Results
The program STABILITYSOFT is online software based on JavaScript and R to calculate several univariate parametric and non‐parametric statistics for various crop traits. These statistics include Plaisted and Peterson's mean variance component (θ i), Plaisted's GE variance component (θ (i)), Wricke's ecovalence stability index (W i 2), regression coefficient (b i), deviation from regression (S di 2), Shukla's stability variance (σ i 2), environmental coefficient of variance (CV i), Nassar and Huhn's statistics (S (1), S (2)), Huhn's equation (S (3) and S (6)), Thennarasu's non‐parametric statistics (NP (i)), and Kang's rank‐sum. These statistics are important in the identification of stable genotypes; hence, this program can compare and select genotypes across multiple environment trials for a given data set. This program supports both the repeated data across environments and matrix data types. The accuracy of the results obtained from this software was tested on several crop plants.
Conclusions
This new software provides a user‐friendly interface to estimate stability statistics accurately for plant scientists, agronomists, and breeders who deal with large volumes of quantitative data. This software can also show ranking patterns of genotypes and describe associations among different statistics with yield performance through a heat map plot. The software is available at https://mohsenyousefian.com/stabilitysoft/.
Keywords: adaptability, phenotypic stability, quantitative traits, ranking method, STABILITYSOFT
Genotype‐by‐environment interactions (GEI) refer to changes in genotypic performance across different environments. The presence of GEI in multi‐environment trials is expressed either as inconsistent responses of different genotypes (relative to others) due to changes in genotypic rank, or as the absolute difference between genotypes without rank change (Crossa, 2012). This effect can be used to assess quantitative traits of economic importance—often investigated in plant and animal breeding, pharmacogenomics, genetic epidemiology, and conservation biology research—including longevity, weight, height, biomass, yield, and even disease resistance. The identification and subsequent selection of superior crop varieties in target environments are important goals of agronomic and plant breeding studies (Ahmadi et al., 2015; Vaezi et al., 2018). To identify superior varieties across multiple environments, plant breeders undertake trials across several years and locations, usually during the final developmental stages of a crop variety. The GEI effect reduces the association observed between genotypic and phenotypic values and complicates the selection of the best variety (Ebdon and Gauch, 2002). Interpreting the GEI effect in multi‐environment trials assists in the selection of stable varieties for a wide range of environments (Vaezi et al., 2017).
Many statistical approaches have been proposed for using stability analyses to interpret GEI, all of which have been based on univariate and multivariate models (Flores et al., 1998). There are two major statistical groups for interpreting GEI by numerical analysis. The first group contains parametric methods such as the regression coefficient (b i; Finlay and Wilkinson, 1963), variance of deviations from the regression (S di 2; Eberhart and Russell, 1966), Wricke's ecovalence stability index (W i 2; Wricke, 1962), Shukla's stability variance (σ i 2; Shukla, 1972), environmental coefficient of variance (CV i; Francis and Kannenberg, 1978), Plaisted and Peterson's mean variance component (θ i; Plaisted and Peterson, 1959), Plaisted's GE variance component (θ (i); Plaisted, 1960), and the yield‐stability index (YS i; Kang, 1991). These parametric statistics are primarily used to assess genotype stability by relating observed genotypic responses (e.g., yield, plant height, seed oil content) to a sample of environmental conditions (e.g., rainfall, temperature, osmotic stress, soil type). Parametric stability statistics have suitable properties under certain statistical assumptions, including a normal distribution and homogeneity of variance of the errors and their interaction effects. However, parametric statistics may not be the best method for assessing genotype stability if the assumptions are not met (Huhn, 1990). The second group of analytical methods includes non‐parametric methods such as Nassar and Huhn's statistics (S (1), S (2); Nassar and Huhn, 1987), Huhn's equation (S (3) and S (6); Huhn, 1990), Thennarasu's statistics (NP (i); Thennarasu, 1995), Kang's rank‐sum (KR or Kang; Kang, 1988), and Fox's top rank (FOX or Top‐rank; Fox et al., 1990). Non‐parametric statistics explain environments and phenotypes relative to both biotic and abiotic factors. Non‐parametric statistics are a feasible alternative to parametric statistics because their performance is based on ranked data (Nassar and Huhn, 1987) and no assumptions are needed about the distribution and homogeneity of the variance of the errors. Because each method has its own merits and weaknesses, most breeding programs now incorporate both parametric and non‐parametric methods for the selection of stable genotypes (Becker and Leon, 1988).
