Abstract
Competition among microbes is a key determinant of successful host colonization and persistence. For Streptococcus pneumoniae, lower than predicted rates of co-colonizing strains suggests a competitive advantage for resident bacteria over newcomers. In light of evolutionary theory, we hypothesised that S. pneumoniae use “owner / intruder” asymmetries to settle contests, leading to the disproportionate success of the initial resident “owner”, regardless of the genetic identity of the “intruder”. We investigated the determinants of within-host competitive success utilizing S. pneumoniae colonization of the infant mouse upper respiratory tract. Within six hours, colonization by the resident inhibited colonization by an isogenic challenger. The competitive advantage of the resident was dependent on quorum sensing via the competence (Com) regulon and downstream choline binding protein D (CbpD) and the competence-induced bacteriocins (CibAB) implicated in fratricide. CbpD and CibAB are highly conserved across pneumococcal lineages, indicating evolutionary advantages for asymmetric competitive strategies within the species. Mathematical modelling supported a significant role for quorum sensing via the Com regulon in competition, even for strains with different competitive advantages. Our study suggests that asymmetric owner/intruder competitive strategies do not require complex cognition and are used by a major human pathogen to determine “ownership” of human hosts.
Competition is a dominant force in microbial communities, evidenced by the widespread inhibitory impacts of microbial lineages on each other1,2. From an evolutionary perspective, both theory and experiments in higher organisms suggest that an “always fight” strategy is not evolutionarily stable, and will be outcompeted by more sophisticated strategies that settle conflicts with less individual cost by responding to resource asymmetries3. In a canonical example, male red deer settle contests via vocal and posture displays of body mass, and only rarely resort to physical fights4. Asymmetries can extend beyond physical size, and include occupancy of territory, or “ownership”5,6. Conflicts between the resident of a territory and newcomer (challenger) are often resolved in favor of the resident without escalation of conflict, even when intrinsic animal fitness is experimentally controlled7. Experiments in robins show that if a resident is removed from its territory and a challenger is introduced, the likelihood of the resident winning upon reintroduction decreased in proportion to the amount of time the newcomer spent establishing itself8. Herein, we show that asymmetric “owner/intruder” competitive strategies do not require complex cognition and are deployed by the major human bacterial pathogen Streptococcus pneumoniae to settle conflicts over host niche dominance.
S. pneumoniae, or pneumococcus, is an opportunistic pathogen that colonizes the nasopharynx asymptomatically, especially during early childhood (when carriage rates can exceed 50%). Simultaneous colonization with multiple strains is observed, but is far less frequent than would be predicted by the high rate of carriage of individual strains9–12. This disparity between low co-colonization rates and high rates of carriage and transmission suggest pneumococci experience direct competition with one another13. In this regard, pneumococci contain multiple bacteriocin-like genes that could mediate competition between strains14.
We previously observed that established resident S. pneumoniae excludes subsequently encountered challenger strains from colonization in an infant mouse model of transmission, suggesting a competitive advantage for resident bacteria over intruders15. We observed asymmetry in pneumococcal competition where an established resident strain always inhibited colonization by an isogenic challenger dependent on Com-regulated CbpD and CibAB that mediate fratricide in vitro16,17.
Methods
Ethics statement
This study followed guidelines as outlined by the National Science Foundation Animal Welfare Requirements and the Public Health Service Policy on the Humane Care and Use of Laboratory Animals. The New York University Medical Center IACUC oversees the welfare, well-being, proper care and use of all vertebrate animals.
Bacterial growth conditions, strains and mutant construction
Strains of S. pneumoniae were grown statically in Tryptic Soy broth (Becton Dickinson) at 37°C. At an optical density (620 nm) of 1.0, bacteria were washed and diluted in sterile PBS to the desired concentration.
All strains used in this study are listed in supplementary Table 1. For different antibiotic resistance markers, we used 23F strains resistant to streptomycin (P139718, P2499), both streptomycin and kanamycin (P2578), as well as 6A strains resistant to spectinomycin (P2397) or kanamycin (P2405)15. P2499 was generated by construction of a PCR product containing the rpsL gene causing streptomycin resistance that was then transformed into P1121. The source of the rpsL is genomic DNA isolated from a streptomycin resistant pneumolysin mutant P172619 using the MasterPure DNA purification kit (Illumina) and primers listed in supplementary Table 2. P2499 was then transformed with genomic DNA isolated from a mutant of P1121 with a kanamycin resistance cassette inserted into the IgA protease gene to make a mutant P2578 resistant to both streptomycin and kanamycin.
All primers utilized for mutant construction are listed in supplementary Table 2. The cbpD (P2500) and cibAB (P2575) mutants were constructed by insertion of the Janus cassette (via a PCR product generated from the genomic DNA of P1121 and a Janus cassette-containing strain P240819) into the desired gene loci of P2499 and P1121, respectively, as previously described20. Mutants were selected for kanamycin resistance. The double mutant cbpD-cibAB- (P2576) was constructed by first making an unmarked, in-frame deletion of cbpD in P2500 as previously described20. This resulted in strain P2516, that was then transformed with genomic DNA from P2575, selecting for kanamycin resistance to generate P2576. To correct mutations in cbpD-cibAB-, P2500 was first transformed with genomic DNA from P2499 to correct the mutation in cbpD, selecting for streptomycin resistance. Genomic DNA from P2575 was then used to transform in the cibAB mutation, selecting for kanamycin resistance. P2499 genomic DNA was used to correct the mutation, generating a streptomycin resistant, corrected mutant P2577. The comM-cbpD- (P2579) mutant was made by insertion of the Janus cassette into the comM gene locus of P2516, selecting for kanamycin resistance. The lytA-lytC- mutant was constructed by inserting the Janus cassette into the lytC locus of P2499, selecting for kanamycin resistance, then transformation with genomic DNA from the lytA mutant P228221 to generate the lytA-lytC- mutant, by selecting for spectinomycin resistance. The comE mutant P2076 was constructed by Janus cassette insertion into the comE gene of P1121 as previously described22. A streptomycin resistant 6A strain was transformed with genomic DNA isolated from a blp- mutant obtained from the laboratory of Dr. Suzanne Dawid and made kanamycin resistant with P2405 DNA to generate P2444. The genotype of each mutant was confirmed by PCR.
Colonization in infant mice
C57BL/6J mice were obtained from The Jackson Laboratory, maintained in a conventional animal facility, and bred as described previously23,24. Pups were housed with a dam (mother) for the entire duration of the study. S. pneumoniae colonization did not impact the weight gain of pups. Animal sample size was chosen based on numbers required to carry out the described statistical tests. In addition, independent experiments were repeated at least twice to ensure reproducibility, and all the data were pooled. In addition, mixed sexes of pups were used and litters were subjected to randomization
4–5 day old pups were colonized with S. pneumoniae suspended in 3uL PBS administered intra-nasally without anesthesia. To determine the impact of in vivo adaptation on the challenger in some experiments, 4–5 day old pups were colonized with the challenger strain (inoculum 103 CFU) for 3 days. Challenger bacteria were collected from the nasal lavage as described below, diluted to the indicated concentration with PBS, and immediately inoculated into a different group of mice already colonized with the resident strain (inoculum 103 CFU). At the indicated time points, pups were euthanized by CO2 asphyxiation followed by cardiac puncture, and nasal lavage samples were collected as previously described19. Briefly, the trachea was cannulated and nasal lavage fluid collected from the nares by instilling 200μl PBS through the trachea. Colonization density in the nasal lavage was assessed by plating serial dilutions of samples on Tryptic Soy Agar supplemented with catalase (6,300 U/plate) (Worthington Biochemical Corporation) and the appropriate antibiotic: neomycin (5μg/ml), streptomycin (200μg/ml), kanamycin (250μg/ml), or spectinomycin (200μg/ml). Plates were incubated overnight with 5% CO2 at 37°C. Unless otherwise stated, P1121 and P1397 were utilized as the resident and challenger strains, respectively, throughout the study. Resident P1121 density was calculated by subtracting the P1397 CFU determined by plating on streptomycin plates from the total bacterial burden cultured on neomycin plates.
To generate heat-killed bacteria, P1121 grown in Tryptic Soy broth and diluted to 105 CFU/3μl in PBS was incubated in a 56°C water bath for 10 min and inoculated into infant mice as described above. Where indicated, infant mice were treated with 3μl of streptomycin (800μg/ml) in PBS intra-nasally. We confirmed that streptomycin treatment cleared colonization of the streptomycin sensitive resident strain without affecting that of the streptomycin resistant challenger.
