Abstract
Somatic mutations can promote malignant transformation of airway epithelial cells, and induce inflammatory responses directed against resultant tumors. Tumor-infiltrating T lymphocytes (TILs) in early stage non-small cell lung cancer (NSCLC) secrete distinct pro-inflammatory cytokines, but the contribution of these TILs to tumor development and metastasis remains unknown. We show here that TILs in early stage NSCLC are biased toward IL17A expression (Th17) when compared to adjacent tumor-free tissue, whereas Th17 cells are decreased in tumor infiltrating loco-regional lymph nodes in advanced NSCLC. Mice in which Pten and Smad4 (Pts4d/d) are deleted from airway epithelial cells develop spontaneous tumors, that share genetic signatures with squamous- (SQ.2b), and adeno- (AD.1) subtypes of human NSCLC. Pts4d/d mice globally lacking in IL17a (Pts4d/dIl17a−/−) showed decreased tumor latency and increased metastasis. Th17 cells were required for recruitment of CD103+ dendritic cells, and adoptive transfer of IL17a-sufficient CD4+ T cells reversed early tumor development and metastasis in Pts4d/dIl17a−/− mice. Together, these findings support a key role for Th17 cells in TILs associated with the Pts4d/d model of NSCLC and suggest therapeutic and biomarker strategies for human SQ2b and AD1 lung cancer.
Introduction
Lung cancer is responsible for over a quarter of all cancer-related deaths worldwide (1). More than two thirds of lung cancer cases are detected at inoperable stages, and among those who undergo lung resection for treatment of early stage cancer, approximately 40% develop metastatic recurrence (2). Although individual genetic susceptibility factors affect lung cancer development, little is known about the underlying mechanisms that regulate tumor latency and metastasis. Cigarette smoke has been shown to promote somatic mutations, double strand DNA breaks, and malignant transformation in airway epithelial cells(3). Despite heightened inflammatory states in the lungs in response to tumor, induction of immune checkpoint molecules (e.g. PD-1, CTLA4) in T cells inhibits recognition of primary or metastatic cells, rendering local immune responses ineffective (4).
IL17A-expressing CD4+ T cells (Th17 cells) are a necessary component of adaptive immune responses in humans and in experimental models of emphysema (5, 6). Although IL17A and Th17 cell have been implicated in human and pre-clinical models of non-small cell lung cancer (NSCLC) (7), their exact activities remain unclear. For instance, immunohistochemical analysis of primary NSCLC tissue showed increased IL17A expression, and genetic polymorphisms that increase its expression are associated with NSCLC (8, 9). Increased serum concentration of IL17A and Th17 cells in peripheral blood of NCLSC have been reported, albeit inconsistently (10). Furthermore, IL17A can induce angiogenesis and promotes proliferation of lung tumor cell lines and metastasis in immunodeficient mice (11). Similarly, protumor and antitumor roles for Th17 cells have also been proposed in other solid tumors(12–14). Specifically, IL17A has been shown to promote hepatocellular carcinoma, and in a model of breast cancer, IL17A has been shown to induce CXCL12 expression and promote metastasis (13, 15). In contrast, in a model of melanoma, IL17A has been shown to promote antitumor responses mediated by activation of T helper 1 (Th1) cells and induction of cytotoxic T cells (CTLs) (12). These divergent (e.g., pro- or antitumor) roles for IL17A might be in part governed by the complex nature of solid organ tumors. However, less is known about the activities of IL17A/Th17 cells in most solid tumors, including NSCLC, in an immune competent host.
NSCLC, the most common histological subtype of lung cancer, is characterized by the presence of an expanding repertoire of oncogenic mutations (e.g., epidermal growth factor receptor) and the loss of tumor suppressors such as KRAS, PTEN, and SMAD4 (16–18). We have previously shown that PTEN and SMAD4 mutations are found in 5.9% and 2.9% of NSCLC respectively(19). Although deletion of many or most gene copies (approximating near total loss by SNP array) for PTEN and SMAD4 occurred in 5.3% and 2.1% of NSCLC cases, respectively, deletion of a few copies (approximating partial copy loss by SNP array) for PTEN and SMAD4 occurred in 36% and 44% of NSCLC cases, respectively(19). We have previously shown that airway-specific deletion of Pten and Smad4 (Pts4d/d) using codon optimized (i)Cre recombinase under the Club Cell Secretory Protein promote (CCSPiCre) in Pten and Smad4 floxed (Pts4f/f) mice results in a single proximal airway tumor (18). At 12 months of age, 100% of Pts4d/d mice develop lung cancer, which spontaneously metastasizes to distant organs, and has histological features consistent with mixed adeno- and squamous cell carcinoma (18).
NSCLC subtypes have therapeutic implications (19, 20), and little is known about the activities of IL17A in early or advanced-staged disease. In this study, we observed that tumor-infiltrating lymphocytes in early stage NSCLC (Stage I–II) patients harbor increased numbers of Th17 cells when compared to tumor-free lung parenchyma, indicating an increased association of this helper subset with lung cancer. In patients with advanced NSCLC (Stage III and IV), Th17 cells present in tumor-positive draining lymph nodes also showed a reverse correlation with PD-1+ CD4+ T cells. Although loss of Il17a has been shown to protect against several tumors driven by Kras mutations, including NSCLC (14), Kras plays an etiologic role in a subset of all lung tumors. We therefore investigated whether IL17a, which is associated with lung inflammation, could alter tumor latency and metastasis in an autochthonous NSCLC model that is independent of Kras mutation.
Here we show that the Pts4d/d-driven mouse model of lung adeno-squamous carcinoma displays genetic signatures characteristic of the SQ.2b, and AD.1 subtypes of human NSCLC. However, Il17a deficiency in the Pts4d/d model resulted in more rapid tumor development and more frequent metastasis. Interrogation of the immune cell profiles within lung tumor, draining lymph nodes, and metastatic organs in Pts4d/dIl17a−/− mice showed that the antitumor immune responses were mediated by IL17a. Together, our findings show that Th17 cells function against the tumor in certain subtypes of NSCLC.
Material and methods
Mice
Ptend/dSmad4d/d mice (Pts4d/d) have been previously described (18). Il17a−/− mice (C57BL/6 background) were obtained from Dr. Chen Dong (The University of Texas MD Anderson Cancer Center, Houston, TX). Pts4 d/d mice were crossed three times with Il17a−/− mice to generate Pts4 d/dIl17a−/− mice. Both male and female mice were used. All mice were bred in the transgenic animal facility at Baylor College of Medicine. All experimental protocols used in this study were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine and followed the National Research Council Guide for the Care and Use of Laboratory Animals.
Reagents
Recombinant murine FLT3L was purchased from eBioscience (San Diego, CA). Recombinant murine GM-CSF, murine IL4, murine and human IL17A was purchased from R&D Systems (Minneapolis, MN). TLR4 ligand lipopolysaccharide LPS and TLR3 ligand poly (I:C) were purchased from InvivoGen (San Diego, CA).
Immune cell isolation from mouse lung, mediastinal lymph node and spleen
Mouse lung, mediastinal lymph node or spleen single-cell suspensions were prepared by mincing whole organs through a 40-μm cell strainer (BD Falcon, San Jose, CA) followed by red blood cell (RBC) lysis (ACK lysis buffer, Sigma-Aldrich) for 3 min. In some experiments, the lung F4/80+ cells were isolated using anti-F4/80 conjugated magnetic beads (Miltenyi Biotec) and were treated with recombinant IL17A as indicated. Alternatively, lungs were fixed with instillation of 4% paraformaldehyde solution via a tracheal cannula at 25-cm H2O pressure followed by paraffin embedding and were sectioned for histopathological studies. Hematoxylin and eosin (H&E) staining was performed as described (21). Immunohistochemical staining was performed as previously described (18). TTF-1 (H190) and CK7 (RCK-105) antibodies were purchased from Santa Cruz (Dallas, TX). SOX2 antibody was purchased from Millipore (Billerica, MA).
Mouse immune cell isolation from mouse stomach
Cells were prepared based on the previously described method for the isolation of small intestinal lamina propria cells (22) with the following modifications. The stomach was cut open into 2 pieces and the contents removed. The tissue was then washed once in a DTT/EDTA solution (1mM DTT, 30mM EDTA and 10 mM HEPES) for10 minutes in a bacterial shaker at 37 degrees and then followed by an additional EDTA wash (30mM EDTA and 10 mM HEPES) for 10 minutes. The tissue was washed with complete RPMI and then cut into 1 to 2 cm pieces, digested for 1 hour in a bacterial shaker, in complete RPMI containing 100 U/ml type VIII collagenases (Sigma) and 150 μg/ml DNase I (Sigma). After digestion, cells were eluted from the tissue by shaking. Live cells were then isolated using a Percoll (GE) gradient.
Human immune cell isolation from lung, lung tumor and lung draining lymph nodes
Current or former smokers with early stage (I–II; n = 17), or advanced stage (II–IV; n = 22) NSCLC serially entered the study; all subjects had a significant (>20 pack-years) history of smoking (Supplementary Table S1). The patients had no history of allergy or asthma, had not received oral or systemic corticosteroids, and were free of acute symptoms suggestive of upper or lower respiratory tract infection during the last 6 weeks.