Parametric and non‐parametric statistics are used by researchers from different fields, but the lack of a user‐friendly statistical package makes these methods unavailable for agronomists and plant breeders. A literature review revealed that other studies have attempted to introduce codes for SAS (Piepho, 1999; Hussein et al., 2000; Akbarpour et al., 2016; Dia et al., 2016) or R (Branco, 2015; Yaseen and Eskridge, 2018) to calculate some of the stability indices. Table 1 compares the available features and capabilities of these codes and packages. Currently, researchers interested in applying stability statistics are required to use several programs to obtain the desired results, and further applications are needed to describe and visualize the correlations between these parameters, which are crucial for the selection of stable varieties. The general lack of platform‐independent software capable of calculating all parametric and nonparametric statistics in one package motivated our work to create an online tool (STABILITYSOFT) that is able to overcome these difficulties by providing a user‐friendly interface for the non‐specialist.
Table 1.
Statistical capacity and available features of STABILITYSOFT relative to other codes and packages
| Statistical capacity/Features | SAS codes | R packages | STABILITYSOFT | ||||
|---|---|---|---|---|---|---|---|
| Piepho (1999) | Hussein et al. (2000) | Akbarpour et al. (2016) | Dia et al. (2016) | Branco (2015) | Yaseen and Eskridge (2018) | ||
| Statistic | |||||||
| Mean variance component | X | X | |||||
| GE variance component | X | X | |||||
| Wricke's ecovalence stability index | X | X | X | X | |||
| Regression coefficient | X | X | X | ||||
| Deviation from regression | X | X | X | X | |||
| Shukla's stability variance | X | X | X | X | X | ||
| Environmental coefficient of variance | X | X | X | X | |||
| Nassar and Huhn's non‐parametric statistics and Huhn's statistics | X | X | X | X | |||
| Thennarasu's non‐parametric statistics | X | X | X | ||||
| Kang's rank‐sum | X | X | X | X | |||
| Correlation coefficients | X | ||||||
| Ranking pattern of genotypes through all statistics | X | ||||||
| Calculation of statistics based on both types of data (row data and matrix mean data) | X | ||||||
| Features | |||||||
| Windows support | X | X | X | X | X | X | X |
| Unix/Linux support | X | X | X | X | X | X | X |
| Mac OS support | X | X | X | ||||
| Portable (without installation) | X | ||||||
| GUI (graphical user interface) | X | ||||||
| Offline usage capability | X | X | X | X | X | X | |
METHODS AND RESULTS
STABLITYSOFT is written in JavaScript at the browser‐side and PHP at the server‐side and is available as a Web application at https://mohsenyousefian.com/stabilitysoft/. Alternately, users can access the source codes and data sets in GitHub (https://github.com/pour-aboughadareh/stabilitysoft). Figure 1 shows the information flow in the STABILITYSOFT program. The software can be used online and is available in R programming language for advanced users, which offers more flexibility. The data used for testing the software are available online and can be used as example files to run the program. The input file is in a standard Excel file format, which is widely supported by other well‐known software. Our program supports two data types: (1) repeated data across environments (year, location, and year × location), with genotype i in year n and location m (or environment j for one year or one location) and replication k, and (2) matrix data that includes genotypes (rows) and environments (columns). The program first calculates the average of the objective trait for each genotype and then provides a data matrix by environment. Based on the data matrix, the software calculates several univariate parametric and non‐parametric statistics, namely Plaisted and Peterson's mean variance component (θ i), Plaisted's GE variance component (θ (i)), Wricke's ecovalence stability index (W i 2), regression coefficient (b i), deviation from regression (S di 2), Shukla's stability variance (σ i 2), environmental coefficient of variance (CV i), Nassar and Huhn's non‐parametric statistics (S (1), Z 1 , S (2), and Z 2), Huhn's nonparametric statistics (S (3) and S (6)), Thennarasu's non‐parametric statistics (NP (i)), and Kang's rank‐sum. For further description and details about these statistics see Appendix 1. The program also calculates ranking patterns of the genotypes, based on each index. After following the instructions outlined on the website, STABILITYSOFT produces a simple Excel output on two separate sheets. The first sheet (named “Statistics”) includes the average of crop yield along with 16 parametric and non‐parametric statistics, and the second sheet (“Ranks”) consists of the ranking of each genotype for each statistic along with sum of ranks (SR), average of sum of ranks (ASR), and standard deviation (SD). It is important to note that the rank of genotypes for the regression coefficient (b i) is not calculated because a significance test (H0: B ≠ 1) must be conducted to determine the stability using this parameter. For further details, see Finlay and Wilkinson (1963). STABILITYSOFT also renders a heat map plot, based on Pearson's correlation coefficients (Pearson, 1895), using Canvas tools (W3C, Cambridge, Massachusetts, USA) to display the interrelationships between the stability statistics and yield performance.