Transmission
7–8 day old pups were intra-nasally inoculated with influenza A virus (IAV) HKx31 strain as previously described to promote bacterial transmission15. After 2 days, each half of the litter was then given either the WT or mutant bacteria, or two different serotypes, as described in Table 1. To determine transmission events, the pups were euthanized 4–6 days later as described above for the collection and culturing of nasal lavage samples. Both the comE- and cbpD-cibAB-mutants were able to transmit to naïve mice.
Table 1:
Competition during transmission in infant mice
| Experiment | Donor Colonization | Recipient Colonization | Number of recipient mice with donor strain | P value* |
|---|---|---|---|---|
| Competition between genetically identical strains | 23F WT | Naive | 5/11 | |
| 23F WT 23F comE- |
23F comE- 23F WT |
7/9 0/10 |
0.0007 | |
| 23F WT 23F cbpD-cibAB- |
23F cbpD-cibAB- 23F WT |
10/11 0/11 |
0.0001 | |
| Competition between genetically different strains | 6A WT | Naive | 9/14 | |
| 6A WT 23F WT |
23F WT 6A WT |
1/11 1/11 |
1.00 | |
| 6A WT 23F comE- |
23F comE- 6A WT |
5/11 0/11 |
0.04 |
WT, wild-type
Two-tailed Fisher’s Exact Test, p<0.05 is considered significant
Competence-stimulating peptide (CSP) stimulation in vitro
P1397 was grown to OD 0.5, diluted 1:10 in THY media pH 6.8, and grown to OD 0.06–0.08. The bacteria were further diluted 1:10 in THY media pH 8.0 and stimulated with a mixture of CSP (AnaSpec), diluted 1:1000 (THY media pH 8.0) for 10–12 minutes. For in vivo experiments, CSP-stimulated bacteria were washed and diluted to the desired CFU and administered to mice in 3μl PBS containing CSP (1:1000 dilution). To assess gene expression, bacteria in culture were immediately treated with RNAprotect Bacterial Reagent (Qiagen) according to the manufacturer’s instructions. Bacterial pellets were stored at −80°C until RNA isolation.
RNA isolation and real-time quantitative PCR
RNAprotect treated bacterial cells were lysed and RNA was isolated using the AllPrep Bacterial DNA/RNA/Protein kit (Qiagen) according to the manufacturer’s instructions. To obtain host RNA, nasal lavages with RLT lysis buffer (Qiagen) were performed and RNA isolated with the RNeasy minikit (Qiagen) as previously described25.
cDNA was reverse transcribed using a high-capacity cDNA kit (Applied Biosystems) following the manufacturer’s instructions. Real-time, quantitative reverse transcriptase PCR was carried out with Power SYBR Green Master Mix (Applied Biosystems) using 10–50 ng cDNA and 0.5 μM primers per reaction. Samples were run in duplicate on a StepOnePlus real-time PCR system (Applied Biosystems). Gene expression was determined using the ΔΔCt method. Target gene expression was first normalized to the housekeeping gene gyrA (for bacteria) or GAPDH (for host) of the same sample and subsequently expressed as fold change over the indicated control group. Primers to gyrA, Gapdh, Il1b, Il6, Cxcl1 and Cxcl2 have been previously published25–27. Primers to comX, comM, cbpD, Tnfa, and Ifnb are listed in supplementary Table 2.
Statistical analysis
All statistical analyses of in vivo data, unless otherwise stated, were carried out using GraphPad Prism 6.0 (GraphPad). Data were analyzed using the Mann-Whitney U test to compare two groups, the Kruskal-Wallis test with Dunn’s post-analysis or one way ANOVA for multiple group comparisons. Statistical tests are two tailed and all measurements are acquired from distinct samples. Unless otherwise stated, median values of datasets are shown on graphs.
Population dynamics model formulation
The Lotka-Volterra model is written as two coupled nonlinear first-order ODEs:
where R and C are the resident and challenger population sizes, rR and rC are the resident and challenger growth rates, K is the carrying capacity (assumed equal for R and C) and αCR and αRC are the competitive effects of challenger on resident and resident on challenger respectively. The analytic solutions at equilibrium are well known28: low values of interspecific competition relative to intraspecific competition promote co-existence, whereas high values of interspecific competition relative to intraspecific competition promote one species going extinct, and the other reaching its carrying capacity.
To include the effect of Com regulon activation in this model, we wrote the competition terms α as a sum of the competitive effect in the absence of Com regulon activation γ (which we set equal to 1 in the isogenic case, to be equal to the inter-specific competition) and a time-dependent Com regulon activation as follows:
Where β is the strength of the effect of Com regulon activation, t = 0 is the point when the resident first arrives, tcom is the time taken from this initial colonization to Com regulon activation switching on, tarrival is the time between resident and challenger arrival, and H is the Heaviside step function:
This formulation of time dependence of Com regulon activation is illustrated with an example in Supplementary Figure 1.
Although Com regulon switching on is quorum-dependent, the population size and arrival time strongly co-vary. In reality, we expect that Com activation will be a more complex function of both arrival time (tcom) and density (Ncom), though we do not have sufficient time points in our data to attempt such a formulation here. Com regulon switching on as a function of time is therefore a convenient alternative formulation when considering the ability to colonize, and the time threshold tcom can easily be converted to a density threshold Ncom. If Ncom were truly fixed, tcom would vary depending on initial inoculum size, potentially leading to an overestimation of tcom compared to other conditions. However, for physiologically relevant conditions of an initial single cell bottleneck this variance is minimized15. This gives an integral in four pieces (which are continuous, but not differentiable at the boundaries): from resident arrival to challenger arrival; from challenger arrival until resident Com regulon activation; from resident Com regulon activation until both are competent; from both are competent onwards. We considered using a function (such as tanh) with a smooth transition in Com regulon switching on. However, given that it is known that translation turns on at a speed much faster than the time to exclusion we observed, we decided not to as it would require fitting an extra parameter operating on a time scale much shorter than the resolution possible from our experimental data.
To solve the ODEs and obtain each population size as a function of time we used numerical integration (odeint in the scipy package).
At low values of interspecific competition γ, although the population of the challenger declined below 1 CFU due to the competitive effect of Com regulon activation of the resident, using ODEs the population will never equal zero, (Supplementary Fig 2). We therefore sought to develop a stochastic version of this model, which has population sizes of zero as an absorbing state29, and thus avoids this issue.
We started with a continuous-time Markov chain (CTMC) representation of the populations, which are then discrete random variables rather than a continuous function of time:
These have transition probabilities between states derived from equation 1:
| Event | Change | Notation | Probability |
|---|---|---|---|
| Resident cell duplication | ΔR = +1 | BR | rRR |
| Challenger cell duplication | ΔC = +1 | BC | rCC |
| Resident cell death | ΔR = −1 | DR | |
| Challenger cell death | ΔC = −1 | DC |
To simulate runs of the chain given parameter values and starting values for R and C we used the Gillespie algorithm30.
We also followed previous work which has related a CTMC formulation of a two population coupled model to derive equivalent stochastic differential equations (SDEs)29,31. This led to coupled Itô equations, where R and C are once again continuous functions of time:
Where the rates BR, DR, BC and DC are as in the CTMC above, and dW are two independent Wiener processes, as in a finite sized population covariance between R and C can be ignored. We solved these equations using the Euler-Maruyama method32, using 2×103 equally spaced evaluation points.
To ensure that the three model formulations were equivalent, we tried running them using the same parameters, finding similar long term behaviour (Supplementary Fig 3). The stochastic formulations solved the issue of population sizes between 0 and 1 bouncing back at long times (Supplementary Fig 2). We used the SDE formulation for medium length runs (used for parameter inference) due to time and robustness to negative populations, and the CTMC formulation with tau-leaping for long runs (used for prediction and plots) due to speed and allowing potentially negative populations to be ignored in averaging.