Studies were approved by the institutional review board (IRB) and in accordance with U.S. Common Rule, at Baylor College of Medicine. Informed consent was obtained from each patient. Human lung single cell suspensions were prepared as previously described (5). Briefly, fresh lung or tumor tissue were minced into 0.1-cm pieces in Petri dishes and treated with 2 mg/ml of collagenase D (Roche Pharmaceuticals, Basel, Switzerland) in HBSS and incubated for 30 to 40 min at 37°C. Single cells were collected using a 40-μm cell strainer (BD Falcon) followed by RBC lysis. Loco-regional and distal lung lymph node cells were collected from lung cancer patients using Endobronchial Ultrasound (EBUS) guided sampling (23), were collected in HBSS, and transported to the lab immediately. Single cells were otherwise harvested using the same protocol used for collection of cells from lung tissue or tumors (described above).
In silico analysis of The Cancer Genome Atlas (TCGA) and Pts4d/d NSCLC
In total, 1023 NSCLC cases from TCGA (which represented the cases assayed on at least three different molecular profiling platforms among RNA sequencing, DNA methylation arrays, miRNA sequencing, Affymetrix SNP arrays, whole-exome sequencing, and RPPA arrays) were included in the analysis. Previously, multiplatform analysis of these TCGA data uncovered nine genomic subtypes of NSCLC. RNA-seq data were obtained from the Broad Institute Firehose pipeline (http://gdac.broadinstitute.org/). Copy-number alteration data (originally obtained using Broad Institute’s Firehose’s “thresholded” calls) and somatic mutation data were obtained (19). Visualization using heat maps was performed using JavaTreeview (version 1.1.6r4) (24). For heat map presentation and for gene signature scoring, gene expression values were log-transformed and then z-normalized to standard deviations from the median across all cancer samples. TCGA RNA-seq profiles were scored based on the manifestation of the Pts4d/d gene signature, using a “t-score” metric as previously described (25), where the overall t-score is high in a profile where the genes up in the Pts4d/d expression signature are relatively high, whereas the genes down in the signature are relatively low. The GEO accession number for the microarray data of Pts4d/d mice in the previous report (18) is GSE57133.
Flow cytometry and antibodies
Flow cytometry was performed with a BD LSR II (BD BioSciences), and data were analyzed with FlowJo software (Tree Star Inc., Ashland, OR). The following antibodies to mouse proteins were purchased from BD Pharmingen and used: Pacific Blue-CD3 (500A2), PE-Cy5-CD4 (RM4-5), FITC-CD4 (RM4-5), APC-Cy7-CD8 (53-6.7), PE-IL17A (TC11-18H10), APC-CD86 (GL-1), PE-CD86 (GL-1) and APC-Cy7-Ly6G/C (RB6-8C5). The following antibodies to mouse proteins were purchased from eBioscience and used: FITC-γδTCR (eBioGL3), eFluro450-B220 (RA3-6B2), PE-CD11b (M1/70), APC-CD11c (N418), APC-CD103 (2E7), PE-Foxp3 (FJK-16s), APC-CD25 (PC61.5), PE-CD217 (IL17 Receptor A) (PAJ-17R). The following antibodies to mouse proteins were purchased from Biolegend (San Diego, CA) and used: PerCP/Cy5.5-F4/80 (BM8), PE-CD206 (C068C2), PerCP/Cy5.5-CD152 (CTLA-4) (UC10-4B9), PE/Cy7-CD279 (PD-1) (29F.1A12), PE/Cy7-CD11b (M1/70), Pacific Blue-CD45 (30-F11), BV510-CD45 (30-F11), APC-CD45.1 (A20), APC-IFNγ (XMG1.2) and PerCP/Cy5.5-CD11c (N418). The following antibodies to human proteins were purchased from BD Pharmingen and used: PE-IL17A (N49-653), APC-IFNγ (B27), FITC-CD4 (RPA-T4), and APC/Cy7-CD8 (SK1). The following antibodies to human proteins were purchased from Biolegend and used: Pacific Blue-CD45 (HI30), APC/Cy7-CD1c (BDCA-1) (L161) and PerCP/Cy5.5-CD279 (PD-1) (EH12.2H7). eFluor450-CD3 (OKT3) was purchased from eBioscience. LIVE/DEAD™ Fixable Dead Cell Stain kit (Thermo Fisher, Waltham, MA) was used to measure the viability of tumor-free adjacent tissue (T-FAT) and tumor infiltrated lymphocytes (TIL) isolated from Stage I–II surgical samples. Anti-mouse CD16/CD32 (eBioscience) or Human TruStain FcX (Biolegend) was added to the cells and were incubated for 5 minutes prior to other antibodies to block Fc receptor. Intracellular cytokine staining was performed by the stimulation of cells with phorbol 12-myristate 13-acetate (PMA, 10 ng/ml; Sigma-Aldrich, St. Louis, MO) and ionomycin (1 μg/ml; Sigma-Aldrich) for overnight supplemented with brefeldin A (10 μg/ml; Sigma-Aldrich) for the last 6 h. Cells were stained for surface markers and then fixed with FACS™ lysing solution (BD BioSciences, San Jose, CA), permeabilized with 0.5% saponin (Sigma-Aldrich), and stained with anti-IFNγ and anti-IL17A.
Micro computed tomography (microCT) detection of lung tumor
Mice were anesthetized and scanned using microCT as previously described (26). Briefly, mice were exposed to etomidate (30 mg/kg) and placed in an animal CT scanner (Gamma Medica, Salem, NH), and completed images of the chest were obtained and analyzed at the Animal Phenotyping Core in Baylor College of Medicine. Amira 3.1.1 software (FEI, Hillsboro, OR) was used to process the images.
Adoptive transfer of CD4+ T cells
Splenic CD4+ T cells from Pts4f/f mice were isolated using anti-CD4 conjugated magnetic beads (Miltenyi Biotec). Pts4d/dIl17a−/− mice were intraperitoneally injected with 15 × 106 isolated CD4+ T cells at 2.5 and 5 months of age. Mice were then euthanized at 7 or 9 months of age and lung tumor and metastasis were analyzed using lung, spleen, liver and stomach histology.
Bone marrow–derived dendritic cell (BMDC) and iCD103 DC culture
Mouse BMDCs were prepared as previously described with some modification (27). Femurs and tibias of 4 to 8 weeks old female mice were isolated and freed from the surrounding tissue. Intact bones were kept in 70% ethanol for 3 min followed by PBS wash. Both ends of the bones were cut with scissors and the marrow was flushed out with RPMI-1640 medium through a syringe with 26.5 needle. RBCs were then removed by ACK lysis buffer and cell debris or tissue clusters were filtered out. Cells from bone marrow were cultured in a 6-well plate with 20 ng/ml recombinant murine GM-CSF and 10 ng/ml recombinant murine IL4 for 5 to 6 days.
Induced CD103+ DCs (iCD103 DC) were generated using bone marrow cells as previously described (28). Briefly, 15×106 bone marrow cells were cultured in 10ml RPMI medium with 10% FBS and penicillin/streptomycin supplemented with 200 ng/ml recombinant murine FLT3L and 5 ng/ml recombinant murine GM-CSF (Complete Medium; CM) for 9 days. New CM was added on day 5 to minimize cell death. Non-adherent cells were harvested on day 9, counted and re-plated at 3×106 cells in 10 ml CM. Differentiated iCD103 DCs were harvested on day 13 or 14.
Bone marrow-derived macrophages (BMDM) culture and macrophage differentiation in vitro
BMDMs were generated as previously described (29). Bone marrow cells were cultured in DMEM with 20% FBS and the supernatant collected from L929 fibroblast cell line culture. After 5 or 6 days of culture, mature macrophages were harvested and stimulated for 24 hours with 10 ng/ml recombinant murine IL4 (M2 macrophage) or with 1 ng/ml LPS (M1 macrophage) differentiation. Recombinant murine IL17A was added in some groups as indicated.
iCD103 DC and T-cell coculture and cytokine measurement
iCD103 DCs collected on day 13 or 14 of culture were treated with indicated amount of poly (I:C) or IL17A for 24 hours, washed, and placed in coculture assays with splenic CD8+ T cells (at 1:10 ratio). CD8+ T cells were isolated using anti-CD8 conjugated magnetic beads (Miltenyi Biotec) and were isolated with an autoMACS cell separator. In some experiments when OT-I cells were used, iCD103 DCs were pretreated with ovalbumin (10 μg/ml) for 3 hours and washed three times with PBS before coculture with OT-I T cells. ELISA (BD BioSciences) was used for the measurement of concentration of IFNγ in the supernatant collected from cultured cells.
In vitro mouse CD4+ T-cell differentiation
Naïve CD4+ T cells were isolated using anti-CD4 conjugated magnetic beads (Miltenyi Biotec) and autoMACS cell separator. Cells were differentiated under Treg polarizing conditions. In brief, 2 to 2.5 × 106/ml cells were activated with 1.5 μg/ml plate-bound anti-CD3 and 1.5 μg/ml soluble anti-CD28 in addition to 10 μg/ml anti-CD4, 10 μg/ml anti-IFNγ, 50 U/ml IL2 and 6 ng/ml TGFβ. Cells were cultured for 3 to 5 days, harvested, and washed for intracellular staining of Foxp3 and the surface staining of CD25 to determine Treg differentiation.
mRNA isolation and quantitative PCR
Cell pellets were treated with TRIzol (Life Technologies), and mRNA was extracted as previously described (30). All probes, mouse Arg1 (Mm00475988_m1), mouse Vegf (Mm00437306_m1) and mouse Il10 (Mm01288386_m1) were purchased from Thermo Fisher. All data were normalized to 18S ribosomal RNA (Hs99999901_s1) expression.