Figure 1.
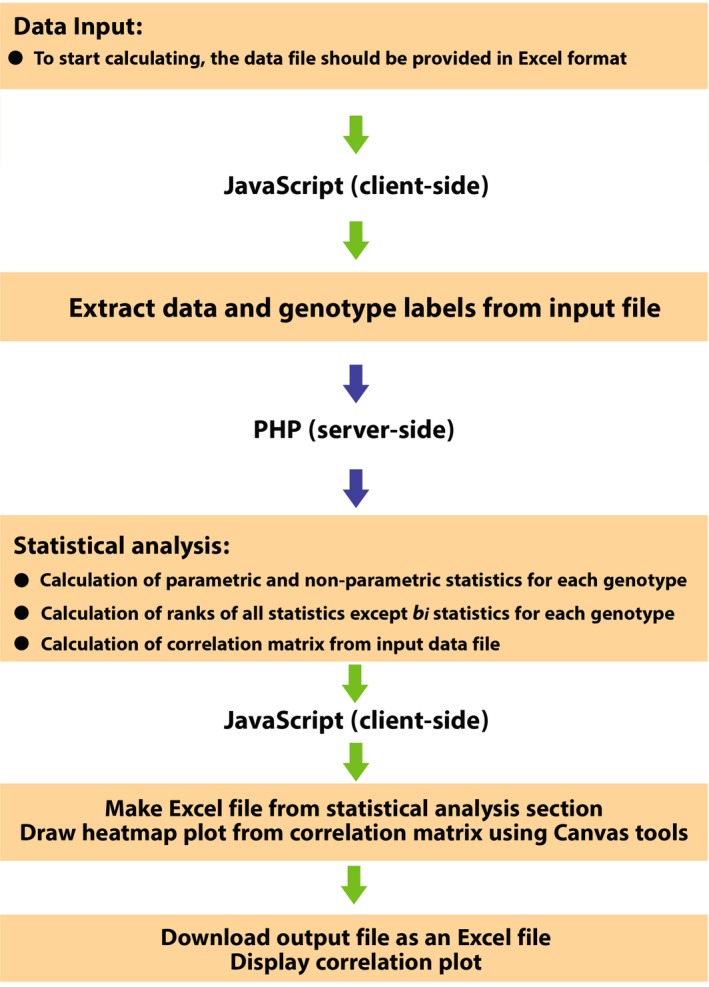
Information flow diagram for the STABILITYSOFT software tool.