Fitting model parameters from experimental data
To fit the model to our isogenic experimental data required estimating the value of the parameters r, K, β and tcom. To find r and K we used the analytic solution to equation 1 when C = 0 ∀ t > 0, which is 3-parameter logistic growth:
We then used lmfit to minimize the squared distance between this and the time series data from Fig 3A, constraining r between 0 and 5, and K between 1 and 105. We used the slope of a linear regression fit and the maximum observed R as starting values for r and K respectively. We obtained values of 4.36×104 CFU ± 3.54×103 CFU for K and 1.03 h−1 ± 0.03 h−1 for r (Supplementary Fig 4).
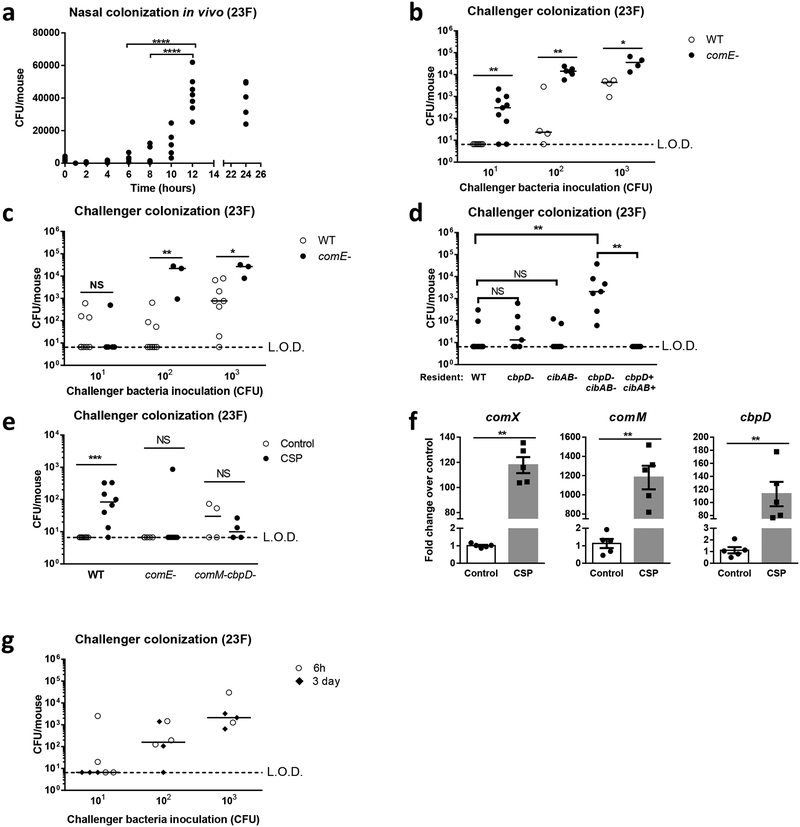
Figure 3: Prevention of challenger colonization is dependent on quorum sensing in the resident and competence-induced fratricide.
a, Day 4–5 old pups were inoculated with 103 CFU of 23F S. pneumoniae. Nasal lavage samples were collected at indicated time points to determine colonization densities by culturing. Bacterial burdens between time points were compared by one-way ANOVA, n=5–7. b-c, Pups were colonized with 103 CFU of 23F WT or comE- resident for 6h (b) or 3 days (c) prior to isogenic challenger inoculation (101-103 CFU). Colonization densities were determined 3 days later. Differences at each challenger inoculation dose were analyzed by two-tailed Mann-Whitney U test, n=3–9. Median values are shown. NS: p=0.8 in (c). d, Pups were colonized with 103 CFU of 23F WT resident, strains deficient in CbpD, CibAB, or a double mutant. A corrected double mutant resident was also included as control. WT challenger bacteria (101 CFU) were introduced 6–8h later and bacterial densities compared by Kruskal–Wallis one-way analysis of variance, n=5–10, NS: p>0.9999. Median values are shown. e, Mice were colonized with 103 CFU 23F resident for 6–8h before introduction of an isogenic challenger (indicated genotype, 101 CFU) with or without Competence Stimulating Peptide (CSP) stimulation. To isolate the effect of ComM, a resident strain deficient in CibAB, instead of WT, was utilized for mice later given the comM-cbpD- challenger. Two-tailed Mann-Whitney U test was carried out for each genotype, n=4–10, comE- control vs CSP NS: p>0.9999, comM-cbpD- control vs CSP NS: p=0.7. Median values are shown. f, CSP-treated WT challenger bacterial RNA was isolated and Com regulon-dependent gene expression was assessed by real-time quantitative RT-PCR and compared to un-stimulated controls by two-tailed Mann-Whitney U test, n=5. Data shown as mean ± SEM. g, Nasal lavages from pups colonized with the challenger strain for 3 days were directly inoculated into a different group of mice already colonized with the resident strain for 6h or 3 days. Challenger colonization density was determined 3 days later, median values are shown, n=7 (101 CFU), n=6 (102 CFU), n=5 (103 CFU). LOD, limit of detection *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. NS, non-significant.
Given the conservation of the mechanism, we assumed β and tcom were equal for all strains. To fit the values of β and tcom from the experimental data in Fig 1, we used approximate Bayesian computation as implemented in ELFI33,34. This treats the model, with given parameters, as a simulator. The distance between the simulated values and observations is then evaluated for different combinations of parameter values, using which a posterior likelihood can be approximated. To integrate out tarrival and the size of the challenger inoculum, in each simulator run we ran the SDE model using the same set of values for these parameters as the 74 experimental conditions tested. As a distance measure, we used the difference in final log(CFU) count of resident and challenger at three days between the simulated and observed values. As there was very strong evidence from the experimental data (Fig 1B) that tcom is between 1 and 6 hrs we picked a prior range bounded by these values. Test simulator runs suggested β would be around 1, so we used a wide prior spanning this value (larger values rapidly excluded all densities of challenger). We therefore used uniform priors, which would not bias the approximate posterior, between 0 and 3 for β and 1 and 6 hrs for tcom. We also tried using a wider uniform prior range, between 0 and 10 for β and 0 and 24 hrs for tcom, to validate our choice of prior. For both simulated and experimental data the approximate posterior did not contain significant density of samples from the edge of the β range, suggesting it correctly spanned the plausible range of values (Supplementary Figures 5 and 6). Additionally, these priors gave similar estimates for both parameters when run on the real data to the more restricted range we report on below.
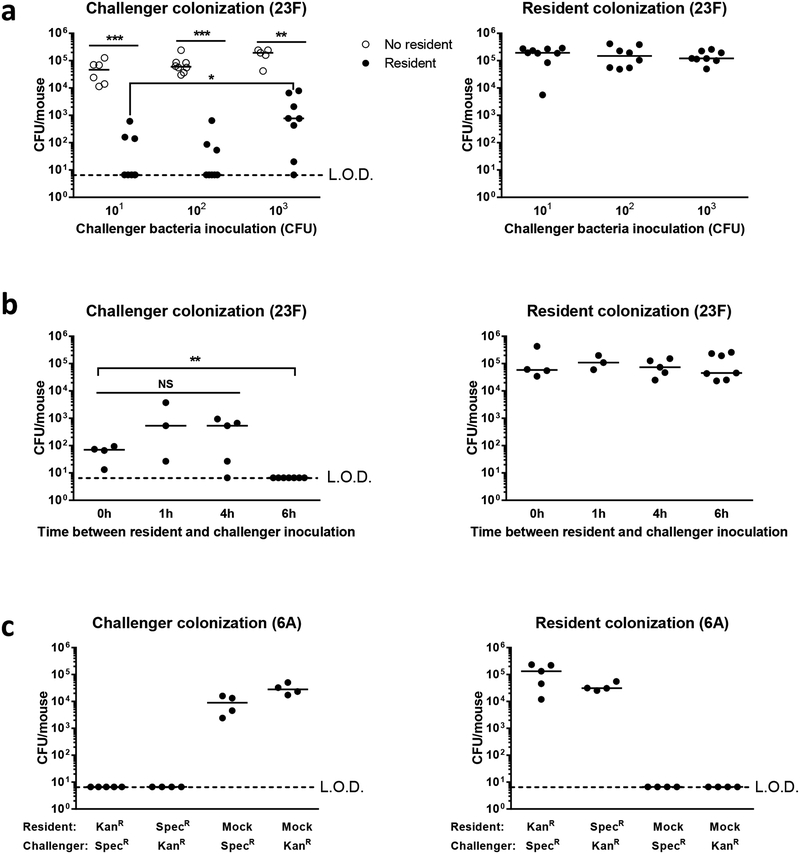
Figure 1: The resident prevails in asymmetric pneumococcal competition for colonization.
a, Day 4–5 old pups were inoculated with a serotype 23F resident strain (103 CFU). After 3 days an isogenic challenger strain was introduced with increasing CFU (101-103). Age-matched naïve mice were inoculated with the challenger only. Nasal lavages were obtained 3 days following challenger inoculation for quantitative culture. Challenger bacterial burdens from pups with or without resident colonization were compared using the two-tailed Mann-Whitney U test, n=5–8. b, The time period the resident was allowed to establish before introduction of 101 CFU challenger was varied. Challenger colonization densities were compared between time points by Kruskal–Wallis one-way analysis of variance (NS: p>0.9999), n=3–7. c, Pups were inoculated with a serotype 6A (103 CFU) kanamycin resistant (KanR) resident for 15h before introduction of a spectinomycin (SpecR) isogenic challenger. Colonization densities were quantified 3 days following challenger inoculation. The two strains were also switched such that the SpecR strain was the resident and the KanR strain was the challenger. Colonization levels by each strain individually in naïve mice are included as controls, n=4–5. LOD, limit of detection, median values are shown for all graphs *p<0.05, **p<0.01, ***p<0.001. NS, non-significant.