Transwell cell migration assay
In vitro cell migration assay was performed as we have described previously with slight modifications (31). Briefly 24-transwell plates with 5μm pore-sized membrane inserts (Corning, Corning, NY) were used to add 105 iCD103 DCs in 100 μl RPMI medium on the top chambers; lung homogenate collected from Pts4d/dIl17a−/− mice or recombinant murine IL17A were added into the bottom chambers. Cells was cultured for indicated times and then the migrated cells in the lower chambers were collected for flow cytometry to detect the cell number of CD103+ DCs.
BEAS-2B cell culture and shRNA experiment
Human broncho-epithelial BEAS-2B cells (ATCC®CRL-9609™) were purchased from ATCC and cultured according to the ATCC culture condition [BEGM medium (CC-3171, LONZA), growth factors (CC-4175, LONZA), 100 IU/ml of penicillin and streptomycin (100 μg/ml)]. Stable shRNA knockdown of PTEN and SMAD4 in BEAS-2B cells are previously described (18).
Transwell invasion and cell viability assay
24-well Matrigel-coated invasion chambers (BD Biosciences) were used to measure cell invasion following the manufacturer’s suggestions and as previously described (18). Briefly, BEAS-2B cells were trypsinized, resuspended in serum-free BEGM medium, and seeded at 1×105 cells per 100 μl per top chamber. Chambers were placed in complete culture medium containing growth factor as chemo-attractants. Following 22 hours of incubation, cells in the top chambers were removed; chambers were then fixed with 4% paraformaldehyde solution and stained with crystal violet for cell counting. BEAS-2B cell viability was measured by CellTiter 96 Aqueous Solution Reagent (Promega, Madison, WI) according to the manufacturer’s instructions. After 1hour incubation, the cell viability was determined by live-dead dye.
Statistical Analysis
For the comparison of immune cell profiles, gene expression and cytokine production in mouse experiments or in vitro cell culture, we used the Student’s t-test, the Paired student’s t-test or one-way analysis of variance (ANOVA) test and Bonferroni’s multiple comparison test. Linear regression was used to evaluate the correlation between Th17 and PD-1+ T cells in lung draining lymph nodes in human subjects. All data shown are the mean ± standard error of the mean (SEM), and all analyses were performed with the Prism software (GraphPad Software).
Results
Tumor infiltrating lymphocytes in early and late stage non-small cell lung cancer (NSCLC)
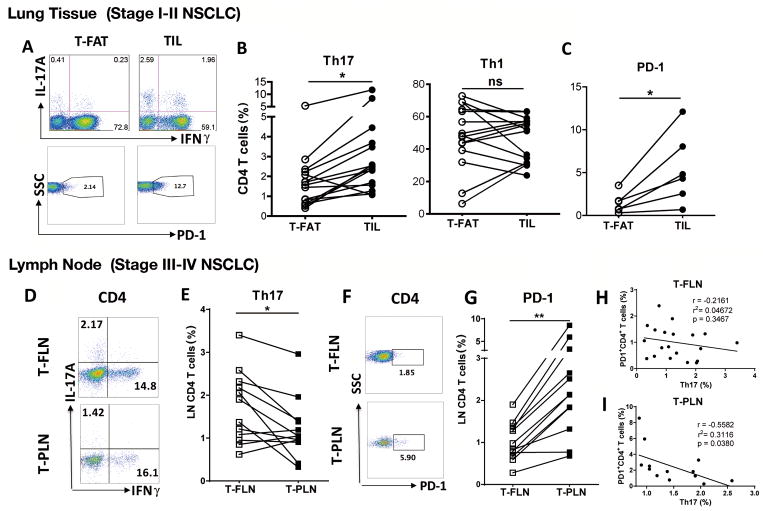
We examined T helper cell subsets in paired samples using tumor infiltrating lymphocytes (TILs) and tumor-free adjacent lung tissue (T-FAT) procured from patients undergoing lung resection for early stages (I–II) of non-small cell lung cancer (NSCLC). Overall there were comparable numbers of IFNγ-producing T helper type 1 (Th1) cells and cytotoxic T lymphocytes (CTLs) among freshly isolated cells in TILs and T-FAT (Supplementary Fig. S1A and B). However, using the same paired samples, we found significantly increased IL17A producing Th17 cells in early stage TILs compared to T-FAT (Fig. 1A and B). As expected, we found increased expression of PD-1, an exhaustion marker on CD4+ T cells within lung tumors (Fig. 1C). Lung cancer is often diagnosed in late stages (III–IV)(1), when surgical resection is not feasible. Therefore, to gain access to fresh lung tissue with advanced tumor, we examined the association between Th17 cells and late stage NSCLC using loco-regional and distal lymph nodes collected in patients undergoing nodal staging with Endobronchial Ultrasound (EBUS) guided sampling (23). Compared with tumor-free lymph nodes (T-FLN), tumor-positive lymph nodes (T-PLN) showed similar abundance of Th1 cells (Supplementary Fig. S1C) whereas Th17 cells were significantly reduced in the latter tissue (Fig. 1D and E). Similarly, in late stage NSCLC, we found increased expression of PD-1 in CD4+ T cells and CTLs in T-PLN when compared to T-FLN (Fig. 1F and G; Supplementary Fig. S1C). Th17 cells and PD-1 expression on CD4+ T cells were negatively correlated in T-PLN samples but not in T-FLN (Fig. 1H and I). Together, these findings suggest that Th17 cells may modulate the immune response to NSCLC in humans.
Figure 1. Th17 cell infiltration in early and late human NSCLC.
(A) Representative intracellular, and (B) cumulative (n = 15) staining for paired analysis of lung CD4+ T cells expressing IL17A (Th17) and IFNγ (Th1) isolated from tumor-free adjacent tissue (T-FAT) and tumor infiltrated lymphocytes (TIL) in early stage (I–II) surgical samples (n = 6). (C) PD-1 cell surface expression in the same cell population (n = 6). (D) Representative intracellular, and (E) cumulative staining for paired analysis of tumor-free (T-FLN) and tumor-positive (T-PLN) lymphocytes expressing IL17A (Th17) and IFNγ (Th1) in sentinel lymph node in late stage (III–IV) NSCLC. (F) Representative PD-1 expression in CD4+ T cells and (G) cumulative (n = 12) in the same cell population. Correlation between PD-1+ CD4+ T cells and Th17 cells in (H) T-FLN (n = 21) or (I) T-PLN samples (n = 14). **P < 0.01, * P < 0.05 as determined by the Paired Student’s t-test. p and r value were obtained by linear regression model. Data are mean ± SEM. Please also see Supplementary Fig. S1.
PTEN and SMAD4 deficiency in human and autochthonous non-Kras model of NSCLC
Genomic studies have identified specific and overlapping molecular subtypes in NSCLC, indicating that cellular heterogeneity could affect clinical outcome (32). Examination of NSCLC cases in TCGA cohort identified three molecular subtypes associated with squamous-carcinoma (e.g. SQ.1, SQ.2a. SQ.2b) and six with adenocarcinomas (e.g. AD.1. AD.2, AD.3, AD.4, AD.5a, and AD.5b) (19). As expected, in the TCGA cohort, NSCLC with KRAS mutations segregated with most adenocarcinoma subtypes, whereas PTEN mutations, copy loss, and lower differential expression was associated with SQ subtypes (Fig. 2A). RNAseq profiles in mice with Club cell specific deletion of Pten and Smad 4 (Pts4d/d), which spontaneously develop lung cancer beginning at 7 months of age (18), showed gene signature patterns associated with SQ.2b and AD.1 types of human NSCLC, indicating the relevance of this autochthonous model of non-Kras NSCLC to human disease (Fig. 2B and C).
Figure 2. Survey of molecular alterations associated with PTEN and SMAD4 in human NSCLC, and Pts4d/d mice.
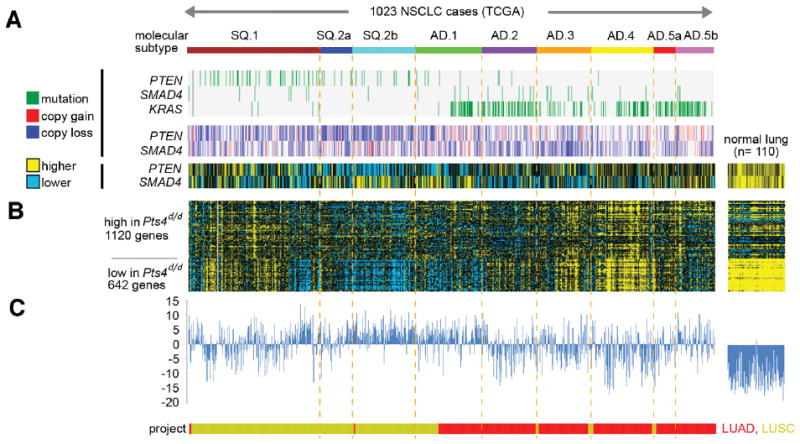
(A) Data from 1023 NSCLC cases in The Cancer Genome Atlas (TCGA) are represented, compared with data from normal lung specimens (n = 110); LUAD, TCGA lung adenocarcinoma specimens; LUSC, TCGA lung squamous specimens. TCGA cases were previously assigned (19) molecular subtypes as indicated (SQ.1/SQ.2, squamous-enriched subtypes; AD.1–5, adenocarcinoma-enriched subtypes). Copy number alterations, somatic mutations, and relative expression for PTEN and SMAD4 genes are shown (along with KRAS mutation). (B) Genes differentially expressed in lung tumors from Pts4d/d mice showing signature patterns of previously-defined set (18). (C) TCGA gene signature was in each case scored according to overall similarity with the Pts4d/d signature pattern.
