Abstract
Polyphosphoinositides (PPIs) are essential phospholipids located in the cytoplasmic leaflet of eukaryotic cell membranes. Despite contributing only a small fraction to the bulk of cellular phospholipids, they make remarkable contributions to practically all aspects of a cell’s life and death. They do so by recruiting cytoplasmic proteins/effectors or by interacting with cytoplasmic domains of membrane proteins at the membrane–cytoplasm interface to organize and mold organelle identity. The present study summarizes aspects of our current understanding concerning the metabolism, manipulation, measurement, and intimate roles these lipids play in regulating membrane homeostasis and vital cell signaling reactions in health and disease.
Introduction
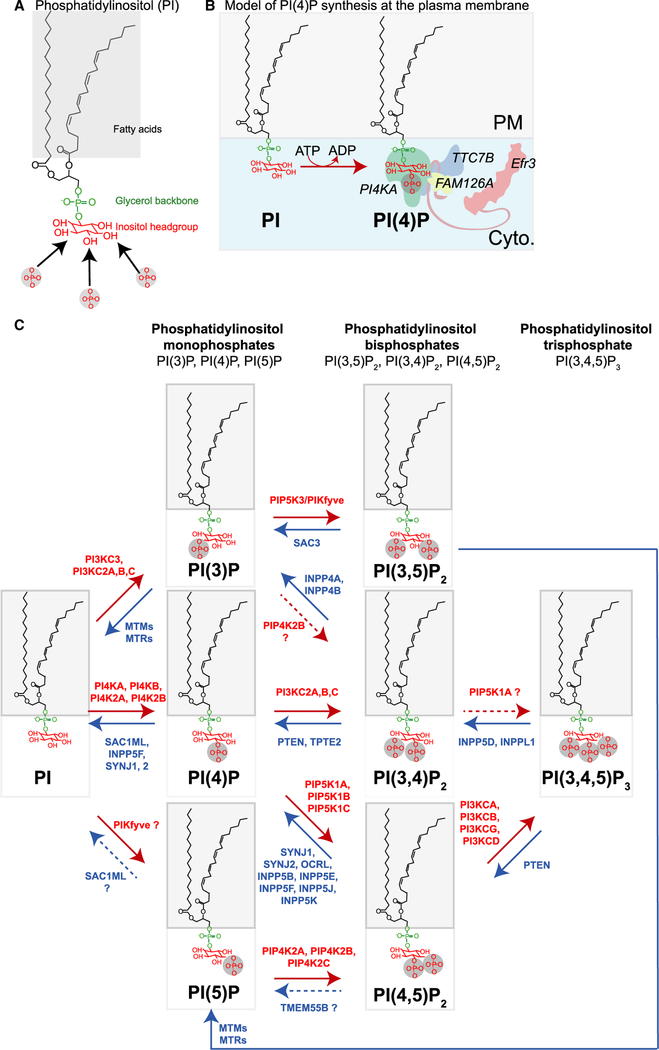
PPIs ( polyphosphoinositides) are reversibly phosphorylated derivatives of the membrane phospholipid phosphatidylinositol (PI, Figure 1A). They are low-abundance lipids of the cytoplasmic leaflet of all eukaryotic cellular membranes. PPIs are amphiphilic, meaning they have a polar inositol head group that faces toward the cytoplasm and non-polar hydrophobic fatty acid tails embedded in the lipid bilayer (Figure 1A). The inositol ring of PI can be phosphorylated by numerous cytoplasmic lipid kinases that add phosphates to the hydroxyl groups on inositol positions 3, 4, or 5, giving polyacidic lipids with a high negative charge [e.g. see PI(4)P synthesis from PI in Figure 1B]. Complementary, lipid phosphatases remove the phosphate. Such reversible phosphorylation gives rise to seven total PPI species (Figure 1C) with dissimilar localizations in the cell. Figure 2A shows a map of the preferred distribution of specific PPIs among the membranes of cellular organelles and the cell surface. We will refer to the PPIs as forming a ZIP code of signature lipids instructing cellular proteins where to bind or where to be active. Nevertheless, the concept of signature lipid should not be taken too strictly. For example, when we describe the Golgi as being a PI(4)P organelle (Figure 2A, blue membrane), this reflects a preponderance of the PI(4)P form but not that PI(4)P is the only species found there. Typically, at least a precursor of the signature lipid will also be present. In addition, cellular activities and signaling may alter the PPI composition of any membrane profoundly but transiently.
Figure 1. PPI biogenesis.
(A) Chemical structure of 1-stearoyl 2-arachidonoyl phosphatidylinositol (PI 38:4). Arrows pointing at the D3, D4, and D5 position on the myo-inositol head group indicate the three reversibly phosphorylatable hydroxyl groups. (B) Model of PI(4)P production from PI at the cytoplasmic (cyto.) interface of the plasma membrane (PM). (C) Diagram summarizing major PPI lipid kinase and phosphatase reaction pathways. Red arrows represent the PPI lipid kinases and blue arrows represent PPI lipid phosphatases. Red and blue labels are the gene name of enzymes capable of catalyzing each reaction. Gene names with question marks (?) and dashed reaction arrows represent enzymes with some ambiguity surrounding their ability to catalyze a specific reaction.
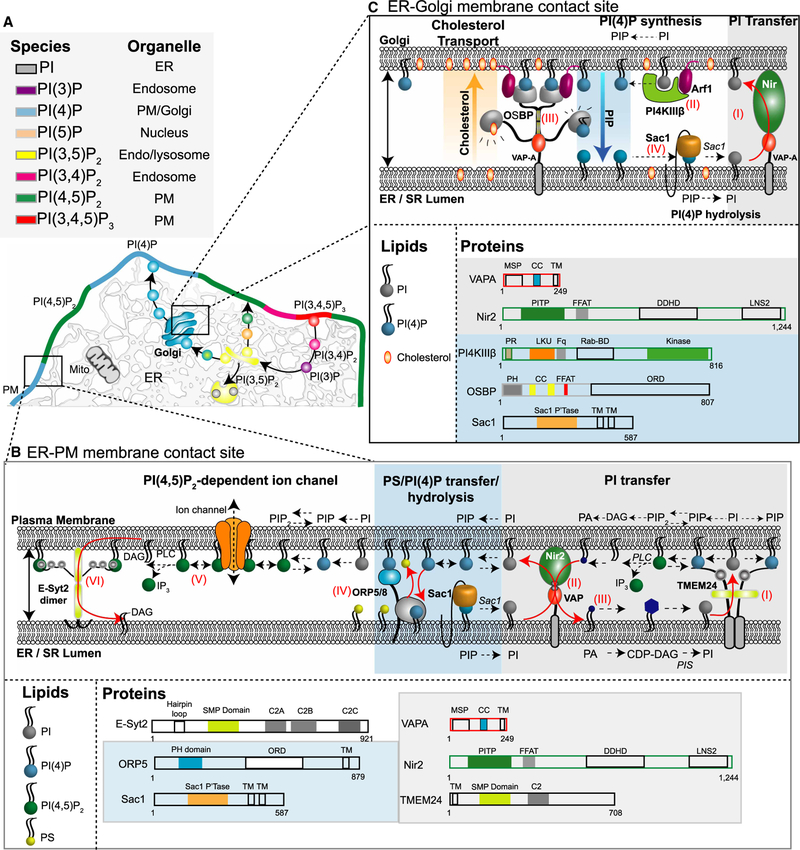
Figure 2. PPI metabolism at MCSs.
(A) Diagrammatic representation of the preferential distribution of PPIs in a eukaryotic cell. (B) Known proteins residing in ER– PM MCSs arranged in a hypothetical scenario. It remains to be fully tested whether each MCS contains this full standardized set of proteins or rather just a subset. For the dynamic equilibrium of PPI metabolism, (I) PI is transferred from the ER to the PM via TMEM24 dimers and this PI is then converted to PI(4)P by PM PI 4-kinases. (II) Under times of VAP-mediated Nir2 recruitment, PI would be transferred to the PM in exchange for phosphatidic acid (PA). (III) PA is subsequently converted to PI in a multi-step reaction. (IV) ORP5/8 countertransports PM PI(4)P to the ER in exchange for phosphatidylserine. Transferred PI (4)P is subsequently dephosphorylated to PI by the ER-resident phosphatase Sac1. (V) PM PI(4,5)P2 supports PM ion channel function and is also the substrate of PLC. Receptor-mediated activation of PLC hydrolyzes PM PI(4,5)P2 into IP3 and DAG. IP3 binds to IP3 receptors on ER membranes and to initiate release of Ca2+ from the ER, while DAG recruits PKC. Excess PM DAG may be cleared from the PM to the ER via extended synaptotagmin 2 (E-Syt2). (C) Known proteins residing in ER-Golgi MCSs. (I) PI is transferred from the ER to Golgi via VAP-A-Nir2 interactions. (II) Golgi PI is the substrate for PI4KIIIβ which generates PI (4)P in an Arf-1-dependent manner. (III) OSBP tethers ER-Golgi membranes through FFAT-mediated interactions with VAP-A on ER membranes and PH domain binding of PI(4)P on Golgi membranes. The ORD (OSBP-related domain) domain of OSBP can bind and transport PI(4)P from Golgi to ER membranes and cholesterol (against its concentration gradient) to Golgi membranes. (IV) Transferred ER PI(4)P is subsequently dephosphorylated into PI by ER Sac1, supporting the steep PI(4)P gradient between the two membranes.
We first discuss PPI biogenesis, before defining the cardinal roles these lipids play in orchestrating cellular signaling cascades, and the devastating consequences that disorders of PPI metabolism have for human health.
Biogenesis of phosphoinositides
PI, the precursor of all phosphoinositides, is generally thought to be synthesized in the endoplasmic reticulum (ER) by conjugation of myo-inositol with CDP-DAG (diacylglycerol) by a PI synthase (PIS) enzyme (Figure 2B). PI produced in the ER is believed to be transported out to requiring membranes via traditional vesicular transport and by non-vesicular lipid transfer protein mechanisms at membrane contact sites (MCSs) (see section ‘Molecular architecture of phosphoinositide transport’). Additionally, one study identified highly mobile PI-synthase-containing vesicles that were postulated to be sites of PI synthesis [1]. These platforms originated from the ER, moved rapidly throughout the cytoplasm, and have been suggested to mediate the delivery of PI to other organelles such as the plasma membrane (PM) or its de novo synthesis [2] at non-ER cellular membranes. Despite the abundance of PI among mammalian phospholipids (∼10–20 mol%), we ironically have the least information regarding its subcellular distribution, as currently there are no reports of PI-specific biosensors. Nevertheless, some information can be gleaned from monophosphorylated PI(3)P and PI(4)P. The knowledge that PI can be converted either to PI(3)P on endosomal membranes or to PI(4)P at the trans-Golgi and PMs suggests that PI is available on these membranes for de novo synthesis of the designated monophosphorylated phosphoinositide. The synthesis, distribution, delivery, and metabolism of PI remains an area with significant questions still to be answered.
Phosphatidylinositol 4-phosphate
(PI(4)P) can be generated in mammalian cells via one of three pathways: (1) phosphorylation of phosphatidylinositol at the 4-position of the inositol ring (Figure 1B), (2) dephosphorylation of PI(4,5)P2 by PI(4,5)P2-5-phosphatases, or (3) dephosphorylation of PI(3,4)P2 by PI(3,4)P2-3-phosphatases (Figure 1C). A combination of cell fractionation, radiolabeling, fluorescent biosensors, and immunofluoresence investigations have confirmed that PI(4)P is found in significant amounts across several organelle compartments, especially the PM and the Golgi (Figure 2A). The PM pool of PI(4)P, which acts as a precursor for PM PI(4,5) P2, appears to be generated mostly by the PI4KA enzyme [3], which is targeted via an evolutionarily conserved complex containing TTC7B, FAM126A and EFR3 [3] (Figure 1B). The majority of Golgi PI(4)P is localized to the trans-Golgi, where PI4KB along with PI4K2A and B are responsible for its synthesis via lipid modifications and Arf1-dependent recruitment, respectively (Figure 2C) [4–7]. Following its production, Golgi PI(4)P is tightly regulated by the ER lipid PI(4)P-4-phosphatase, Sac1 (Figure 2B,C). (Like many other names of PPI-metabolizing enzymes, the name Sac1 comes from yeast genetics.) Whether Sac1 dephosphorylates its substrate in the cis configuration [acting on PI(4)P in the ER] or the trans configuration [reaching out and acting on PI(4)P on another membrane], or both, has been a topic of discussion. Crystal structures suggest that the long flexible linker between the catalytic domain and the transmembrane domain could allow Sac1 to hydrolyze PI(4)P in either configuration [8]. Currently, there appears more evidence for Sac1 working in cis [9–11] to dephosphorylate PI(4)P on ER membranes following its transfer by OSBP at ER-Golgi membrane junctions (Figure 2C; see ‘ER-Golgi sites’).
Phosphatidylinositol-3-phosphate
(PI(3)P) is a key player in membrane dynamics and trafficking. Based on biosensor information, the majority of cellular PI(3)P is found on early endosomal autoantigen (EEA1)-positive vesicles, suggesting that this monophosphorylated PPI is most abundant in an early endosome compartment. In vitro, PI(3)P can be generated following the hydrolysis of PI(3,4)P2 by INPP4A and INPP4B [12,13]. Cellular experiments further underscore the concept that INPP4A can produce PI(3)P with its overexpression rescuing the enlarged endosome morphology caused by PI(3)P deficiency [14]. Alternatively, phosphorylation of PI on the 3-position of the inositol ring by the activity of the class III PI 3-kinase (PIK3C3) [15], with additional contributions coming from the class II 3-kinase [16,17], produces PI(3)P. In humans, PIK3C3 seems to exist in at least two heterotetrametric complexes: complex I (containing PIK3C3, p150, Beclin1, and ATG14L) functioning in autophagy, and complex II (containing PIK3C3, p150, Beclin1, and UVRAG) participating in endocytic sorting [18,19]. The turnover of PI(3)P is regulated by myotubularin family phosphatases that dephosphorylate it to PI. Additionally, the PI(3)P 5-kinase, PIKfyve, can convert PI(3)P into PI(3,5)P2 as endosomes mature into late endosomes/multivesicular bodies.