As each simulator run took around 30s, we wished to use a method that required fewer than ~104 simulator evaluations. We therefore used BOLFI35, which uses a Gaussian process regression to leverage information from previous distance estimates to optimize future picks of parameter values, and thereby reducing the number of simulator evaluations. We used default values for the BOLFI fit, using 200 evidence points, and taking the log of the above distance to minimize the effect of large discrepancies. This inference took 6.1h on a single core. After approximating the posterior we used Hamiltonian Monte Carlo36 to take samples from it. We took 2000 samples, discarding the first half as burn-in and averaging over four chains using to estimate the marginal distributions for β and tcom.
To test that there was sufficient evidence in this scheme to estimate the parameters, we first simulated observations from the model across the experimental conditions for known values β = 0.1 and tcom = 5hrs (Supplementary Fig 7). While this gave reasonable estimates of both parameters, there appeared to be covariance of tcom and β in the approximate posterior samples, as well as a posterior for tcom that was close to its prior. We therefore simulated data from the model for a grid of values for tcom and β to test whether both parameters could be correctly and separately recovered (Supplementary Table 3). While we had some ability to estimate β in this range, we found we were not able to separately extract tcom. We are only able to measure the final colonization density at the end of the experiment (after 48 hours) and not the at a higher time resolution due to the impractically large number of competition experiments this would require. This is a limitation of fitting this complex model to our in vivo data, which has a relative scarcity of data points. The best quality fit we were able to obtain was therefore by imposing the above experimentally justified prior on tcom, and then attempting to extract a joint estimate of β using our in vivo data.
Performing this fit of these parameters to the real data we estimated mean posterior values of β = 1.48 (0.39 – 2.91 95% HPD) and tcom = 3.76 hrs (2.19 – 5.92 hrs 95% HPD) (Supplementary Fig 8 and 9). This tcom corresponded to a colonization density of 48 CFU (8.9 – 530 CFU 95% HPD) with an initial inoculum of a single CFU.
We did not vary the growth rate r in the model, both because it is likely to be similar between strains, and it makes no difference to the long term stability of the LV model.
Population genomics
To determine the conservation of competition related genes in the natural population, we used a core genome alignment of 616 S. pneumoniae isolates which had been taken from the nasopharynx of asymptomatically carrying children in Massachusetts37,38. We used the top blastp bitscore match between the annotated clusters of orthologous genes (COGs)39,40 to relate genes to their TIGR4 annotations41. We analysed the five genes of interest in table 2 and any COGs annotated as bacteriocins or immunity proteins (Supplementary table 4).
Table 2:
Conservation of genes involved in Com regulon -dependent competition
| Gene | TIGR4 ID* | CLS ID# | Isolates present (/616) | dN/dS | |
|---|---|---|---|---|---|
| All serotypes | cbpD | SP_2201 | CLS00029 | 616 | 0.35 |
| comM | SP_1945 | CLS01685 | 616 | 0.19 | |
| cibA | SP_0125 | CLS00190 | 605 | 0.59 | |
| cibB | SP_0124 | CLS00189 | 605 | 0.52 | |
| cibC | SP_0122 | CLS00187 | 616 | 0.27 |
ID in the TIGR4 reference genome.
Cluster ID refers to a previously described annotation40.
To quantify conservation, which we first checked the frequency of each COG in the whole population. We then calculated dN/dS for each COG as follows. We first aligned each COG at the codon level using RevTrans v1.1042, and at the amino acid level using MUSCLE v3.8.3143. We used the nucleotide alignment to generate a maximum likelihood tree using a GTR rate model with IQ-TREE v1.5.544. These alignments and the tree were then input into HYPHY v2.3, where we used Single-Likelihood Ancestor Counting45 to calculate dN/dS for each COG. For the bacteriocins we also calculated Tajima’s D using our own C++ program.
To determine conservation of Com regulon genes in serotype 3 we combined the 11 serotype 3 genomes from the above population, and 82 serotype 3 genomes previously found to have negligible recombination levels46. We took a single representative sequence from each COG in the Massachusetts population to create a reference panel, and then clustered the genes of these isolates first with CD-HIT47, then with blastp. These matches were used to efficiently annotate the serotype 3 genes consistently with the Massachusetts samples. We analysed the five genes of interest in table 2 and any COGs annotated as being involved in Com regulon activation, using the same methods as above (Supplementary table 5).
Results
The resident prevails in asymmetric pneumococcal competition for colonization
We used isogenic resident and challenger strains to eliminate genetic differences as a factor during competition. Bacteria were inoculated intra-nasally into infant mice and bacterial density in the nasal lavage fluid was assessed. The resident and challenger strains were distinguished by insertion of different antibiotic resistance markers and selective plating.
Infant mice are highly susceptible to S. pneumoniae colonization. In naïve mice, maximal colonization levels were established with an inoculum as low as 101 CFU of a 23F serotype challenger strain (Fig 1A). In contrast, when an isogenic resident strain was first established in the host for 3 days, colonization by the challenger was inhibited. Increasing the inoculation dose of the challenger partially restored its colonization without impacting resident strain density. We then varied the time period between inoculation of the resident and challenger (Fig 1B). The resident was able to completely exclude challenger colonization (101 CFU inoculation) when given 6h prior, but not at earlier time points. Resident colonization levels were not impacted (Fig 1B). Competition was also not strain specific (Fig 1C). While two serotype 6A isogenic strains both colonized naïve mice, establishment of either strain as the resident excluded subsequent colonization by the other strain. These data suggest that competition for pneumococcal colonization is asymmetric: a resident strain established at 6h prior always prevails even when the challenger is isogenic.