Th17 cells in animal model of NSCLC driven by Pten and Smad4 deficiency
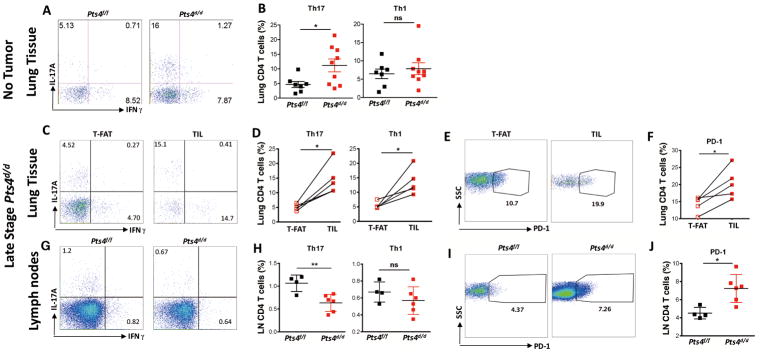
To examine the role of Th17 cells in NSCLC, we immunophenotyped inflammatory cells recruited to early- and late-stage lung cancer using the Pts4dd/-driven model of lung cancer (18). At 5 months of age, Pts4d/d mice exhibit lung epithelium hyperplasia without evidence of tumor (18), but showed abnormal expansion of Th17 cells in the lung tissue as compared to wild type (Pts4f/f) mice (Fig. 3A and B; Supplementary Fig. S2A). Further, at 11 to 12 months of age, when nearly all mice exhibit lung tumor with distant metastasis (18), we found increased Th17, PD-1+ T cells, and CTLs in TILs when compared to T-FAT (Fig. 3C–F; Supplementary Fig. S2B). Consistent with advanced stage NSCLC in humans, mediastinal lymph nodes also showed decreased Th17 cells and increased PD-1+ T cells in Pts4d/d mice when compared to Pts4f/f mice (Fig. 3G–J; Supplementary Fig. S2C). In this pre-neoplastic stage, we also detected increased IL6 and IL1β in the lungs of Pts4d/d mice, indicating that the lung microenvironment is conditioned to promote Th17 cells (Supplementary Fig. S3). These findings further demonstrate an association between advanced NSCLC and the presence of Th17 cells in the lung.
Figure 3. Infiltration of Th17 cells in pre- and late stage Ptsd/d model of NSCLC.
Representative (A) and (B) cumulative intracellular cytokine (ICC) to detect lung CD4+ T cells expressing IL17A (Th17) and IFNγ (Th1) in Ptenf/fSmad4f/f (Pts4f/f) and Ptend/dSmad4d/d (Pts4d/d) prior to tumor development (5 months; n = 7 or 9 per group). Representative (C) and (D) cumulative ICC detection of Th17 and Th1 cells in tumor-free adjacent tissue (T-FAT) and tumor infiltrated lymphocytes (TIL) in late stage Pts4d/d NSCLC (11 to 12 months; n = 5). Representative (E) and cumulative (F) PD-1 expression in CD4+ T cells in the same cell population described in C and D. (G) Representative, and (H) cumulative ICC detection of T cells expressing IL17A (Th17) and IFNγ (Th1) in mediastinal lymph nodes in late stage Pts4d/d NSCLC (11 to 12 months n = 6), and aged matched Pts4f/f mice (n = 4). Representative (I) and cumulative (J) PD-1 expression in CD4+ T cells in the same cell population described in G and H. Data are mean ± SEM and representative of two independent experiments. ** P < 0.01, * P < 0.05 as determined by the Paired Student’s t-test. Please also see Supplementary Figs. S2 and S3.
Il17a deficiency reduces lung tumor latency and promotes metastasis in Pts4d/d mice
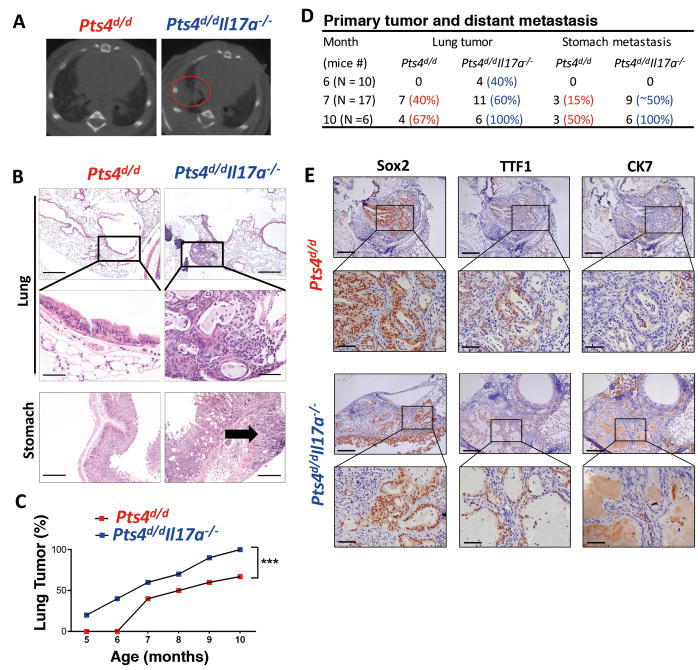
IL17A expression has been implicated in the pathogenesis of several different tumors in humans and in animal models of cancer (12, 14). We next crossed Pts4d/d with Il17a−/− mice, to investigate a link between upregulation of the cytokine Il17a and lung tumor invasion capacity. We found that compared to IL17a-sufficient Pts4d/d mice, global loss of Il17a−/− (Pts4d/dIl17a−/−) resulted in earlier development of tumors (Fig. 4A–C) and more metastasis (Fig. 4D). Although deletion of Il17a resulted in more aggressive tumors, histological examination of tumors showed similar expressional pattern of tumor markers (Fig. 4E). Further, IL17A failed to increase invasiveness (Supplementary Fig. S4A and B) or viability (Supplementary Fig. S4C) of human epithelial cells lacking PTEN and SMAD4, indicating that this cytokine does not directly play a role in transformed epithelial cell function.
Figure 4. Global loss of IL17a in Pts4d/d mice reduces primary tumor latency and increases metastasis.
(A) Representative images of microCT scan mid thorax in 6 month-old Pts4d/d, and Pts4d/dIl17a−/− mice. Red circle identifies detection of tumor in the right lobe. (B) Representative H&E of the lungs (top two panels), and 4X magnifications (depicted in rectangles) detect epithelial hyperplasia in Pts4d/d mice, and adenosquamous tumor in Pts4d/dIl17a−/− mice. Bottom panel show representative of stomach H&E, shows no evidence of metastasis in Pts4d/d mice, while tumor invasion is localized to stomach parenchyma in Pts4d/dIl17a−/− mice (arrow). scale bar = 200 μm. (C) Primary adenosquamous lung cancer detection (%) in Pts4d/d and Pts4d/dIl17a−/− mice. (D) Cumulative analyses of the frequency of lung tumor and stomach metastases of Pts4d/d and Pts4d/dIl17a−/− mice at indicated ages. (E) Representative lung tissue immumohistochemical analysis of Sox2, TTF1, and CK7 in Pts4d/d and Pts4d/dIl17a−/− mice. *** P < 0.001 as determined by the paired Student’s t-test. Please also see Supplementary Fig. S4.
Suppressive myeloid cells reduce tumor latency in Pts4d/dIl17a−/− mice
Our data thus far indicated that TILs in early stage NSCLC were enriched in Th17 cells, and Il17a deficiency decreased tumor latency and increased metastasis in the Pts4d/d model of lung cancer indicating a potential protective effect of this cytokine in tumor progression. Therefore, we next examined the possible mechanism for IL17A-mediated tumor suppression in the lung tumor microenvironment.
Because we have previously shown that Pts4d/d mice develop lung cancer at approximately 7 months of age, we comprehensively surveyed lung immune phenotypes during pre- and peri- lung tumor development (e.g., 5- and 7-month old) in this model of NSCLC. Compared with wild type (Pts4f/f) mice, Pts4d/d mice at 5 months showed more tumor associated macrophages (TAMs), marked by CD11b/CD206 expression, and regulatory T cells (Tregs) in the lungs (Supplementary Fig. S5A–D). However, examination of single cells isolated from the whole lung in Pts4d/dIl17a−/− mice showed more TAMs and Tregs when compared to Pts4d/d mice of the same age group (Supplementary Fig. S5A–D).