To test the program, we are providing two examples and data sets gathered from five yield trials in grass pea and barley (advanced and doubled haploid lines) taken from Ahmadi et al. (2015), Khalili and Pour‐Aboughadareh (2016), and Vaezi et al. (2018). In the first example, we used grain yield (kg ha−1) for 14 advanced grass pea lines grown in three Iranian semi‐arid regions (Kermanshah, Gachsaran, and Lorestan) during four consecutive years (2005–2008) (see Ahmadi et al., 2015 for further details on growing conditions and experimental design). In this example, only the averages of grain yield across replications were used for calculations. The averages of grain yield, along with 16 parametric and non‐parametric statistics, are shown in Appendices S1 and S2 together with significance tests for S (1) and S (2) statistics, Z1 and Z2, respectively. According to Nassar and Huhn (1987), if Z1 and Z2 are less than the critical value of χ2, the results show non‐significant differences in rank stability among the studied genotypes grown in the test environments (Huhn, 1990). In the second sheet of the output file, which is named “Ranks,” the rank of each genotype for each statistic is calculated (Appendix S2). The calculated values indicate there are significant differences between tested lines in terms of grain yield, and five lines (G3, G9, G5, G7, and G12) could be identified with high‐yield performance across nine diverse environments. The calculated S (1), S (2), S (3), S (6), NP (3), NP (4), and θ i statistics showed G3 to be the most stable line, but according to other parameters (S (3), S (6), NP (1), NP (2), NP (3), NP (4), θ (i), W i 2, b i, S di 2, σ i 2, and CV i), line G6 was also shown to possess desired stability traits. In these cases, the heat map plot function of STABILITYSOFT based on Pearson's correlation can be used to further investigate the interrelationships among different stability statistics. This showed that yield performance only positively correlates with the regression slope (b i) (Appendix S3). Because identifying stable lines based on grain yield and sole parameters could be problematic, as shown in this example, our program provides an estimation of the average of the sum ranks (ASR) for all statistics to select potentially superior stable lines. Accordingly, a genotype with low ASR value can be selected as a superior stable genotype. Based on our results (Appendix S2), lines G6 (ASR = 3.44; SD = 3.01) followed by G3 (ASR = 5.69; SD = 5.13) and G11 (ASR = 4.50; SD = 4.50) could be selected as stable lines for cultivation in the semi‐arid regions of Iran.
In the second example, we tested the software using grain yield data from a two‐year (2016–2017) barley trial using 18 genotypes grown at four semi‐arid regions in Iran (Gonbad, Gachsaran, Moghan, and Lorestan). At each location, the experimental layout was a randomized complete block design with four replications. Each experimental plot consisted of six rows with 17.5‐cm row spacing. Each location received optimal agricultural practices, with total grain yield for each genotype estimated at harvest. The data file is available on GitHub as “Example5.xlsx,” and the results are summarized in Appendix S4. Grain yield (Y) ranged from 1820 to 2415 kg ha−1 (average 1985 kg ha−1), which was used as the first statistic for assessing genotypes. G2 had the highest yield performance, followed by G13, G5, G1, and G18 (2415, 2246, 2125, 2054, and 2053 kg ha−1, respectively). Two parametric statistics (W i 2 and σ i 2) showed that genotypes G6 and G18 had the lowest values and are, therefore, deemed stable lines. According to S (1), S (2), S (3), and S (6), genotypes G2, G6, G7, and G13 were selected as desirable genotypes. Two statistics (NP (3) and NP (4)) showed a similar trend, such that G2 and G13 were selected as stable genotypes. Selection based on the KR statistic orders the remaining genotypes as G18 > G13 > G1 > G16. The heat map plot based on Pearson's correlation revealed that S (1), S (2), S (3), and S (6) were positively and significantly correlated with each other and with NP (1), NP (3), and NP (4) (Appendix S5), and grain yield had a significant positive correlation with b i and CV i. Moreover, grain yield also had a positive association with θ i, W i 2, and σ i 2, but a negative association with NP (2), NP (3), NP (4), S (1), S (2), S (3), S (6), and KR. Two statistics (W i 2 and σ i 2) were negatively associated with θ (i). Based on these associations and the use of the ranking method integrated in STABILITYSOFT, we were able to identify the six most stable genotypes (G18, G13, G12, G6, G2, and G1), which had the lowest ASR values (4.81, 4.88, 5.13, 5.63, 6.75, and 7.69, respectively; Appendix S6). With respect to yield performance, two high‐yielding genotypes (G1 [2054 kg ha−1] and G2 [2415 kg ha−1]) with relatively low ASR values are ideally suited for introduction to desirable growth environments, whereas two genotypes (G13 [2246 kg ha−1] and G18 [2053 kg ha−1]) with high and middle yield performance along with low ASR values can be introduced to semi‐arid or similar regions of Iran. Furthermore, genotypes G6 (1840 kg ha−1) and G12 (1923 kg ha−1), which have acceptable ASR values and low grain yields, were identified as low‐yielding genotypes, hence these genotypes can be introduced to marginal cultivation environments.
