Abstract
Plasmacytoid dendritic cells (pDCs) are a unique sentinel cell type that can detect pathogen-derived nucleic acids and respond with rapid and massive production of type I interferon. This review summarizes our current understanding of pDC biology, including transcriptional regulation, heterogeneity, role in antiviral immune responses, and involvement in immune pathology, particularly in autoimmune diseases, immunodeficiency, and cancer. We also highlight the remaining gaps in our knowledge and important questions for the field, such as the molecular basis of unique interferon-producing capacity of pDCs. A better understanding of cell type-specific positive and negative control of pDC function should pave the way for translational applications focused on this immune cell type.
Plasmacytoid dendritic cells (pDCs) are innate immune sentinels that play important roles in immunity to infection and autoimmunity. This review by Boris Reizis highlights recent progress and emerging areas of interest in pDC biology as well as translational applications.
Main Text
Introduction
2019 marks the 20th anniversary of the definitive description of plasmacytoid dendritic cells (pDCs) as a unique cell type by the Liu and Colonna groups (Cella et al., 1999, Siegal et al., 1999). This description was based on extensive prior research by multiple groups (reviewed in Colonna et al., 2004, Liu, 2005) that collectively defined the essential properties of pDCs: secretory “plasmacytoid” morphology akin to that of plasma cells, rapid and massive production of type I interferons (IFN-I) in response to viruses, and the ability to differentiate into conventional dendritic cells (cDCs) in vitro. The last 20 years produced extraordinary insights into all aspects of pDC biology, including their development, mechanism of activity, and role in the immune system. At the same time, this progress brought us to fundamental unanswered questions about the role and mechanism of pDC function. Solving these questions would hasten the translation of accumulated knowledge into clinical applications focused on pDCs.
Accordingly, the goal of this review is both to summarize the current understanding and to highlight the gaps in our knowledge of pDC biology. Rather than an extensive listing of all pDC-related topics, it focuses on several key areas and concepts and also emphasizes the studies of the last several years that have not been covered in the previous reviews. I sincerely apologize to the researchers whose important original contributions could not be cited due to the limited time frame and scope of this review.
Development and Transcriptional Regulation of pDCs
pDCs primarily reside in and recirculate through lymphoid organs, where they typically comprise 0.1%–0.5% of nucleated cells. They have the round morphology of a secretory lymphocyte and express low levels of MHC class II that can be upregulated upon activation. pDCs express high levels of endosomal nucleic acid-sensing Toll-like receptors (TLRs) TLR7 and TLR9, which recognize single-stranded RNA and unmethylated CpG motif-containing DNA, respectively. pDCs respond to these nucleic acids with massive secretion of IFN-I, which commences within 1–3 h and involves IFN-β and most IFN-α subtypes. pDCs also produce type III interferon (IFN-λ or IL-28/IL-29) and additional cytokines (e.g., TNF-α) and chemokines (Gilliet et al., 2008, Reizis et al., 2011, Swiecki and Colonna, 2015). The speed, magnitude, and broad spectrum of interferon production by pDCs are reflected in the original definition of these cells as “natural interferon-producing cells” (Colonna et al., 2004, Liu, 2005). This and other key genetic and functional features of pDCs are shared between humans and mice (Crozat et al., 2010, Guilliams et al., 2016), highlighting evolutionary conservation of this immune cell type. This review will primarily focus on IFN-I production as the most prominent, unique, and well-established feature of pDCs, even though other aspects of pDC function such as antigen presentation are undoubtedly important (Villadangos and Young, 2008).
pDCs are continuously produced in the bone marrow (BM) and emerge as mature cells into the periphery, where they remain non-proliferative and have a relatively short lifespan of several days (Zhan et al., 2016). Similar to cDCs, pDC express cytokine receptor Flt3 (CD135) and are strictly dependent on its ligand Flt3L for their development. Conversely, Flt3L in the absence of other signals is sufficient to drive the development of pDCs and cDCs from myeloid and lymphoid progenitors (Sathe et al., 2013), consistent with the potential of both progenitor types to produce pDCs in vivo (Shigematsu et al., 2004). The myeloid pathway to pDCs includes a potential common dendritic cell (DC) progenitor (CDP) (Naik et al., 2007, Onai et al., 2007) and pDC-biased DC progenitors (Onai et al., 2013, Schlitzer et al., 2011). The identity of lymphoid progenitors of pDCs has been recently studied in greater detail (Herman et al., 2018, Rodrigues et al., 2018). Notably, relative rarity and quiescence of pDC-committed lymphoid progenitors (Rodrigues et al., 2018) are not consistent with these cells being a major source of much more abundant pDCs. Indeed, lineage tracing using the CDP marker Csf1r showed that the vast majority of pDCs became labeled (Loschko et al., 2016); in contrast, recently deposited results of B cell progenitor tracing suggest that a minor distinct population of pDCs is derived from these progenitors (Dekker et al., 2018). Furthermore, pDCs develop from stem cells in vivo with the same kinetics as myeloid cells including cDCs (Sawai et al., 2016), and progenitors with transcriptomic features of pDCs emerge prior to lymphoid progenitors (Upadhaya et al., 2018). Collectively, the results so far suggest that pDCs develop primarily through a Flt3L-driven pathway shared with cDCs, with a potential additional contribution from lymphoid progenitors. Importantly, clonal tracing studies of transplanted or cultured progenitors (Dursun et al., 2016, Lee et al., 2017, Lin et al., 2018, Naik et al., 2013) suggest that the commitment to produce pDC and/or cDC subsets may be determined at early stages of differentiation, i.e., prior to the emergence of phenotypically defined DC progenitors.
The fact that Flt3L alone is sufficient for pDC development suggests the existence of a robust transcriptional program that drives spontaneous pDC differentiation and pDC versus cDC lineage bifurcation, unless subverted by competing stimuli. Transcription factors such as PU.1 control the entire Flt3L-driven program of DC development (Carotta et al., 2010), whereas the subsequent specification of pDCs requires the E protein transcription factor TCF4 (E2-2) (Cisse et al., 2008, Nagasawa et al., 2008). TCF4 is expressed in all hematopoietic progenitors and can amplify its own expression in developing pDCs through a BRD protein-dependent feedback loop, underlying a self-sustained program of pDC development (Grajkowska et al., 2017). The pDC expression program appears to be initiated in progenitors that express IRF8 (Upadhaya et al., 2018), which is required for pDC development in vitro (although surprisingly, not in vivo [Sichien et al., 2016]). TCF4 acts jointly with its protein cofactor MTG16 (Ghosh et al., 2014) and additional factors such as BCL11A (Ippolito et al., 2014, Wu et al., 2013) to promote pDC development (Figure 1 A). Conversely, E protein inhibitor ID2 may “break” the TCF4-driven transcriptional program to channel the development into cDCs, particularly the ID2-dependent cDC1 subset. Thus, the loss of MTG16 or of transcriptional repressor ZEB2 (Scott et al., 2016, Wu et al., 2016) causes de-repression of ID2 and impairs pDC development, whereas the development of cDC1 is enhanced. A milder reduction of TCF4 activity by the deletion of pDC-specific “long” isoform TCF4L (Grajkowska et al., 2017) reduces pDC development but facilitates the development of the non-canonical DC subset discussed below. TCF4 is a close homolog of other E protein transcription factors TCF3 (E2A) and TCF12 (HEB), the key regulators of lymphocyte development. All three E proteins bind to the same consensus sequence and share target genes; accordingly, TCF4 in pDCs binds to and activates many genes that are activated by E2A/HEB in developing lymphocytes (Ceribelli et al., 2016, Ghosh et al., 2014). This unique feature of transcriptional control distinguishes pDCs from cDCs and likely accounts for their transcriptomic similarity to lymphoid progenitors (Herman et al., 2018). For instance, Dntt (TdT) and Ccr9 are canonical targets of E proteins in lymphoid progenitors and are also among the most TCF4-dependent genes in murine pDCs (Cisse et al., 2008), even though Dntt has no obvious function in these cells. Furthermore, deletion of TCF4 from mature pDCs induces the switch to dendritic morphology and expression of cDC gene signature (Ceribelli et al., 2016, Ghosh et al., 2010). Additional “resolution” between pDC and cDC expression programs is provided by transcription factor ETV6, which represses the cDC gene signature in pDCs and vice versa (Lau et al., 2018). Altogether, these studies underscore a close relationship between pDC and the cDC lineages and reveal a unique transcriptional program that maintains pDCs in a “lymphoid-like” state and opposes alternative cDC fates.
Figure 1.
Transcriptional Control of pDC Lineage
Transcription factors with enriched expression in pDCs, cDCs, or all DCs are shown in red, blue, or violet, respectively.
(A) Transcription factors that regulate pDC specification in the bone marrow. Lineage splits leading to alternative cell fates including cDC1s and non-canonical DCs are highlighted. TCF4L denotes the pDC-specific long isoform of TCF4.
(B) Transcription factors that regulate the differentiation of committed pDCs in the bone marrow and pDC functionality in the periphery.
Following the initial lineage commitment, pDCs undergo an elaborate differentiation program that ensures their functionality (Figure 1B). Thus, transcription factor SPIB facilitates the retention of immature pDCs in the BM (Sasaki et al., 2012); conversely, RUNX2 facilitates the exit of mature pDCs from BM to the periphery (Chopin et al., 2016, Sawai et al., 2013). The IFN-I-producing capacity of pDCs is dependent on interferon response family (IRF) transcription factors including IRF7 (Honda et al., 2005b), IRF5 (Dai et al., 2011, Yasuda et al., 2013), and IRF8 (Sichien et al., 2016). Consistent with the key role of IRF7, several factors were shown to regulate its activity in pDCs: for instance, SPIB (Sasaki et al., 2012) and NFATC3 (Bao et al., 2016) interact with IRF7 and facilitate the activation of IFN-I genes. Furthermore, RUNX2 was proposed to directly activate IRF7 expression (Chopin et al., 2016) whereas MYC may repress it to limit IFN-I production (Kim et al., 2016). Finally, pDC-enriched epigenetic regulator CXXC5 together with DNA demethylase Tet2 was shown to promote Irf7 expression by maintaining the hypomethylation of its promoter in pDC (Ma et al., 2017). Collectively, these studies begin to reveal a complex transcriptional network that maintains the capacity of pDCs for rapid response while limiting unwanted hyperactivation. Key parts of this network, including epigenetic mechanisms and cis-regulatory elements that facilitate pDC functionality, still remain to be identified.