Phosphatidylinositol 5-phosphate
(PI(5)P) synthesis and cellular functions remain poorly understood. Similar to PI(3)P, it is present only in low amounts in mammalian cells. Once considered an intermediate in the synthesis of phosphatidylinositol bisphosphonates or trisphosphate, PI(5)P has been reported to be present at the PM and endomembranes, as well as the nucleus. At these diverse cellular locations it appears to have roles in Akt/mTOR signaling [20], apoptosis [21], and as a nuclear transducer of stress signaling [22], respectively. The biochemical route for PI(5)P generation in mammalian cells remains controversial. The most ‘direct’ route is generation by phosphorylation of PI by a PI 5-kinase (such as PIKfyve or Type I PI5K enzymes), with an ‘indirect’ route via dephosphorylation of PI(3,5)P2 by myotubularin phosphatases [23] (Figure 1). Interestingly, during the early phases of an infection by Salmonella, PI(5)P is produced from PI(4,5)P2 [24] via the actions of type I and II PI(4,5)P2 4-phosphatases [25,26]. In contrast, the type II P(5)P 4-kinase can transform PI(5)P into PI(4,5)P2. In resting cells, it appears that the amount of PI(4,5)P2 synthesized from PI(5)P is minor; however, it cannot be ruled out that local PM PI(5)P pools could serve as precursors for the generation of PM PI(4,5)P2.
Phosphatidylinositol 4,5-bisphosphate
(PI(4,5)P2), the most abundant phosphorylated PPI, is the signature phosphoinositide of the PM (Figure 2A). It is produced primarily through the sequential phosphorylation of PI by PI 4-kinases [27] to generate PI(4)P and type I PI(4)P 5-kinases to generate PI(4,5)P2. The three type I PI 5-kinases (α, β, and γ) are localized to the PM and involved in the conversion of PI(4)P into PI(4,5)P2 [28]. In human cells, the type I PIP5Kβ forms homo- and hetero-dimers with the PIP5Kγ that appear essential for enzymatic activity and localization to the PM [29]. Additionally, PI(4,5)P2 is also produced through dephosphorylation of PI(3,4,5)P3 by PI(3,4,5)P3-3-phosphatases, such as PTEN, TPIPα, β, and γ [30]. PI(4,5)P2 can be down-regulated at the PM via two main pathways; (1) receptor-mediated hydrolysis by various subtypes of phospholipase C (PLC), and (2) dephosphorylation by members of the PI(4,5)P2-5-phosphatase family (SYNJ1, SYNJ2, OCRL, INPP5B, INPP5E, INPP5J, INPP5K: see Figure 1C). The importance of 5-phosphatase-mediated regulation of cellular PI(4,5)P2 concentrations is highlighted by the broad spectrum of diseases and disorders, including various cancers, obesity, type 2 diabetes, neurodegenerative diseases, and rare genetic conditions, which result from mutations or alterations in the expression of these enzymes (see section ‘Defective phosphoinositide metabolism in disease’).
Phosphatidylinositol 3,4-bisphosphate
(PI(3,4)P2) represents <0.1% of total PPI (Table 1) in human cells under basal conditions and mainly localizes to the PM and endocytic compartments (Figure 2A). The majority of PI(3,4)P2 seems to be formed downstream of receptors that activate PI3K, consistent with its generation from PI(3,4,5)P3 [31,32] by INPP5D (SHIP1) and INPPL1 (SHIP2) phosphatases (Figure 1C). An alternative pathway that could potentially increase PI(3,4)P2 at the PM is phosphorylation of PI(4)P via class II PI3K lipid kinases [33]. In addition, the PPI 5-phosphatases INPP5D, INPPL1, OCRL1, INPP5B, as well as synaptojanins 1 and 2 can dephosphorylate PI(3,4,5)P3 into PI(3,4)P2 (Figure 1C), it remains to be fully understood if these enzymes participate in its generation in vivo. PI(3,4)P2 is under a further layer of regulation through the metabolic phosphatase actions of INPP4A/B [34] and PTEN [35], that act on their substrates to generate PI(3)P and PI(4)P, respectively. Thus, the current view is that PI(3,4)P2 is transiently elevated at the PM following stimulation of growth factor receptors and persists through and contributes towards endocytotic events, before being trafficked to more distal membrane compartments.
Table 1.
Abundance, location, measurement, and roles of PPI lipids in eukaryotic cells
| Lipid | Abundance | Distribution | Assays for measurement | Cellular role |
|---|---|---|---|---|
| Phosphatidylinositol (a.k.a. PI, PtdIns) | ∼80 mol% of total cellular PPIs | Abundant in ER but potentially all membranes | TLC, HPLC-MS/MS, [3H] or [32P] radiolabeling to equilibrium | Precursor for PI(3)P and PI(4)P |
| Phosphatidylinositol 4-phosphate (a.k.a. PI (4)P, PtdIns4P, PI4P) | ∼2–5 mol% of total cellular PPI | PM, endosomes, trans-Golgi | TLC, HPLC-MS/MS, [3H] or [32P] radiolabeling to equilibrium, immunofluorescence, fluorescent biosensor (P4M) | Precursor for PI(4,5) P2, recruitment of clathrin adaptors, lipid binding to OSBP, ORP, CERT, FAPP2. |
| Phosphatidylinositol 3-phosphate (a.k.a. PI (3)P, PtdIns3P, PI3P) | ∼0.2–0.5 mol % of total cellular PPI | Early endosomes | TLC, [3H] or [32P] radiolabeling to equilibrium, fluorescent biosensors (FYVE) | Involved in endocytic vesicle fusion and trafficking, autophagy |
| Phosphatidylinositol 5-phosphate (a.k.a. PI (5)P, PtdIns5P, PI5P) | ∼0.01 mol% of total cellular PPI | PM, endosomes, nuclear envelope | TLC, [3H] or [32P] radiolabeling to equilibrium | Stress signal, apoptosis, Akt/ mTOR signal pathway |
| Phosphatidylinositol 4,5-bisphosphate (a.k. a. PI(4,5)P2, PtdIns4,5P2, PI4,5P2, ‘PIP2’) | 2–5 mol% of total PPI | PM, recycling endosomes, lysosomes | TLC, HPLC-MS/MS, [3H] or [32P] radiolabeling to equilibrium, immunofluorescence, genetically encoded fluorescent biosensor (PH domain from PLCδ1 for PM labeling), ion channel currents. | Intimately involved in key PM events, including regulation of endo- and exocytosis, phagocytosis, cell motility, signal transduction, and ion channel function |
| Phosphatidylinositol 3,4-bisphosphate (a.k. a. PI(3,4)P2, PtdIns3,4P2, PI3,4P2) | <0.1mol% of total PPI | PM, early endosomes | TLC, HPLC-MS/MS, [3H] or [32P] radiolabeling to equilibrium, immunofluorescence, genetically encoded fluorescent biosensor (TAPP1 PH domain) | PI3K/Akt signaling pathway, clathrin-coated vesicle maturation, endocytic trafficking |
| Phosphatidylinositol 3,5-bisphosphate (a.k. a. PI(3,5)P2, PtdIns3,5P2, PI3,5P2) | <∼2 mol% of total PPI | Late endosomes and Lysosomes | TLC, HPLC-MS/MS, [3H] or [32P] radiolabeling to equilibrium, immunofluorescence, genetically encoded fluorescent biosensor (Fyve) | Protein sorting at the late endosomes/ multivesicular bodies |
| Phosphatidylinositol 3,4,5 trisphosphate (a. k.a. PI(3,4,5)P3, PtdIns3,4,5P3, PI3,4,5P3, PIP3) | <0.05% of total PPI | PM, some endocytic compartments | Thin layer chromatography (TLC), HPLC-MS/MS, [3H] or [32P] radiolabeling to equilibrium, immunofluorescence, genetically encoded fluorescent biosensor (AKT, BTK biosensors). | Cell proliferation and cell survival, cytoskeleton dynamics, cell motility, membrane trafficking and apoptosis |
Abbreviations: ER, endoplasmic reticulum; HPLC, High-pressure liquid chromatography; MS, mass spectrometry; PM, plasma membrane; PPI, Polyphosphoinositides; TLC, Thin layer chromatography.
Phosphatidylinositol 3,5-bisphosphate
(PI(3,5)P2) is the signature PPI of late endosomal membranes. It regulates endosomal fission and fusion to maintain endomembrane homeostasis and exchange of membrane cargo. To date, there have been no reports of a PPI 4-phosphatase hydrolyzing PI(3,4,5)P3 to generate PI(3,5)P2 in cells. Therefore, the only currently known route for PI(3,5)P2 production in humans is through synthesis catalyzed by the PI(3)P 5-kinase, PIKfyve [36] (Figure 1C). As in yeast cells, VAC14 serves as a platform regulating PI(3,5)P2 synthesis by interacting directly with PIKfyve, FIG4/SAC3, and VAC7 to fine tune the regulation of PI(3,5)P2 levels [37,38]. At endosomes, production of PI(3,5)P2 induces the release of cortactin from the endosomal, branched actin network via direct interaction between the actin filament-binding region of cortactin and PI(3,5)P2. This regulation is important for membrane trafficking since actin cytoskeleton dynamics regulate membrane curvature and transport of vesicles [39]. Finally, the importance of regulated PI(3,5)P2 abundance for cellular processes is highlighted by the embryonic lethality of PIKfyve−/− knock-out mice [40].
Phosphatidylinositol 3,4,5 trisphosphate
(PI(3,4,5)P3) represents <0.05% of total PPI in human cells, and in quiescent cells it is almost undetectable. The class I PI3Ks, of which there are four isoforms in mammalian cells (α, β, δ, and γ), can be activated by a variety of cell-surface receptors to increase PI(3,4,5)P3 levels rapidly and transiently up to 100-fold in the inner leaflet of the PM [41,42]. The major reason that this minor lipid abundance is so tightly regulated is that it plays a central role in key signaling pathways: cell proliferation, migration, growth, survival, and cancer. For example, the pleckstrin homology (PH) domains of proteins such as Akt/PKB, PDK1, and Btk1 interact electrostatically with PI(3,4,5)P3. This means that receptor-mediated elevations in PI(3,4,5)P3 recruit these kinases to the PM to shape cellular signaling cascades. In many cell types, the production of PI(3,4,5)P3 and subsequent recruitment of PH domain-containing proteins to the cytoplasmic leaflet results in local actin polymerization, important for cell migration and division. The many downstream consequences of PI(3,4,5)P3 production highlight the oncogenic potential of such a powerful anabolic signal and the essential need for regulated metabolism of this minor lipid. A major regulator is the PTEN phosphatase, which catalyzes the dephosphorylation of PI(3,4,5)P3 on the 3-position to regenerate PI(4,5)P2. PTEN has been characterized extensively as a tumor suppressor [30], and mutations in the PTEN gene are found in many cancers [43]. Indeed, loss of PTEN phosphatase activity, coupled with the very common mutations in the PIK3CA isoform, makes enhanced PI3K signaling the most common oncogenic event [44].
Molecular architecture of phosphoinositide transport
We have been discussing enzymatic pathways and molecular mechanisms underlying the generation and destruction of each PPI species in a membrane-specific way. This is an oversimplification. Indeed, cellular membranes communicate extensively with one another via two distinct cellular processes: by traditional membrane trafficking and by lipid exchange at MCSs. PPIs co-ordinate and regulate intracellular trafficking by acting as molecular beacons and effectors to co-ordinate and recruit important trafficking machinery. Furthermore, the continuous membrane flow between organelle compartments mediated by vesicular traffic, coupled with precise spatial and temporal localization of PPI-metabolizing enzymes, is an essential process that ensures the heterogeneous distribution of PPIs throughout eukaryotic cells. Underscoring the symbiotic relationship between trafficking, PPI enzyme localization, and PPI distribution, local changes in PPI levels mediate fine-tuning of key cellular events, such as vesicular budding and membrane fusion, whereas gross manipulation of PPI abundance at the Golgi [45–47], lysosomes [48], or endosomes [49] results in disruption of trafficking organelles and downstream reorganization of the PPI zip code. It is clear that co-ordinated vesicular trafficking is required for normal PPI distribution.
The second class of transport mechanisms used to transport PPIs are MCSs. These sites are platforms of information transfer akin to synapses where two intracellular membranes come into close proximity to one another (∼15 nm) and allow lipids (among other things) to be exchanged. The concept of close proximity between two organelle membranes without membrane fusion is important. Unlike traditional vesicular trafficking that transports cargo (lipids and protein) from one organelle to another via membrane fusion, the exchange of lipids at MCSs occurs via protein-mediated transport or exchange (Figure 2B,C). The lipid transfer proteins form a protected conduit — a bridge — for lipid transfer. Thus, organelle identity and architecture are preserved as molecules are transferred. Despite great heterogeneity in the molecular elements defining each MCS, it is striking that the majority of these MCSs complexes contain and transport several PPIs and catalyze their metabolism.
ER–PM contact sites
The best characterized MCS in terms of PPI metabolism is that between the ER and the peripheral PM (Figure 2B). First characterized in muscle cells in the 1950s, an essential role for ER–PM contact sites is the regulation of cytoplasmic calcium during excitation–contraction coupling in striated muscle cells [50–52]. More recently, it has become increasingly evident that ER–PM MCSs also provide a rapid non-vesicular transport pathway for PPIs, phosphatidylserine, and sterols to be transferred and modified at the ER and PM.
Transfer of PI at MCSs:
The precursor for all PPIs is PI. PI can be transferred to the PM from the ER via the ER-resident membrane protein TMEM24 [53] (Figure 2A). The TMEM24-mediated transfer of PI is enabled by C2-domain interactions with the PM and by a lipid transport module of the synaptotagmin-like, mitochondrial, and lipid-binding protein (SMP) family, which binds glycerolipids, including PI. The binding of ER TMEM24 ‘in trans’ with the PM occurs in a phosphorylation- and Ca2+ -dependent manner, allowing the protein to disconnect transiently from the PM as the Ca2+ concentration spikes and then reassociate upon dephosphorylation [53]. TMEM24 is thought to be important in neuro-endocrine cells for regulated insulin secretion [53]. Another mechanism of PI transfer at ER–PM MCSs involves the phosphoinositide transfer proteins (PITPs), Nir2 and Nir3 (Figure 2B). Following Gq-coupled receptor stimulation, PM PI(4,5)P2 is hydrolyzed and Nir2 and Nir3, are recruited to ER–PM MCSs (Figure 2B) [54–57]. This recruitment is triggered through the combined actions of phosphatidic acid and DAG [54,56] and mediated through an interaction with the FFAT motif on the ER vesicle-associated membrane protein (VAMP)-associated proteins A and B (VAP-A and VAP-B) [58] (Figure 2B). The stimulated recruitment of Nir2 and Nir3 at ER–PM junctions delivers PI to the PM as a precursor to replenish PM PPIs, while concurrently returning phosphatidic acid to the ER for future PI regeneration [56].