CONCLUSIONS
We developed an online software to calculate several parametric and non‐parametric stability statistics that are important in the identification of stable crop genotypes. Some statistical programs are available for stability analyses, but unlike our program they are not platform independent and cannot calculate all the required statistics. In addition to the favorable features listed in Table 1, STABILITYSOFT has the following advantages over other R and SAS packages: (1) it directly calculates different parametric and non‐parametric statistics along with Pearson's correlation with high accuracy; (2) it is a cross‐platform software that needs no additional downloads or installation, calculations are performed on PHP servers, and users are not limited to the processing power of their computers when using large data sets; (3) unlike other codes based on SAS and R packages, which require additional user knowledge of these packages, STABILITYSOFT has a web‐based user interface; and (4) it is compatible with all major browsers (e.g., Google Chrome, Mozilla Firefox). In conclusion, we expect that this software will be useful for analyzing essential data for stability studies related to agronomy and plant breeding. Our program is also able to visualize the interrelationships between different indices, which is crucial for selecting stable varieties. STABILITYSOFT will be helpful for agronomists and plant breeders who deal with large volumes of quantitative data and require user‐friendly software to explore GEI and accurately calculate stability parameters.
DATA ACCESSIBILITY
The source code used to develop STABILITYSOFT is available on GitHub (https://github.com/pour-aboughadareh/stabilitysoft). STABILITYSOFT is available at https://mohsenyousefian.com/stabilitysoft/.
Supporting information
APPENDIX S1. Parametric and non‐parametric stability statistics calculated with STABILITYSOFT for grain yield (kg ha−1) of 14 grass pea advanced lines across nine different environments in Iran.
APPENDIX S2. Ranks of parametric and non‐parametric stability statistics calculated with STABILITYSOFT for grain yield (kg ha−1) of 14 grass pea advanced lines across nine different environments in Iran.
APPENDIX S3. Heat map plot rendered based on Pearson’s correlation analysis for Example 1. See Appendix 1 for full definitions of statistics. θ i = mean variance component; θ (i) = GE variance component; W i 2 = Wricke’s ecovalence stability index; b i = regression coefficient; S di 2 = deviation from regression; σ i 2 = Shukla’s stability variance; CV i = environmental coefficient of variance; S (1) and S (2) = Nassar and Huhn’s non‐parametric statistics; S (3) and S (6) = Huhn’s non‐parametric statistics; NP (1–4) = Thennarasu’s non‐parametric statistics; KR = Kang’s rank‐sum; Y = yield.
APPENDIX S4. Parametric and non‐parametric stability statistics calculated with STABILITYSOFT for grain yield (kg ha−1) of 18 barley genotypes across four different environments in Iran.
APPENDIX S5. Heat map plot rendered based on Pearson’s correlation analysis for Example 2. See Appendix 1 for full definitions of statistics. θ i = mean variance component; θ (i) = GE variance component; W i 2 = Wricke’s ecovalence stability index; b i = regression coefficient; S di 2 = deviation from regression; σ i 2 = Shukla’s stability variance; CV i = environmental coefficient of variance; S (1) and S (2) = Nassar and Huhn’s non‐parametric statistics; S (3) and S (6) = Huhn’s non‐parametric statistics; NP (1–4) = Thennarasu’s non‐parametric statistics; KR = Kang’s rank‐sum; Y = yield.
APPENDIX S6. Ranks of parametric and non‐parametric stability statistics calculated with STABILITYSOFT for grain yield grain yield (kg ha−1) of 18 barley genotypes across four different environments in Iran.
ACKNOWLEDGMENTS
The authors thank Dr. Mohsen Shekarbaigi (Department of Mathematics, Imam Khomeini International University) for his assistance with the dissection of mathematical formulae, Prof. Jafar Ahmadi and Dr. Valiollah Yousefi (Department of Genetics and Plant Breeding, Imam Khomeini International University) for their suggestions, and Dr. Behroz Vaezi and Dr. Marouf Khalili for providing the data sets for testing this software.