Heterogeneity and Post-activation Fate of pDCs
Recent single-cell analysis of human DC population revealed a subset with some phenotypic features of pDCs that was incapable of IFN-I production but efficiently primed T cells (Alcántara-Hernández et al., 2017, See et al., 2017, Villani et al., 2017). Together with other reports on human pDC heterogeneity (Matsui et al., 2009, Zhang et al., 2017), these results suggest the existence of a “non-canonical” DC population that is intermediate between the canonical pDCs and cDCs. Further analysis of this population, defined by several markers including AXL, revealed that it is heterogeneous and comprises “pDC-like” as well as “cDC-like” cells (Alcántara-Hernández et al., 2017, Villani et al., 2017). Importantly, “non-canonical” DC subsets have been described previously in the mouse, most notably the distinct Cx3cr1+ subset of CD8+ cDCs (Bar-On et al., 2010, Lau et al., 2016). These cells manifest the surface phenotype and functionality of cDCs, lack IFN-I production, yet have a transcriptional profile related to pDCs and require TCF4 for development. Furthermore, a recently deposited study proposed a distinct minor subset of Axl+ murine pDCs that are derived from lymphoid progenitors, lack IFN-I production, but show increased antigen-presenting capacity (Dekker et al., 2018). Preliminary comparison of expression profiles and/or phenotypes of these subsets suggests a concordance between human and murine “non-canonical” pDC and cDC subsets, which potentially comprise an evolutionarily conserved spectrum (Figure 2 ). Notably, non-canonical pDCs and cDCs co-express TCF4 and ID2, consistent with their intermediate position between TCF4hi pDCs and ID2hi cDCs. This proposed cross-species alignment of DCs is supported by high-dimensional DC phenotyping in humans and mice (J. Idoyaga, personal communication) and remains to be comprehensively tested in future studies. Most importantly, the developmental origin and function of non-canonical subsets need to be elucidated in both species.
Figure 2.
The Proposed Spectrum of Dendritic Cell Subsets
Shown on this hypothetical scheme are canonical pDCs and cDCs (including cDC1 and cDC2) and the intermediate “non-canonical” populations including non-canonical pDCs (nc-pDC) and cDCs (nc-cDC). Shown are functional properties including the IFN-I production capacity and antigen presentation capacity in the steady state, expression of key transcription factors, and surface markers in the human (Alcántara-Hernández et al., 2017, Villani et al., 2017) and mouse (Bar-On et al., 2010, Lau et al., 2016, Dekker et al., 2018). Italicized genes and markers denote expression based on reported transcript levels.
Within the canonical mature pDC subset, heterogeneity is evident from the expression of surface markers such as murine CD4, CD8 (O’Keeffe et al., 2002), or Ccr2 (Sawai et al., 2013), yet its significance remains unclear. The question of pDC heterogeneity is directly tied to a broader question of pDC function and eventual fate upon TLR-mediated activation. While activated pDCs are capable of cytokine production as well as antigen presentation and cross-presentation, these processes may either occur sequentially in the same cells or represent separate fates of different pDCs. The latter appears more likely because only a fraction of pDCs produce IFN-I irrespective of the stimulus, as confirmed using IFN-I reporter strains (Bauer et al., 2016, Kumagai et al., 2007, Tomasello et al., 2018). Furthermore, pDC activation results in the massive pDC apoptosis caused by IFN-I (Swiecki et al., 2011). It is therefore conceivable that the majority of IFN-producing pDCs die in vivo, whereas the surviving ones remain capable of antigen presentation. Indeed, in vitro activated human pDCs generate distinct populations with preferential IFN-producing or antigen-presenting capacities (Alculumbre et al., 2018). These populations were proposed to diversify spontaneously through paracrine cytokine signaling; however, pre-existing commitment toward these fates within the pDC population cannot be ruled out. Importantly, the observed differentiation into cDC-like cells represents an intrinsic property of activated pDCs (Grouard et al., 1997) that cannot be explained by contamination with AXL+ DCs (Alcántara-Hernández et al., 2017, Alculumbre et al., 2018). The analysis of such activation-induced differentiation in vivo, as well as the broad characterization of pDC cell fate during immune responses, represent important challenges for future studies.
Mechanism of Interferon Production by pDCs
Signaling by endosomal TLRs through the adaptor MyD88 and transcription factor IRF7 appears to be a major (albeit not exclusive) mechanism of pDC activation that results in IFN-I production (Honda et al., 2005a, Honda et al., 2005b). Another consequence of TLR/MyD88 signaling is the activation of NF-κB and the ensuing induction of cytokines (such as TNF-α) and costimulatory molecules; this pathway is very important but not unique to pDCs and will not be further discussed here. Non-endosomal MyD88-dependent TLRs such as TLR2 may also be functional in pDCs in response to certain stimuli (Dasgupta et al., 2014). The cGAS/STING and RIG-I pathways of intracellular sensing may be functional in pDCs in response to replicating DNA viruses (Li et al., 2013) and RNA viruses (Bruni et al., 2015, Kumagai et al., 2009), respectively. Although it has been suggested that intracellular sensing pathways such as cGAS/STING and MDA5/MAVS may restrict IFN-I response of pDCs (Yu et al., 2016), this model is not supported by other studies (Spaulding et al., 2016). Importantly, endosomal TLR-induced IFN-I production represents a very specific feature of pDCs: for example, in vivo administration of TLR9 ligand CpG induces a rapid systemic IFN-I response that is mediated exclusively by pDCs (Asselin-Paturel et al., 2003, Blasius et al., 2004, Cervantes-Barragan et al., 2012).
This, however, opens up a truly challenging question: what makes the TLR7/9-induced IFN-I response so specific for pDCs? The expression of TLR7 and TLR9 is a distinct but not unique property that is shared with B cells and several myeloid cell types (particularly in the mouse). Additional important mechanisms of TLR-induced IFN-I production include the PI3K/mTOR pathway activation (Cao et al., 2008, Guiducci et al., 2008), prolonged retention of TLR ligands in early endosomes (Guiducci et al., 2006, Honda et al., 2005a), and regulation of endosomal TLR signaling by the AP-3 adaptor complex and the AP-3-interacting cation transporter Slc15a4 (Blasius et al., 2010, Sasai et al., 2010). However, none of these mechanisms or molecules is specific for pDCs; conversely, candidate pDC-specific signaling mediators such as endocytic adaptor Pacsin1 contribute to IFN-I response but are not essential for it (Esashi et al., 2012). Finally, fundamental differences may exist in the response of pDCs to synthetic TLR agonists and to viruses: for instance, AP-3 was recently shown to be required for the former but not the latter (Tomasello et al., 2018).
In most cell types, the expression of IRF7 is normally low or absent and is induced by the first wave of IFN-β production through an IFN-I receptor (IFNAR)-mediated feedback loop. Quiescent pDCs express high levels of IRF7 and therefore were proposed to secrete IFN-I rapidly and independently of the IFNAR-based feedback signaling (Barchet et al., 2002, Dalod et al., 2002). Indeed, IFNAR was shown to be dispensable for pDC response to viruses such as vesicular stomatitis virus (VSV) (Barchet et al., 2002) and murine cytomegalovirus (MCMV) (Tomasello et al., 2018). Surprisingly, IFNAR-deficient pDCs manifested normal response to MCMV despite expressing low baseline levels of IRF7 (Tomasello et al., 2018). On the other hand, IFNAR appears necessary for full-strength IFN-I response to TLR ligands in vivo (Asselin-Paturel et al., 2005, Blasius et al., 2010, Tomasello et al., 2018) and to certain viruses in vitro (Kumagai et al., 2009), and for optimal functionality of human pDC in vitro (Laustsen et al., 2018). These data suggest that IFNAR-independent signaling does not require high IRF7 levels and may promote pDC responses to certain viruses, although full pDC functionality still requires intact IFNAR signaling.