Asymmetric competition is dependent on direct interactions between resident and challenger pneumococci
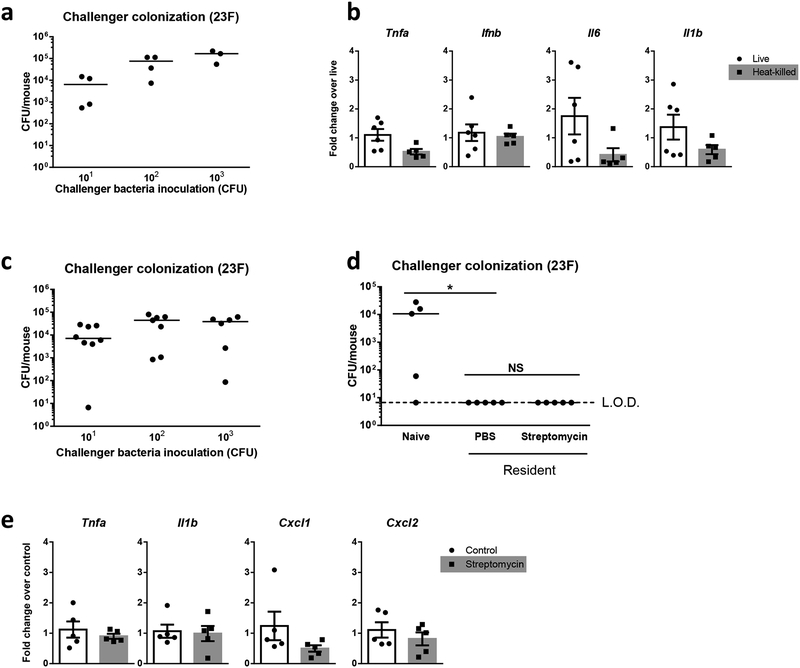
We investigated if the resident acted directly on the challenger, or if challenger exclusion occurred as a consequence of a host environment altered by the resident. Heat-killed resident given 6h prior to challenger inoculation was unable to prevent challenger colonization (Fig 2A), especially at low dose challenger-inoculation where we previously observed complete exclusion in the presence of live resident. Given that live and heat-killed resident induced similar expression levels of early inflammatory markers of pneumococcal colonization48, including tumour necrosis factor (TNF)-α, interferon (IFN)-β, interleukin (IL)-6, and IL-1β, it is unlikely that the challenger was excluded through a resident-elicited inflammatory response (Fig 2B). Eradication of the resident by streptomycin also rescued colonization by a streptomycin resistant challenger (Fig 2C). In contrast, when the same experiment was repeated with a streptomycin resistant resident, streptomycin treatment did not rescue challenger colonization (Fig 2D). We controlled for any impact of streptomycin treatment on non-pneumococcal components of the nasal microbiota by using resident S. pneumoniae that are sensitive or resistant to streptomycin. Additionally, streptomycin treatment did not alter expression levels of cytokines induced at 3 days post-pneumococcal colonization19 (Fig 2E). Any impact of the resident on the host environment was insufficient to exclude the challenger if the resident was no longer present. This shows that the resident acts directly on the challenger to attenuate its colonization during asymmetric competition.
Figure 2: Asymmetric competition is dependent on direct interactions between resident and challenger pneumococci.
a, Day 4–5 old pups were inoculated with a heat-killed serotype 23F resident strain (105 CFU) for 6h before introduction of the live isogenic challenger strain with increasing CFU (101-103). Nasal lavages were cultured 3 days later to determine challenger colonization densities, n=3–4. Median values are shown. b, Pups were inoculated with live (103 CFU) or heat-killed (105 CFU) resident strain for 6h and nasal lavages with RLT lysis buffer were obtained to isolate host RNA. Early inflammatory cytokine expression levels were assessed by SYBR green real-time quantitative RT-PCR. Cytokine expression induced by heat-killed S. pneumoniae were shown as fold change relative to that elicited by live bacteria, n=5–6. Data are shown as mean ± SEM. c-d, Pups were inoculated with either a streptomycin sensitive (c) or resistant (d) resident strain for 3 days before intranasal administration of streptomycin. An isogenic streptomycin resistant challenger was inoculated 6h later and its colonization density determined after 3 days. Median values are shown. Colonization levels by 101 CFU challenger alone, and in resident-colonized pups treated with either phosphate buffered saline (PBS) or streptomycin, were compared by Kruskal–Wallis one-way analysis of variance in (d), n=4–8, *p<0.05, NS: p>0.9999, LOD, limit of detection. e, RLT nasal lavages were collected from pups after 3-day colonization with the streptomycin sensitive resident 6h following PBS control or streptomycin treatment for RNA analysis, n=5. Data are shown as mean ± SEM. NS, non-significant.
Prevention of challenger colonization is dependent on quorum sensing in the resident and competence-induced fratricide
We tested the hypothesis that the resident reaches maximal colonization by 6h, excluding challenger colonization due to competition for space (i.e. the carrying capacity has already been reached). However, when we assessed the in vivo colonization density of the resident over time, it was far below maximal colonization levels until 12h post-inoculation (Fig 3A). These data suggested a time-dependent mechanism of bacterial interference that occurs before full occupancy of the niche.
A common mechanism of bacterial-bacterial interactions involves quorum sensing (QS), where bacterial cells communicate with one another based on the cell density-dependent accumulation of signalling molecules in the surrounding environment that induces coordinated responses in the overall population49. In S. pneumoniae, there are two well-studied QS systems that could contribute to competition, the bacteriocin-like peptides (Blp) and the Com regulon, with cross-talk recently reported to occur between the two systems50,51. We ruled out a role for Blp as a blp- resident still excluded challenger colonization (Supplementary Fig 10). However, when the resident was deficient in ComE, the master response regulator for Com regulon activation, challenger colonization was restored at both 6h and 3 days post-resident inoculation, although a higher inoculum was required to restore challenger colonization at 3 days (Fig 3B and 3C).
The murein hydrolase choline binding protein D (CbpD) and competence-induced bacteriocins A and B (CibAB) are up-regulated by the pheromone competence-stimulating peptide (CSP) in a ComE-dependent manner and have been reported to mediate the killing of non-competent pneumococci (referred to as fratricide) in vitro16,17. We constructed mutants of cbpD and cibAB and assessed their ability as the resident to exclude WT challenger colonization (Fig 3D). There was no effect of single mutations in cbpD or cibAB. Rather, challenger colonization was restored only when the expression of both CbpD and CibAB were disrupted in the resident. We corrected both mutations and confirmed exclusion of challenger colonization similar to WT resident. Downstream of comE, the autolysin lytA and lysozyme lytC also contributes to fratricide in vitro52. However, we did not observe differences in challenger colonization using a lytA-lytC- resident (Supplementary Fig 11).
Since competent cells protect themselves from the effect of CbpD and CibAB through the cognate immunity factors ComM and CibC, respectively17,53, we hypothesized Com regulon activation in the challenger should confer protection against the resident via immunity factor expression. CSP stimulation of the challenger prior to host inoculation significantly increased challenger colonization (Fig 3E). This effect was QS-dependent as CSP stimulation did not rescue colonization of a comE- challenger. To determine if the protective effect of CSP is due to immunity factor expression, we utilized a CibAB deficient resident to isolate the contribution of CbpD and ComM. The challenger in this case was deficient in both CbpD and ComM to prevent self-killing. Protection induced by CSP stimulation in the challenger was lost in the absence of ComM. Furthermore, RNA levels of comX, a marker of Com regulon activation, comM, and cbpD were all up-regulated by CSP treatment of the WT challenger in vitro (Fig 3F). To determine if the challenger retains Com regulon activation during host-to-host transit, we directly inoculated pneumococci from the nasal lavage of challenger-colonized mice into mice already colonized with the resident for 6h or 3 days (Fig 3G). The challenger was still excluded from colonization despite prior in vivo adaptation, suggesting that the Com regulon was quickly switched off such that immunity factor expression was lost upon entry into the new host environment. These data showed that the resident excluded challenger colonization through QS via the expression of CbpD and CibAB downstream of Com regulon activation. This mechanism is cell density-dependent, taking around 6h to take effect in the host, and is sustained, lasting at least 3 days. Upon arrival in the host, the challenger does not reach the population size necessary to activate the Com regulon, and is thus unable to express corresponding immunity factors in time to avoid being killed by the resident.
Since competition would naturally occur following transmission events when bacteria are establishing in new hosts, we assessed the role of QS in asymmetric competition in a transmission model. The 23F WT strain transmits to naïve mice (Table 1). When 23F WT or comE- mutant colonized mice were co-housed together, the WT bacteria transmitted to hosts colonized with resident comE- mutant. However, none of the WT-colonized mice acquired the comE- mutant. Similarly, transmission of WT bacteria only occurred with a cbpD-cibAB- resident. We concluded that QS promotes competition for colonization during transmission.
Genes involved in Com regulon-dependent competition are highly conserved in S. pneumoniae
To assess whether induction of the Com regulon could be a general mechanism for competition, we evaluated the conservation of Com regulon-dependent CbpD, CibAB and immunity factors across pneumococcal isolates of different genetic backgrounds. For each gene, we counted its frequency in a population sample of 616 S. pneumoniae genomes collected from asymptomatic carriage in children and found them to be present in almost all isolates (Table 2). We also calculated the proportion of non-synonymous to synonymous changes (dN/dS) from a codon alignment of each gene to assess amino acid sequence conservation. The dN/dS of all genes listed in table 2 are below 1 and close to that of the mean dN/dS of all pneumococcal core genes (mean 0.28, median 0.22, 95% quantiles 0.058–0.88), and accessory genes present in over 5% of isolates (mean 1.29, median 0.38, 95% quantiles 0.076–2.51), suggesting purifying selection and therefore high conservation across strains40. Since the Com regulon has been studied primarily for its role in extracellular DNA uptake and incorporation, this raised the question whether its induction contributes to fitness through competition, genetic transformation, or both. We examined a large collection of serotype 3 isolates of S. pneumoniae previously shown to have a negligible DNA recombination rate, possibly because of steric hindrance by its unusually thick capsule46. We reasoned that if the genes in table 2, as well as the Com regulon machinery in general, were conserved in this non-recombining lineage then this would provide further evidence that there is an evolutionarily conserved function of Com regulon activation besides DNA uptake. Indeed, we found no difference in Com regulon gene frequency (χ2 = 0.12; df = 21; two-sided p = 1.0) or sequence conservation (median dN/dS 0.27 vs. 0.31 W = 248.5, two-sided p = 0.8879, Wilcox test) between serotype 3 and the overall population (Supplementary Table 5). Thus, the coupling of DNA acquisition may have evolved to maximize benefit during competition in most strains.