APPENDIX 1. Description of stability statistics calculated for the crop traits by STABILITYSOFT.
| Statistic | Symbol | Definition |
|---|---|---|
| Mean variance component | θ i | Plaisted and Peterson (1959) proposed the variance component of genotype‐by‐environment interactions (GEI) for interactions between each of the possible pairs of genotypes. This statistic considers the average of the estimate for all combinations with a common genotype to be a measure of stability. Accordingly, the genotypes that show a lower value for θ i are considered more stable. |
| GE variance component | θ (i) | This statistic is a modified measure of stability parameter. In this approach, the ith genotype is deleted from the entire set of data and the GEI variance from this subset is the stability index for the ith genotype. According to this statistic, the genotypes that show higher values for the (i) are considered more stable. |
| Wricke's ecovalence stability index | W i 2 | Wricke (1962) proposed the concept of ecovalence as the contribution of each genotype to the GEI sum of squares. The ecovalence (W i) of the ith genotype is its interaction with the environments, squared and summed across environments. Thus, genotypes with low values have smaller deviations from the mean across environments and are more stable. |
| Regression coefficienta | b i | The regression coefficient (b i) is the response of the genotype to the environmental index that is derived from the average performance of all genotypes in each environment (Finlay and Wilkinson, 1963). If b i does not significantly differ from 1, then the genotype is adapted to all environments. A b i > 1 indicates genotypes with higher sensitivity to environmental change and greater specificity of adaptability to high‐yielding environments, whereas a b i < 1 describes a measure of greater resistance to environmental change, thereby increasing the specificity of adaptability to low‐yielding environments. |
| Deviation from regression | S di 2 | In addition to the regression coefficient, variance of deviations from the regression (S di 2) has been suggested as one of the most‐used parameters for the selection of stable genotypes. Genotypes with an S di 2 = 0 would be most stable, while an S di 2 > 0 would indicate lower stability across all environments. Hence, genotypes with lower values are the most desirable (Eberhart and Russell, 1966). |
| Shukla's stability variance | σ i 2 | Shukla (1972) suggested the stability variance of genotype i as its variance across environments after the main effects of environmental means have been removed. According to this statistic, genotypes with minimum values are intended to be more stable. |
| Environmental coefficient of variance | CV i | The coefficient of variation is suggested by Francis and Kannenberg (1978) as a stability statistic through the combination of the coefficient of variation, mean yield, and environmental variance. Genotypes with low CV i, low environmental variance (EV), and high mean yield are considered to be the most desirable. |
| Nassar and Huhn's non‐parametric statistics and Huhn's statisticsb |
S
(1)
S (2) S (3) S (6) |
Huhn (1990) and Nassar and Huhn (1987) suggested four non‐parametric statistics: (1) S (1), the mean of the absolute rank differences of a genotype over all tested environments; (2) S (2), the variance among the ranks over all tested environments; (3) S (3), the sum of the absolute deviations for each genotype relative to the mean of ranks; and (4) S (6), the sum of squares of rank for each genotype relative to the mean of ranks. To compute these statistics, the mean yield data have to be transformed into ranks for each genotype and environment, and the genotypes are considered stable if their ranks are similar across environments. The lowest value for each of these statistics reveals high stability for a certain genotype. |
| Thennarasu's non‐parametric statistics |
NP
(1)
NP (2) NP (3) NP (4) |
Four NP (1–4) statistics are a set of alternative non‐parametric stability statistics defined by Thennarasu (1995). These parameters are based on the ranks of adjusted means of the genotypes in each environment. Low values of these statistics reflect high stability. |
| Kang's rank‐sum | Kang or KR | Kang's rank‐sum (Kang, 1988) uses both yield and σ i 2 as selection criteria. This parameter gives a weight of 1 to both yield and stability statistics to identify high‐yielding and stable genotypes. The genotype with the highest yield and lower σ i 2 is assigned a rank of 1. Then, the ranks of yield and stability variance are added for each genotype, and the genotypes with the lowest rank‐sum are the most desirable. |
To determine stability using this parameter, the significance test (H0: B ≠ 1) must be conducted. For more detail, see Finlay and Wilkinson (1963).
In addition to S (i) statistics, two significance tests for S (1) and S (2), namely Z 1 and Z 2, are calculated.