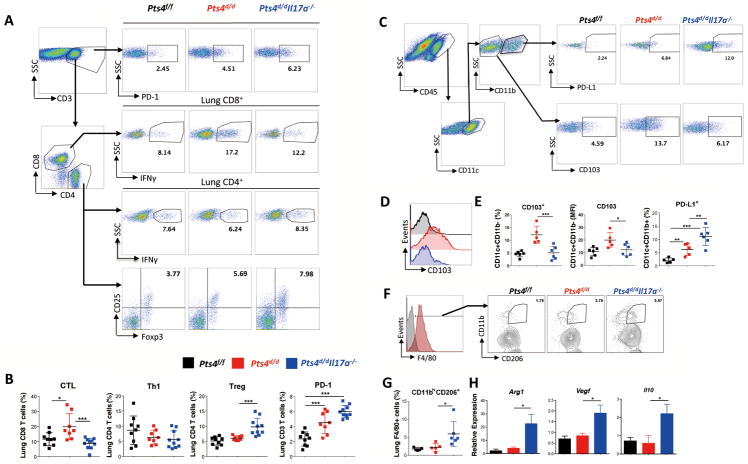
At 7 months of age, when approximately 40% of Pts4d/d and 60% of Pts4d/dIl17a−/− mice develop lung tumors, we found increases in PD-1 expression and Treg numbers, and decreases in IFNγ+ expression and CD8+ cytotoxic T-cell (CTL) numbers in Pts4d/dIl17a−/− mice, when compared with Pts4d/d and Pts4f/f mice (Fig. 5A and B), indicating an immunosuppressive tumor microenvironment in the absence of IL17A. Immunosuppressive cells did not affect IFNγ expression in CD4+ T cells because Th1 responses remained comparable in all three genotypes (Fig. 5A and B). In contrast, CD103+CD11blowmyeloid DCs, which cross-present antigen and prime CD8+ T cells (33, 34), were decreased in Pts4d/dIl17a−/− when compared to Pts4d/d mice (Fig. 5C–E). In addition to the increased abundance of PD-1 expression in T cells, PD-L1-expressing CD11c+ CD11bhighmyeloid DCs were more abundant in Pts4d/dIl17a−/− mice (Fig. 5C and E). Together these data suggest IL17A is required for initiation of antitumor responses in this model of NSCLC, possibly through upregulation of immunosuppressive PD-1/PD-L1. Further, consistent with an immunosuppressive microenvironment, we also found significantly increased TAMs (CD11bhighCD206+) in Pts4d/dIl17a−/− mice (Fig. 5F and G) that expressed significantly more arginase-1 (Arg1), VEGF (Vegf) and IL10 (Il10), factors that are known to promote tumor growth (Fig. 5H) (35). Immunosuppressive myeloid derived suppressor cells (MDSCs) marked by CD11b/Gr-1 were not affected by lack of IL17A in this model (Supplementary Fig. S5 E and F). These findings suggest that IL17A deficiency promotes TAMs and PD-L1+ DCs and limits abundance of CD8-priming CD103+ DCs.
Figure 5. Comprehensive immunophenotype in 7-month old Pts4d/dIl17a−/− mice.
Representative flow gating, and ICC (A) and cumulative analysis (B) for lung IFNγ expressing CD8+ cells(CTLs), CD4+ (Th1), foxp3+ expressing (Tregs), and PD-1 expression in 7-month old age matched mice: WT (Pts4f/), Pts4d/d, and Pts4d/dIl17a−/− (data pooled from two independent experiments; n = 9 per group) (C) Representative flow gating and relative abundance of lung CD103+ DCs in CD45+CD11c+CD11b− lung cells and PD-L1expression in CD11b+ DCs in the same groups of mice. (D) Representative histogram and (E) cumulative relative abundance of CD103 expression, and MFI, and PD-L1 expression in CD11c+CD11b− DC subset described in panel C. (n = 5 or 6 per group) (F) Representative histogram and (G) cumulative expression of CD206 on F4/80 tumor-associated macrophages (TAM) in whole lung single cells homogenates in the indicated groups of mice. (H) Quantitative expression of Arg1, Vegf and Il10 gene using qPCR in BAL cells isolated from the same groups of mice. (n = 5 or 6 per group). Data are mean ± SEM and representative of three independent experiments; *** P < 0.001, ** P < 0.01, * P < 0.05 as determined by the one-way ANOVA and Bonferroni’s Multiple Comparison test. Please also see Supplementary Figs. S5, S6, S7, S8, and S9.
We next assessed the phenotype of mediastinal tumor-associated sentinel lymph nodes, in the same group of mice (7-month old). We found significantly fewer CTLs and Th1 cells in Pts4d/dIl17a−/− when compared with IL17A-sufficient Pts4d/d mice (Supplementary Fig. S6A and B). Additionally, CD44, a marker of effector T cells, was upregulated in both CD4+ and CD8+ T cells in Pts4d/d when compared to Pts4d/dIl17a−/− mice (Supplementary Fig. S6C and D), indicating defective T-cell activation in lung tumor sentinel lymph nodes in the absence of IL17A.
We found CTL and Th1 cell responses in metastatic sites (e.g. stomach) in Pts4d/d mice; similar T cells were fewer in Pts4d/dIl17a−/− mice (Supplementary Fig. S6E and F). CD103+CD11blow DCs that are capable of efficient antigen cross-presentation and are present in metastatic tissues in Pts4d/dIl17a−/− mice were reduced when compared to IL17A-sufficient controls (Supplementary Fig. S6G and H). These findings suggest an immunosuppressive microenvironment in the lungs, tumor draining lymph nodes, and distant metastatic sites in Pts4d/dIl17a−/− mice.
IL17A inhibits alternative macrophage differentiation
Our findings thus far pointed to a lack of activated endogenous immune cells in the lungs of Pts4d/dIl17a−/− mice that resulted in decreased tumor latency when compared to Il17a-sufficient Pts4d/d mice. Recombinant (r)IL17A did not directly affect Treg differentiation (Supplementary Fig. S7) or CD8+ T-cell proliferation and activation in vitro (36). However, because we had found an increase of CD206+ M2 macrophage-like TAM from tumor-bearing Pts4d/dIl17a−/− mice, we next examined whether IL17A directly affects differentiation in this cell population. We found that IL4-mediated differentiation of bone marrow-derived macrophages (BMDM) to CD206+ M2 cells was inhibited by IL17A in a dose-dependent manner (Supplementary Fig. S8A and B). Moreover, lung F4/80+ macrophages from Pts4d/dIl17a−/− mice exhibited less differentiation of CD206+ cells when treated with IL17A in vitro (Supplementary Fig. S8C and D). Expression of the M2 macrophage signature genes Arg1 and Vegf were also inhibited by IL17A treatment (Supplementary Fig. S8E). Il17a-deficient bone marrow cells did not display an intrinsic defect in M2 differentiation in vitro (Supplementary Fig. S9). We conclude that IL17A suppressed genes associated with M2-like macrophage differentiation, because its absence resulted in abundant TAM and exaggerated lung tumor progression in Pts4d/d mice.
IL17A recruits CD103+ DCs critical in CTL activation
Next, we examined whether IL17A could induce differentiation and function of CD103+ (iCD103) DCs (Supplementary Fig. S10A). Exogenous addition of rIL17A to bone marrow derived cells failed to promote iCD103 differentiation and/or activation as determined by Clec9a expression, a functional marker for DC-mediated CTL cross-priming (Supplementary Fig. S10B) (37). Because of reduced CD103+ DCs in the lungs of Pts4d/dIl17a−/− mice, we next examined whether IL17A might act as a chemoattractant and recruit CD103+ DCs into the lungs. Using a transwell assay system, we found that IL17A in a dose-dependent manner enhanced iCD103 DC migration (Fig. 6A). Moreover, iCD103 DCs showed increased migration when Pts4d/dIl17a−/− lung homogenates were supplemented with rIL17A (Fig. 6B). These findings support the notion that IL17A may be required for efficient recruitment of CD103+ DCs into the lungs of Pts4d/d mice. In an autocrine/paracrine manner, rIL17A treatment induced expression of IL17A receptor in iCD103 DCs (Fig. 6C, D). Costimulation molecule CD86 was also upregulated in CD103+ DCs treated with rIL17A (Fig. 6C and E; Supplementary Fig. S11).
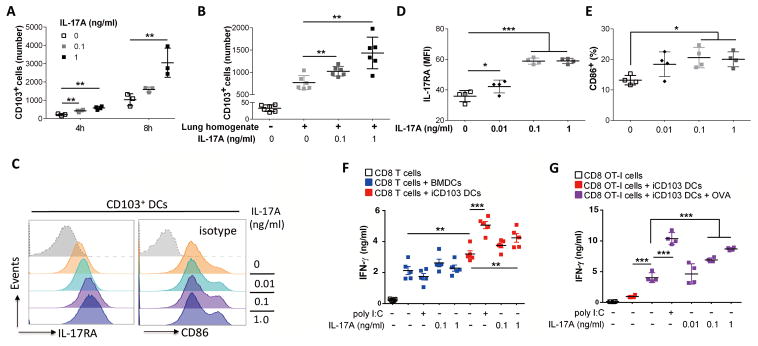
Figure 6. IL17A recruits CD103+ DC and potentiates CD8+ T-cell activation.