As bacteriocins are a well-known competition mechanism in the pneumococcus, we broadly assessed Com-mediated competitive mechanisms as a distinct strategy by examining bacteriocin population-level characteristics (gene frequencies and dN/dS). Our analysis of bacteriocins in the same population as above (Supplementary Table 4) found that they were variably present (mean frequency 40%), had higher levels of sequence diversity than core genes (median dN/dS 0.5; mean = 16.5), and higher levels of allelic diversity maintained in the population (median Tajima’s D = −0.23; mean = 0.11; for population, median = 0.76, mean=0.54). It has been shown that there is frequency dependent selection on these genes39,54, meaning they are present at intermediate frequencies in the pneumococcal population55. An example of this is a ‘rock-paper-scissors’ interaction: a strain can encode a bacteriocin, its immunity protein or neither, and its fitness depends on the proportion of other strategies in the population56. The diversifying and frequency-dependent selection shows that bacteriocins serve as a general strategy for conditional competition between strains. In contrast, we found that the Com regulon genes in Table 2 are conserved and consistently present in a high proportion of the population. This genetically-blind and quorum-dependent mechanism is therefore likely a distinct competitive strategy from the bacteriocins.
Mathematical model of Com regulon-mediated asymmetric competition
We wished to generalize our conclusions from our in vivo experiments with isogenic challenger and resident strains: how much of an effect does Com regulon activation have on competition amongst colonizing pneumococci more generally? The large diversity of the pneumococcal population makes comprehensive experiments between many different strains of resident and challenger impossible. We therefore formulated a mathematical model to describe the phenomenon we observed between an isogenic resident and challenger which could then be generalized to residents and challengers with different genetic backgrounds. We modelled the population dynamics of resident and challenger strains using a two-species, three-parameter Lotka-Volterra (LV) model, which has previously been used to describe the dynamics of co-infection57. The two strains compete with themselves for resources (intraspecific competition leading to carrying capacity K), and with each other at potentially different strengths depending on the strains in question (interspecific competition terms αRC and αCR).
We modified this model to include a time-dependent effect of Com regulon activation with strength β which turns on in the resident after time tcom. and created a stochastic formulation of the model to correctly deal with small population sizes when changing competition strengths over time (see methods). Using two approaches, we then fitted the four free parameters of the resulting model to our experimental data (Fig 4). We were unable to accurately estimate tcom from our in vivo data, so instead relied on an experimentally informed prior distribution for this parameter. Though the relative scarcity of data points available to fit the model also limited the accuracy of estimation of β, the overall combined effect of the parameters β and tcom over their estimated range was a strong effect for the Com regulon-mediated competition.
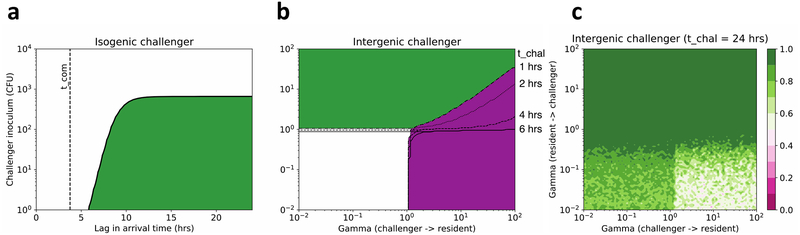
Figure 4: Predictions of winners between resident and challenger from the mathematical model.
a, For a range of lags in arrival time between an isogenic resident and challenger, and inoculum sizes the green region is that in which ‘the resident always wins’ in the deterministic model. The fitted value of tcom is shown as a vertical dashed line. b, For a range of strengths of competition γ between different resident and challenger strains, regions where at long times in the deterministic model: the resident wins (green); the challenger wins (purple); there is co-existence (white). Marked contours show how the area where the resident wins increases with increasing lag times marked on the y-axis. c, At long times between arrival (lag time 24hrs) the probability of being a long-term winner in the stochastic model: from resident always wins (green) through co-existence (white) to challenger always wins (pink). Averaged over twenty runs.
So, by using our best estimate for the fitted values of these parameters in our model, we aimed to predict when the resident wins against an isogenic challenger across a range of times and challenger inoculum sizes (Fig 4A). The green region shows when ‘the resident always wins’ for the deterministic model. This was necessarily consistent with our experimental data: for small inoculum sizes exclusion occurs between 4 and 6h, and a large inoculum (103 CFU) can restore challenger colonization. The stochastic model gave very similar conclusions (Supplementary Fig 12).
After fitting our model to an isogenic challenger, we then varied the relative strength of resident and challenger’s inter-specific competition γ to test the global effect of the development of Com regulon activation on exclusion of the challenger. For equal arrival times this replicates the known results of the standard LV model, however as the lag between arrival times increases the challenger can only win if it exerts a competitive advantage over the resident, and the resident does not effectively compete with the challenger (Fig 4B). Again, the stochastic model gave similar results (Supplementary Fig 13). After 24h, our model predicts that the effect of Com regulon activation is complete, and that the challenger will normally be excluded, regardless of its intrinsic competitive ability in a head to head race (Fig 4C). Our model therefore predicts that Com regulon activation plays a critical role in competition for colonization even between pneumococcal strains with different genetic backgrounds, and that its effect occurs rapidly (in less than 24h).
Validation of mathematical model with genetically different resident and challenger strains
To validate the prediction made by our mathematical model that Com regulon activation can have a significant advantage even between strains with different competition strengths, we performed transmission experiments as before, but using two genetically different S. pneumoniae strains. As with serotype 23F WT, the serotype 6A WT strain transmits to naïve mice (Table 1). When serotype 23F or 6A WT colonized mice were co-housed together, both strains blocked transmission of the other serotype. We did see a small probability for co-colonization of the two strains (1/11 mice). This could be explained by the transmission event occurring soon enough after inoculation such that the resident had not yet activated the Com regulon (less than 6h). However, when we co-housed mice colonized with either the serotype 6A WT strain or the serotype 23F comE- mutant, we observed that the serotype 6A WT bacteria can now transmit to mice colonized with the serotype 23F comE- mutant. In contrast, none of the mice colonized with the serotype 6A WT strain acquired the serotype 23F comE- mutant. This Com regulon dependent effect on competition between different strains is consistent with the predictions of our model.
Discussion
We report that a pneumococcal resident (owner) excluded a challenger (intruder) dependent on QS via Com regulon activation. While owner/intruder biases in competitive outcomes have been widely observed across animal model systems7,8, the molecular and ecological mechanisms of owner advantage have largely remained obscure. From an ecological perspective, owner advantage can follow from (1) quality asymmetry: enrichment of superior competitors in ownership positions58; (2) convention asymmetry: use of ownership convention or ‘social norms’ to minimize costly fights1; and (3) value asymmetry: higher valuation of habitat to the owner8. Our experimental design using isogenic competitors rules out quality asymmetry. The evolutionarily stable use of convention asymmetry requires that the expected costs of escalated fights exceed the expected benefits of territory ownership, and has received experimental support in butterfly contests where alternate sunspot territories are readily accessible and physical contests are costly7. In the context of pneumococcal competition over host territory, the probability of an ‘intruder’ clone to avoid conflict by switching to an alternate host is likely to be both low and out of the clone’s strategic control. In contrast, there is a clear basis for value asymmetry in the context of the pneumococcus, mediated by clonal population size. By virtue of an earlier arrival time, the resident clone is able to build greater cellular resources through within-host growth, and is also able to access and respond to information on its reproductive value via Com regulon QS.