Pour‐Aboughadareh, A. , Yousefian M., Moradkhani H., Poczai P., and Siddique K. H. M.. 2019. STABILITYSOFT: A new online program to calculate parametric and non‐parametric stability statistics for crop traits. Applications in Plant Sciences 7(1): e1211.
LITERATURE CITED
- Ahmadi, J. , Vaezi B., Shaabani A., Khademi K., Fabriki‐Ourang S., and Pour‐Aboughadareh A.. 2015. Non‐parametric measures for yield stability in grass pea (Lathyrus sativus L.) advanced lines in semi warm regions. Journal of Agricultural Science and Technology 17: 1825–1838. [Google Scholar]
- Akbarpour, O. A. , Dehghani H., Dorkhi‐Lalelo B., and Kang M. S.. 2016. A SAS macro for computing statistical tests for two‐way table and stability indices of nonparametric method from genotype‐by‐environment interaction. Acta Scientiarum. Agronomy 38: 35–50. [Google Scholar]
- Becker, H. C. , and Leon J.. 1988. Stability analysis in plant breeding. Plant Breeding 101: 1–23. [Google Scholar]
- Branco, L. C. 2015. Nonparametric stability analysis (phenability R package). Website https://cran.r-project.org/web/packages/phenability/phenability.pdf [accessed 12 August 2018].
- Crossa, J. 2012. From genotype × environment interaction to gene × environment interaction. Current Genomics 13: 225–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dia, M. , Wehner T. C., and Arellano C.. 2016. Analysis of genotype × environment interaction (GE) using SAS programming. Agronomy Journal 108: 1838–1852. [Google Scholar]
- Ebdon, J. S. , and Gauch H. G.. 2002. Additive main effect and multiplicative interaction analysis of national turf grass performance trials: I. Interpretation of genotype × environment interaction. Crop Science 42: 489–496. [Google Scholar]
- Eberhart, S. A. T. , and Russell W. A.. 1966. Stability parameters for comparing varieties. Crop Science 6: 36–40. [Google Scholar]
- Finlay, K. W. , and Wilkinson G. N.. 1963. Adaptation in a plant breeding programme. Australian Journal of Agricultural Research 14: 742–754. [Google Scholar]
- Flores, F. , Moreno M. T., and Cubero J. I.. 1998. A comparison of univariate and multivariate methods to analyze environments. Field Crops Research 56: 271–286. [Google Scholar]
- Fox, P. , Skovmand B., Thompson B., Braun H. I., and Cormier R.. 1990. Yield and adaptation of hexaploid spring triticale. Euphytica 47: 57–64. [Google Scholar]
- Francis, T. R. , and Kannenberg L. W.. 1978. Yield stability studies in short‐season maize: I. A descriptive method for grouping genotypes. Canadian Journal of Plant Science 58: 1029–1034. [Google Scholar]
- Huhn, M. 1990. Nonparametric measures of phenotypic stability. Part 1: Theory. Euphytica 47: 189–194. [Google Scholar]
- Hussein, M. A. , Bjornstad A., and Aastveit H.. 2000. SASG×ESTAB: A SAS program for computing genotype × environment stability statistics. Agronomy Journal 92: 454–459. [Google Scholar]
- Kang, M. S. 1988. A rank‐sum method for selecting high‐yielding, stable corn genotypes. Cereal Research Communication 16: 113–115. [Google Scholar]
- Kang, M. S. 1991. Modified rank‐sum method for selecting high yielding, stable crop genotypes. Cereal Research Communication 19: 361–364. [Google Scholar]
- Khalili, M. , and Pour‐Aboughadareh A.. 2016. Parametric and non‐parametric measures for evaluating yield stability and adaptability in barley doubled haploid lines. Journal of Agricultural Science and Technology 18: 789–803. [Google Scholar]
- Nassar, R. , and Huhn M.. 1987. Studies on estimation of phenotypic stability: Tests of significance for nonparametric measures of phenotypic stability. Biometrics 43: 45–53. [Google Scholar]
- Pearson, K. 1895. Notes on regression and inheritance in the case of two parents. Proceedings of the Royal Society of London 58: 240–242. [Google Scholar]
- Piepho, H. S. 1999. Stability analysis using the SAS system. Agronomy Journal 91: 154–160. [Google Scholar]
- Plaisted, R. L. 1960. A shorter method for evaluating the ability of selections to yield consistently over locations. American Potato Journal 37: 166–172. [Google Scholar]
- Plaisted, R. I. , and Peterson L. C.. 1959. A technique for evaluating the ability of selection to yield consistently in different locations or seasons. American Potato Journal 36: 381–385. [Google Scholar]
- Shukla, G. K. 1972. Some statistical aspects of partitioning genotype‐environmental components of variability. Heredity 29: 237–245. [DOI] [PubMed] [Google Scholar]
- Thennarasu, K. 1995. On certain non‐parametric procedures for studying genotype‐environment interactions and yield stability. PhD thesis, PJ School, Indian Agricultural Research Institute, New Delhi, India.