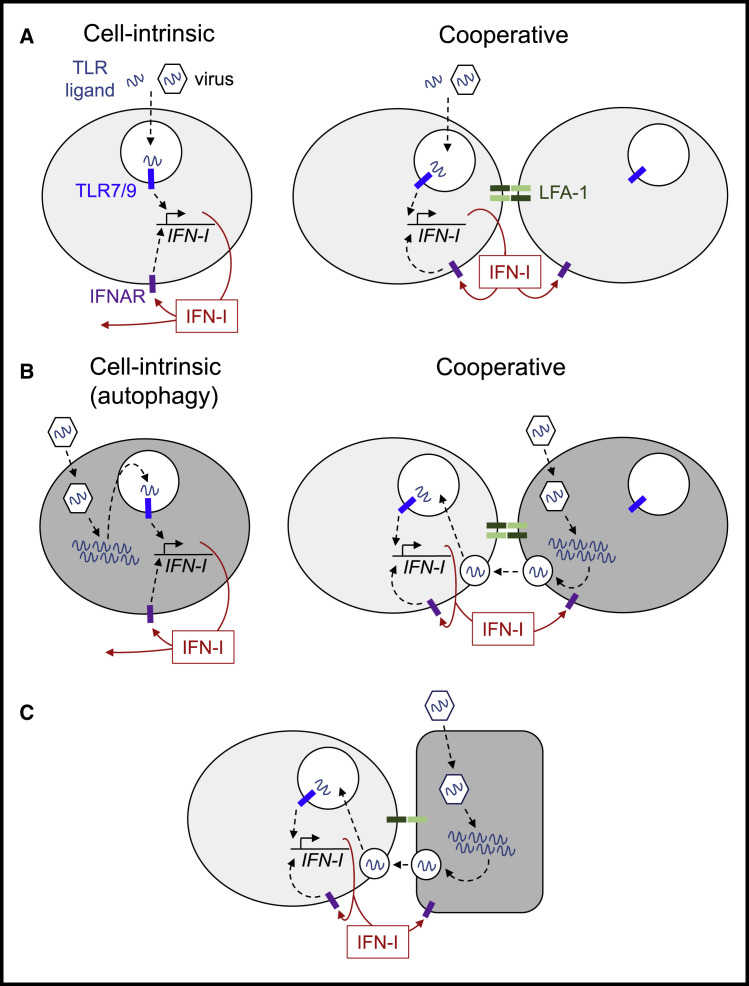
In contrast to the traditional “cell-intrinsic” view of pDC activation, recent studies highlighted its unexpectedly cooperative nature (Figure 3 A). It has been observed early on that pDC form tight clusters within hours of activation by TLR ligands in vivo (Asselin-Paturel et al., 2005), although the functional significance of such clustering remained unclear. In vitro, the magnitude of IFN-I response to TLR ligands was shown to be facilitated by cell density (Kim et al., 2014), as confirmed recently by single-cell activation assay (Wimmers et al., 2018). These studies ascribed the observed cooperativity to autocrine/paracrine IFN-I signaling, which may indeed be important in these settings as discussed above. However, another potentially important mechanism is cell-cell interaction and the associated polarization of IFN-I production and sensing, as observed upon pDC activation in vitro (Saitoh et al., 2017). This study showed that pDC response to TLR ligands and to the influenza virus is impaired in the absence of integrin LFA-1 and of downstream regulators of intracellular trafficking that localize TLRs and secreted IFN-I to the sites of cell-cell contact (Saitoh et al., 2017). The contribution of LFA-1 expressed on pDCs to IFN-I production in response to MCMV and to TLR9 ligand has been recently confirmed in vivo (Tomasello et al., 2018). These recent studies highlight cell-cell interactions (including homotypic clustering of pDCs) and the associated profound polarization of TLR sensing, IFN-I production, and IFN-I-mediated feedback signaling as important features of pDC response. Collectively, the unique IFN-I-producing capacity of pDCs cannot be easily explained by any single known mechanism or molecule. Instead, it appears to integrate multiple pathways and cellular processes, in a manner that still remains to be fully elucidated.
Figure 3.
The Mechanism of pDC Activation
(A) Activation by TLR ligands or viruses that do not infect pDCs. Shown is the traditional “cell-intrinsic” model and the emerging “cooperative” model that highlights homotypic interaction between pDCs and directional signaling and IFN secretion.
(B) Activation by viruses that infect and replicate in pDCs. Shown is the “cell-intrinsic” model based on autophagy-mediated TLR signaling, and the proposed “cooperative” model whereby virus replication and TLR signaling occur in different cells, and TLR ligands are transferred via exosomes or viral particles. An infected cell is highlighted in dark gray.
(C) Activation by viruses that infect cells other than pDCs. TLR ligands are transferred from infected cells (highlighted in dark gray) to pDCs during polarized contact via exosomes or viral particles.
Negative Regulation of IFN-I Production by pDCs
The IFN-I-producing capacity and “trigger-happy” state of pDC must have necessitated the need for counterbalancing inhibitory mechanisms. Some of these act through broadly expressed surface receptors such as CXCR4 and CD44, which inhibit pDC activation in response to natural monoamines (Smith et al., 2017) and galectin-9 (Panda et al., 2018), respectively. Several inhibitory receptors are shared with lymphocytes and seem to be “re-purposed” for inhibitory function in pDCs, such as murine Lag3 (Workman et al., 2009) and CD28 (Macal et al., 2016). Yet other receptors are relatively specific for pDCs, such as murine SiglecH and human BDCA-2 (CD303) and ILT-7 (CD85). Additional members of this class are likely to exist, as illustrated by the recent description of mouse pDC-specific inhibitory receptor CD300c (Kaitani et al., 2018). These receptors inhibit pDC response when engaged with agonistic antibodies and are thought to signal through a common pathway involving ITAM-containing adaptors such as Fc receptors and DAP12 (Gilliet et al., 2008). SiglecH is a sialic acid-binding lectin whose exact ligands are poorly understood, although its deletion causes enhanced IFN-I response by pDCs (Puttur et al., 2013, Schmitt et al., 2016). SiglecH-deficient mice also develop autoimmunity several weeks after viral infection (Schmitt et al., 2016), although it is unclear whether this striking phenotype emanates from pDCs. Unless some pDCs acquire a long lifespan after infection, the delayed autoimmunity is likely mediated by long-lived SiglecH+ cells such as macrophages. BDCA-2 is a C-type lectin that binds multiple glycoproteins including serum immunoglobulins (Kim et al., 2018), raising the possibility that antibody responses inhibit pDC activation through BDCA-2-mediated homeostatic mechanism. ILT-7 is an immunoglobulin-like receptor whose single known ligand is BST2 (tetherin), an interferon-inducible antiviral membrane protein that may provide a negative feedback signal to IFN-I-producing pDC (Cao et al., 2009). Murine pDCs lack an obvious ortholog of ILT-7 but specifically express Bst2 (Blasius et al., 2006b), which paradoxically facilitates IFN-I production (Swiecki et al., 2012) and may contribute to pDC resistance to infection.
The species-specific nature of above-mentioned inhibitory pathways likely reflects separate evolutionary pressures from pathogens. On the other hand, a distinct and evolutionarily conserved pDC-specific pathway involves a receptor-type protein tyrosine phosphatase PTPRS, which is expressed in multiple tissues but is highly pDC-specific within the immune system of humans and mice (Bunin et al., 2015). The deletion or ligation of PTPRS (along with the homologous pDC-specific phosphatase PTPRF in the mouse) respectively enhanced or inhibited pDC activation. The ligands of PTPRS likely involve diverse heparan sulfate proteoglycans, suggesting a potential way for pDCs to monitor the integrity of extracellular matrix. The intracellular substrates of PTPRS are unknown, but the inhibitory activity of a tyrosine phosphatase mirrors the recently identified positive roles of tyrosine kinases SYK (Aouar et al., 2016), LYN, and FYN (Dallari et al., 2017) in pDC activation. Global or dendritic cell-specific deletion of PTPRS/PTPRF in the mouse caused spontaneous IFN-I production and colitis (Bunin et al., 2015), highlighting the inherent danger posed by even mild pDC hyperactivation. Overall, negative regulation of pDC responses appears fundamental to pDC biology and is responsible for the most prominent cell type-specific molecular features, including the hallmark surface markers such as SiglecH and BDCA-2. Further analysis of these pathways may provide attractive opportunities for therapeutic targeting of pDC activation, e.g., inhibition in autoimmune diseases or activation in cancer.
The Nature of Virus Recognition by pDCs
pDCs were defined by their ability to respond to the purified influenza virus added to cultures, and this certainly represents an important mode of pathogen recognition by these cells (Figure 3A). Direct recognition of internalized virions through endosomal TLRs is indeed strongly supported by the ability of pDCs to respond to viruses that have been rendered replication-deficient, e.g., by physical inactivation (Asselin-Paturel et al., 2001, Deal et al., 2010, Kumagai et al., 2009, Lund et al., 2003). It is also consistent with the relative resistance of pDCs to infection by many viruses to which they are capable of responding with IFN-I production.
Other viruses such as VSV elicit IFN-I production only when replication-active (Hornung et al., 2004, Lee et al., 2007), and productive infection of pDCs by IFN-I-inducing viruses has been described in vitro (Cervantes-Barragan et al., 2007, Dai et al., 2011, Manuse et al., 2010) and in vivo (Macal et al., 2012). Notably, these experimental settings utilized high viral doses in vitro, and the resulting infection of pDCs was still lower than of other cell types. Autophagy may represent one mechanism whereby a virus replicating inside the pDC would get access to the endosomal compartment and induce IFN-I production (Figure 3B; Lee et al., 2007). On the other hand, single-cell analysis of response to several viruses (including VSV) demonstrated that viral replication and IFN-I production occurs in different cells within the pDC population (Deal et al., 2010, Döring et al., 2014, Frenz et al., 2014). This may reflect the heterogeneity of IFN-I response by pDCs as noted above—in this scenario, IFN-I-producing pDCs would suppress viral replication while those lacking IFN-I production would sustain it. Another possibility, however, is that pDCs spontaneously “divide the labor”—some get infected and others recognize the virus replicating in the infected pDCs (Figure 3B). This scenario is consistent with the highly cooperative nature of pDC activation as discussed above, as well as with the emerging unique feature of pDCs: the ability to recognize virus-infected cells.
This new paradigm of virus recognition by pDCs (Figure 3C) was introduced in a study of hepatitis C virus (HCV), an RNA virus with a strict tropism for hepatocytes. It showed that pDCs produce IFN-I by interacting with and recognizing HCV-infected hepatocytes rather than the free HCV (Takahashi et al., 2010). This recognition is TLR7 dependent, requires physical contact between pDCs and live infected cells, and was initially proposed to occur via the short-range transfer of viral RNA in exosomes (Dreux et al., 2012, Takahashi et al., 2010). Subsequent studies have demonstrated the same phenomenon for multiple diverse RNA viruses including retroviruses (Lepelley et al., 2011, Rua et al., 2012), lymphocytic choriomeningitis virus (LCMV) (Wieland et al., 2014), hepatitis A virus (Feng et al., 2015), Dengue and West Nile viruses (Décembre et al., 2014), and yellow fever vaccine virus (Bruni et al., 2015). These studies also showed that RNA of a replicating virus may be transferred between infected cells and pDCs within immature viral particles or enveloped virions. In cells latently infected with the DNA herpesvirus Epstein-Barr virus (EBV), EBV-derived RNA can be transferred to pDCs through exosomes and can elicit the expression of IFN-I and interferon-stimulated genes (ISG) (Baglio et al., 2016). In addition to human pDCs, this phenomenon was also observed in murine (Frenz et al., 2014) and porcine (García-Nicolás et al., 2016) pDCs. It may also be applicable to non-viral pathogens, as suggested by the close interactions of pDCs with macrophages during TLR7-mediated activation of pDCs in malaria-infected mice (Spaulding et al., 2016).