Bone marrow cells were treated with FLT3L (200 ng/ml) and GM-CSF (5 ng/ml) for 13 to 14 days to induce CD103+ DCs (iCD103 DCs). (A) Number of iCD103 DCs migrated to lower transwell chamber after 4 or 8 hours in response to increasing concentration of mouse rIL17A (0, 0.1, and 1ng/ml). (B) Number of iCD103 DCs migrated in transwell chamber after 3 hrs in response to 100 ul of lung homogenate freshly collected from Pts4d/dIl17a−/− mice at 7 months of age in the presence or absence of increasing dose of mouse rIL17A (0, 0.1, and 1ng/ml). (C) Representative histogram, and cumulative quantification of (D) IL17A receptor A (IL17RA), and (E) CD86 expression in iCD103 DCs treated with mouse rIL17A. (F) IFNγ concentration detected by ELISA after 72 hr coculture of splenic CD8+ T cells and iCD103 DCs or BMDCs treated with poly I:C (5 μg/ml) or IL17A at indicated concentration in the presence of anti-CD3 (1 μg/ml). (G) IFNγ concentration detected by ELISA after 72 hr coculture of splenic OT-1 CD8+ T cells and iCD103 DCs treated with poly I:C (5 μg/ml) or mouse rIL17A at indicated concentration with or without pretreatment of OVA. Data are mean ± SEM and representative of two independent experiments; *** P < 0.001, ** P < 0.01, * P < 0.05 as determined by the one-way ANOVA and Bonferroni’s Multiple Comparison test. Please also see Supplementary Figs. S10 and S11.
Toll-like receptor (TLR)3 activation is known to promote DC maturation and prime CD8+ T cells (38), therefore, we compared IL17A–mediated activation of DCs relative to TLR3 stimulation. Autologous splenic CD8+ T cells, cocultured with the TLR3 agonist poly I:C, activated iCD103 DCs and induced CTL responses (Fig. 6F). Treatment with rIL17A in a dose-dependent manner also resulted in iCD103-mediated enhanced CD8+ T-cell priming in both a non-specific (Fig. 6F) and an ovalbumin-specific transgenic T-cell (OT-I) model (Figure 6.G), consistent with IL17A-induced activation of iCD103 DCs. Together, these data suggest that the mechanism for increased IL17A-mediated antitumor responses may be recruitment and activation of CD103+ DCs to the primary and metastatic tumors and promotion of CTL development.
IL17A-sufficient CD4+ T cells prevent lung tumor progression and metastasis
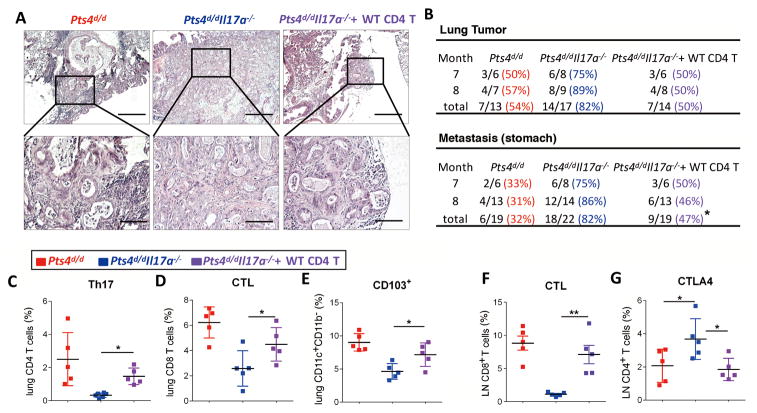
Although Th17 cells are considered a potent source of IL17A, other cells (e.g. γδ T cells, monocytes and neutrophils) also express IL17A under homeostatic or activated conditions. To determine whether Th17 cells specifically contribute to the antitumor immune response in the Pts4−/− model of NSCLC, CD4+ T cells isolated from splenocytes of WT (Pts4f/f) mice were adoptively transferred to Pts4d/dIl17a−/− mice at 2.5 and 5 months of age. IL17A-sufficient T cells persisted over 5 months and significantly reduced lung tumor incidence and stomach metastasis in Pts4d/dIl17a−/− mice at 7 to 8 months of age (Fig. 7A–C). Furthermore, CTL responses and CD103+ DCs were upregulated in Pts4d/dIl17a−/− mice after adoptive transfer of IL17A-sufficient CD4+ T cells as compared with control (Fig. 7D and E). Consistently, CTL responses were stronger in lung mediastinal nodes in response to CD4+ T-cell transfer whereas CTLA-4 expression, a marker of T-cell exhaustion, was reduced (Fig. 7F and G). Together these findings support a homeostatic effect of IL17A-expressing T cells that is required to activate CTLs and suppress lung tumor progression and metastasis in Pts4d/dIl17a−/− mice.
Figure 7. Adoptive transfer of IL17a sufficient CD4+ T cells reverses decreased tumor latency and metastasis in Pts4d/dIl17a−/− mice.
15 × 106 wild type CD4+ T cells were adoptively transferred to Pts4d/dIl17a−/− mice at 2.5 and 5 months of age. (A) Representative H&E staining of lung tumors in the indicated groups of mice (inset 4× magnification). scale bar = 100 μm. (B) Frequency of lung tumor and stomach metastases in Pts4d/d and Pts4d/dIl17a−/−, Pts4d/dIl17a−/− (+WT CD4) mice at 7 and 8 months. (C) Relative abundance of Th17 cells, (D) ICC detection of IFNγ and IL17A in lung CD4+ and CD8+ T cells (CTLs) respectively. (E) Relative expression of CD103 in lung DCs (CD11c+CD11b−) in the same groups of mice. (F) ICC detection of IFNγ in lung CD8+ T cells (CTLs) in mediastinal lymph nodes in the same groups of mice at 7 months of age. (G) Relative expression of CTLA in CD4+ T cells in mediastinal lymph nodes of the same groups of mice. Data are mean ± SEM and representative of two independent experiments (C – G); ** P < 0.01, * P < 0.05 as determined by the Chi-square test (B) or the one-way ANOVA and Bonferroni’s Multiple Comparison test (C-G).
Discussion
Th1 and Th17 cells are associated with emphysema development (5), which is linked to increased risk of lung cancer (39). Carcinogens in cigarette smoke promote large numbers of somatic mutations that result in production of altered proteins in premalignant and transformed airway epithelial cells (40). Studies of tumor survivors and animal models(41) show that CTLs that target mutated tumor antigens are more abundant and are sustained longer than CTLs that target non-mutated tumor antigens (42). However, how accumulation of altered proteins shapes early protective CTL tumor responses remains unclear. CD4+ T cells within TILs are thought to play a role in development of effective CTLs (43). To assess their role, we examined the relative abundance of different subset of CD4+ and CD8+ T cells within and outside the lung tumor microenvironment in early and late stage in NSCLC. We found increased relative abundance of Th17 cells in early stage NSCLC and an inverse relation between the T-cell exhaustion marker (PD-1) and Th17 cells in late stage tumor, indicating a role for Th17 cells in human lung cancer. This premise was further supported in our pre-clinical model of adeno-squamous lung cancer that shares features with human NSCLC (18).
We have previously shown that loss of Pten in mice with a codon-optimized Cre recombinase results in hyperplasic airway epithelia, whereas concurrent loss of Smad4 in these mice (Pts4d/d), results in proximal airway adeno-squamous lung tumors(18), and promotes recruitment of acquired immune cells. Lung tumors in Pts4d/d mice share characteristics with the human SQ2b and AD.1 subtypes of NSCLC as determined by RNAseq signature. Using the TCGA database, we have previously shown that the fraction of estimated mutations varies for mutational signatures and by smoking status in NSCLC molecular subtypes (19). Compared with other subtypes, SQ.2b, and AD.1 show the highest proportion of patients who were active- or short-term former-smokers, and have the highest manifestation of the SI4 transversion signature, a smoking-related signature of cytosine to adenine (C > A) transversions (19). Our work here underlines the importance of antitumor immunity with respect to tumor molecular diversity (19). Prior work has shown that global loss of Il17a can prevent tumor formation in the Kras-driven model of NSCLC(14). Here we show an opposite response, whereby Pts4d/d/Il17a−/− mice display reduced tumor latency and increased metastasis. We found that IL17A expression is upregulated early in response to tumor in the lungs and constitutes an immune response to hyperplastic and premalignant airway epithelial cells in NSCLC distinct molecular subtype. These observations are supported by our findings that global loss of Il17a results in earlier malignant transformation and increased distant organ metastasis when compared to Il17a sufficient Pts4d/d mice. Adoptive transfer of Il17a-sufficient CD4+ T cells to Pts4d/d/Il17a−/− mice reversed early tumor development and metastasis, indicating the antitumor function of Th17 cells.
Several studies have identified protumor roles for Th17 cells and IL17A in preclinical models and in human of NSCLC (14, 44), whereas some have implicated anti-malignant function (10). Although pre-clinical studies have examined the activity of IL17A and Th17 cells in the context of mice with immune deficiency (45), human NSCLC develops under immune competent conditions. Studies in mouse models and human breast (46), colon (47), and pancreatic (48) cancers have suggested that IL17A activates several transcriptional factors and can promote tumor growth and metastasis. In our study, however, we found no evidence for IL17A to directly induce cell proliferation or an invasion phenotype of human airway epithelial cells that lack PTEN and SMAD4. However, consistent with the pro-inflammatory nature of pten/smad4-driven tumors, we have previously found suppressed TGFβ1 signaling (18) and increased recruitment of Th17 cells prior to tumor development the lungs of Pts4d/d mice, suggesting that the endogenous immune system suppresses lung tumors.