- Vaezi, B. , Pour‐Aboughadareh A., Mohammadi R., Armion M., Mehraban A., Hossein‐Pour T., and Dorri M.. 2017. GGE biplot and AMMI analysis of barley yield performance in Iran. Cereal Research Communications 45: 500–511. [Google Scholar]
- Vaezi, B. , Pour‐Aboughadareh A., Mehraban A., Hossein‐Pour T., Mohammadi R., Armion M., and Dorri M.. 2018. The use of parametric and non‐parametric measures for selecting stable and adapted barley lines. Archives of Agronomy and Soil Science 64: 597–611. [Google Scholar]
- Wricke, G. 1962. Übereine Methode zur Erfassung der ökologischen Streubreite in Feldversuchen. Zeitschrift für Pflanzenzüchtung 47: 92–96. [Google Scholar]
- Yaseen, M. , and Eskridge K. M.. 2018. Stability analysis of genotype by environment interaction (GEI) (stability R package). Website https://cran.r-project.org/web/packages/stability/stability.pdf [accessed 12 August 2018].
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
APPENDIX S1. Parametric and non‐parametric stability statistics calculated with STABILITYSOFT for grain yield (kg ha−1) of 14 grass pea advanced lines across nine different environments in Iran.
APPENDIX S2. Ranks of parametric and non‐parametric stability statistics calculated with STABILITYSOFT for grain yield (kg ha−1) of 14 grass pea advanced lines across nine different environments in Iran.
APPENDIX S3. Heat map plot rendered based on Pearson’s correlation analysis for Example 1. See Appendix 1 for full definitions of statistics. θ i = mean variance component; θ (i) = GE variance component; W i 2 = Wricke’s ecovalence stability index; b i = regression coefficient; S di 2 = deviation from regression; σ i 2 = Shukla’s stability variance; CV i = environmental coefficient of variance; S (1) and S (2) = Nassar and Huhn’s non‐parametric statistics; S (3) and S (6) = Huhn’s non‐parametric statistics; NP (1–4) = Thennarasu’s non‐parametric statistics; KR = Kang’s rank‐sum; Y = yield.
APPENDIX S4. Parametric and non‐parametric stability statistics calculated with STABILITYSOFT for grain yield (kg ha−1) of 18 barley genotypes across four different environments in Iran.
APPENDIX S5. Heat map plot rendered based on Pearson’s correlation analysis for Example 2. See Appendix 1 for full definitions of statistics. θ i = mean variance component; θ (i) = GE variance component; W i 2 = Wricke’s ecovalence stability index; b i = regression coefficient; S di 2 = deviation from regression; σ i 2 = Shukla’s stability variance; CV i = environmental coefficient of variance; S (1) and S (2) = Nassar and Huhn’s non‐parametric statistics; S (3) and S (6) = Huhn’s non‐parametric statistics; NP (1–4) = Thennarasu’s non‐parametric statistics; KR = Kang’s rank‐sum; Y = yield.
APPENDIX S6. Ranks of parametric and non‐parametric stability statistics calculated with STABILITYSOFT for grain yield grain yield (kg ha−1) of 18 barley genotypes across four different environments in Iran.
Data Availability Statement
The source code used to develop STABILITYSOFT is available on GitHub (https://github.com/pour-aboughadareh/stabilitysoft). STABILITYSOFT is available at https://mohsenyousefian.com/stabilitysoft/.