Transfer of PI(4)P at ER–PM MCS:
There are two major pools of PI(4)P within cells, in the PM and in the Golgi. The majority of PM PI(4)P appears to be generated by PI4KIIIα and its associated factors (Figure 1B,C). Recently, two integral ER membrane proteins, oxysterol-binding protein (OSBP)-related protein 5 (ORP5) and 8 (ORP8), have been identified as additional key regulators of PM PI(4)P [59,60]. ORP5 and 8 tether ER–PM MCSs through the action of a hydrophobic tail sequence that is anchored in the ER membrane and a PH domain that interacts with PM PI(4)P and PI(4,5)P2 [59]. These ORP junctions facilitate the countertransport of phosphatidylserine (PS) for PI(4)P. Transferred PI(4)P is then dephosphorylated to PI by the PI(4) P-4-phosphatase Sac1 on ER membranes. The major distinction between ORP5 and 8 is their binding affinities towards PI(4)P and PI(4,5)P2, with ORP5 capable of engaging PM PPIs under resting conditions, whereas ORP8 requires higher levels of each substrate. These differences in PPI affinity present two modes of action for ORP5 and 8. Mode one involves ORP5 binding and transferring PI(4)P to the ER under resting levels of PI(4) P. Here, decreases in PM PI(4)P or PI(4,5)P2 will uncouple ORP5 from the PM to aid replenishment of each PPI pool. Mode two involves ORP8, which binds PI(4)P and PI(4,5)P2 under periods of excess PI(4)P and PI (4,5)P2 production. Here, increases in PM PI(4)P and PI(4,5)P2 would enhance PM ORP8 binding to facilitate increased transfer of PI(4)P to the ER, reducing PM PI(4)P and PM PI(4,5)P2. In both scenarios, ORP5 and 8 proteins serve as a rheostat regulating PM PI(4)P levels.
ER-PM contact sites are under a further layer of regulation from the extended synaptotagmin (E-Syt) family of proteins [61]. These three (E-Syt 1, 2, 3) ER-resident proteins interact with PM PI(4,5)P2, via their C2 domains, to tether ER-PM membrane contact sites. In the context of PPI metabolism, E-Syt1 and E-Syt2 appear important for the extraction and transfer of DAG from the PM to the ER during periods of Gq coupled receptor activation (Figure 2B) [61]. Given this role, it seems likely that E-Syt’s act in cooperation with the aforementioned Nir2 pathway to stabilize ER-PM contact sites and control the accumulation of PI(4,5)P2 metabolites, while providing precursor elements for future PI synthesis.
ER-Golgi contact sites
Transfer of PI(4)P at ER-Golgi MCSs:
At ER-Golgi MCSs three lipid transfer proteins with FFAT domains bind ER-localized VAP proteins: CERT, Nir2, and OSBP. OSBP1 contains an ORD and PH domain to bind and transfer both sterols and PI(4)P, respectively. Under conditions of high Golgi PI(4)P, paired with the presence of VAP-A and -B on ER membranes, OSBP is recruited and tethers ER-Golgi junctions (Figure 2C). Once positioned at ER-Golgi MCSs the ‘OSBP cycle’ proceeds wherein cholesterol on ER membranes is exchanged for PI (4)P on trans-Golgi membranes [11,62]. The energy required for this cycle is supported by the steep PI(4)P gradient between Golgi and ER. There is continuous dephosphorylation of PI(4)P by the ER-localized Sac1 and continuous production of PI(4)P at the TGN by PI4-kinases, notably PI4KIIIβ located close to OSBP in an Arf1-dependent manner. Accordingly, in the OSBP cycle, PI(4)P serves as a lipid form of free energy to support the unidirectional transport of cholesterol from the ER (site of its synthesis) to the Golgi, against its concentration gradient. In this sense, OSBP function is analogous to that of the Na+/Ca2+ exchanger that uses the energy stored in the electrochemical gradient of Na+ to drive Ca2+ extrusion out of the cell against its concentration gradient.
ER–endosome contact sites
Recent studies have revealed multiple MCSs between the ER and endosomes [63–66]. Two of them have a PPI as a central component to promote microtubule-dependent endosomal transport [67] or retromer- and WASH-dependent budding from endosomes [68]. For PPI-dependent, ER–endosome-mediated transport, the ER protein protrudin forms contact sites with late endosomes through electrostatic interactions with PI(3)P in a Rab7-GTP-dependent manner. This coincidence binding on late endosome membranes promotes MCS formation and enables hand-over of kinesin-1 from protrudin to the adaptor protein FYCO1 on late endosomes. Consequently, late endosomes are transported via plus-end-directed transport along microtubules into growing neurites to promote protrusion formation and neurite outgrowth.
Contacts between the ER and endosomes also influence vesicle budding through changes in lipid composition that affect actin dynamics. These MCSs are formed by VAPA/B binding to the retromer subunit SNX2 and to OSBP, which binds PI(4)P on the endosomal membrane via its PH domain. In WT cells, transient accumulation of PI(4)P on endosomes via type II PI4Ks is coupled to a transient burst of WASH-dependent actin nucleation to facilitate retromer function [68]. In cells that lack VAP, excessive and persistent PI(4)P accumulation on endosomal membranes causes disruption of retromer-dependent budding and disruption of trafficking between endosomes and the Golgi complex.
It is worth noting that ER–endosome contact sites containing either protrudin or VAP-retromer expand the role of MCS from stages of lipid and calcium transfer to hubs of protein docking and sorting that control critical cellular events.
Tools for exploring phosphoinositide pools
Part of the reason that we know so much about the biogenesis and distribution of these rare lipids is development of new tools. Indeed, outstanding cell biologists who excel in logic, biochemistry, genetics, microscopy, and molecular biology have pioneered development of biosensors for detecting PPIs and other tools that alter PPI abundance. Using these complementary approaches allows investigators to visualize the PPI species in question and to manipulate that species to determine its potential function. We now discuss techniques and strategies for the detection and manipulation of these lipids. Such outstanding tools, expanded upon below, have been instrumental in revealing the important cellular roles of PPIs.
Detecting the PPIs
Historically, the most practical approach to monitor pools of cellular PPIs and their dynamics is via classical biochemical approaches that depend on chemical analysis of lipid extracts from biological samples. These methodologies measure the aggregate PPI content of populations of cells rather than the content of a specific cellular membrane or organelle. Although it is possible to assay subcellular fractions, the important phosphorylation state of PPIs may change during procedures lasting many minutes, requiring fastidious attention to slowing the actions of endogenous lipid phosphatases and kinases. The chemical methods often need >100 000 cells, and they are static, with each assay resolving only one time point. One of the first methods was equilibrating preincubation of cells with [32P]phosphate or [3H]inositol for several days, acid extraction of lipids into an organic phase, and reading the counts in the various peaks resolved by thin-layer or other chromatography. In such labeling experiments, it is possible with different elution solvents to discriminate most of the major PPI classes and to determine their relative amounts. With briefer incubations with [32P]phosphate instead, it is possible to observe fluxes, the rates of synthesis of PPIs. A more elaborate method without labeling, separates the isolated glyceryl-headgroups on anion exchange HPLC with detection by conductivity [69]. Becoming more common now is ultra-high-pressure liquid chromatography coupled to tandem mass spectrometry (HPLC-MS/MS). This still-evolving method can resolve large numbers of lipid species, and permits lipidomics. For PPI lipids, the negative phosphate groups in the sample need to be methylated to make them neutral [70]. The analysis of ionized fragments yields the atomic mass, readily making possible a determination of the number of phosphates, the total chain length of attached fatty acid side chains, and their degree of unsaturation [71–73]. With care for sensitivity, more than a dozen different fatty acid side-chain combinations are readily resolved for each major PPI class. In the pools of PI and PPIs of native mammalian cells the dominant (∼70%) fatty acid composition is 1-stearoyl 2-arachidonoyl (symbolized 38:4 for 38 carbons and 4 double bonds; Figure 1A), but in cultured mammalian cells the arachidonoyl content is lower [71–73]. The fatty acid content is roughly the same in all classes of PPI in a single sample as if they form a dynamic and interconverting metabolic pool. Methods are being developed to distinguish some of the regio-isomers in a PPI family [e.g. separating PI(3,4)P2 from PI(3,5)P2] [73], but this is still not yet routine. Mass spectrometry can be calibrated with internal standards to yield the absolute numbers of molecules of each lipid per cell.
Another static method for visualizing distribution and abundance of PPIs is immunofluorescence. Commercial antibodies have been raised against PI(4)P and PI(4,5)P2. They are used in conjunction with cell fixation and permeabilization protocols tailored to preserve PPI lipids and specific membranes of interest. The cells are stained with primary and secondary antibodies to give fluorescence localization under confocal microscopy [74]. Such results are qualitative, but they have two important advantages over the other static methods: The observations are made on single cells, and they provide subcellular localization at the light-microscope level.
Genetically encoded biosensors are versatile tools that allow dynamic optical, real-time monitoring of PPIs in single cells [75]. They take advantage of the lipid-binding modules that many cytoplasmic proteins have evolved to interact with specific membrane PPIs [76]. Recently, several excellent reviews carefully detail methods used to determine the lipid binding specificity of individual biosensors, the limitations of these methods, and the challenges when interpreting the resulting data [75,77–79]. To illustrate the general utility in using biosensors, we provide the example of a binding domain from phospholipase Cδ1 (PLCδ1). PLCδ1 has a PH domain that selectivity recognizes PI(4,5)P2. Fusion proteins formed between the 120-residue PH domain sequence and fluorescent proteins serve as genetically expressible fluorescent labels that indicate where PI(4,5) P2 is in cells [80,81]. In normal cells, the fluorescent diffusible PH-PLCδ1 probes localize to the PM, virtually the only membrane with PI(4,5)P2 (Figure 2), but if the lipid is depleted by receptor-activated PLC, the probes move quickly into the cytoplasm, having lost their binding partners. They return to the membrane as PI(4,5)P2 is resynthesized from PI. Probe movements can be monitored as translocation in confocal or total internal reflection microscopy or by changes in fluorescence resonance energy transfer between the probe and another suitable fluorophore. A large repertoire of such lipid-binding modules has been developed as PPI probes for live-cell experiments [79]. Considerations when using such biosensors include how selective they are for a particular PPI species and that some biosensors may not report every membrane containing their target lipid but rather act like coincidence detectors that require other cofactors such as Arf-GTPases to recognize a membrane [76]. Illustrating these two points, PH-PLCδ1 domains can bind IP3 as well as PI(4,5)P2, and a more selective choice for monitoring PI(4,5)P2 might be the Tubby domain, whereas the PH domain from OSH1 requires the coincidence of PI4P and Arf1 for targeting to trans-Golgi network membranes. Despite these caveats and the fact they do not report absolute amounts of lipids, these genetically expressible probes have great advantages of versatility, speed, low cost, and continuous real-time monitoring in living cells. They are readily detected in single cells or cell populations. They do require the use of a fluorescence microscope and expression of proteins in the cells.
Manipulating PPIs
Many good tools exist for manipulating the pools of PPIs during chronic or acute experiments in living cells. Most involve manipulation of enzymes of PPI metabolism: the lipid kinases, the lipid phosphatases, and the phospholipases. These enzymes have been knocked out and knocked down genetically, they have been overexpressed by transfection, they have been activated by receptors or by voltage, they have been blocked pharmacologically, and they have been recruited from cytoplasm to membranes by chemical- or light-mediated dimerization. Although much basic and important PPI biology was discovered through classical genetics and molecular biology in yeast and then mice, we focus here on other cell biological approaches.
Receptor-activated lipid perturbation has been limited to two families of enzymes, PLC (activated by Gq-coupled receptors or growth-factor receptors) and PI-3-kinases (also activated by growth-factor receptors) (see section ‘Physiological roles of phosphoinositides’). Receptor-linked lipid phosphatases seem uncommon. Nevertheless, PI(4,5)P2-5-phosphatases and PI(4)P-4-phosphatases have been engineered to make several powerful genetically expressed tools. The idea is to attach the enzyme catalytic domain without regulatory or localization sequences to a platform that can be recruited to the PM or to an intracellular membrane. The recruitment needs another genetically expressed partner, the anchor, already attached to the chosen target membrane. The platform with its enzyme is engineered to be dispersed in the cytoplasm where the enzyme does not encounter its substrate. Heterodimerization of the platform with the anchor then is achieved chemically as with a rapamycin dimerization system [82,83] or by light-activated dimerization systems [84,85] translocating the enzyme to the desired target. Translocatable platforms combining two enzymes, e.g. both PI(4,5) P2-5-phosphatase and PI(4)P-4-phosphatase together [86], or just PI(4)P-5-kinase or a 3-kinase [82] also have been useful. These tools when directed to a known membrane serve to localize subcellular PPI pools and to document the effects of perturbing one pool on another [86–88]. These tools are convenient and easily used. The translocation and the actions of the enzymes on specific PPI pools are completed in 10–20 s. An even faster depletion of PI(4,5)P2 can be accomplished with voltage-sensing phosphatases (VSPs) that, depending on the animal species of origin, have different ranges of voltage sensitivity [89,90]. The principal enzymatic activity is a lipid 5-phosphatase, but the enzyme also removes 3-phosphates at a lower rate [91,92]. This tool requires patch-clamp electrophysiology to make depolarizing pulses. Since any applied depolarization is limited to the PM, it acts very specifically on PM PPIs. With zebrafish voltage-sensing phosphatase (DrVSP), a plasma membrane depolarization to +110 mV can dephosphorylate the PI(4,5)P2 pool to PI(4)P in <1 s [93]. The main advantages of the method are speed, and the possibility of spatial and temporal control of the amount of PI (4,5)P2 depletion.
Several other tools are convenient for PPI manipulation. The simplest is a pharmacological block of lipid kinases or phosphatases. The two most widely used are wortmannin, which blocks lipid 3-kinases at <10 nM and some 4-kinases at >100 nM concentrations [94,95], and phenylarsine oxide, which reacts with vicinal cysteines and blocks 4-kinases as well as tyrosine kinases [81]. Other, more specific inhibitors of PPI-metabolizing enzymes are being developed, including inhibitors against PI4KA [96], PI4KB [97], PIP5KA [98], and PIP5KC [99]. For the class 1 PI3Ks, there is a large and expanding body of data establishing the target selectivity of numerous inhibitors. These efforts were made specifically to improve on older molecules such as wortmannin that have many off-target effects. Illustrative examples of relatively selective drugs include Idelalisib, the class I PI3K delta selective inhibitor approved for B cell malignancies [100], or pan-class I inhibitors such as Pictilisib/GDC0941 [101] or Buparlisib/BKM120 [102] currently in clinical studies for the treatment of patients with advanced solid tumors and breast cancer, respectively. Despite the growing list of compounds that can alter PPI species it would be valuable to develop others. Another approach requires exposing the cytoplasmic face of the PM as is done in inside-out patch clamping. While recording ion channel function, the effective membrane PPI concentration then can be reduced by bathing the cytoplasmic face with PPI-chelating polycations such as polylysine or polyamines or with anti-PPI antibodies. PPIs can be restored by perfusing the membrane patch with water-soluble dioctanoyl-PPI analogs such as diC8-PI(4,5)P2. For ion channel studies, the use of diC8 compounds has been very successful to estimate relative affinities from concentration–response experiments [103,104]. Recently, it has also become possible to apply PPIs to intact cells from the outside as membrane-permeable acetoxymethyl esters and even as membrane-permeable, light-releasable caged PPIs [105].