The recognition of virus-infected cells by pDCs has multiple potential advantages. First, it helps pDCs detect the virus during the main intracellular events of its life cycle including replication and possibly even latency, rather than during the often brief and rare extracellular transit. Indeed, the recognition of a free virus such as VSV, even at a high multiplicity of infection, appears less efficient than the recognition of cells infected with the same virus (Frenz et al., 2014). Second, it makes pDCs resistant to multiple evasion mechanisms used by viruses to prevent recognition and IFN-I secretion by the infected cell. Third, it allows the detection of viruses with a very narrow tropism (e.g., HCV) and facilitates precise tissue localization of responses toward infected cells. This mode of recognition also offers a potential explanation for the above-mentioned cases where pDCs themselves get infected: in this scenario, infected pDCs may elicit TLR-driven production of IFN-I from uninfected pDCs (Figure 3B). Thus, pDCs appear to have evolved a unique ability to monitor the intracellular compartment in a manner resembling other sentinel cells such as NK cells and cytotoxic T cells.
Fundamental questions still remain about this emerging mechanism of virus recognition by pDCs. Is the recognition of virus-infected cells a special case of certain viruses recognized through TLR7, or a “default” mode applicable to the majority of RNA and DNA viruses? TLR9-mediated recognition of infected cells has not been demonstrated yet, although the transfer of the virus and cellular material from cells infected with the DNA virus herpes simplex virus (HSV) to pDCs has been described (Megjugorac et al., 2007). Recent histological analysis of IFN-I-producing pDCs during MCMV infection suggests that they are indeed localized in proximity to virus-infected cells (Tomasello et al., 2018). However, the recognition of primary infected cells by pDCs still remains to be documented in vivo, ideally by intravital microscopy. Perhaps most importantly, we need to understand how the virus-derived material is being transferred and why it can be recognized so efficiently compared to the free virus. The role of LFA-1-mediated adhesion in the process would be consistent with the analysis of homotypic pDC interactions (Saitoh et al., 2017) and is supported by blocking studies in vitro (Assil et al., 2018, García-Nicolás et al., 2016) and by genetic deletion in vivo (Tomasello et al., 2018). These and other data support the formation of integrin-mediated “interferogenic synapse” between the infected cell and a pDC, as proposed in a recently deposited study (Assil et al., 2018). Future studies should elucidate molecular mechanisms that mediate the formation and function of this synapse at both sides, particularly those responsible for its specificity for pDCs.
The Role of pDCs in Immune Responses to Infections
Studies of pDC activation in vitro typically demonstrate satisfyingly robust and specific responses of pDCs to the majority of viruses. However, in vivo testing of pDC function during infections has been frequently disappointing: for instance, pDCs are not strictly required for the in vivo control of such “model” murine viruses as influenza, VSV, MCMV, or acute LCMV (Cervantes-Barragan et al., 2012, Swiecki et al., 2010, Wolf et al., 2009). These results may reflect the notoriously multilayered nature of immune responses—for instance, conventional DC were shown to compensate for the absence of pDCs in infections with ectromelia virus (Kaminsky et al., 2015) or MCMV (Puttur et al., 2016). Additional reasons may include peculiarities of laboratory mice as a model, such as the mutations of key antiviral ISG in common strains. Conversely, the role of pDCs has to be rigorously assessed by genetic ablation, as depleting or inactivating antibodies (Table 1 ) are not fully pDC specific: for instance, the commonly used depletion target Bst2 (120G8, CD317, mPDCA-1) is also expressed on plasmacytes in the steady state and is induced on many cell types during infection (Blasius et al., 2006b). Ultimately, genetic models using human BDCA2-driven diphtheria toxin receptor (DTR) for acute depletion (Swiecki et al., 2010) or Tcf4 targeting for constitutive pDC loss (Cervantes-Barragan et al., 2012) (Table 1) revealed several viral infections in which pDCs play a major non-redundant role. Although these tools implicated pDCs in response to certain non-viral pathogens such as bacteria (Crother et al., 2012) and apicomplexan parasites (Spaulding et al., 2016, Yu et al., 2016), the net activity of pDCs in these models appears complex and incompletely understood.
Table 1.
In vivo Depletion and Functional Modulation of pDCs
| Driver/Target | Targeting Strategy | Advantages | Caveats | References |
|---|---|---|---|---|
| Genetic (Mouse) | ||||
| Tcf4 (E2-2) | DC-specific deletion using Itgax-Cre | constitutive specific depletion of peripheral pDCs | complex allele combination; depletion incomplete on C57BL/6 background | Cisse et al., 2008, Cervantes-Barragan et al., 2012 |
| Tcf4 (E2-2) | monoallelic germline deletion | constitutive functional impairment of pDCs; single allele | partial reduction of pDC numbers and functionality; effects in non-pDCs formally possible | Cisse et al., 2008, Sisirak et al., 2014 |
| Ikzf1 (Ikaros) | germline hypomorphic mutation on Rag2-null background | specific absence of peripheral pDCs | complex allele combination; requires lymphocyte reconstitution; effects in non-pDCs formally possible | Allman et al., 2006, Guillerey et al., 2012 |
| CLEC4C (BDCA-2) | human transgene driving DTR | DT-inducible depletion; efficient; repeated DT injections used for several weeks; single allele | transient; side effects of DT | Swiecki et al., 2010, Rowland et al., 2014 |
| SiglecH | knock-in of DTR | DT-inducible depletion; efficient; based on endogenous mouse gene | nonspecific effects in non-pDCs demonstrated; concomitant SiglecH deletion; side effects of DT | Takagi et al., 2011, Swiecki et al., 2014 |
| Antibody-Mediated (Mouse) | ||||
| Bst2 (CD317) | depleting mAb (clones 120G8, mPDCA-1, 927) | rapid depletion | transient; can be used only in the steady state; targets other cells (e.g., plasma cells) | Asselin-Paturel et al., 2003, Krug et al., 2004, Blasius et al., 2006b |
| SiglecH | agonistic mAb (clone 440c) | rapid functional impairment | transient; may affect other cells (e.g., macrophages) | Blasius et al., 2004, Blasius et al., 2006a |
| Antibody-Mediated (Human, Primate) | ||||
| IL-3Rα (CD123) | depleting mAb (clone CSL362) | efficient depletion | depletes other CD123+ cells (e.g., basophils); blocks IL-3 signaling | Oon et al., 2016 |
| CLEC4C (BDCA-2, CD303) | agonistic mAb (clone 24F4A) | specific; causes functional impairment | efficiency to be demonstrated in human subjects | Pellerin et al., 2015 |
Abbreviations: DT, diphtheria toxin; DTR, diphtheria toxin receptor; mAb, monoclonal antibody.
Mouse hepatitis virus (MHV) causes an acute infection that is normally controlled in a TLR7- and IFNAR-dependent manner. A strict requirement for pDCs in the control of MHV infection has been demonstrated using both antibody-mediated and genetic pDC depletion (Cervantes-Barragan et al., 2007, Cervantes-Barragan et al., 2012). Within 2 days post-infection, pDCs produce nearly all detectable IFN-I, which is critical for the survival of cDCs and macrophages (Cervantes-Barragán et al., 2009). Another prominent example of pDC dependence is systemic (but not local) HSV infection, in which pDC are required for early production of IFN-I within 8–12 h, NK cell activation, and ultimately for survival (Swiecki et al., 2013). Using a complementary genetic approach in which IRF7-dependent IFN-I production was restricted to pDCs, it was shown that pDCs are sufficient to control acute infection with dengue and chikungunya viruses (Webster et al., 2018). In all these models, a very rapid production of IFN-I by pDCs appears critical for protection against cytopathic effects of the virus, NK cell activation, amplification of cytokine response, and overall innate control of viral replication. Notably, systemic IFN-I production is not always detectable (Webster et al., 2018), suggesting that close-range IFN-I production by pDCs may be particularly important. Given that coronaviruses, herpesviruses, and flaviviruses (exemplified by MHV, HSV, and dengue virus, respectively) exerted major evolutionary pressures on mammalian immune systems, these results emphasize the critical role of pDCs in immunity and explain their emergence and evolutionary conservation.
In addition to their innate protective function, pDCs also play a key role in shaping the adaptive responses to viruses. This was first illustrated by impaired survival and accumulation of VSV-specific cytotoxic T cells upon pDC depletion (Swiecki et al., 2010). Moreover, pDC-deficient mice are unable to control persistent LCMV infection due to impaired CD4+ T cell priming, which in turn impairs CD8+ T cell response to LCMV (Cervantes-Barragan et al., 2012). The role of pDCs in T cell priming may be mediated at least in part through their cooperation with cDC (Rogers et al., 2017), particularly the recruitment of XCR1+ cDC1s that mediate CD8+ T cell cross-priming (Brewitz et al., 2017). The effect of pDCs on T cell responses is not limited to cytotoxic T cell priming and may exert broader effects on virus-induced pathology. For example, pDCs contribute to IFN-I response and control of respiratory syncytial virus (RSV) infection in the lung (Davidson et al., 2011, Lynch et al., 2018). In addition, they dampen RSV-induced Th2 cell responses, lung inflammation, and asthma (Cormier et al., 2014), at least in part through the induction of regulatory T (Treg) cells (Lynch et al., 2018). Likely reflecting the role of pDCs in long-term adaptive responses, persistent viruses such as LCMV impair the IFN-I producing capacity of pDCs (Lee et al., 2009, Zuniga et al., 2008). This “exhausted” state of pDCs was recently shown to result from a combination of impaired development, enhanced self-renewal in the periphery, and persistent signaling through TLR7 (Macal et al., 2018). The role of pDCs in human persistent viral infections has been largely studied in the HIV infection, where pDCs mount vigorous IFN-I response that is protective at early stages but may drive immune pathology later in the course of infection (Aiello et al., 2018). Collectively, these results highlight the critical dual function of pDCs in antiviral responses: (1) rapid protection through innate mechanisms such as cytoprotective effects and NK cell activation; (2) fine-tuning of adaptive responses through enhanced T cell differentiation. Both phenomena are likely to involve IFN-I production (both local and systemic) as well as other mechanisms such as production of additional cytokines and chemokines and possibly antigen presentation; elucidating these mechanisms represents an important challenge for years to come.