The pneumococcal Com regulon has been studied primarily as a mechanism for DNA acquisition and genetic transformation. There is heterogeneity in the population during QS such that not all cells activate the Com regulon in response to CSP at the same time, and pneumococcal fratricide, a process whereby competent cells target non-competent cells for lysis and DNA acquisition, occurs59. The release of DNA and virulence factors, such as pneumolysin, are thought to be advantageous for the remaining population. DNA transformation can maintain bacterial fitness, including defending against the spread of parasitic mobile genetic elements, while pneumolysin facilitates invasiveness and transmission16,17,19,52,60.
The murein hydrolase CbpD and bacteriocins CibAB have been extensively studied in vitro, although reports of their relative importance to competence-induced fratricide have been conflicting depending on the assay utilized16,17,52. CbpD is an extracellular, cell wall anchored murein hydrolase that binds to peptidylglycan of target cells61. CibAB, on the other hand, have double glycine leaders and are secreted bacteriocins that are thought to cause lysis by insertion into the cell membrane of target cells17,62. Our in vivo data show a redundancy in the system where either CbpD or CibAB alone was sufficient for competition during colonization, further highlighting the evolutionary importance of this mechanism. Lysis, the result of competition, subsequently provides a source of external DNA for genetic transformation. Activation of the Com regulon likely also impact closely related microbiome members as CbpD from S. pneumoniae has been reported to target non-competent S. oralis and S. mitis for lysis in vitro63. Whether pneumococcal CbpD and CibAB have activity against distantly related bacteria is unknown.
Given the significance of Com regulon activation during competition, we wondered whether its effects are more constitutive in the host environment when compared to in vitro conditions where Com regulon-induced genes are only upregulated for a short period, and expression declines within minutes64. We observed that challenger colonization was significantly rescued when the resident was deficient in ComE even at 3 days post-resident inoculation. The long term effect of Com regulon activation in vivo could be due to factors specific to the nasopharyngeal environment, including a lower temperature, limited nutrients, interactions with the host epithelium, and biofilm formation65. Consistent with our conclusions, constitutive induction of comX and comD relative to planktonic culture, for up to 3 days, was observed in an in vitro biofilm assay65. Our study suggests a narrow window for co-colonization with different pneumococci strains that occur before Com regulon activation by the resident, and possibly during clearance of colonizing strains when the quorum is below the threshold for Com regulon activation, accounting for the lower than predicted levels of co-colonization observed in humans11.
It may be costly to maintain constitutive activation of the Com regulon despite an advantage for competition. We speculate that the bacteria sense population density and environmental factors to activate the Com regulon only when conditions favor maximum effectiveness. Perhaps the most well-known factor to impact Com regulon activation is the initial cell density, with lower bacterial inoculums taking hours longer to activate the Com regulon than higher density populations in vitro66. Indeed, toxin production by bacterial populations have been suggested to be favored by a high density of self-cells, as to ensure maximum effectiveness67. We observed that although the challenger still established colonization in infant mice when given at the same time as the resident, its final colonization density was proportional to its inoculation dose (Supplementary Fig 14). We speculate that when the resident was given at a higher inoculum than the challenger, it activated the Com regulon faster and was able to target a portion of the challenger population that has not yet activated the Com regulon, resulting in lower challenger colonization due to a lack of immunity factor expression. These results support that the threshold of CSP concentration was achieved more quickly with a higher starting cell density. Com regulon activation is also shaped by conditions of the host environment, with the synchronization of cells affected by growth conditions, pH and exposure to antibiotics66. Additionally, environmental factors affecting other QS systems, such as LuxS, may also impact competition through the Com regulon-dependent expression of CbpD68. Lastly, a host factor asparagine, released by infected host cells, was recently shown to activate QS and bacteriocin-mediated competition in group A Streptococcus69. Whether there are any host factors that activate the Com regulon or other QS systems in S. pneumoniae remains unknown.
The general stability of the microbiome composition over time suggests a competitive advantage for established resident bacteria over newcomers. We showed competition amongst bacteria to be asymmetric using S. pneumoniae. Bacteria adopt an evolutionarily stable strategy resembling value asymmetry during competition similar to that described in higher level organisms. In value asymmetry, the resident invests time and energy in establishing its territory before it can outcompete the challenger8. Rather than behavioral cues, our study shows that microbes employ QS to detect population asymmetries and modulate competition. Mathematical modelling supports the idea that Com regulon activation has a significant role in competition for colonization, even for strains with very different competitive advantages, and that its effects manifest quickly (in less than 24h). Although it plays a crucial role in competition, the relative contribution of the Com regulon in competition may vary depending on differences in bacteriocin expression between different genotypes. QS is common among microbes, including pathogens, and may be a general mechanism for asymmetric bacterial competition during colonization.
Supplementary Material
Acknowledgements
We wish to thank Simon Frost and Jukka Corander for helpful advice on the formulation of the stochastic model and likelihood-free model fitting respectively.
Footnotes
Data availability
The in vivo measurements, formatted as used to fit the competition model, are available on figshare with doi:10.6084/m9.figshare.7166432. All code used to run and fit the population dynamics model, and draw the associated plots is available on github: https://github.com/johnlees/competition_model (Apache 2.0 license). The code we used to calculate Tajima’s D is available on github: https://github.com/johnlees/tajima-D (GPL 2.0 license). The data that support the findings of this study are available from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
References
- 1.Foster KR & Bell T Competition, not cooperation, dominates interactions among culturable microbial species. Curr. Biol 22, 1845–1850 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Chassaing B & Cascales E Antibacterial Weapons: Targeted Destruction in the Microbiota. Trends in Microbiology 26, 329–338 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Davies NB, Krebs JR & West SA An introduction to behavioural ecology, 4th edition. An Introduction to Behavioural Ecology (2012). doi: 10.1037/026600 [DOI] [Google Scholar]
- 4.Clutton-Brock TH & Albon SD The Roaring of Red Deer and the Evolution of Honest Advertisement. Behaviour 69, 145–170 (1979). [Google Scholar]
- 5.Smith JM & Parker GA The logic of asymmetric contests. Anim. Behav 24, 159–175 (1976). [Google Scholar]
- 6.Leimar O & Enquist M Effects of asymmetries in owner-intruder conflicts. J. Theor. Biol 111, 475–491 (1984). [Google Scholar]
- 7.Davies NB Territorial defence in the speckled wood butterfly (Pararge aegeria): The resident always wins. Anim. Behav 26, 138–147 (1978). [Google Scholar]
- 8.Tobias J Asymmetric territorial contests in the European robin: The role of settlement costs. Anim. Behav 54, 9–21 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Regev-Yochay G et al. Nasopharyngeal carriage of Streptococcus pneumoniae by adults and children in community and family settings. Clin. Infect. Dis 38, 632–639 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Kamng’ona AW et al. High multiple carriage and emergence of Streptococcus pneumoniae vaccine serotype variants in Malawian children. BMC Infect. Dis 15, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Numminen E, Cheng L, Gyllenberg M & Corander J Estimating the Transmission Dynamics of Streptococcus pneumoniae from Strain Prevalence Data. Biometrics 69, 748–757 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Lees JA et al. Genome-wide identification of lineage and locus specific variation associated with pneumococcal carriage duration. Elife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pessoa D et al. Comparative analysis of Streptococcus pneumoniae transmission in Portuguese and Finnish day-care centres. BMC Infect. Dis 13, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Javan RR, Tonder AJ Van King JP, Harrold CL & Brueggemann AB Genome sequencing reveals that Streptococcus pneumoniae possesses a large and diverse repertoire of antimicrobial toxins Corresponding author Running title Keywords 1–36 (2017). [Google Scholar]