We and others have previously shown that cigarette smoke induces Th17 cell-mediated lung inflammation, and IL17A and its receptor are critical for emphysema development in mice (5, 26, 49). Although induction of smoke-induced Th17 cells seemly counters the antitumor function of IL17A as shown here, several carcinogens in cigarette smoke activate pro-inflammatory cytokines, in particular IL6, which induces tumor growth(50)−(51). Thus, we propose that induction of Th17 cells may represent an immune response to tumor subtypes and to the protumorigenic effect of inflammatory cytokines. Using a non-Kras–driven pre-clinical model of lung cancer, here, we have provided insight into the role of Th17 cell–bias in a subtype of NSCLC. Our findings show that Th17 cells increase early in human NSCLC, and that Th17 cells correlate negatively with an immune suppressive microenvironment in tumors. Adoptive transfer of T cells isolated from TILs to patients with metastatic disease has shown promising results, although such immune targeted therapy is limited to solid tumors (7, 52). Our findings here suggest that an induced bias of TILs towards Th17 cells in non-Kras driven tumors may boost adoptive T-cell therapeutic functions in NSCLC.
Current adjuvant cancer therapies that target immune checkpoint molecules (e.g. PD-1/PD-L1, CTLA-4) can elicit durable, albeit low (20%), clinical responses in NSCLC (53). Although expression of these markers in the tumor microenvironment does not correlate with clinical responses to immune checkpoint inhibition in humans (54), increased somatic mutation, which can promote expression of mutated proteins and their presentation by activating CD103+ DCs in the tumor landscape, has been a reliable predictor of favorable response to immune adjuvant therapy (55). High frequency of failed clinical response to immune checkpoint inhibitors is thought to be due to the presence of TAMs, MDSCs and cancer-associated fibroblasts that mediate T-cell exclusion in CD4+ T cells and CTLs in solid tumors (4). We showed here that IL17A can inhibit TAM differentiation and promote activation of CD103 DCs, which are capable of antigen cross presentation, alter the immunosuppressive tumor microenvironment and promote a more effective antitumor immune response. We showed that IL17A, in a dose-dependent manner, inhibited Arg-1 and Vegf expression in TAMs, Th17 cells restored tumor latency in Pts4d/dIl17a−/− mice comparable to Pts4d/d mice. Moreover, we showed that Th17 cells recruited CTL-priming CD103+ DCs to the lungs, and activated CD8+ T cells, necessary for antitumor responses in vivo.
In summary, we show that the immune phenotype of CD4+ T cells in early and advanced stage human NSCLC varies, and that PD-1 expression in CD4+ T cells and Th17 cells show an inverse correlation. Using Pts4d/d mice, a model of NSCLC that shares with human NSCLC a propensity for spontaneous distant metastasis, we show that IL17A functions in early endogenous antitumor responses. Global deficiency in Il17a in Pts4d/d mice resulted in early and more invasive lung tumors, indicating a protective role for this cytokine in NSCLC. The mechanisms for Th17 cell-mediated antitumor activities are in part through inhibition of TAMs, whereby CD103+ DCs can be activated and recruited in the lungs. The immunosuppressive role of TAM and CTL-priming role of CD103 DCs have been reported in other cancers (56) and may also extend to the Pts4d/d model and/or human NSCLC. Adoptive transfer of IL17A-sufficient CD4+ T cells reduced lung cancer and metastasis in IL17-deficient Pts4d/d mice. Although IL17A has been shown to promote tumors associated with Kras mutations, our studies here support a role for Th17 cells in antitumor immune responses in tumors independent of the Kras mutation. Thus, molecular phenotyping of lung cancer should be considered prior to immune therapy. We propose that Th17-based immune therapy could be considered in some cases of NSCLC. Adoptive transfer of TILs differentiated to express IL17A in NSCLC patients with metastatic disease could provide a more effective result in immune-targeted cancer therapy.
Supplementary Material
Acknowledgments
This work was supported in part by the R01HL117181-01, (FK, DBC), CX000104 (FK) and BX002221 (DBC) VA Merit Awards.
This work was supported in part by the R01HL117181-01, (FK, DBC) CX000104 (FK) and BX002221 (DBC) VA Merit Awards. This project was supported by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the NIH (AI036211, CA125123, and RR024574), Mouse Phenotyping Core at Baylor College of Medicine with funding from the NIH (UM1HG006348), and the expert assistance of Joel M. Sederstrom and Dr. Corey Reynolds. This project was done in collaboration with the Adrienne Helis Malvin Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with Baylor College of Medicine and the Immune Therapy in a Novel Humanized Model of Lung Cancer Program (to F.J.D. and F.K.) and the Natural Killer Cell Immunotherapy to Cure Lung Cancer (to S.P, and F.K). CJC was supported in part by NIH grant CA125123 and Cancer Prevention and Research Institute of Texas grant RP120713.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Howlader NNA, Krapcho M, Neyman N, Aminou R, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Chen HS, Feuer EJ, Cronin KA, editors. SEER Cancer Statistics Review, 1975–2013. Bethesda, MD: National Cancer Institute; 2016. [Google Scholar]
- 2.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 3.Huang YT, Lin X, Liu Y, Chirieac LR, McGovern R, Wain J, et al. Cigarette smoking increases copy number alterations in nonsmall-cell lung cancer. Proc Natl Acad Sci U S A. 2011;108(39):16345–50. doi: 10.1073/pnas.1102769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015;348(6230):74–80. doi: 10.1126/science.aaa6204. [DOI] [PubMed] [Google Scholar]
- 5.Shan M, Cheng HF, Song LZ, Roberts L, Green L, Hacken-Bitar J, et al. Lung myeloid dendritic cells coordinately induce TH1 and TH17 responses in human emphysema. Sci Transl Med. 2009;1(4):4ra10. doi: 10.1126/scitranlsmed.3000154. [DOI] [PubMed] [Google Scholar]
- 6.Roos A, Sanden C, Mori M, Bjermer L, Stampfli M, Erjefalt J. IL-17A Is Elevated in End-Stage Chronic Obstructive Pulmonary Disease and Contributes to Cigarette Smoke–induced Lymphoid Neogenesis. American Journal of Respiratory and Critical Care Medicine Volume. 2015;191(11):1232–41. doi: 10.1164/rccm.201410-1861OC. [DOI] [PubMed] [Google Scholar]
- 7.Sheng SY, Gu Y, Lu CG, Tang YY, Zou JY, Zhang YQ, et al. The Characteristics of Naive-like T Cells in Tumor-infiltrating Lymphocytes From Human Lung Cancer. J Immunother. 2017;40(1):1–10. doi: 10.1097/CJI.0000000000000147. [DOI] [PubMed] [Google Scholar]
- 8.Cheng S, Shao Z, Liu X, Guo L, Zhang X, Na Q, et al. Interleukin 17A polymorphism elevates gene expression and is associated with increased risk of nonsmall cell lung cancer. DNA Cell Biol. 2015;34(1):63–8. doi: 10.1089/dna.2014.2628. [DOI] [PubMed] [Google Scholar]
- 9.Ma QY, Chen J, Wang SH, Wu N, Hao ZH, Chen XF. Interleukin 17A genetic variations and susceptibility to non-small cell lung cancer. APMIS. 2015;123(3):194–8. doi: 10.1111/apm.12341. [DOI] [PubMed] [Google Scholar]
- 10.Zhao L, Yang J, Wang HP, Liu RY. Imbalance in the Th17/Treg and cytokine environment in peripheral blood of patients with adenocarcinoma and squamous cell carcinoma. Med Oncol. 2013;30(1):461. doi: 10.1007/s12032-013-0461-7. [DOI] [PubMed] [Google Scholar]
- 11.Wei L, Wang H, Yang F, Ding Q, Zhao J. Interleukin-17 potently increases non-small cell lung cancer growth. Mol Med Rep. 2016;13(2):1673–80. doi: 10.3892/mmr.2015.4694. [DOI] [PubMed] [Google Scholar]
- 12.Nunez S, Saez JJ, Fernandez D, Flores-Santibanez F, Alvarez K, Tejon G, et al. T helper type 17 cells contribute to anti-tumour immunity and promote the recruitment of T helper type 1 cells to the tumour. Immunology. 2013;139(1):61–71. doi: 10.1111/imm.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma S, Cheng Q, Cai Y, Gong H, Wu Y, Yu X, et al. IL-17A produced by gammadelta T cells promotes tumor growth in hepatocellular carcinoma. Cancer Res. 2014;74(7):1969–82. doi: 10.1158/0008-5472.CAN-13-2534. [DOI] [PubMed] [Google Scholar]