The Role of pDC Activation in Autoimmunity
Soon after pDCs were conclusively identified as a distinct IFN-I-producing cell type, their potential role in excessive cytokine production in autoimmune disease was proposed (Rönnblom and Alm, 2001). In subsequent years, excessive or aberrant pDC activity has been implicated in almost every autoimmune or inflammatory disease. As in the case of infections, rigorous testing in genetic models appears critical to define immune pathologies with a particularly prominent role of pDCs. For instance, genetic ablation of pDCs revealed only a mild stage-specific role of pDCs in a genetic model of psoriasis (Glitzner et al., 2014) and no involvement in a chemically induced model (Wohn et al., 2013). In other examples, genetic impairment of pDCs had no effect in two genetic models of colitis (Sawai et al., 2018) and exacerbated the disease in an antibody-mediated model of arthritis (Nehmar et al., 2017). Even genetic approaches occasionally produce conflicting results as illustrated by studies in atherosclerosis, where constitutive or inducible pDC ablation was shown to impair (Sage et al., 2014) or promote (Yun et al., 2016) the disease, respectively. On the other hand, recent genetic studies strongly support the pathogenic role of pDCs in several autoimmune diseases.
The role of pDCs in type I diabetes was proposed based on the expansion of pDCs at the onset of human diabetes (Allen et al., 2009) and pDC infiltration into islets in the NOD mouse model (Diana et al., 2013). Indeed, constitutive genetic depletion of pDCs was shown to ameliorate insulitis and significantly reduce the incidence of diabetes in NOD mice (Hansen et al., 2015). In scleroderma (systemic sclerosis, SSc), an antibody-mediated autoimmune disease that targets connective tissue including the skin, patients manifest the elevated expression of multiple ISGs (“interferon signature”) and the pathogenic immune complexes induce IFN-I production in pDCs (Eloranta et al., 2010, Kim et al., 2008). Moreover, pDCs from SSc patients produce high levels of proinflammatory hemokine CXCL4, which correlates with the risk and progression of the disease (van Bon et al., 2014). The most recent study connected the aberrant CXCL4 production to increased IFN-I expression in pDCs from SSc patients and showed that transient pDC depletion could prevent and even revert skin fibrosis in a chemically induced animal model (Ah Kioon et al., 2018). In both diabetes and SSc, pDC likely act locally at the sites of inflammation (pancreatic islets and skin, respectively) and/or in the regional lymphoid organs to promote inflammation through the production of IFN-I and other mediators such as CXCL4.
Perhaps the strongest case for the pathogenic role of pDCs has been built in systemic lupus erythematosus (SLE), a systemic disease driven by immune complexes of autoantibodies and nucleic acid-containing nuclear antigens (reviewed in Panda et al., 2017). More than half of SLE patients manifest interferon signature that correlates with clinical disease, and IFNAR deletion ameliorates SLE in experimental models. Clinical SLE is associated with the reduction of pDCs in the peripheral blood and their concomitant accumulation in tissue lesions. pDCs can be induced to produce IFN-I by multiple SLE-associated molecular features including nucleic acid-containing immune complexes, complexes of DNA with antimicrobial peptides, neutrophil extracellular traps (NETs), and as shown most recently, oxidized mitochondrial DNA (Caielli et al., 2016, Lood et al., 2016). Consistent with the predominant occurrence of SLE in females, female pDCs have higher IFN-I-producing capacity than male pDCs, due to both cell-intrinsic and -extrinsic factors (Griesbeck et al., 2015, Laffont et al., 2014). Importantly, even a transient DTR-based ablation of pDCs ameliorated SLE in genetic models (Davison and Jørgensen, 2015, Rowland et al., 2014). Furthermore, a constitutive impairment of pDCs by monoallelic deletion of Tcf4 strongly reduced autoantibody production and all disease manifestations in two different spontaneous models of SLE (Sisirak et al., 2014). The magnitude of the effect was particularly striking, given that Tcf4 haplodeficiency causes only a partial reduction of pDC numbers and IFN-I-producing capacity. These data confirmed an essential role of pDCs in SLE pathogenesis as well as revealed its systemic nature: pDCs are required not only for inflammation triggered by immune complexes, but also for the production of autoantibodies. This emerging role of pDCs in both innate and adaptive immune responses to self is reminiscent of their dual role in antiviral responses as discussed above, and poses major questions about its mechanism. It remains to be determined what initial molecular signals cause pDC hyperactivation, and how they are sensed; the issue is particularly challenging because TLR9, the proposed major sensor of self-DNA in pDCs, appears to have a net tolerogenic activity in experimental SLE. Interestingly, the mechanism of sensing in some cases may resemble the recognition of virus-infected cells, e.g., the recognition of microRNA transferred via exosomes (Salvi et al., 2018). Another question is whether pDCs act primarily through IFN-I secretion (local or systemic), the production of other soluble mediators such as IL-6 (Jego et al., 2003), or other functions such as antigen presentation. Finally, pDCs may act directly on the differentiation or maintenance of autoreactive B cells and/or promote autoreactivity indirectly through T cells or other cell types; the former possibility is supported by studies in human cells in vitro (Jego et al., 2003, Menon et al., 2016) and remains to be confirmed and analyzed in vivo.
The Role of Impaired pDC Activity in Immunodeficiency and Cancer
Mirroring pDC hyperactivation in autoimmunity, impaired pDC activity has been implicated in immunodeficient states or inefficient immune responses, e.g., to tumors. pDCs in neonates have a significantly reduced capacity to produce IFN-I in response to viruses such as RSV (Cormier et al., 2014, Zhang et al., 2014) and rhinovirus (Barlow-Anacker et al., 2017). This reduced pDC function may contribute to increased susceptibility to infections in neonates and in particular may underline the ability of RSV to promote lung inflammation and asthma later in life (Lynch et al., 2018). On the other end of the age spectrum, pDCs from aged mice showed a significant impairment of IFN-I production in vitro and during viral infection in vivo, potentially due to increased oxidative stress (Stout-Delgado et al., 2008); a similarly impaired function was observed in pDCs from elderly humans (Jing et al., 2009). Together with pDC impairment by persistent viruses as discussed above, the likely cell-extrinsic effect of aging on pDCs may contribute to impaired responses to infections and vaccines in the elderly.
Finally, studies in cancer patients showed a profoundly aberrant and/or hypofunctional state of tumor-infiltrating pDCs: in particular, pDCs in breast and ovarian tumors produce IFN-I poorly yet have an increased capacity to induce Treg cell differentiation (Conrad et al., 2012, Labidi-Galy et al., 2011, Sisirak et al., 2012). This aberrant function may be imparted by tumor-derived soluble factors such as TGF-β (Sisirak et al., 2013) and is thought to promote tumor growth as suggested by animal models (Le Mercier et al., 2013). The role of TGF-β and the tumor-promoting properties of pDCs have been recently supported by genetic pDC depletion in mice (Terra et al., 2018). Although activated pDCs can be administered to elicit anti-tumor T cell responses in vivo (Liu et al., 2008, Tel et al., 2013), the data so far suggest a net tumor-promoting effect of endogenous pDCs that manifest impaired IFN-I production and immunosuppressive properties. The molecular basis of pDC reprogramming in the tumor environment remains to be fully elucidated and may represent a target for immunotherapy.
Concluding Remarks
Based on the knowledge of pDC biology accumulated over the last two decades, it should now be possible to address major outstanding issues, such as the molecular basis of IFN-I-producing capacity of pDCs, the nature and consequences of pathogen recognition, and the mechanisms whereby pDCs contribute to protective and pathogenic immune responses. Moreover, the field seems to be rapidly crossing the “bottleneck” toward translation of the basic knowledge into clinically relevant applications. For example, the study of pDC development helped elucidate a highly aggressive leukemia type, the blastic plasmacytoid dendritic cell neoplasm (BPDCN), from its initial identification as the leukemic counterpart of pDCs (Chaperot et al., 2001) to better diagnostic tools and potential therapeutic approaches targeting pDC-specific transcriptional machinery (Ceribelli et al., 2016). Furthermore, the analysis of pDC function led to the development of antibodies that deplete pDCs or inhibit their function (Table 1). Further dissection of pDC regulation should facilitate the development of non-depleting reagents to modulate pDC function in the long term, e.g., dampen pDC hyperactivation in autoimmunity or reverse their dysfunction in cancer. The observations that even a partial functional modulation (Hansen et al., 2015, Sisirak et al., 2014) or a transient ablation (Ah Kioon et al., 2018, Rowland et al., 2014) of pDCs have a dramatic effect on chronic disease bode well for the utility of this cell type as a therapeutic target.
Acknowledgments
Supported by the NIH grants AI072571 and AI128949, the Lupus Research Alliance, and the Colton Center for Autoimmunity.