A tool of quite a different kind is mathematical modeling. Especially, when working with living cells, trying to dissect out and predict the importance of a particular enzyme of lipid metabolism is complicated by the ongoing actions of many other enzymes and by the dynamics of traffic and exchange reactions between organelles. Qualitative discussions often fail to recognize the emergent properties of such background integrated metabolism. Realistic modeling that includes many enzymes greatly helps in interpreting the activity and consequences of stimulating or inhibiting one of them [92,93,106–110].
Physiological roles of phosphoinositides
Many functions for phosphoinositides have been revealed during the last 50 years, in parallel with growing understanding of their chemical structures and metabolism. Virtually all functions fall loosely under the heading of signaling. Here, we emphasize signaling by the phosphoinositides themselves, but first we mention briefly two important examples where a phosphoinositide is the precursor of a signal. One early major discovery was realization in the 1970s and early 1980s that PI(4,5)P2 is the principal substrate of receptor-activated PLC. This enzyme cleaves PI(4,5)P2 to yield two potent second messengers, inositol trisphosphate IP3 and DAG. In this signaling cascade, the IP3 releases calcium from intracellular stores, and the DAG activates several proteins, notably protein kinase C (PKC) [111,112]. Thus, many hormones and modulatory neurotransmitters acting through Gq-coupled receptors or through some tyrosine-kinase receptors can initiate intracellular calcium signaling and protein phosphorylation by PKC. Virtually every animal cell receives regulatory inputs acting through this important pathway. Another major discovery in the late 1980s was lipid 3-kinases acting on several phosphoinositides that, among other reactions, convert PM PI(4,5)P2 to PI(3,4,5)P3. This very low abundance and transient phosphoinositide recruits protein kinases (Akt, BTK, etc.) and downstream signaling cascades to the membrane. Thus, insulin and many growth hormones that signal through tyrosine-kinase receptors to a PPI 3-kinase initiate metabolic changes, cell proliferation, and growth programs. Defects in such signaling may lead to cell transformation and are common in cancers. We refer the reader to reviews on PI 3-kinase signaling [44,113].
Hilgemann hypothesis for intrinsic membrane proteins
As we have seen, organellar membranes and the PM present a phosphoinositide ZIP code unique to that compartment. After discovering the first examples of an ion channel and of a transporter that each required PI(4,5) P2 for function [114], Donald Hilgemann proposed ‘thus ion channels and transporters that are activated by PIP2 will probably be inactive during the processing and passage through the secretory pathway’ [115]. Such a membrane protein that is synthesized in the ER, traffics through the Golgi, and is sent in vesicles to the PM would remain silent until it arrives ‘home’ at the PI(4,5)P2-containing PM, and if it is subsequently recycled by internalization, it would be silenced again. We call this the Hilgemann hypothesis. By generalization, there could be other membrane proteins that need to be silenced at the PM and are active only at another particular compartment. That could be accomplished by requiring a different phosphoinositide for function. Indeed, two ion channels of endolysosomal compartments (TRPML1 and TPC1) require the endolysosomal signature lipid PI(3,5)P2 and are not activated or are even inhibited by PI(4,5)P2 [116,117]. Finally, there are ion channels whose PPI preference is less strict; they can be activated by more than one phosphoinositide [118]. This might permit them to perform their functions in several specific organellar membranes. In general, Hilgemann’s hypothesis still needs broader testing, but it seems to us to offer a powerful explanation for why many intrinsic membrane proteins have evolved strong preferences for specific phosphoinositides and a neat cell biological principle for functional compartmentalization of membrane proteins.
Regulation of peripheral membrane proteins
Membranes isolated by cell fractionation include proteins that are loosely bound in addition to the transmembrane proteins and proteins that are firmly anchored by lipid modification. These are the peripheral membrane proteins that may be associated by electrostatic bonds to membrane components. Frequently at least one of their interaction partners is a charged lipid head group, especially PPIs. Peripheral proteins can be released by high salt solutions or by low pH. Polybasic domains of the protein associate reversibly with the acidic phosphates of the head group and dissociate to find new partners as the membrane lipids change during membrane trafficking and signaling. In this way, a long list of cytoplasmic proteins including, for example, PLC, cPLA2, pleckstrin, spectrin, ankyrin, AP-2, mSOS1, Rab35, Rho, Arf6, ARNO, and CYTH, transiently associate with the PM through PH domains and other lipid-interaction modules [76]. The emphasis is that this association is dynamic and regulated in part by the PPI phosphorylation pattern on the membrane. This invites the important question as to how one PPI species might regulate many different effector proteins independantly? One general hypothesis is that local diffusional barriers structurally organize or concentrate specific PPI pools, essentially creating segregated PPI signaling domains (reviewed [119]).
Dynamic regulation of intrinsic membrane proteins by Gq-coupled receptors
Brown and Adams [120] discovered a potassium current of sympathetic neurons that was suppressed by muscarinic agonists and by other GPCR agonists. These agonists were subsequently shown to activate Gq and PLC. The current originally called M-current to reflect its muscarinic regulation was eventually shown to be carried by members of the KCNQ (KV7) ion channel family. Since these K+ channels are partially open at rest, they dampen the electrical excitability of the neurons. Receptor-induced closure leads to heightened electrical activity, behavioral arousal in central circuits, contraction of smooth muscle, and vasoconstriction [121–124]. More than 20 years after the discovery of M-current, closure of the channels was finally found to be due to receptor-mediated depletion of PI(4,5)P2 [125,126]. Such current suppression by PLC can be mimicked by stimulating a depolarization-activated voltage-sensing lipid phosphatase [93,127] or light-activated optogenetic lipid 5-phosphatase [85], enzymes that deplete PI(4,5)P2 by dephosphorylation to PI(4)P.
The hypothesis that KCNQ channels require PI(4,5)P2 to function was initially unexpected. Although cleavage of PI(4,5)P2 was the well-known enzymatic activity of PLC, it had not been fully appreciated that PI(4,5)P2 is at such low concentration and is synthesized sufficiently slowly that it could be used up by stimulating endogenous PLC. Subsequently, it was shown by HPLC and by tandem HPLC-mass spectrometry that activating transfected or endogenous Gq-coupled receptors in cell lines and in native cells can deplete nearly the entire cellular pool (∼90%) of PI(4,5)P2 with a time constant as short as 7–10 s. The depletion was perfectly synchronous with the translocation to the cytoplasm of PLCδ1 PH domains and with the loss of KCNQ channel current [72,128,129]. The total cellular PI(4)P pool fell by more than 70% in tens of seconds as well, reflecting the tight metabolic coupling of cellular PI(4,5)P2 and PI(4)P pools despite their partial separation in space. In sympathetic neurons and in expression systems, the same receptor-mediated PI(4,5)P2 depletion also decreased current in two types of CaV1 (L-type) and in two types of CaV2 (P/Q and N-type) voltage-gated Ca2+ channels [130–133]. In these cases, channel gating was modulated, but 40–70% of the original peak current still remained as if PI(4,5)P2 was regulatory but not absolutely essential for Ca2+ channels to open. Such receptor-mediated Ca2+ channel modulation would decrease the release of neurotransmitter at synapses. Since the Ca2+ currents could also be down-regulated by activating voltage-sensing or light-activated lipid 5-phosphatases that generate PI(4)P, we conclude that for both KCNQ channels and CaV channels monophosphorylated PI(4)P is not a good substitute for PI(4,5)P2.
How general is such rapid depletion of lipid by receptors and consequent regulation of membrane protein function? Only a few studies have shown depletion using direct chemical measurements of PI lipids in native cells [72,128,134], but indirect evidence comes from studies of ion channels. Suppression of KCNQ and CaV channel currents occurs in a broad variety of neurons and smooth muscles via a wide range of different endogenous PLC-coupled receptors, including M1 and M3 muscarinic, tachykinin, bradykinin, endothelin, vasopressin, 5-HT2, AT1 angiotensin II, α1-adrenergic, H1 histamine, P2Y purinergic, and GnRH [123,135–140]. When an endogenous PLC-coupled GPCR is present at only low density, it may still evoke sufficient production of IP3 and DAG for signaling but might not activate PLC strongly enough to deplete PI(4,5)P2.This is the case for purinergic and bradykinin receptors in sympathetic ganglia [141,142] and endogenous purinergic receptors in tsA cell lines [108,109]. Nevertheless, many native cells are equipped with a high enough density of PLC-coupled GPCRs and of PLC to experience quick and extensive receptor-mediated depletion of PI(4,5)P2.
PI(4,5)P2-sensitive intrinsic membrane proteins
What membrane proteins are sensitive to PI(4,5)P2? The first two to be recognized were a KATP channel and a Na+–Ca2+ exchanger [114], two multi-pass transmembrane proteins that move ions across the membrane. After two more decades, 80 ion channels and 6 transporters have known PI(4,5)P2 requirements [143–145]. For channels, today’s list includes nearly every TRP channel, the five KCNQ channels, four CaV channels, all of the GIRK channels, many other inward rectifier K+ channels, and numerous others. For transporters, the list includes notably the dopamine and the serotonin transporters [146].
Three questions come immediately to mind that we now proceed to answer: (i) Why are so many channels and transporters sensitive to PPIs? (ii) Why not other kinds of proteins? (iii) How do these proteins interact with the lipids and how does this regulate their activities? The first two questions might be easy to speculate about. We suggest that the list of channels and transporters is so long because all of them are regulated by the PI ZIP codes they encounter during membrane traffic — the Hilgemann hypothesis. This would prevent them from leaking or transporting ions across the wrong compartment membranes, reserving that job for other channels and transporters functionally adapted for that compartment. Alternatively, perhaps the list is long to allow many channels and transporters to be regulated co-ordinately by PLC-coupled receptors. Such a model becomes more reasonable when one considers that not every cell has the full gamut of PI(4,5)P2-sensitive channels and transporters, thus precise spatial distribution of select PLC-coupled receptors, channels, and transporters could allow local changes in electrical activity in specialized cells like neurons. The second question asks why the list contains few transmembrane enzymes, receptors, and other proteins. We suggest here that these proteins present a greater challenge for the experimentalist to study, and quite probably with time many receptors and intrinsic enzymes will appear on the list. Supporting such a statement, recent evidence suggests G-protein-coupled receptors interact with PPIs to stabilize their active state and enhance selectivity of G-protein coupling [147], while β-arrestin binding to PPIs seems required for its trafficking to clathrin-coated pits [148]. Channels have been relatively easy to study because the patch-clamp assay gives an instantaneous before–during–after readout as the lipids are manipulated in living cells by the many tools available. These experiments take only some seconds to perform. For other classes of membrane proteins, we lack rapid read-outs in living cells. An alternative explanation of the paucity of other proteins on the list might be that cellular physiology is not profoundly perturbed if these proteins continue to be functional as they traffic among different compartments or perhaps they are shut off adequately by other factors like low vesicular pH.
We turn now to the mechanism of these interactions. Channels differ in their apparent lipid affinity, in their PPI lipid specificity, and in the effect of the lipid on them. The KCNQ channel exemplifies low-affinity binding, which means that in normal cells the KCNQ channels are not fully saturated with lipid and respond readily to decreases in PI(4,5)P2. The lifetime of the KCNQ channel-lipid complex is < 0.5 s, perhaps much less [93]. The effects of lipid can be described as a large increase in channel open probability, and, with a very low level of the lipid, the channel may be shut off entirely. CaV channels also seem to form low-affinity complexes with PI(4,5)P2. The lifetime of the complex is <100 ms, but standard means of depletion of PI(4,5)P2 still leave 40–70% of the current [133]. At another extreme, Kir2.1 inwardly rectifying K+ channels are nearly saturated with PI(4,5)P2. They form a high-affinity complex with the lipid that requires extensive lipid depletion to dissociate and might not be much affected by typical physiological receptor-mediated PI(4,5)P2 reductions [103,149,150]. KCNQ channels are PI(4,5)P2 preferring but can be activated by other PPI lipids and related compounds (not IP3) presented to an excised membrane patch [104]. Even more promiscuously, GIRK1/ GIRK4 channels need, but do not distinguish between, PI(3,4)P2, PI(3,5)P2, PI(4,5)P2, or PI(3,4,5)P3 [103].
Typical neuronal KCNQ channels are heterotetramers of KCNQ2 and KCNQ3 subunits. In single-channel recordings, careful titration with increasing concentrations of dioctanoyl PI(4,5)P2 reveals a two-component activation curve for the KCNQ2/KCNQ3 heteromers. The higher affinity component attributed to the KCNQ3 subunits has a midpoint at 1.3 µM and the lower affinity attributed to KCNQ2 subunits, at 76 µM [104]. These apparent affinities correspond well with the affinities seen with the two types of homomeric channels. Such experiments show that the same channel tetramer can bind PI(4,5)P2 at more than one site. A simple presumption is that each of the four protein subunits binds one lipid molecule, and the functional results imply that a channel can open when only two sites are occupied, although with a lower open probability than with four.