- 15.Kono M et al. Single Cell Bottlenecks in the Pathogenesis of Streptococcus pneumoniae. PLoS Pathog 12, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kausmally L, Johnsborg O, Lunde M, Knutsen E & Håvarstein LS Choline-binding protein D (CbpD) in Streptococcus pneumoniae is essential for competence-induced cell lysis. J. Bacteriol 187, 4338–4345 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guiral S, Mitchell TJ, Martin B & Claverys JP Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci U S A 102, 8710–8715 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zangari T, Wang Y & Weiser JN Streptococcus pneumoniae transmission is blocked by type-specific immunity in an infant mouse model. MBio 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zafar MA, Wang Y, Hamaguchi S & Weiser JN Host-to-Host Transmission of Streptococcus pneumoniae Is Driven by Its Inflammatory Toxin, Pneumolysin. Cell Host Microbe 21, 73–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sung CK, Li H, Claverys JP & Morrison DA An rpsL Cassette, Janus, for Gene Replacement through Negative Selection in Streptococcus pneumoniae. Appl. Environ. Microbiol 67, 5190–5196 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeBardeleben HK, Lysenko ES, Dalia AB & Weiser JN Tolerance of a phage element by Streptococcus pneumoniae leads to a fitness defect during colonization. J. Bacteriol 196, 2670–2680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowalko JE & Sebert ME The Streptococcus pneumoniae competence regulatory system influences respiratory tract colonization. Infect. Immun 76, 3131–3140 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hergott CB et al. Bacterial exploitation of phosphorylcholine mimicry suppresses inflammation to promote airway infection 125, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Clarke TB & Weiser JN Cellular effectors mediating Th17-dependent clearance of pneumococcal colonization in mice. J Clin Invest 119, 1899–1909 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemon JK, Miller MR & Weiser JN Sensing of IL-1 cytokines during Streptococcus pneumoniae colonization contributes to macrophage recruitment and bacterial clearance. Infect. Immun IAI.00224–15 (2015). doi: 10.1128/IAI.00224-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roche AM, Richard AL, Rahkola JT, Janoff EN & Weiser JN Antibody blocks acquisition of bacterial colonization through agglutination. Mucosal Immunol 8, 176–185 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegel SJ, Tamashiro E & Weiser JN Clearance of Pneumococcal Colonization in Infants Is Delayed through Altered Macrophage Trafficking. PLoS Pathog 11, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blat J & Brown KJ Bifurcation of steady-state solutions in predator-grey and competition systems. Proc. R. Soc. Edinburgh Sect. A Math 97, 21–34 (1984). [Google Scholar]
- 29.Allen LJS A primer on stochastic epidemic models: Formulation, numerical simulation, and analysis. Infect. Dis. Model 2, 128–142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gillespie DT Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem 81, 2340–2361 (1977). [Google Scholar]
- 31.Allen EJ, Allen LJS, Arciniega A & Greenwood PE Construction of Equivalent Stochastic Differential Equation Models. Stoch. Anal. Appl 26, 274–297 (2008). [Google Scholar]
- 32.Maruyama G Continuous Markov processes and stochastic equations. Rend. Circ. Mat. Palermo 4, 48 (1955). [Google Scholar]
- 33.Lintusaari J, Gutmann MU, Dutta R, Kaski S & Corander J Fundamentals and Recent Developments in Approximate Bayesian Computation. Syst. Biol 66, e66–e82 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lintusaari J et al. ELFI: Engine for Likelihood Free Inference. arXiv [stat.ML] (2017). [Google Scholar]
- 35.Gutmann MU & Corander J Bayesian Optimization for Likelihood-free Inference of Simulator-based Statistical Models. J. Mach. Learn. Res 17, 4256–4302 (2016). [Google Scholar]
- 36.Homan MD & Gelman A The No-U-turn Sampler: Adaptively Setting Path Lengths in Hamiltonian Monte Carlo. J. Mach. Learn. Res 15, 1593–1623 (2014). [Google Scholar]
- 37.Croucher NJ et al. Population genomics of post-vaccine changes in pneumococcal epidemiology. Nat. Genet 45, 656–663 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Croucher NJ et al. Population genomic datasets describing the post-vaccine evolutionary epidemiology of Streptococcus pneumoniae. Sci. Data 2, 150058 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corander J et al. Frequency-dependent selection in vaccine-associated pneumococcal population dynamics. Nat. Ecol. Evol 1 (2017). doi: 10.1038/s41559-017-0337-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Croucher NJ et al. Diverse evolutionary patterns of pneumococcal antigens identified by pangenome-wide immunological screening. Proc. Natl. Acad. Sci. U. S. A (2017). doi: 10.1073/pnas.1613937114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tettelin H et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science (80-.) 293, 498–506 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Wernersson R & Pedersen AG RevTrans: Multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res 31, 3537–3539 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen L-T, Schmidt HA, von Haeseler A & Minh BQ IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kosakovsky Pond SL & Frost SDW Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol 22, 1208–1222 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Croucher NJ et al. Dominant role of nucleotide substitution in the diversification of serotype 3 pneumococci over decades and during a single infection. PLoS Genet 9, e1003868 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu L, Niu B, Zhu Z, Wu S & Li W CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dorrington MG et al. MARCO Is Required for TLR2- and Nod2-Mediated Responses to Streptococcus pneumoniae and Clearance of Pneumococcal Colonization in the Murine Nasopharynx. J Immunol 190, 250–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whiteley M, Diggle SP & Greenberg EP Progress in and promise of bacterial quorum sensing research. Nature 551, 313–320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shanker E & Federle MJ Quorum sensing regulation of competence and bacteriocins in Streptococcus pneumoniae and mutans. Genes 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wholey WY, Kochan TJ, Storck DN & Dawid S Coordinated Bacteriocin Expression and Competence in Streptococcus pneumoniae Contributes to Genetic Adaptation through Neighbor Predation. PLoS Pathog 12, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eldholm V, Johnsborg O, Haugen K, Ohnstad HS & Havastein LS Fratricide in Streptococcus pneumoniae: Contributions and role of the cell wall hydrolases CbpD, LytA and LytC. Microbiology 155, 2223–2234 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Håvarstein LS, Martin B, Johnsborg O, Granadel C & Claverys JP New insights into the pneumococcal fratricide: Relationship to clumping and identification of a novel immunity factor. Mol. Microbiol 59, 1297–1307 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Azarian T et al. The impact of serotype-specific vaccination on phylodynamic parameters of Streptococcus pneumoniae and the pneumococcal pan-genome. PLOS Pathog 14, e1006966 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller EL, Abrudan MI, Roberts IS & Rozen DE Diverse Ecological Strategies Are Encoded by Streptococcus pneumoniae Bacteriocin-Like Peptides. Genome Biol. Evol 8, 1072–1090 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kerr B, Riley MA, Feldman MW & Bohannan BJM Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418, 171–174 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Eswarappa SM, Estrela S & Brown SP Within-host dynamics of multi-species infections: facilitation, competition and virulence. PLoS One 7, e38730 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krebs JR Territory and Breeding Density in the Great Tit, Parus Major L. Ecology 52, 2–22 (1971). [Google Scholar]
- 59.Claverys JP, Martin B & Håvarstein LS Competence-induced fratricide in streptococci. Molecular Microbiology 64, 1423–1433 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Croucher NJ et al. Horizontal DNA Transfer Mechanisms of Bacteria as Weapons of Intragenomic Conflict. PLoS Biol 14, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eldholm V et al. Pneumococcal CbpD is a murein hydrolase that requires a dual cell envelope binding specificity to kill target cells during fratricide. Mol. Microbiol 76, 905–917 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Peterson SN et al. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol 51, 1051–1070 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Johnsborg O, Eldholm V, Bjørnstad ML & Håvarstein LS A predatory mechanism dramatically increases the efficiency of lateral gene transfer in Streptococcus pneumoniae and related commensal species. Mol. Microbiol 69, 245–253 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Peterson S, Cline RT, Tettelin H, Sharov V & Morrison DA Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J. Bacteriol 182, 6192–6202 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marks LR, Reddinger RM & Anders P High Levels of Genetic Recombination during Nasopharyngeal Carriage and Biofilm Formation in Streptococcus pneumoniae. MBio 3, e00200–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moreno-Gámez S et al. Quorum sensing integrates environmental cues, cell density and cell history to control bacterial competence. Nat. Commun 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cornforth DM & Foster KR Competition sensing: The social side of bacterial stress responses. Nature Reviews Microbiology 11, 285–293 (2013). [DOI] [PubMed] [Google Scholar]
- 68.Trappetti C, Potter AJ, Paton AW, Oggioni MR & Paton JC LuxS mediates iron-dependent biofilm formation, competence, and fratricide in Streptococcus pneumoniae. Infect Immun 79, 4550–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hertzog BB et al. A Sub-population of Group A Streptococcus Elicits a Population-wide Production of Bacteriocins to Establish Dominance in the Host. Cell Host Microbe 23, 312–323.e6 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.