- 14.Chang SH, Mirabolfathinejad SG, Katta H, Cumpian AM, Gong L, Caetano MS, et al. T helper 17 cells play a critical pathogenic role in lung cancer. Proc Natl Acad Sci U S A. 2014;111(15):5664–9. doi: 10.1073/pnas.1319051111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roy LD, Sahraei M, Schettini JL, Gruber HE, Besmer DM, Mukherjee P. Systemic neutralization of IL-17A significantly reduces breast cancer associated metastasis in arthritic mice by reducing CXCL12/SDF-1 expression in the metastatic niches. BMC Cancer. 2014;14:225. doi: 10.1186/1471-2407-14-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15(24):3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.TCGA. Cancer Genome Atlas Research N. Hammerman PS, Hayes DN, Wilkerson MD, Schultz N, Bose R, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012 doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J, Cho SN, Akkanti B, Jin N, Mao J, Long W, et al. ErbB2 Pathway Activation upon Smad4 Loss Promotes Lung Tumor Growth and Metastasis. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen F, Zhang Y, Parra E, Rodriguez J, Behrens C, Akbani R, et al. Multiplatform-based molecular subtypes of non-small-cell lung cancer. Oncogene. 2017;36(10):1384–93. doi: 10.1038/onc.2016.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang T, Guo L, Creighton CJ, Lu Q, Gibbons DL, Yi ES, et al. A genetic cell context-dependent role for ZEB1 in lung cancer. Nat Commun. 2016;7:12231. doi: 10.1038/ncomms12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goswami S, Angkasekwinai P, Shan M, Greenlee KJ, Barranco WT, Polikepahad S, et al. Divergent functions for airway epithelial matrix metalloproteinase 7 and retinoic acid in experimental asthma. Nature Immunology. 2009;10(5):496–503. doi: 10.1038/ni.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valdez Y, Diehl GE, Vallance BA, Grassl GA, Guttman JA, Brown NF, et al. Nramp1 expression by dendritic cells modulates inflammatory responses during Salmonella Typhimurium infection. Cellular Microbiology. 2008;10(8):1646–61. doi: 10.1111/j.1462-5822.2008.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casal RF, Lazarus DR, Kuhl K, Nogueras-Gonzalez G, Perusich S, Green LK, et al. Randomized trial of endobronchial ultrasound-guided transbronchial needle aspiration under general anesthesia versus moderate sedation. Am J Respir Crit Care Med. 2015;191(7):796–803. doi: 10.1164/rccm.201409-1615OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 25.Gibbons DL, Lin W, Creighton CJ, Zheng S, Berel D, Yang Y, et al. Expression signatures of metastatic capacity in a genetic mouse model of lung adenocarcinoma. PLoS One. 2009;4(4):e5401. doi: 10.1371/journal.pone.0005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shan M, Yuan X, Song LZ, Roberts L, Zarinkamar N, Seryshev A, et al. Cigarette smoke induction of osteopontin (SPP1) mediates T(H)17 inflammation in human and experimental emphysema. Sci Transl Med. 2012;4(117):117ra9. doi: 10.1126/scitranslmed.3003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lutz MB, Kukutsch N, Ogilvie AL, Rossner S, Koch F, Romani N, et al. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods. 1999;223(1):77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
- 28.Mayer CT, Ghorbani P, Nandan A, Dudek M, Arnold-Schrauf C, Hesse C, et al. Selective and efficient generation of functional Batf3-dependent CD103+ dendritic cells from mouse bone marrow. Blood. 2014;124(20):3081–91. doi: 10.1182/blood-2013-12-545772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millien VO, Lu W, Shaw J, Yuan X, Mak G, Roberts L, et al. Cleavage of fibrinogen by proteinases elicits allergic responses through Toll-like receptor 4. Science. 2013;341(6147):792–6. doi: 10.1126/science.1240342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shan M, You R, Yuan X, Frazier MV, Porter P, Seryshev A, et al. Agonistic induction of PPAR reverses cigarette smoke-induced emphysema. Journal of Clinical Investigation. 2014;124(3):1371–81. doi: 10.1172/JCI70587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan X, Shan M, You R, Frazier MV, Hong MJ, Wetsel RA, et al. Activation of C3a receptor is required in cigarette smoke-mediated emphysema. Mucosal Immunol. 2015;8(4):874–85. doi: 10.1038/mi.2014.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, et al. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell. 2017;169(4):750–65. e17. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bedoui S, Whitney PG, Waithman J, Eidsmo L, Wakim L, Caminschi I, et al. Cross-presentation of viral and self antigens by skin-derived CD103+ dendritic cells. Nat Immunol. 2009;10(5):488–95. doi: 10.1038/ni.1724. [DOI] [PubMed] [Google Scholar]
- 34.Broz M, Binnewies M, Boldajipour B, Nelson A, Pollack J, Erle D, et al. Dissecting the Tumor Myeloid Compartment Reveals Rare Activating Antigen-Presenting Cells Critical for T Cell Immunity. Cancer Cell. 2014;26(v):1–15. doi: 10.1016/j.ccell.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 35.Quatromoni JG, Eruslanov E. Tumor-associated macrophages: function, phenotype, and link to prognosis in human lung cancer. Am J Transl Res. 2012;4(4):376–89. [PMC free article] [PubMed] [Google Scholar]
- 36.Xu S, Han Y, Xu X, Bao Y, Zhang M, Cao X. IL-17A-producing gammadeltaT cells promote CTL responses against Listeria monocytogenes infection by enhancing dendritic cell cross-presentation. J Immunol. 2010;185(10):5879–87. doi: 10.4049/jimmunol.1001763. [DOI] [PubMed] [Google Scholar]
- 37.Zelenay S, Keller AM, Whitney PG, Schraml BU, Deddouche S, Rogers NC, et al. The dendritic cell receptor DNGR-1 controls endocytic handling of necrotic cell antigens to favor cross-priming of CTLs in virus-infected mice. J Clin Invest. 2012;122(5):1615–27. doi: 10.1172/JCI60644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Desch AN, Randolph GJ, Murphy K, Gautier EL, Kedl RM, Lahoud MH, et al. CD103+ pulmonary dendritic cells preferentially acquire and present apoptotic cell-associated antigen. J Exp Med. 2011;208(9):1789–97. doi: 10.1084/jem.20110538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de-Torres JP, Wilson DO, Sanchez-Salcedo P, Weissfeld JL, Berto J, Campo A, et al. Lung cancer in patients with chronic obstructive pulmonary disease. Development and validation of the COPD Lung Cancer Screening Score. American Journal of Respiratory & Critical Care Medicine. 2015;191(3):285–91. doi: 10.1164/rccm.201407-1210OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150(6):1121–34. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu YC, Robbins PF. Cancer immunotherapy targeting neoantigens. Semin Immunol. 2016;28(1):22–7. doi: 10.1016/j.smim.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borewicz K, Pragman AA, Kim HB, Hertz M, Wendt C, Isaacson RE. Longitudinal analysis of the lung microbiome in lung transplantation. FEMS Microbiology Letters. 2013;339(1):57–65. doi: 10.1111/1574-6968.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sheng SY, Gu Y, Lu CG, Zou JY, Hong H, Wang R. The distribution and function of human memory T cell subsets in lung cancer. Immunol Res. 2017;65(3):639–50. doi: 10.1007/s12026-016-8882-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Numasaki M, Watanabe M, Suzuki T, Takahashi H, Nakamura A, McAllister F, et al. IL-17 enhances the net angiogenic activity and in vivo growth of human non-small cell lung cancer in SCID mice through promoting CXCR-2-dependent angiogenesis. J Immunol. 2005;175(9):6177–89. doi: 10.4049/jimmunol.175.9.6177. [DOI] [PubMed] [Google Scholar]
- 46.Cochaud S, Giustiniani J, Thomas C, Laprevotte E, Garbar C, Savoye AM, et al. IL-17A is produced by breast cancer TILs and promotes chemoresistance and proliferation through ERK1/2. Sci Rep. 2013;3:3456. doi: 10.1038/srep03456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang K, Kim MK, Di Caro G, Wong J, Shalapour S, Wan J, et al. Interleukin-17 receptor a signaling in transformed enterocytes promotes early colorectal tumorigenesis. Immunity. 2014;41(6):1052–63. doi: 10.1016/j.immuni.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McAllister F, Leach SD. Targeting IL-17 for pancreatic cancer prevention. Oncotarget. 2014;5(20):9530–1. doi: 10.18632/oncotarget.2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen K, Pociask DA, McAleer JP, Chan YR, Alcorn JF, Kreindler JL, et al. IL-17RA is required for CCL2 expression, macrophage recruitment, and emphysema in response to cigarette smoke. PLoS ONE. 2011;6(5) doi: 10.1371/journal.pone.0020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.You R, Lu W, Shan M, Berlin JM, Samuel EL, Marcano DC, et al. Nanoparticulate carbon black in cigarette smoke induces DNA cleavage and Th17-mediated emphysema. Elife. 2015;4:e09623. doi: 10.7554/eLife.09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Caetano MS, Zhang H, Cumpian AM, Gong L, Unver N, Ostrin EJ, et al. IL6 Blockade Reprograms the Lung Tumor Microenvironment to Limit the Development and Progression of K-ras-Mutant Lung Cancer. Cancer Res. 2016;76(11):3189–99. doi: 10.1158/0008-5472.CAN-15-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, et al. T-Cell Transfer Therapy Targeting Mutant KRAS in Cancer. N Engl J Med. 2016;375(23):2255–62. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Topalian SL, Drake CG, Pardoll DM. Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor immunity. Curr Opin Immunol. 2012;24(2):207–12. doi: 10.1016/j.coi.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 55.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–9. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruffell B, Chang-Strachan D, Chan V, Rosenbusch A, Ho CM, Pryer N, et al. Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell. 2014;26(5):623–37. doi: 10.1016/j.ccell.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.