References
- Ah Kioon M.D., Tripodo C., Fernandez D., Kirou K.A., Spiera R.F., Crow M.K., Gordon J.K., Barrat F.J. Plasmacytoid dendritic cells promote systemic sclerosis with a key role for TLR8. Sci. Transl. Med. 2018;10:10. doi: 10.1126/scitranslmed.aam8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello A., Giannessi F., Percario Z.A., Affabris E. The involvement of plasmacytoid cells in HIV infection and pathogenesis. Cytokine Growth Factor Rev. 2018;40:77–89. doi: 10.1016/j.cytogfr.2018.03.009. [DOI] [PubMed] [Google Scholar]
- Alcántara-Hernández M., Leylek R., Wagar L.E., Engleman E.G., Keler T., Marinkovich M.P., Davis M.M., Nolan G.P., Idoyaga J. High-Dimensional Phenotypic Mapping of Human Dendritic Cells Reveals Interindividual Variation and Tissue Specialization. Immunity. 2017;47:1037–1050.e6. doi: 10.1016/j.immuni.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alculumbre S.G., Saint-André V., Di Domizio J., Vargas P., Sirven P., Bost P., Maurin M., Maiuri P., Wery M., Roman M.S. Diversification of human plasmacytoid predendritic cells in response to a single stimulus. Nat. Immunol. 2018;19:63–75. doi: 10.1038/s41590-017-0012-z. [DOI] [PubMed] [Google Scholar]
- Allen J.S., Pang K., Skowera A., Ellis R., Rackham C., Lozanoska-Ochser B., Tree T., Leslie R.D., Tremble J.M., Dayan C.M., Peakman M. Plasmacytoid dendritic cells are proportionally expanded at diagnosis of type 1 diabetes and enhance islet autoantigen presentation to T-cells through immune complex capture. Diabetes. 2009;58:138–145. doi: 10.2337/db08-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allman D., Dalod M., Asselin-Paturel C., Delale T., Robbins S.H., Trinchieri G., Biron C.A., Kastner P., Chan S. Ikaros is required for plasmacytoid dendritic cell differentiation. Blood. 2006;108:4025–4034. doi: 10.1182/blood-2006-03-007757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouar B., Kovarova D., Letard S., Font-Haro A., Florentin J., Weber J., Durantel D., Chaperot L., Plumas J., Trejbalova K. Dual Role of the Tyrosine Kinase Syk in Regulation of Toll-Like Receptor Signaling in Plasmacytoid Dendritic Cells. PLoS ONE. 2016;11:e0156063. doi: 10.1371/journal.pone.0156063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselin-Paturel C., Boonstra A., Dalod M., Durand I., Yessaad N., Dezutter-Dambuyant C., Vicari A., O’Garra A., Biron C., Brière F., Trinchieri G. Mouse type I IFN-producing cells are immature APCs with plasmacytoid morphology. Nat. Immunol. 2001;2:1144–1150. doi: 10.1038/ni736. [DOI] [PubMed] [Google Scholar]
- Asselin-Paturel C., Brizard G., Pin J.J., Brière F., Trinchieri G. Mouse strain differences in plasmacytoid dendritic cell frequency and function revealed by a novel monoclonal antibody. J. Immunol. 2003;171:6466–6477. doi: 10.4049/jimmunol.171.12.6466. [DOI] [PubMed] [Google Scholar]
- Asselin-Paturel C., Brizard G., Chemin K., Boonstra A., O’Garra A., Vicari A., Trinchieri G. Type I interferon dependence of plasmacytoid dendritic cell activation and migration. J. Exp. Med. 2005;201:1157–1167. doi: 10.1084/jem.20041930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assil S., Coléon S., Décembre E., Sherry L., Allatif O., Webster B., Dreux M. Antiviral Response by Plasmacytoid Dendritic Cells via Interferogenic Synapse with Infected Cells. bioRxiv. 2018 doi: 10.1101/374496. [DOI] [PubMed] [Google Scholar]
- Baglio S.R., van Eijndhoven M.A., Koppers-Lalic D., Berenguer J., Lougheed S.M., Gibbs S., Léveillé N., Rinkel R.N., Hopmans E.S., Swaminathan S. Sensing of latent EBV infection through exosomal transfer of 5'pppRNA. Proc. Natl. Acad. Sci. USA. 2016;113:E587–E596. doi: 10.1073/pnas.1518130113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao M., Wang Y., Liu Y., Shi P., Lu H., Sha W., Weng L., Hanabuchi S., Qin J., Plumas J. NFATC3 promotes IRF7 transcriptional activity in plasmacy--toid dendritic cells. J. Exp. Med. 2016;213:2383–2398. doi: 10.1084/jem.20160438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-On L., Birnberg T., Lewis K.L., Edelson B.T., Bruder D., Hildner K., Buer J., Murphy K.M., Reizis B., Jung S. CX3CR1+ CD8alpha+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. USA. 2010;107:14745–14750. doi: 10.1073/pnas.1001562107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barchet W., Cella M., Odermatt B., Asselin-Paturel C., Colonna M., Kalinke U. Virus-induced interferon alpha production by a dendritic cell subset in the absence of feedback signaling in vivo. J. Exp. Med. 2002;195:507–516. doi: 10.1084/jem.20011666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow-Anacker A., Bochkov Y., Gern J., Seroogy C.M. Neonatal immune response to rhinovirus A16 has diminished dendritic cell function and increased B cell activation. PLoS ONE. 2017;12:e0180664. doi: 10.1371/journal.pone.0180664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J., Dress R.J., Schulze A., Dresing P., Ali S., Deenen R., Alferink J., Scheu S. Cutting Edge: IFN-β Expression in the Spleen Is Restricted to a Subpopulation of Plasmacytoid Dendritic Cells Exhibiting a Specific Immune Modulatory Transcriptome Signature. J. Immunol. 2016;196:4447–4451. doi: 10.4049/jimmunol.1500383. [DOI] [PubMed] [Google Scholar]
- Blasius A., Vermi W., Krug A., Facchetti F., Cella M., Colonna M. A cell-surface molecule selectively expressed on murine natural interferon-producing cells that blocks secretion of interferon-alpha. Blood. 2004;103:4201–4206. doi: 10.1182/blood-2003-09-3108. [DOI] [PubMed] [Google Scholar]
- Blasius A.L., Cella M., Maldonado J., Takai T., Colonna M. Siglec-H is an IPC-specific receptor that modulates type I IFN secretion through DAP12. Blood. 2006;107:2474–2476. doi: 10.1182/blood-2005-09-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius A.L., Giurisato E., Cella M., Schreiber R.D., Shaw A.S., Colonna M. Bone marrow stromal cell antigen 2 is a specific marker of type I IFN-producing cells in the naive mouse, but a promiscuous cell surface antigen following IFN stimulation. J. Immunol. 2006;177:3260–3265. doi: 10.4049/jimmunol.177.5.3260. [DOI] [PubMed] [Google Scholar]
- Blasius A.L., Arnold C.N., Georgel P., Rutschmann S., Xia Y., Lin P., Ross C., Li X., Smart N.G., Beutler B. Slc15a4, AP-3, and Hermansky-Pudlak syndrome proteins are required for Toll-like receptor signaling in plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. USA. 2010;107:19973–19978. doi: 10.1073/pnas.1014051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewitz A., Eickhoff S., Dähling S., Quast T., Bedoui S., Kroczek R.A., Kurts C., Garbi N., Barchet W., Iannacone M. CD8+ T Cells Orchestrate pDC-XCR1+ Dendritic Cell Spatial and Functional Cooperativity to Optimize Priming. Immunity. 2017;46:205–219. doi: 10.1016/j.immuni.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruni D., Chazal M., Sinigaglia L., Chauveau L., Schwartz O., Desprès P., Jouvenet N. Viral entry route determines how human plasmacytoid dendritic cells produce type I interferons. Sci. Signal. 2015;8:ra25. doi: 10.1126/scisignal.aaa1552. [DOI] [PubMed] [Google Scholar]
- Bunin A., Sisirak V., Ghosh H.S., Grajkowska L.T., Hou Z.E., Miron M., Yang C., Ceribelli M., Uetani N., Chaperot L. Protein Tyrosine Phosphatase PTPRS Is an Inhibitory Receptor on Human and Murine Plasmacytoid Dendritic Cells. Immunity. 2015;43:277–288. doi: 10.1016/j.immuni.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caielli S., Athale S., Domic B., Murat E., Chandra M., Banchereau R., Baisch J., Phelps K., Clayton S., Gong M. Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J. Exp. Med. 2016;213:697–713. doi: 10.1084/jem.20151876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W., Manicassamy S., Tang H., Kasturi S.P., Pirani A., Murthy N., Pulendran B. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat. Immunol. 2008;9:1157–1164. doi: 10.1038/ni.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W., Bover L., Cho M., Wen X., Hanabuchi S., Bao M., Rosen D.B., Wang Y.H., Shaw J.L., Du Q. Regulation of TLR7/9 responses in plasmacytoid dendritic cells by BST2 and ILT7 receptor interaction. J. Exp. Med. 2009;206:1603–1614. doi: 10.1084/jem.20090547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotta S., Dakic A., D’Amico A., Pang S.H., Greig K.T., Nutt S.L., Wu L. The transcription factor PU.1 controls dendritic cell development and Flt3 cytokine receptor expression in a dose-dependent manner. Immunity. 2010;32:628–641. doi: 10.1016/j.immuni.2010.05.005. [DOI] [PubMed] [Google Scholar]
- Cella M., Jarrossay D., Facchetti F., Alebardi O., Nakajima H., Lanzavecchia A., Colonna M. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat. Med. 1999;5:919–923. doi: 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- Ceribelli M., Hou Z.E., Kelly P.N., Huang D.W., Wright G., Ganapathi K., Evbuomwan M.O., Pittaluga S., Shaffer A.L., Marcucci G. A Druggable TCF4- and BRD4-Dependent Transcriptional Network Sustains Malignancy in Blastic Plasmacytoid Dendritic Cell Neoplasm. Cancer Cell. 2016;30:764–778. doi: 10.1016/j.ccell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L., Züst R., Weber F., Spiegel M., Lang K.S., Akira S., Thiel V., Ludewig B. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood. 2007;109:1131–1137. doi: 10.1182/blood-2006-05-023770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragán L., Kalinke U., Züst R., König M., Reizis B., López-Macías C., Thiel V., Ludewig B. Type I IFN-mediated protection of macrophages and dendritic cells secures control of murine coronavirus infection. J. Immunol. 2009;182:1099–1106. doi: 10.4049/jimmunol.182.2.1099. [DOI] [PubMed] [Google Scholar]