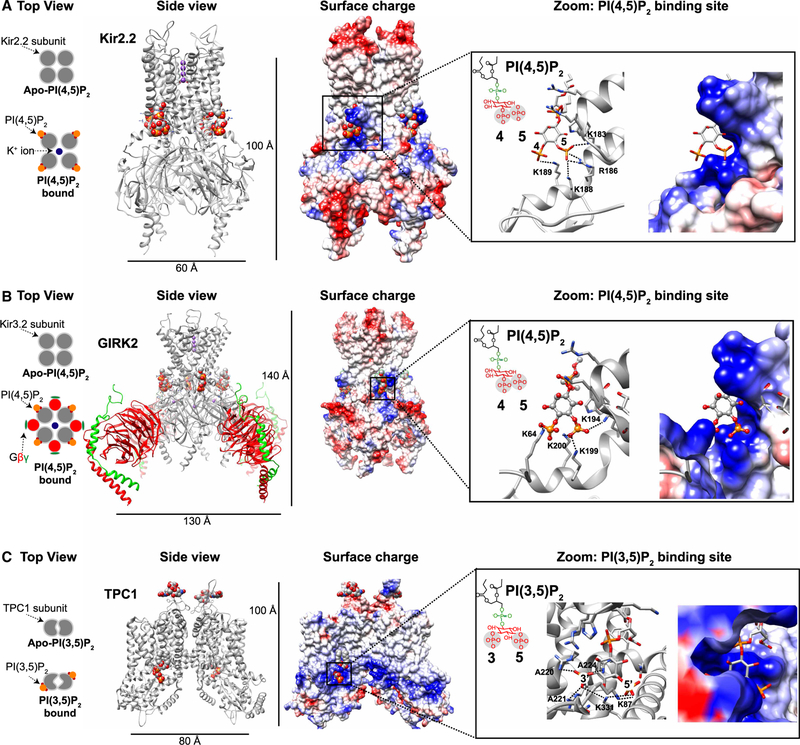
Putative binding sites for PI(4,5)P2 on many ion channels have been identified by a combination of mutagenesis, molecular modeling, and docking [151]. Invariably they contain numerous arginine and lysine residues as basic counterions for the five negative charges on the lipid. More definitively, three cryo-EM structures at atomic resolution show PI(4,5)P2 bound to K+ channels: a Kir2.2 channel [152] (Figure 3A), a GIRK2 channel [153] (Figure 3B), a KATP channel complex [154], and another pair of structures compare an apo-TPC1 channel with a PI(3,5)P2-bound TPC1 channel [155] (Figure 3C). Each tetrameric channel has four bound lipid molecules and the dimeric TPC1 channel has only two. The lipids are oriented with respect to the membrane as one might expect for phospholipids in a membrane with lipid tails extending away from the cytoplasm and the head group lying at the level of the membrane–cytoplasm interface. However, the lipid is mostly encased by hydrophobic regions (blue area, ‘surface charge’, Figure 3) of the pore-forming protein, and the head group phosphates are interacting in a cup with arginines, lysines, a histidine, glutamine, and asparagine of the cytoplasmic domain of the channel protein. Particularly in the Kir2.2 structure (Figure 3A), one might say that the encased lipid acts like a rivet securing the cytoplasmic domain to the membrane domain of the channel in a functional configuration [152]. In each example, the basic residues of the binding site come from well-separated sequences along the main carbon chain that are folded to bring them together. There is no consensus fold such as is found in PH domains. We can expect further direct structure determination for many other channels in the future.
Figure 3. Structural basis of PIP2 interactions with several ion channels.
In each case, the lipid is the diC8 version of a PIP2. (A) Top view: schematic representation of Kir2.2 subunits before and after PI(4,5)P2 binding. Side view: X-ray crystal structures of the Kir2.2 homotetrameric channel in presence of PI(4,5)P2 (gray, PDB: 3SPI [152]). Each channel subunit is in complex with a single PI(4,5)P2 molecule represented as spheres and colored according to atom type: carbon, gray; phosphorous, orange; and oxygen, red. Surface charge: electrostatic map of Kir2.2 homotetrameric channel structure (blue is positive; red negative; white neutral). The PI(4,5)P2 binding site comprises numerous basic residues (blue) that interact electrostatically with the negatively charged phosphates of PI(4,5)P2. Zoom: PI(4,5)P2 binding site. Note that positively charged residues in the protein structure are closely apposed to and predicted to interact with the negatively charged phosphate groups at the D4 and D5 positions of PI(4,5)P2. (B) Top view: schematic representation of Kir3.2 (GIRK2) subunits before and after PI(4,5)P2 binding. Side view: X-ray crystal structure of the Kir3.2 homotetrameric channel with accompanying Gβγ subunits (gray, PDB: 3SYA [153]). Surface charge: electrostatic map of Kir3.2 homotetrameric channel structure. Zoom: PI (4,5)P2 binding site. (C) Top view: schematic representation of TPC1 subunits before and after PI(3,5)P2 binding. Side view: cryo-EM structure of the endolysosomal TPC1 homodimeric channel in the PI(3,5)P2-bound state (gray, PDB: 6C9A [155]). Note that similar to Kir2.1 and Kir3.2, one PI(3,5)P2 molecule binds to each TPC1 subunit. Surface charge: electrostatic map of TPC1 homodimeric channel structure. Zoom: PI(3,5)P2 binding site.
Other physiological roles for the PPIs
We now briefly mention other important roles of PPIs in exo-/endocytosis, tethering, fusion, cell migration, cell division, and autophagy. During calcium-regulated synaptic vesicle release, PI(4,5)P2 is required to attract the C2B domain of vesicular synaptotagmin to the PM active-zone for docking and fusion [156], then to switch on the clathrin adapter protein AP2 to form clathrin-coated pits [157], and later to attract dynamin for pinching off the endocytosing vesicle and synaptojanin for dephosphorylating the PPI for its next roles and releasing AP-2 [158]. In cell migration, the chemoattractant sets up a PPI polarity by activating PI3K at the leading edge to produce PI(3,4,5)P3 and by localizing the 3-phosphatase PTEN to the trailing edge favoring PI (4,5)P2 in the rear [159,160]. A similar scheme might apply to the leading edge of the myelinating Schwann cell as it forms myelin by advancing spirally to wrap around an axon [161]. In cell division, PPIs are implicated in spindle orientation, mitotic cell shape, and bridge stability (see [162] for review), with PI(3,4,5)P3 levels during pro- and metaphase important for spindle positioning, PI(4,5)P2 accumulation during telophase essential for coordinating assembly of the contractile actomyosin ring [163], and PI(3)P participating in abscission during cytokinesis [164]. For autophagy, PI(3)P is the major PPI controlling cellular homeostasis. During the initial phases of autophagosome formation, PI(3)P is produced by the VPS34 kinase complex [165,166]. By recruiting different proteins having PI(3)P-specific binding domains, such as FYVE domains (DFCP1, PIKFYVE), PX domains (Phospholipase D1), or WD40 repeats (WIPI1), PI(3)P promotes autophagosome nucleation and expansion, as well as cargo recruitment and autophagosome maturation.
Defective phosphoinositide metabolism in disease
The spatial and temporal distribution of PPIs is established through the complex activities of ∼50 PPI-metabolizing enzymes (34 phosphatases and 20 kinases) [167]. The dynamic equilibrium of their combined activities results in the PI ZIP code map presented in Figure 1C. Adding to this complexity, a recent proteomics study identified over 400 PPI-interacting proteins that serve to influence membrane identity and cellular function [168]. Given the sheer number of proteins involved in the PPI-interactome, it is not surprising that mutations in the enzymes in Figure 1C result in a wide range of devastating and heterogeneous disorders. Here, we focus on a few, more common monogenetic disorders; for a deeper review of more than 20 different diseases, see Staiano et al. [167].
Oculocerebrorenal syndrome of Lowe/Lowe’s syndrome/OCRL
This X-linked (almost exclusively found in males) recessive disorder primarily affects the brain, eyes, and kidneys. Patients typically present clinically first with congenital cataracts and other visual impairments, followed by cognitive and behavorial impairments. Behavioral problems and seizures have also been reported in children with this rare (∼1 : 500 000 frequency) condition. Genetically, this disease is caused by a loss of function mutation in the OCRL gene, which encodes a PI(4,5)P2 5-phosphatase. In health, the enzyme is predominantly localized to endocytic compartments, Golgi, and PM where its role is to dephosphorylate PI(4,5)P2 to produce PI(4)P. Intuitively, loss of OCRL catalytic activity leads to accumulation of PI(4,5)P2 on endosomal membranes, which is subsequently responsible for cytoskeletal abnormalities [169]. Recently, some patients diagnosed with proximal tubule dysfunction in another X-linked genetic disorder called Dent disease were also found to have mutations in the OCRL gene. More commomly, Dent disease results from mutations in a gene that encodes a chloride/proton exchanger, thus patients presenting with OCRL mutations have been defined as suffering from Dent-2 disease.
Charcot–Marie–Tooth neuropathy
Charcot–Marie–Tooth disease (CMT) is one of the most common inherited neurological disorders, affecting ∼1 in 2500 people in the U.S.A. CMT is a hereditary motor and sensory neuropathy that affects peripheral nerves. Although levels of many different proteins are abnormal in different forms of CMT disease, all the mutations affect the normal structure and function of either the peripheral nerve axon or the myelin sheath. CMT affects both motor and sensory nerves and can be caused by mutations in PPI phosphatases (myotubularin family members, FIG4, and synaptojanin1, SYNJ1) and PPI kinases (PIP5Ks).
Ciliopathies
Joubert and MORM syndrome are ciliopathies that share a dysfunction of primary cilia. Joubert syndrome is a rare autosomal recessive disease whose diagnostic hallmark is a unique cerebellar and brainstem malformation and cerebellar ataxia. It can be caused by mutations in 21 different genes [170]. One of these is the catalytic domain of INPP5E, a 5-phosphatase that dephosphorylates PI(3,4,5)P3 and PI(4,5)P2 into PI(3,4)P2 and PI(4) P, respectively, aiding in the development and stability of the primary cilium [171]. Similar to Joubert syndrome, the related MORM ciliopathy also results from a defect in INPP5E function. Interestingly, the mutation in this disorder results in a truncated protein rather than a catalytically inactive one; it retains PPI phosphatase activity but loses the ability to be correctly targeted to the ciliary axoneme [172].
Alzheimer’s and Parkinson’s
This short list does not underscore sufficiently the importance of PPIs for human health. As noted by Balla ‘with some efforts every human disease can be linked to altered inositol lipid metabolism’ [173]. Indeed, PPIs are responsible not only for the genetic disorders (some rare) mentioned above, but also are implicated in more common disorders such as cancer, obesity, diabetes, Alzheimer’s, and Parkinson’s. In the case of Alzheimer’s disease (AD), there is much evidence linking alterations in PPI levels to disease progression. This includes numerous reports of alterations in PPI abundance and metabolism in diseased brains [174–178] and the correlation between cellular PI(4,5)P2 levels and the abundance of 42-residue amyloid β (Aβ) peptide in familial AD-associated presenilin mutations [179]. Furthermore, binding of Aβ to critical PPI-metabolizing enzymes, such as synaptojanin [176] and PI4K2A [180], significantly alters catalytic activities. Finally, highlighting the potential role for PPIs in controlling amyloid protein processing, genetic polymorphisms or mutations in genes encoding PPI-metabolizing enzymes such as INPP5D and SYNJ1 are risk factors for late-onset AD [181,182].
For Parkinson’s disease, a homozygous missense R258Q mutation within the Sac domain of synaptojanin 1 has been identified in patients with an autosomal recessive early-onset Parkinson’s. This mutation is designated PARK20. A murine model carrying this mutation develops neurological manifestations similar to those of human patients, with synapses of these mice displaying endocytic defects and accumulation of clathrin-coated intermediates, reinforcing the Sac domain’s importance in endocytosis and establishing a link between dysfunction in early endocytic traffic and Parkinson’s disease [183].
Future outlook and concluding remarks
This article has discussed some of the broader issues relating to PPI metabolism in health and disease. It is clear there is significant information still to determine. The rapidly expanding toolkit, now including super-resolution imaging modalities and mass-spectrometry, to interrogate PPI metabolism with nanoscale and quantitative precision promises greater understanding of how PPIs can simultaneously regulate many biological processes. Concerning PPI manipulation, gene editing (CRISPR/Cas9) promises to have a revolutionary impact on the field. For example, the generation of endogenously tagged proteins at physiological levels offers the unique opportunity to identify the preferred patterning and dynamics of PPI-enzymes. Some of the broader questions that will hopefully inspire future research endeavors include: How can we visualize these low-abundance lipids directly, without the need for binding-domains or effectors? What are the spatial and temporal characteristics of PI metabolism? For MCSs, is there heterogeneity in terms of proteins or transfer activities at the same membrane-membrane interface? If so, what governs such signaling diversity? Perhaps most importantly, how can we leverage our current knowledge of PPIs and the extensive coverage of the cellular PPI-interactome to influence the onset or progression of human disease? This latter question may be farthest from our grasp in part because of the relative lack of information on PPI metabolism in native primary cells compared with expression systems, such as HEK cells, and the paucity of specific pharmacological reagents that interfere with the generation and destruction of these lipids. Despite this long list of information still to determine, the past 80 years of PPI research, especially the explosion within the last twenty years impressively underscores how essential these minor lipids are for the fundamental regulation of all aspects of membrane dynamics in eukaryotic cells.
Acknowledgements
We thank Dr Gerald R.V. Hammond for critical reading and helpful comments and suggestions.
Funding
This work was supported by NIH grants R01GM127513 (E.J.D.), R37-NS08174-50 (B.H.) and the Wayne E. Crill Endowed Professorship (B.H.).