- Cervantes-Barragan L., Lewis K.L., Firner S., Thiel V., Hugues S., Reith W., Ludewig B., Reizis B. Plasmacytoid dendritic cells control T-cell response to chronic viral infection. Proc. Natl. Acad. Sci. USA. 2012;109:3012–3017. doi: 10.1073/pnas.1117359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaperot L., Bendriss N., Manches O., Gressin R., Maynadie M., Trimoreau F., Orfeuvre H., Corront B., Feuillard J., Sotto J.J. Identification of a leukemic counterpart of the plasmacytoid dendritic cells. Blood. 2001;97:3210–3217. doi: 10.1182/blood.v97.10.3210. [DOI] [PubMed] [Google Scholar]
- Chopin M., Preston S.P., Lun A.T.L., Tellier J., Smyth G.K., Pellegrini M., Belz G.T., Corcoran L.M., Visvader J.E., Wu L., Nutt S.L. RUNX2 Mediates Plasmacytoid Dendritic Cell Egress from the Bone Marrow and Controls Viral Immunity. Cell Rep. 2016;15:866–878. doi: 10.1016/j.celrep.2016.03.066. [DOI] [PubMed] [Google Scholar]
- Cisse B., Caton M.L., Lehner M., Maeda T., Scheu S., Locksley R., Holmberg D., Zweier C., den Hollander N.S., Kant S.G. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. doi: 10.1016/j.cell.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M., Trinchieri G., Liu Y.J. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 2004;5:1219–1226. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- Conrad C., Gregorio J., Wang Y.H., Ito T., Meller S., Hanabuchi S., Anderson S., Atkinson N., Ramirez P.T., Liu Y.J. Plasmacytoid dendritic cells promote immunosuppression in ovarian cancer via ICOS costimulation of Foxp3(+) T-regulatory cells. Cancer Res. 2012;72:5240–5249. doi: 10.1158/0008-5472.CAN-12-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier S.A., Shrestha B., Saravia J., Lee G.I., Shen L., DeVincenzo J.P., Kim Y.I., You D. Limited type I interferons and plasmacytoid dendritic cells during neonatal respiratory syncytial virus infection permit immunopathogenesis upon reinfection. J. Virol. 2014;88:9350–9360. doi: 10.1128/JVI.00818-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crother T.R., Ma J., Jupelli M., Chiba N., Chen S., Slepenkin A., Alsabeh R., Peterson E., Shimada K., Arditi M. Plasmacytoid dendritic cells play a role for effective innate immune responses during Chlamydia pneumoniae infection in mice. PLoS ONE. 2012;7:e48655. doi: 10.1371/journal.pone.0048655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat K., Guiton R., Guilliams M., Henri S., Baranek T., Schwartz-Cornil I., Malissen B., Dalod M. Comparative genomics as a tool to reveal functional equivalences between human and mouse dendritic cell subsets. Immunol. Rev. 2010;234:177–198. doi: 10.1111/j.0105-2896.2009.00868.x. [DOI] [PubMed] [Google Scholar]
- Dai P., Cao H., Merghoub T., Avogadri F., Wang W., Parikh T., Fang C.M., Pitha P.M., Fitzgerald K.A., Rahman M.M. Myxoma virus induces type I interferon production in murine plasmacytoid dendritic cells via a TLR9/MyD88-, IRF5/IRF7-, and IFNAR-dependent pathway. J. Virol. 2011;85:10814–10825. doi: 10.1128/JVI.00104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallari S., Macal M., Loureiro M.E., Jo Y., Swanson L., Hesser C., Ghosh P., Zuniga E.I. Src family kinases Fyn and Lyn are constitutively activated and mediate plasmacytoid dendritic cell responses. Nat. Commun. 2017;8:14830. doi: 10.1038/ncomms14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalod M., Salazar-Mather T.P., Malmgaard L., Lewis C., Asselin-Paturel C., Brière F., Trinchieri G., Biron C.A. Interferon alpha/beta and interleukin 12 responses to viral infections: pathways regulating dendritic cell cytokine expression in vivo. J. Exp. Med. 2002;195:517–528. doi: 10.1084/jem.20011672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S., Erturk-Hasdemir D., Ochoa-Reparaz J., Reinecker H.C., Kasper D.L. Plasmacytoid dendritic cells mediate anti-inflammatory responses to a gut commensal molecule via both innate and adaptive mechanisms. Cell Host Microbe. 2014;15:413–423. doi: 10.1016/j.chom.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson S., Kaiko G., Loh Z., Lalwani A., Zhang V., Spann K., Foo S.Y., Hansbro N., Uematsu S., Akira S. Plasmacytoid dendritic cells promote host defense against acute pneumovirus infection via the TLR7-MyD88-dependent signaling pathway. J. Immunol. 2011;186:5938–5948. doi: 10.4049/jimmunol.1002635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison L.M., Jørgensen T.N. Sialic acid-binding immunoglobulin-type lectin H-positive plasmacytoid dendritic cells drive spontaneous lupus-like disease development in B6.Nba2 mice. Arthritis Rheumatol. 2015;67:1012–1022. doi: 10.1002/art.38989. [DOI] [PubMed] [Google Scholar]
- Deal E.M., Jaimes M.C., Crawford S.E., Estes M.K., Greenberg H.B. Rotavirus structural proteins and dsRNA are required for the human primary plasmacytoid dendritic cell IFNalpha response. PLoS Pathog. 2010;6:e1000931. doi: 10.1371/journal.ppat.1000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Décembre E., Assil S., Hillaire M.L., Dejnirattisai W., Mongkolsapaya J., Screaton G.R., Davidson A.D., Dreux M. Sensing of immature particles produced by dengue virus infected cells induces an antiviral response by plasmacytoid dendritic cells. PLoS Pathog. 2014;10:e1004434. doi: 10.1371/journal.ppat.1004434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J.D., Rhee C., Hu Z., Lee B.K., Lee J., Iyer V.R., Ehrlich L.I.R., Georgiou G., Tucker H.O., Ippolito G.C. Lymphoid origin of a lineage of intrinsically activated plasmacytoid dendritic cell in mice and humans. bioRxiv. 2018 doi: 10.1101/310680. [DOI] [Google Scholar]
- Diana J., Simoni Y., Furio L., Beaudoin L., Agerberth B., Barrat F., Lehuen A. Crosstalk between neutrophils, B-1a cells and plasmacytoid dendritic cells initiates autoimmune diabetes. Nat. Med. 2013;19:65–73. doi: 10.1038/nm.3042. [DOI] [PubMed] [Google Scholar]
- Döring M., Lessin I., Frenz T., Spanier J., Kessler A., Tegtmeyer P., Dağ F., Thiel N., Trilling M., Lienenklaus S. M27 expressed by cytomegalovirus counteracts effective type I interferon induction of myeloid cells but not of plasmacytoid dendritic cells. J. Virol. 2014;88:13638–13650. doi: 10.1128/JVI.00216-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreux M., Garaigorta U., Boyd B., Décembre E., Chung J., Whitten-Bauer C., Wieland S., Chisari F.V. Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe. 2012;12:558–570. doi: 10.1016/j.chom.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dursun E., Endele M., Musumeci A., Failmezger H., Wang S.H., Tresch A., Schroeder T., Krug A.B. Continuous single cell imaging reveals sequential steps of plasmacytoid dendritic cell development from common dendritic cell progenitors. Sci. Rep. 2016;6:37462. doi: 10.1038/srep37462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloranta M.L., Franck-Larsson K., Lövgren T., Kalamajski S., Rönnblom A., Rubin K., Alm G.V., Rönnblom L. Type I interferon system activation and association with disease manifestations in systemic sclerosis. Ann. Rheum. Dis. 2010;69:1396–1402. doi: 10.1136/ard.2009.121400. [DOI] [PubMed] [Google Scholar]
- Esashi E., Bao M., Wang Y.H., Cao W., Liu Y.J. PACSIN1 regulates the TLR7/9-mediated type I interferon response in plasmacytoid dendritic cells. Eur. J. Immunol. 2012;42:573–579. doi: 10.1002/eji.201142045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Li Y., McKnight K.L., Hensley L., Lanford R.E., Walker C.M., Lemon S.M. Human pDCs preferentially sense enveloped hepatitis A virions. J. Clin. Invest. 2015;125:169–176. doi: 10.1172/JCI77527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenz T., Graalmann L., Detje C.N., Döring M., Grabski E., Scheu S., Kalinke U. Independent of plasmacytoid dendritic cell (pDC) infection, pDC triggered by virus-infected cells mount enhanced type I IFN responses of different composition as opposed to pDC stimulated with free virus. J. Immunol. 2014;193:2496–2503. doi: 10.4049/jimmunol.1400215. [DOI] [PubMed] [Google Scholar]
- García-Nicolás O., Auray G., Sautter C.A., Rappe J.C., McCullough K.C., Ruggli N., Summerfield A. Sensing of Porcine Reproductive and Respiratory Syndrome Virus-Infected Macrophages by Plasmacytoid Dendritic Cells. Front. Microbiol. 2016;7:771. doi: 10.3389/fmicb.2016.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh H.S., Cisse B., Bunin A., Lewis K.L., Reizis B. Continuous expression of the transcription factor e2-2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity. 2010;33:905–916. doi: 10.1016/j.immuni.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh H.S., Ceribelli M., Matos I., Lazarovici A., Bussemaker H.J., Lasorella A., Hiebert S.W., Liu K., Staudt L.M., Reizis B. ETO family protein Mtg16 regulates the balance of dendritic cell subsets by repressing Id2. J. Exp. Med. 2014;211:1623–1635. doi: 10.1084/jem.20132121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilliet M., Cao W., Liu Y.J. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 2008;8:594–606. doi: 10.1038/nri2358. [DOI] [PubMed] [Google Scholar]