Abbreviations
- AD
Alzheimer’s disease
- CaV
voltage-gated Ca2+ channel
- CMT
Charcot–Marie–Tooth
- DAG
diacylglycerol
- diC8-PIP2
a dioctanoyl version of the lipid
- ER
endoplasmic reticulum
- E-Syt2
extended synaptotagmin 2
- Kir
inwardly rectifying K+ channel
- KV
voltage-gated K+ channel
- MCS
membrane contact site
- ORD
OSBP-related domain
- ORP
OSBP-related protein
- OSBP
oxysterol-binding protein
- PH
pleckstrin homology
- PI
phosphatidylinositol
- PKC
protein kinase C
- PLC
phospholipase C
- PM
plasma membrane
- PPI
polyphosphoinositide
Footnotes
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Kim YJ, Guzman-Hernandez ML and Balla T (2011) A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev. Cell 21, 813–824 10.1016/j.devcel.2011.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baskin JM, Wu X, Christiano R, Oh MS, Schauder CM, Gazzerro E et al. (2015) The leukodystrophy protein FAM126A (hyccin) regulates PtdIns(4)P synthesis at the plasma membrane. Nat. Cell Biol 18, 132–138 10.1038/ncb3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakatsu F, Baskin JM, Chung J, Tanner LB, Shui G, Lee SY et al. (2012) Ptdins4p synthesis by PI4KIIIα at the plasma membrane and its impact on plasma membrane identity. J. Cell Biol 199, 1003–1016 10.1083/jcb.201206095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Matteis M, Godi A and Corda D (2002) Phosphoinositides and the Golgi complex. Curr. Opin. Cell Biol 14, 434–447 10.1016/S0955-0674(02)00357-5 [DOI] [PubMed] [Google Scholar]
- 5.Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C et al. (1999) ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat. Cell Biol 1, 280–287 10.1038/12993 [DOI] [PubMed] [Google Scholar]
- 6.Wong K, Meyers dd R and Cantley LC (1997) Subcellular locations of phosphatidylinositol 4-kinase isoforms. J. Biol. Chem 272, 13236–13241 10.1074/jbc.272.20.13236 [DOI] [PubMed] [Google Scholar]
- 7.Nakagawa T, Goto K and Kondo H (1996) Cloning, expression, and localization of 230-kDa phosphatidylinositol 4-kinase. J. Biol. Chem 271, 12088–12094 10.1074/jbc.271.20.12088 [DOI] [PubMed] [Google Scholar]
- 8.Manford A, Xia T, Saxena AK, Stefan C, Hu F, Emr SD et al. (2010) Crystal structure of the yeast Sac1: implications for its phosphoinositide phosphatase function. EMBO J 29, 1489–1498 10.1038/emboj.2010.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zewe JP, Wills RC, Sangappa S, Goulden BD and Hammond GR (2018) SAC1 degrades its lipid substrate PtdIns4P in the endoplasmic reticulum to maintain a steep chemical gradient with donor membranes. eLife 7, e35588 10.7554/eLife.35588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mesmin B, Bigay J, Polidori J, Jamecna D, Lacas-Gervais S and Antonny B (2017) Sterol transfer, PI4P consumption, and control of membrane lipid order by endogenous OSBP. EMBO J 36, 3156–3174 10.15252/embj.201796687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G and Antonny B (2013) A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell 155, 830–843 10.1016/j.cell.2013.09.056 [DOI] [PubMed] [Google Scholar]
- 12.Norris FA, Auethavekiat V and Majerus PW (1995) The isolation and characterization of cDNA encoding human and rat brain inositol polyphosphate 4-phosphatase. J. Biol. Chem 270, 16128–16133 10.1074/jbc.270.27.16128 [DOI] [PubMed] [Google Scholar]
- 13.Norris FA and Majerus PW (1994) Hydrolysis of phosphatidylinositol 3,4-bisphosphate by inositol polyphosphate 4-phosphatase isolated by affinity elution chromatography. J. Biol. Chem 269, 8716–8720 [PubMed] [Google Scholar]
- 14.Ivetac I, Gurung R, Hakim S, Horan KA, Sheffield DA, Binge LC et al. (2009) Regulation of PI(3)K/Akt signalling and cellular transformation by inositol polyphosphate 4-phosphatase-1. EMBO Rep 10, 487–493 10.1038/embor.2009.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marat AL and Haucke V (2016) Phosphatidylinositol 3-phosphates — at the interface between cell signalling and membrane traffic. EMBO J 35, 561–579 10.15252/embj.201593564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devereaux K, Dall’Armi C, Alcazar-Roman A, Ogasawara Y, Zhou X, Wang F et al. (2013) Regulation of mammalian autophagy by class II and III PI 3-kinases through PI3P synthesis. PLoS ONE 8, e76405 10.1371/journal.pone.0076405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franco I, Gulluni F, Campa CC, Costa C, Margaria JP, Ciraolo E et al. (2014) PI3K class II α controls spatially restricted endosomal PtdIns3P and Rab11 activation to promote primary cilium function. Dev. Cell 28, 647–658 10.1016/j.devcel.2014.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marat AL, Wallroth A, Lo WT, Muller R, Norata GD, Falasca M et al. (2017) mTORC1 activity repression by late endosomal phosphatidylinositol 3,4-bisphosphate. Science 356, 968–972 10.1126/science.aaf8310 [DOI] [PubMed] [Google Scholar]
- 19.Rostislavleva K, Soler N, Ohashi Y, Zhang L, Pardon E, Burke JE et al. (2015) Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science 350, aac7365 10.1126/science.aac7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta A, Toscano S, Trivedi D, Jones DR, Mathre S, Clarke JH et al. (2013) Phosphatidylinositol 5-phosphate 4-kinase (PIP4K) regulates TOR signaling and cell growth during Drosophila development. Proc. Natl Acad. Sci. U.S.A 110, 5963–5968 10.1073/pnas.1219333110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emerling BM, Hurov JB, Poulogiannis G, Tsukazawa KS, Choo-Wing R, Wulf GM et al. (2013) Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell 155, 844–857 10.1016/j.cell.2013.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viaud J, Boal F, Tronchere H, Gaits-Iacovoni F and Payrastre B (2014) Phosphatidylinositol 5-phosphate: a nuclear stress lipid and a tuner of membranes and cytoskeleton dynamics. BioEssays 36, 260–272 10.1002/bies.201300132 [DOI] [PubMed] [Google Scholar]
- 23.Zolov SN, Bridges D, Zhang Y, Lee WW, Riehle E, Verma R et al. (2012) In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc. Natl Acad. Sci. U.S.A 109, 17472–17477 10.1073/pnas.1203106109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mason D, Mallo GV, Terebiznik MR, Payrastre B, Finlay BB, Brumell JH et al. (2007) Alteration of epithelial structure and function associated with PtdIns(4,5)P2 degradation by a bacterial phosphatase. J. Gen. Physiol 129, 267–283 10.1085/jgp.200609656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ungewickell A, Hugge C, Kisseleva M, Chang SC, Zou J, Feng Y et al. (2005) The identification and characterization of two phosphatidylinositol-4,5-bisphosphate 4-phosphatases. Proc. Natl Acad. Sci. U.S.A 102, 18854–18859 10.1073/pnas.0509740102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zou J, Marjanovic J, Kisseleva MV, Wilson M and Majerus PW (2007) Type I phosphatidylinositol-4,5-bisphosphate 4-phosphatase regulates stress-induced apoptosis. Proc. Natl Acad. Sci. U.S.A 104, 16834–16839 10.1073/pnas.0708189104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brockerhoff H and Ballou CE (1962) Phosphate incorporation in brain phosphionositides. J. Biol. Chem 237, 49–52 [PubMed] [Google Scholar]
- 28.Ishihara H, Shibasaki Y, Kizuki N, Wada T, Yazaki Y, Asano T et al. (1998) Type I phosphatidylinositol-4-phosphate 5-kinases. Cloning of the third isoform and deletion/substitution analysis of members of this novel lipid kinase family. J. Biol. Chem 273, 8741–8748 10.1074/jbc.273.15.8741 [DOI] [PubMed] [Google Scholar]
- 29.Hu J, Yuan Q, Kang X, Qin Y, Li L, Ha Y et al. (2015) Resolution of structure of PIP5K1A reveals molecular mechanism for its regulation by dimerization and dishevelled. Nat. Commun 6, 8205 10.1038/ncomms9205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y and Bankaitis VA (2010) Phosphoinositide phosphatases in cell biology and disease. Prog. Lipid Res 49, 201–217 10.1016/j.plipres.2009.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens LR, Hughes KT and Irvine RF (1991) Pathway of phosphatidylinositol(3,4,5)-trisphosphate synthesis in activated neutrophils. Nature 351, 33–39 10.1038/351033a0 [DOI] [PubMed] [Google Scholar]
- 32.Hawkins PT, Jackson TR and Stephens LR (1992) Platelet-derived growth factor stimulates synthesis of Ptdlns(3,4,5)P3 by activating a Ptdlns(4,5) P2 3-OH kinase. Nature 358, 157–159 10.1038/358157a0 [DOI] [PubMed] [Google Scholar]
- 33.Misawa H, Ohtsubo M, Copeland NG, Gilbert DJ, Jenkins NA and Yoshimura A (1998) Cloning and characterization of a novel class II phosphoinositide 3-kinase containing C2 domain. Biochem. Biophys. Res. Commun 244, 531–539 10.1006/bbrc.1998.8294 [DOI] [PubMed] [Google Scholar]
- 34.Sasaki J, Kofuji S, Itoh R, Momiyama T, Takayama K, Murakami H et al. (2010) The PtdIns(3,4)P2 phosphatase INPP4A is a suppressor of excitotoxic neuronal death. Nature 465, 497–501 10.1038/nature09023 [DOI] [PubMed] [Google Scholar]
- 35.Malek M, Kielkowska A, Chessa T, Anderson KE, Barneda D, Pir P et al. (2017) PTEN regulates PI(3,4)P2 signaling downstream of Class I PI3K. Mol. Cell 68, 566–580.e510 10.1016/j.molcel.2017.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shisheva A (2008) PIKfyve: partners, significance, debates and paradoxes. Cell Biol. Int 32, 591–604 10.1016/j.cellbi.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikonomov OC, Sbrissa D, Fligger J, Delvecchio K and Shisheva A (2010) ArPIKfyve regulates Sac3 protein abundance and turnover: disruption of the mechanism by Sac3I41T mutation causing Charcot–Marie–Tooth 4J disorder. J. Biol. Chem 285, 26760–26764 10.1074/jbc.C110.154658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin N, Chow CY, Liu L, Zolov SN, Bronson R, Davisson M et al. (2008) VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P2 in yeast and mouse. EMBO J 27, 3221–3234 10.1038/emboj.2008.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong NH, Qi A and Weaver AM (2015) PI(3,5)P2 controls endosomal branched actin dynamics by regulating cortactin-actin interactions. J. Cell Biol 210, 753–769 10.1083/jcb.201412127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ikonomov OC, Sbrissa D, Delvecchio K, Xie Y, Jin JP, Rappolee D et al. (2011) The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve−/− embryos but normality of PIKfyve+/− mice. J. Biol. Chem 286, 13404–13413 10.1074/jbc.M111.222364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milne SB, Ivanova PT, DeCamp D, Hsueh RC and Brown HA (2005) A targeted mass spectrometric analysis of phosphatidylinositol phosphate species. J. Lipid Res 46, 1796–1802 10.1194/jlr.D500010-JLR200 [DOI] [PubMed] [Google Scholar]
- 42.Clark J, Anderson KE, Juvin V, Smith TS, Karpe F, Wakelam MJ et al. (2011) Quantification of PtdInsP3 molecular species in cells and tissues by mass spectrometry. Nat. Methods 8, 267–272 10.1038/nmeth.1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chalhoub N and Baker SJ (2009) PTEN and the PI3-kinase pathway in cancer. Annu. Rev. Pathol 4, 127–150 10.1146/annurev.pathol.4.110807.092311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC and Abraham RT (2017) The PI3K pathway in human disease. Cell 170, 605–635 10.1016/j.cell.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Matteis MA and Godi A (2004) PI-loting membrane traffic. Nat. Cell Biol 6, 487–492 10.1038/ncb0604-487 [DOI] [PubMed] [Google Scholar]
- 46.Godi A, Di Campli A, Konstantakopoulos A, Di Tullio G, Alessi DR, Kular GS et al. (2004) FAPPs control Golgi-to-cell-surface membrane traffic by binding to ARF and PtdIns(4)P. Nat. Cell Biol 6, 393–404 10.1038/ncb1119 [DOI] [PubMed] [Google Scholar]
- 47.Szentpetery Z, Varnai P and Balla T (2010) Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc. Natl Acad. Sci. U.S.A 107, 8225–8230 10.1073/pnas.1000157107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mironova YA, Lenk GM, Lin JP, Lee SJ, Twiss JL, Vaccari I et al. (2016) PI(3,5)P2 biosynthesis regulates oligodendrocyte differentiation by intrinsic and extrinsic mechanisms. eLife 5, e13023 10.7554/eLife.13023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoetzel C, Bar S, De Craene JO, Scheidecker S, Etard C, Chicher J et al. (2016) A mutation in VPS15 (PIK3R4) causes a ciliopathy and affects IFT20 release from the cis-Golgi. Nat. Commun 7, 13586 10.1038/ncomms13586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moore DH and Ruska H (1957) Electron microscope study of mammalian cardiac muscle cells. J. Biophys. Biochem. Cytol 3, 261–268 10.1083/jcb.3.2.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Block BA, Imagawa T, Campbell KP and Franzini-Armstrong C (1988) Structural evidence for direct interaction between the molecular components of the transverse tubule/sarcoplasmic reticulum junction in skeletal muscle. J. Cell Biol 107, 2587–2600 10.1083/jcb.107.6.2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franzini-Armstrong C and Protasi F (1997) Ryanodine receptors of striated muscles: a complex channel capable of multiple interactions. Physiol. Rev 77, 699–729 10.1152/physrev.1997.77.3.699 [DOI] [PubMed] [Google Scholar]
- 53.Lees JA, Messa M, Sun EW, Wheeler H, Torta F, Wenk MR et al. (2017) Lipid transport by TMEM24 at ER-plasma membrane contacts regulates pulsatile insulin secretion. Science 355, eaah6171 10.1126/science.aah6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim YJ, Guzman-Hernandez ML, Wisniewski E and Balla T (2015) Phosphatidylinositol-phosphatidic acid exchange by Nir2 at ER-PM contact sites maintains phosphoinositide signaling competence. Dev. Cell 33, 549–561 10.1016/j.devcel.2015.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang CL, Hsieh TS, Yang TT, Rothberg KG, Azizoglu DB, Volk E et al. (2013) Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep 5, 813–825 10.1016/j.celrep.2013.09.038 [DOI] [PubMed] [Google Scholar]
- 56.Kim S, Kedan A, Marom M, Gavert N, Keinan O, Selitrennik M et al. (2013) The phosphatidylinositol-transfer protein Nir2 binds phosphatidic acid and positively regulates phosphoinositide signalling. EMBO Rep 14, 891–899 10.1038/embor.2013.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang CL and Liou J (2015) Phosphatidylinositol 4,5-bisphosphate homeostasis regulated by Nir2 and Nir3 proteins at endoplasmic reticulum-plasma membrane junctions. J. Biol. Chem 290, 14289–14301 10.1074/jbc.M114.621375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amarilio R, Ramachandran S, Sabanay H and Lev S (2005) Differential regulation of endoplasmic reticulum structure through VAP-Nir protein interaction. J. Biol. Chem 280, 5934–5944 10.1074/jbc.M409566200 [DOI] [PubMed] [Google Scholar]
- 59.Sohn M, Korzeniowski M, Zewe JP, Wills RC, Hammond GRV, Humpolickova J et al. (2018) PI(4,5)P2 controls plasma membrane PI4P and PS levels via ORP5/8 recruitment to ER-PM contact sites. J. Cell Biol 217, 1797–1813 10.1083/jcb.201710095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chung J, Torta F, Masai K, Lucast L, Czapla H, Tanner LB et al. (2015) PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science 349, 428–432 10.1126/science.aab1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saheki Y et al. (2016) Control of plasma membrane lipid homeostasis by the extended synaptotagmins. Nat. Cell Biol 10.1038/ncb3339 [DOI] [PMC free article] [PubMed]
- 62.Antonny B, Bigay J and Mesmin B (2018) The oxysterol-binding protein cycle: burning off PI(4)P to transport cholesterol. Annu. Rev. Biochem 87, 809–837 10.1146/annurev-biochem-061516-044924 [DOI] [PubMed] [Google Scholar]
- 63.Wilhelm LP, Wendling C, Védie B, Kobayashi T, Chenard MP, Tomasetto C et al. (2017) STARD3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites. EMBO J 36, 1412–1433 10.15252/embj.201695917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao K and Ridgway ND (2017) Oxysterol-binding protein-related protein 1L regulates cholesterol egress from the endo-lysosomal system. Cell Rep 19, 1807–1818 10.1016/j.celrep.2017.05.028 [DOI] [PubMed] [Google Scholar]
- 65.Wilhelm LP, Tomasetto C and Alpy F (2016) Touche! STARD3 and STARD3NL tether the ER to endosomes. Biochem. Soc. Trans 44, 493–498 10.1042/BST20150269 [DOI] [PubMed] [Google Scholar]
- 66.Wijdeven RH, Jongsma ML, Neefjes J and Berlin I (2015) ER contact sites direct late endosome transport. BioEssays 37, 1298–1302 10.1002/bies.201500095 [DOI] [PubMed] [Google Scholar]
- 67.Raiborg C, Wenzel EM, Pedersen NM, Olsvik H, Schink KO and Schultz SW (2015) Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature 520, 234–238 10.1038/nature14359 [DOI] [PubMed] [Google Scholar]
- 68.Dong R, Saheki Y, Swarup S, Lucast L, Harper JW and De Camilli P (2016) Endosome-ER contacts control actin nucleation and retromer function through VAP-dependent regulation of PI4P. Cell 166, 408–423 10.1016/j.cell.2016.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nasuhoglu C, Feng S, Mao J, Yamamoto M, Yin HL, Earnest S et al. (2002) Nonradioactive analysis of phosphatidylinositides and other anionic phospholipids by anion-exchange high-performance liquid chromatography with suppressed conductivity detection. Anal. Biochem 301, 243–254 10.1006/abio.2001.5489 [DOI] [PubMed] [Google Scholar]
- 70.Kielkowska A, Niewczas I, Anderson KE, Durrant TN, Clark J, Stephens LR et al. (2014) A new approach to measuring phosphoinositides in cells by mass spectrometry. Adv. Biol. Regul 54, 131–141 10.1016/j.jbior.2013.09.001 [DOI] [PubMed] [Google Scholar]
- 71.Wenk MR, Lucast L, Di Paolo G, Romanelli AJ, Suchy SF, Nussbaum RL et al. (2003) Phosphoinositide profiling in complex lipid mixtures using electrospray ionization mass spectrometry. Nat. Biotechnol 21, 813–817 10.1038/nbt837 [DOI] [PubMed] [Google Scholar]
- 72.Traynor-Kaplan A, Kruse M, Dickson EJ, Dai G, Vivas O, Yu H et al. (2017) Fatty-acyl chain profiles of cellular phosphoinositides. Biochim. Biophys. Acta 1862, 513–522 10.1016/j.bbalip.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang C, Palavicini JP, Wang M, Chen L, Yang K, Crawford PA et al. (2016) Comprehensive and quantitative analysis of polyphosphoinositide species by shotgun lipidomics revealed their alterations in db/db mouse brain. Anal. Chem 88, 12137–12144 10.1021/acs.analchem.6b02947 [DOI] [PubMed] [Google Scholar]
- 74.Hammond GR, Schiavo G and Irvine RF (2009) Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5) P2. Biochem. J 422, 23–35 10.1042/BJ20090428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hammond GRV and Balla T (2015) Polyphosphoinositide binding domains: key to inositol lipid biology. Biochim. Biophys. Acta 1851, 746–758 10.1016/j.bbalip.2015.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moravcevic K, Oxley CL and Lemmon MA (2012) Conditional peripheral membrane proteins: facing up to limited specificity. Structure 20, 15–27 10.1016/j.str.2011.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Idevall-Hagren O and De Camilli P (2014) Detection and manipulation of phosphoinositides. Biochim. Biophys. Acta 1851, 736–745 10.1016/j.bbalip.2014.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wills RC, Goulden BD and Hammond GRV (2018) Genetically encoded lipid biosensors. Mol. Biol. Cell 29, 1526–1532 10.1091/mbc.E17-12-0738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Varnai P, Gulyas G, Toth DJ, Sohn M, Sengupta N and Balla T (2017) Quantifying lipid changes in various membrane compartments using lipid binding protein domains. Cell Calcium 64, 72–82 10.1016/j.ceca.2016.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stauffer TP, Ahn S and Meyer T (1998) Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol 8, 343–346 10.1016/S0960-9822(98)70135-6 [DOI] [PubMed] [Google Scholar]
- 81.Várnai P and Balla T (1998) Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol 143, 501–510 10.1083/jcb.143.2.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Suh BC, Inoue T, Meyer T and Hille B (2006) Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science 314, 1454–1457 10.1126/science.1131163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Inoue T, Heo WD, Grimley JS, Wandless TJ and Meyer T (2005) An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods 2, 415–418 10.1038/nmeth763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Benedetti L, Barentine AES, Messa M, Wheeler H, Bewersdorf J and De Camilli P (2018) Light-activated protein interaction with high spatial subcellular confinement. Proc. Natl Acad. Sci. U.S.A 115, E2238–E2245 10.1073/pnas.1713845115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Idevall-Hagren O, Dickson EJ, Hille B, Toomre DK and De Camilli P (2012) Optogenetic control of phosphoinositide metabolism. Proc. Natl Acad. Sci. U.S.A 109, E2316–E2323 10.1073/pnas.1211305109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hammond GR, Fischer MJ, Anderson KE, Holdich J, Koteci A, Balla T et al. (2012) PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science 337, 727–730 10.1126/science.1222483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hammond GR, Machner MP and Balla T (2014) A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J. Cell Biol 205, 113–126 10.1083/jcb.201312072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dickson EJ, Jensen JB and Hille B (2014) Golgi and plasma membrane pools of PI(4)P contribute to plasma membrane PI(4,5)P2 and maintenance of KCNQ2/3 ion channel current. Proc. Natl Acad. Sci. U.S.A 111, E2281–E2290 10.1073/pnas.1407133111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Okamura Y, Murata Y and Iwasaki H (2009) Voltage-sensing phosphatase: actions and potentials. J. Physiol 587, 513–520 10.1113/jphysiol.2008.163097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Murata Y, Iwasaki H, Sasaki M, Inaba K and Okamura Y (2005) Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature 435, 1239–1243 10.1038/nature03650 [DOI] [PubMed] [Google Scholar]
- 91.Kurokawa T, Takasuga S, Sakata S, Yamaguchi S, Horie S, Homma KJ et al. (2012) 3’ phosphatase activity toward phosphatidylinositol 3,4-bisphosphate [PI(3,4)P2] by voltage-sensing phosphatase (VSP). Proc. Natl Acad. Sci. U.S.A 109, 10089–10094 10.1073/pnas.1203799109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Keum D, Kruse M, Kim DI, Hille B and Suh BC (2016) Phosphoinositide 5- and 3-phosphatase activities of a voltage-sensing phosphatase in living cells show identical voltage dependence. Proc. Natl Acad. Sci. U.S.A 113, E3686–E3695 10.1073/pnas.1606472113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Falkenburger BH, Jensen JB and Hille B (2010) Kinetics of PIP2 metabolism and KCNQ2/3 channel regulation studied with a voltage-sensitive phosphatase in living cells. J. Gen. Physiol 135, 99–114 10.1085/jgp.200910345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meyers R and Cantley LC (1997) Cloning and characterization of a wortmannin-sensitive human phosphatidylinositol 4-kinase. J. Biol. Chem 272, 4384–4390 10.1074/jbc.272.7.4384 [DOI] [PubMed] [Google Scholar]
- 95.Wymann MP and Schultz C (2012) The chemical biology of phosphoinositide 3-kinases. Chembiochem 13, 2022–2035 10.1002/cbic.201200089 [DOI] [PubMed] [Google Scholar]
- 96.Bojjireddy N, Botyanszki J, Hammond G, Creech D, Peterson R, Kemp DC et al. (2014) Pharmacological and genetic targeting of the PI4KA enzyme reveals its important role in maintaining plasma membrane phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate levels. J. Biol. Chem 289, 6120–6132 10.1074/jbc.M113.531426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Toth B, Balla A, Ma H, Knight ZA, Shokat KM and Balla T (2006) Phosphatidylinositol 4-kinase IIIβ regulates the transport of ceramide between the endoplasmic reticulum and Golgi. J. Biol. Chem 281, 36369–36377 10.1074/jbc.M604935200 [DOI] [PubMed] [Google Scholar]
- 98.Semenas J, Hedblom A, Miftakhova RR, Sarwar M, Larsson R, Shcherbina L et al. (2014) The role of PI3K/AKT-related PIP5K1α and the discovery of its selective inhibitor for treatment of advanced prostate cancer. Proc. Natl Acad. Sci. U.S.A 111, E3689–E3698 10.1073/pnas.1405801111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wright BD, Loo L, Street SE, Ma A, Taylor-Blake B, Stashko MA et al. (2014) The lipid kinase PIP5K1C regulates pain signaling and sensitization. Neuron 82, 836–847 10.1016/j.neuron.2014.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fruman DA and Cantley LC (2014) Idelalisib–a PI3Kδ inhibitor for B-cell cancers. N. Engl. J. Med 370, 1061–1062 10.1056/NEJMe1400055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sarker D, Ang JE, Baird R, Kristeleit R, Shah K, Moreno V et al. (2015) First-in-human phase I study of pictilisib (GDC-0941), a potent pan-class I phosphatidylinositol-3-kinase (PI3K) inhibitor, in patients with advanced solid tumors. Clin. Cancer Res 21, 77–86 10.1158/1078-0432.CCR-14-0947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Di Leo A, Johnston S, Lee KS, Ciruelos E, Lonning PE, Janni W et al. (2018) Buparlisib plus fulvestrant in postmenopausal women with hormone-receptor-positive, HER2-negative, advanced breast cancer progressing on or after mTOR inhibition (BELLE-3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 19, 87–100 10.1016/S1470-2045(17)30688-5 [DOI] [PubMed] [Google Scholar]
- 103.Rohács T, Chen J, Prestwich GD and Logothetis DE (1999) Distinct specificities of inwardly rectifying K+ channels for phosphoinositides. J. Biol. Chem 274, 36065–36072 10.1074/jbc.274.51.36065 [DOI] [PubMed] [Google Scholar]
- 104.Telezhkin V, Reilly JM, Thomas AM, Tinker A and Brown DA (2012) Structural requirements of membrane phospholipids for M-type potassium channel activation and binding. J. Biol. Chem 287, 10001–10012 10.1074/jbc.M111.322552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Walter AM, Muller R, Tawfik B, Wierda KD, Pinheiro PS, Nadler A et al. (2017) Phosphatidylinositol 4,5-bisphosphate optical uncaging potentiates exocytosis. eLife 6, e30203 10.7554/eLife.30203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu C, Watras J and Loew LM (2003) Kinetic analysis of receptor-activated phosphoinositide turnover. J. Cell Biol 161, 779–791 10.1083/jcb.200301070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Brown SA, Morgan F, Watras J and Loew LM (2008) Analysis of phosphatidylinositol-4,5-bisphosphate signaling in cerebellar Purkinje spines. Biophys. J 95, 1795–1812 10.1529/biophysj.108.130195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dickson EJ, Falkenburger BH and Hille B (2013) Quantitative properties and receptor reserve of the IP3 and calcium branch of Gq-coupled receptor signaling. J. Gen. Physiol 141, 521–535 10.1085/jgp.201210886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Falkenburger BH, Dickson EJ and Hille B (2013) Quantitative properties and receptor reserve of the DAG and PKC branch of Gq-coupled receptor signaling. J. Gen. Physiol 141, 537–555 10.1085/jgp.201210887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Olivenca DV, Uliyakina I, Fonseca LL, Amaral MD, Voit EO and Pinto FR (2018) A mathematical model of the phosphoinositide pathway. Sci. Rep 8, 3904 10.1038/s41598-018-22226-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Berridge MJ (2016) The inositol trisphosphate/calcium signaling pathway in health and disease. Physiol. Rev 96, 1261–1296 10.1152/physrev.00006.2016 [DOI] [PubMed] [Google Scholar]
- 112.Berridge MJ and Irvine RF (1984) Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature 312, 315–321 10.1038/312315a0 [DOI] [PubMed] [Google Scholar]
- 113.Manning BD and Cantley LC (2007) AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274 10.1016/j.cell.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hilgemann DW and Ball R (1996) Regulation of cardiac Na+,Ca2+ exchange and KATP potassium channels by PIP2. Science 273, 956–959 10.1126/science.273.5277.956 [DOI] [PubMed] [Google Scholar]
- 115.Hilgemann DW, Feng S and Nasuhoglu C (2001) The complex and intriguing lives of PIP2 with ion channels and transporters. Sci. STKE 2001, re19 10.1126/stke.2001.111.re19 [DOI] [PubMed]
- 116.Dong XP, Shen D, Wang X, Dawson T, Li X, Zhang Q et al. (2010) PI(3,5)P2 controls membrane trafficking by direct activation of mucolipin Ca2+ release channels in the endolysosome. Nat. Commun 1, 38 10.1038/ncomms1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang X, Li X and Xu H (2012) Phosphoinositide isoforms determine compartment-specific ion channel activity. Proc. Natl Acad. Sci. U.S.A 109, 11384–11389 10.1073/pnas.1202194109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ufret-Vincenty CA, Klein RM, Collins MD, Rosasco MG, Martinez GQ and Gordon SE (2015) Mechanism for phosphoinositide selectivity and activation of TRPV1 ion channels. J. Gen. Physiol 145, 431–442 10.1085/jgp.201511354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Trimble WS and Grinstein S (2015) Barriers to the free diffusion of proteins and lipids in the plasma membrane. J. Cell Biol 208, 259–271 10.1083/jcb.201410071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Brown DA and Adams PR (1980) Muscarinic suppression of a novel voltage-sensitive K+ current in a vertebrate neurone. Nature 283, 673–676 10.1038/283673a0 [DOI] [PubMed] [Google Scholar]
- 121.Joshi S, Sedivy V, Hodyc D, Herget J and Gurney AM (2009) KCNQ modulators reveal a key role for KCNQ potassium channels in regulating the tone of rat pulmonary artery smooth muscle. J. Pharmacol. Exp. Ther 329, 368–376 10.1124/jpet.108.147785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brown DA and Passmore GM (2009) Neural KCNQ (Kv7) channels. Br. J. Pharmacol 156, 1185–1195 10.1111/j.1476-5381.2009.00111.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mani BK, O’Dowd J, Kumar L, Brueggemann LI, Ross M and Byron KL (2013) Vascular KCNQ (Kv7) potassium channels as common signaling intermediates and therapeutic targets in cerebral vasospasm. J. Cardiovasc. Pharmacol 61, 51–62 10.1097/FJC.0b013e3182771708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vivas O, Kruse M and Hille B (2014) Nerve growth factor sensitizes adult sympathetic neurons to the proinflammatory peptide bradykinin. J. Neurosci 34, 11959–11971 10.1523/JNEUROSCI.1536-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Suh BC and Hille B (2002) Recovery from muscarinic modulation of M current channels requires phosphatidylinositol 4,5-bisphosphate synthesis. Neuron 35, 507–520 10.1016/S0896-6273(02)00790-0 [DOI] [PubMed] [Google Scholar]
- 126.Zhang H, Craciun LC, Mirshahi T, Rohacs T, Lopes CM, Jin T et al. (2003) PIP2 activates KCNQ channels, and its hydrolysis underlies receptor-mediated inhibition of M currents. Neuron 37, 963–975 10.1016/S0896-6273(03)00125-9 [DOI] [PubMed] [Google Scholar]
- 127.Murata Y and Okamura Y (2007) Depolarization activates the phosphoinositide phosphatase Ci-VSP, as detected in Xenopus oocytes coexpressing sensors of PIP2. J. Physiol 583, 875–889 10.1113/jphysiol.2007.134775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kruse M, Vivas O, Traynor-Kaplan A and Hille B (2016) Dynamics of phosphoinositide-dependent signaling in sympathetic neurons. J. Neurosci 36, 1386–1400 10.1523/JNEUROSCI.3535-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Horowitz LF, Hirdes W, Suh BC, Hilgemann DW, Mackie K and Hille B (2005) Phospholipase C in living cells: activation, inhibition, Ca2+ requirement, and regulation of M current. J. Gen. Physiol 126, 243–262 10.1085/jgp.200509309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bernheim L, Beech DJ and Hille B (1991) A diffusible second messenger mediates one of the pathways coupling receptors to calcium channels in rat sympathetic neurons. Neuron 6, 859–867 10.1016/0896-6273(91)90226-P [DOI] [PubMed] [Google Scholar]
- 131.Hille B, Beech DJ, Bernheim L, Mathie A, Shapiro MS and Wollmuth LP (1995) Multiple G-protein-coupled pathways inhibit N-type Ca channels of neurons. Life Sci 56, 989–992 10.1016/0024-3205(95)00038-8 [DOI] [PubMed] [Google Scholar]
- 132.Delmas P, Coste B, Gamper N and Shapiro MS (2005) Phosphoinositide lipid second messengers: new paradigms for calcium channel modulation. Neuron 47, 179–182 10.1016/j.neuron.2005.07.001 [DOI] [PubMed] [Google Scholar]
- 133.Suh BC, Leal K and Hille B (2010) Modulation of high-voltage activated Ca2+ channels by membrane phosphatidylinositol 4,5-bisphosphate. Neuron 67, 224–238 10.1016/j.neuron.2010.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li Y, Gamper N, Hilgemann DW and Shapiro MS (2005) Regulation of Kv7 (KCNQ) K+ channel open probability by phosphatidylinositol 4,5-bisphosphate. J. Neurosci 25, 9825–9835 10.1523/JNEUROSCI.2597-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Adams PR, Brown DA and Constanti A (1982) Pharmacological inhibition of the M-current. J. Physiol 332, 223–262 10.1113/jphysiol.1982.sp014411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hawryluk JM, Moreira TS, Takakura AC, Wenker IC, Tzingounis AV and Mulkey DK (2012) KCNQ channels determine serotonergic modulation of ventral surface chemoreceptors and respiratory drive. J. Neurosci 32, 16943–16952 10.1523/JNEUROSCI.3043-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Haick JM, Brueggemann LI, Cribbs LL, Denning MF, Schwartz J and Byron KL (2017) PKC-dependent regulation of Kv7.5 channels by the bronchoconstrictor histamine in human airway smooth muscle cells. Am. J. Physiol 312, L822–Ll834 10.1152/ajplung.00567.2016 [DOI] [PubMed] [Google Scholar]
- 138.Delmas P and Brown DA (2005) Pathways modulating neural KCNQ/M (Kv7) potassium channels. Nat. Rev. Neurosci 6, 850–862 10.1038/nrn1785 [DOI] [PubMed] [Google Scholar]
- 139.Mathie A, Bernheim L and Hille B (1992) Inhibition of N- and L-type calcium channels by muscarinic receptor activation in rat sympathetic neurons. Neuron 8, 907–914 10.1016/0896-6273(92)90205-R [DOI] [PubMed] [Google Scholar]
- 140.Zaika O, Lara LS, Gamper N, Hilgemann DW, Jaffe DB and Shapiro MS (2006) Angiotensin II regulates neuronal excitability via phosphatidylinositol 4,5-bisphosphate-dependent modulation of Kv7 (M-type) K+ channels. J. Physiol 575, 49–67 10.1113/jphysiol.2006.114074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cruzblanca H, Koh DS and Hille B (1998) Bradykinin inhibits M current via phospholipase C and Ca2+ release from IP3-sensitive Ca2+ stores in rat sympathetic neurons. Proc. Natl Acad. Sci. U.S.A 95, 7151–7156 10.1073/pnas.95.12.7151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zaika O, Zhang J and Shapiro MS (2011) Combined phosphoinositide and Ca2+ signals mediating receptor specificity toward neuronal Ca2+ channels. J. Biol. Chem 286, 830–841 10.1074/jbc.M110.166033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hille B, Dickson EJ, Kruse M, Vivas O and Suh B-C (2015) Phosphoinositides regulate ion channels. Biochim. Biophys. Acta, Mol. Cell Biol 1851, 844–856 10.1016/j.bbalip.2014.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Suh BC and Hille B (2005) Regulation of ion channels by phosphatidylinositol 4,5-bisphosphate. Curr. Opin. Neurobiol 15, 370–378 10.1016/j.conb.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 145.Logothetis DE, Petrou VI, Adney SK and Mahajan R (2010) Channelopathies linked to plasma membrane phosphoinositides. Pflugers Arch 460, 321–341 10.1007/s00424-010-0828-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Razavi AM, Khelashvili G and Weinstein H (2018) How structural elements evolving from bacterial to human SLC6 transporters enabled new functional properties. BMC Biol 16, 31 10.1186/s12915-018-0495-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yen H-Y, Hoi KK, Liko I, Hedger G, Horrell MR, Song W et al. (2018) Ptdins(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling. Nature 559, 423–427 10.1038/s41586-018-0325-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Eichel K, Jullie D, Barsi-Rhyne B, Latorraca NR, Masureel M, Sibarita JB et al. (2018) Catalytic activation of beta-arrestin by GPCRs. Nature 557, 381–386 10.1038/s41586-018-0079-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Rohács T, Lopes CMB, Jin T, Ramdya PP, Molnár Z and Logothetis DE (2003) Specificity of activation by phosphoinositides determines lipid regulation of Kir channels. Proc. Natl Acad. Sci. U.S.A 100, 745–750 10.1073/pnas.0236364100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kruse M, Hammond GR and Hille B (2012) Regulation of voltage-gated potassium channels by PI(4,5)P2. J. Gen. Physiol 140, 189–205 10.1085/jgp.201210806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tang QY, Larry T, Hendra K, Yamamoto E, Bell J, Cui M et al. (2015) Mutations in nature conferred a high affinity phosphatidylinositol 4,5-bisphosphate-binding site in vertebrate inwardly rectifying potassium channels. J. Biol. Chem 290, 16517–16529 10.1074/jbc.M115.640409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hansen SB, Tao X and MacKinnon R (2011) Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature 477, 495–498 10.1038/nature10370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Whorton MR and MacKinnon R (2011) Crystal structure of the mammalian GIRK2 K+ channel and gating regulation by G proteins, PIP2, and sodium. Cell 147, 199–208 10.1016/j.cell.2011.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li N, Wu JX, Ding D, Cheng J, Gao N and Chen L (2017) Structure of a pancreatic ATP-sensitive potassium channel. Cell 168, 101–110.e110 10.1016/j.cell.2016.12.028 [DOI] [PubMed] [Google Scholar]
- 155.She J, Guo J, Chen Q, Zeng W, Jiang Y and Bai X-C (2018) Structural insights into the voltage and phospholipid activation of the mammalian TPC1 channel. Nature 556, 130 10.1038/nature26139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Perez-Lara A, Thapa A, Nyenhuis SB, Nyenhuis DA, Halder P, Tietzel M et al. (2016) Ptdinsp2 and PtdSer cooperate to trap synaptotagmin-1 to the plasma membrane in the presence of calcium. eLife 5, e15886 10.7554/eLife.15886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kelly BT, Graham SC, Liska N, Dannhauser PN, Honing S, Ungewickell EJ et al. (2014) Clathrin adaptors. AP2 controls clathrin polymerization with a membrane-activated switch. Science 345, 459–463 10.1126/science.1254836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Saheki Y and De Camilli P (2012) Synaptic vesicle endocytosis. Cold Spring Harb. Perspect. Biol 4, a005645 10.1101/cshperspect.a005645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Devreotes P and Janetopoulos C (2003) Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J. Biol. Chem 278, 20445–20448 10.1074/jbc.R300010200 [DOI] [PubMed] [Google Scholar]
- 160.Nichols JM, Veltman D and Kay RR (2015) Chemotaxis of a model organism: progress with dictyostelium. Curr. Opin. Cell Biol 36, 7–12 10.1016/j.ceb.2015.06.005 [DOI] [PubMed] [Google Scholar]
- 161.Noseda R, Belin S, Piguet F, Vaccari I, Scarlino S, Brambilla P et al. (2013) DDIT4/REDD1/RTP801 is a novel negative regulator of Schwann cell myelination. J. Neurosci 33, 15295–15305 10.1523/JNEUROSCI.2408-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cauvin C and Echard A (2015) Phosphoinositides: lipids with informative heads and mastermind functions in cell division. Biochim. Biophys. Acta 1851, 832–843 10.1016/j.bbalip.2014.10.013 [DOI] [PubMed] [Google Scholar]
- 163.Brill JA, Wong R and Wilde A (2011) Phosphoinositide function in cytokinesis. Curr. Biol 21, R930–R934 10.1016/j.cub.2011.10.001 [DOI] [PubMed] [Google Scholar]
- 164.Nezis IP, Sagona AP, Schink KO and Stenmark H (2010) Divide and ProsPer: the emerging role of PtdIns3P in cytokinesis. Trends Cell Biol 20, 642–649 10.1016/j.tcb.2010.08.010 [DOI] [PubMed] [Google Scholar]
- 165.Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H and Ohsumi Y (2013) Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci 126, 2534–2544 10.1242/jcs.122960 [DOI] [PubMed] [Google Scholar]
- 166.Funderburk SF, Wang QJ and Yue Z (2010) The Beclin 1-VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell Biol 20, 355–362 10.1016/j.tcb.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Staiano L, De Leo MG, Persico M and De Matteis MA (2015) Mendelian disorders of PI metabolizing enzymes. Biochim. Biophys. Acta 1851, 867–881 10.1016/j.bbalip.2014.12.001 [DOI] [PubMed] [Google Scholar]
- 168.Jungmichel S, Sylvestersen KB, Choudhary C, Nguyen S, Mann M and Nielsen ML (2014) Specificity and commonality of the phosphoinositide-binding proteome analyzed by quantitative mass spectrometry. Cell Rep 6, 578–591 10.1016/j.celrep.2013.12.038 [DOI] [PubMed] [Google Scholar]
- 169.Nandez R, Balkin DM, Messa M, Liang L, Paradise S, Czapla H et al. (2014) A role of OCRL in clathrin-coated pit dynamics and uncoating revealed by studies of Lowe syndrome cells. eLife 3, e02975 10.7554/eLife.02975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Romani M, Micalizzi A and Valente EM (2013) Joubert syndrome: congenital cerebellar ataxia with the molar tooth. Lancet Neurol 12, 894–905 10.1016/S1474-4422(13)70136-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hardee I, Soldatos A, Davids M, Vilboux T, Toro C, David KL et al. (2017) Defective ciliogenesis in INPP5E-related Joubert syndrome. Am. J. Med. Genet. A 173, 3231–3237 10.1002/ajmg.a.38376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jacoby M, Cox JJ, Gayral S, Hampshire DJ, Ayub M, Blockmans M et al. (2009) INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse. Nat. Genet 41, 1027–1031 10.1038/ng.427 [DOI] [PubMed] [Google Scholar]
- 173.Balla T (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev 93, 1019–1137 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhu L, Zhong M, Elder GA, Sano M, Holtzman DM, Gandy S et al. (2015) Phospholipid dysregulation contributes to ApoE4-associated cognitive deficits in Alzheimer’s disease pathogenesis. Proc. Natl Acad. Sci. U.S.A 112, 11965–11970 10.1073/pnas.1510011112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bennett SA, Valenzuela N, Xu H, Franko B, Fai S and Figeys D (2013) Using neurolipidomics to identify phospholipid mediators of synaptic (dys) function in Alzheimer’s disease. Front. Physiol 4, 168 10.3389/fphys.2013.00168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Berman DE, Dall’Armi C, Voronov SV, McIntire LB, Zhang H, Moore AZ et al. (2008) Oligomeric amyloid-beta peptide disrupts phosphatidylinositol-4,5-bisphosphate metabolism. Nat. Neurosci 11, 547–554 10.1038/nn.2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ferrari-DiLeo G and Flynn DD (1993) Diminished muscarinic receptor-stimulated [3H]-PIP2 hydrolysis in alzheimer’s disease. Life Sci 53, PL439–PL444 10.1016/0024-3205(93)90037-4 [DOI] [PubMed] [Google Scholar]
- 178.Stokes CE and Hawthorne JN (1987) Reduced phosphoinositide concentrations in anterior temporal cortex of Alzheimer-diseased brains. J. Neurochem 48, 1018–1021 10.1111/j.1471-4159.1987.tb05619.x [DOI] [PubMed] [Google Scholar]
- 179.Landman N, Jeong SY, Shin SY, Voronov SV, Serban G, Kang MS et al. (2006) Presenilin mutations linked to familial Alzheimer’s disease cause an imbalance in phosphatidylinositol 4,5-bisphosphate metabolism. Proc. Natl Acad. Sci. U.S.A 103, 19524–19529 10.1073/pnas.0604954103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wu B, Kitagawa K, Liu B, Zhang NY, Xiong ZM and Inagaki C (2006) Attenuation of amyloid beta (Abeta)-induced inhibition of phosphatidylinositol 4-kinase activity by Abeta fragments, Abeta20–29 and Abeta31–35. Neurosci Lett 396, 148–152 10.1016/j.neulet.2005.11.026 [DOI] [PubMed] [Google Scholar]
- 181.Karch CM and Goate AM (2015) Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol. Psychiatry 77, 43–51 10.1016/j.biopsych.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhu L, Zhong M, Zhao J, Rhee H, Caesar I, Knight EM et al. (2013) Reduction of synaptojanin 1 accelerates Aβ clearance and attenuates cognitive deterioration in an Alzheimer mouse model. J. Biol. Chem 288, 32050–32063 10.1074/jbc.M113.504365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Cao M, Wu Y, Ashrafi G, McCartney AJ, Wheeler H, Bushong EA et al. (2017) Parkinson sac domain mutation in synaptojanin 1 impairs clathrin uncoating at synapses and triggers dystrophic changes in dopaminergic axons. Neuron 93, 882–896.e885 10.1016/j.neuron.2017.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]