- Glitzner E., Korosec A., Brunner P.M., Drobits B., Amberg N., Schonthaler H.B., Kopp T., Wagner E.F., Stingl G., Holcmann M., Sibilia M. Specific roles for dendritic cell subsets during initiation and progression of psoriasis. EMBO Mol. Med. 2014;6:1312–1327. doi: 10.15252/emmm.201404114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajkowska L.T., Ceribelli M., Lau C.M., Warren M.E., Tiniakou I., Nakandakari Higa S., Bunin A., Haecker H., Mirny L.A., Staudt L.M., Reizis B. Isoform-Specific Expression and Feedback Regulation of E Protein TCF4 Control Dendritic Cell Lineage Specification. Immunity. 2017;46:65–77. doi: 10.1016/j.immuni.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbeck M., Ziegler S., Laffont S., Smith N., Chauveau L., Tomezsko P., Sharei A., Kourjian G., Porichis F., Hart M. Sex Differences in Plasmacytoid Dendritic Cell Levels of IRF5 Drive Higher IFN-α Production in Women. J. Immunol. 2015;195:5327–5336. doi: 10.4049/jimmunol.1501684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grouard G., Rissoan M.C., Filgueira L., Durand I., Banchereau J., Liu Y.J. The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)-3 and CD40-ligand. J. Exp. Med. 1997;185:1101–1111. doi: 10.1084/jem.185.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiducci C., Ott G., Chan J.H., Damon E., Calacsan C., Matray T., Lee K.D., Coffman R.L., Barrat F.J. Properties regulating the nature of the plasmacytoid dendritic cell response to Toll-like receptor 9 activation. J. Exp. Med. 2006;203:1999–2008. doi: 10.1084/jem.20060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiducci C., Ghirelli C., Marloie-Provost M.A., Matray T., Coffman R.L., Liu Y.J., Barrat F.J., Soumelis V. PI3K is critical for the nuclear translocation of IRF-7 and type I IFN production by human plasmacytoid predendritic cells in response to TLR activation. J. Exp. Med. 2008;205:315–322. doi: 10.1084/jem.20070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillerey C., Mouriès J., Polo G., Doyen N., Law H.K., Chan S., Kastner P., Leclerc C., Dadaglio G. Pivotal role of plasmacytoid dendritic cells in inflammation and NK-cell responses after TLR9 triggering in mice. Blood. 2012;120:90–99. doi: 10.1182/blood-2012-02-410936. [DOI] [PubMed] [Google Scholar]
- Guilliams M., Dutertre C.A., Scott C.L., McGovern N., Sichien D., Chakarov S., Van Gassen S., Chen J., Poidinger M., De Prijck S. Unsupervised High-Dimensional Analysis Aligns Dendritic Cells across Tissues and Species. Immunity. 2016;45:669–684. doi: 10.1016/j.immuni.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L., Schmidt-Christensen A., Gupta S., Fransén-Pettersson N., Hannibal T.D., Reizis B., Santamaria P., Holmberg D. E2-2 Dependent Plasmacytoid Dendritic Cells Control Autoimmune Diabetes. PLoS ONE. 2015;10:e0144090. doi: 10.1371/journal.pone.0144090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman J.S., Sagar, Grün D. FateID infers cell fate bias in multipotent progenitors from single-cell RNA-seq data. Nat. Methods. 2018;15:379–386. doi: 10.1038/nmeth.4662. [DOI] [PubMed] [Google Scholar]
- Honda K., Ohba Y., Yanai H., Negishi H., Mizutani T., Takaoka A., Taya C., Taniguchi T. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434:1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Hornung V., Schlender J., Guenthner-Biller M., Rothenfusser S., Endres S., Conzelmann K.K., Hartmann G. Replication-dependent potent IFN-alpha induction in human plasmacytoid dendritic cells by a single-stranded RNA virus. J. Immunol. 2004;173:5935–5943. doi: 10.4049/jimmunol.173.10.5935. [DOI] [PubMed] [Google Scholar]
- Ippolito G.C., Dekker J.D., Wang Y.H., Lee B.K., Shaffer A.L., 3rd, Lin J., Wall J.K., Lee B.S., Staudt L.M., Liu Y.J. Dendritic cell fate is determined by BCL11A. Proc. Natl. Acad. Sci. USA. 2014;111:E998–E1006. doi: 10.1073/pnas.1319228111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jego G., Palucka A.K., Blanck J.P., Chalouni C., Pascual V., Banchereau J. Plasmacytoid dendritic cells induce plasma cell differentiation through type I interferon and interleukin 6. Immunity. 2003;19:225–234. doi: 10.1016/s1074-7613(03)00208-5. [DOI] [PubMed] [Google Scholar]
- Jing Y., Shaheen E., Drake R.R., Chen N., Gravenstein S., Deng Y. Aging is associated with a numerical and functional decline in plasmacytoid dendritic cells, whereas myeloid dendritic cells are relatively unaltered in human peripheral blood. Hum. Immunol. 2009;70:777–784. doi: 10.1016/j.humimm.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaitani A., Izawa K., Maehara A., Isobe M., Takamori A., Matsukawa T., Takahashi M., Yamanishi Y., Oki T., Yamada H. Leukocyte mono-immunoglobulin-like receptor 8 (LMIR8)/CLM-6 is an FcRγ-coupled receptor selectively expressed in mouse tissue plasmacytoid dendritic cells. Sci. Rep. 2018;8:8259. doi: 10.1038/s41598-018-25646-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky L.W., Sei J.J., Parekh N.J., Davies M.L., Reider I.E., Krouse T.E., Norbury C.C. Redundant Function of Plasmacytoid and Conventional Dendritic Cells Is Required To Survive a Natural Virus Infection. J. Virol. 2015;89:9974–9985. doi: 10.1128/JVI.01024-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Peck A., Santer D., Patole P., Schwartz S.M., Molitor J.A., Arnett F.C., Elkon K.B. Induction of interferon-alpha by scleroderma sera containing autoantibodies to topoisomerase I: association of higher interferon-alpha activity with lung fibrosis. Arthritis Rheum. 2008;58:2163–2173. doi: 10.1002/art.23486. [DOI] [PubMed] [Google Scholar]
- Kim S., Kaiser V., Beier E., Bechheim M., Guenthner-Biller M., Ablasser A., Berger M., Endres S., Hartmann G., Hornung V. Self-priming determines high type I IFN production by plasmacytoid dendritic cells. Eur. J. Immunol. 2014;44:807–818. doi: 10.1002/eji.201343806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.W., Hong S., Lin Y., Murat E., Joo H., Kim T., Pascual V., Liu Y.J. Transcriptional Repression of IFN Regulatory Factor 7 by MYC Is Critical for Type I IFN Production in Human Plasmacytoid Dendritic Cells. J. Immunol. 2016;197:3348–3359. doi: 10.4049/jimmunol.1502385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.W., Budzak J., Liu Y., Jégouzo S.A.F., Drickamer K., Taylor M.E. Identification of serum glycoprotein ligands for the immunomodulatory receptor blood dendritic cell antigen 2. Glycobiology. 2018;28:592–600. doi: 10.1093/glycob/cwy050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug A., French A.R., Barchet W., Fischer J.A., Dzionek A., Pingel J.T., Orihuela M.M., Akira S., Yokoyama W.M., Colonna M. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity. 2004;21:107–119. doi: 10.1016/j.immuni.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Kumagai Y., Takeuchi O., Kato H., Kumar H., Matsui K., Morii E., Aozasa K., Kawai T., Akira S. Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity. 2007;27:240–252. doi: 10.1016/j.immuni.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Kumagai Y., Kumar H., Koyama S., Kawai T., Takeuchi O., Akira S. Cutting Edge: TLR-Dependent viral recognition along with type I IFN positive feedback signaling masks the requirement of viral replication for IFN-alpha production in plasmacytoid dendritic cells. J. Immunol. 2009;182:3960–3964. doi: 10.4049/jimmunol.0804315. [DOI] [PubMed] [Google Scholar]
- Labidi-Galy S.I., Sisirak V., Meeus P., Gobert M., Treilleux I., Bajard A., Combes J.D., Faget J., Mithieux F., Cassignol A. Quantitative and functional alterations of plasmacytoid dendritic cells contribute to immune tolerance in ovarian cancer. Cancer Res. 2011;71:5423–5434. doi: 10.1158/0008-5472.CAN-11-0367. [DOI] [PubMed] [Google Scholar]
- Laffont S., Rouquié N., Azar P., Seillet C., Plumas J., Aspord C., Guéry J.C. X-Chromosome complement and estrogen receptor signaling independently contribute to the enhanced TLR7-mediated IFN-α production of plasmacytoid dendritic cells from women. J. Immunol. 2014;193:5444–5452. doi: 10.4049/jimmunol.1303400. [DOI] [PubMed] [Google Scholar]
- Lau C.M., Nish S.A., Yogev N., Waisman A., Reiner S.L., Reizis B. Leukemia-associated activating mutation of Flt3 expands dendritic cells and alters T cell responses. J. Exp. Med. 2016;213:415–431. doi: 10.1084/jem.20150642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C.M., Tiniakou I., Perez O.A., Kirkling M.E., Yap G.S., Hock H., Reizis B. Transcription factor Etv6 regulates functional differentiation of cross-presenting classical dendritic cells. J. Exp. Med. 2018;215:2265–2278. doi: 10.1084/jem.20172323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laustsen A., Bak R.O., Krapp C., Kjær L., Egedahl J.H., Petersen C.C., Pillai S., Tang H.Q., Uldbjerg N., Porteus M. Interferon priming is essential for human CD34+ cell-derived plasmacytoid dendritic cell maturation and function. Nat. Commun. 2018;9:3525. doi: 10.1038/s41467-018-05816-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Mercier I., Poujol D., Sanlaville A., Sisirak V., Gobert M., Durand I., Dubois B., Treilleux I., Marvel J., Vlach J. Tumor promotion by intratumoral plasmacytoid dendritic cells is reversed by TLR7 ligand treatment. Cancer Res. 2013;73:4629–4640. doi: 10.1158/0008-5472.CAN-12-3058. [DOI] [PubMed] [Google Scholar]
- Lee H.K., Lund J.M., Ramanathan B., Mizushima N., Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- Lee L.N., Burke S., Montoya M., Borrow P. Multiple mechanisms contribute to impairment of type 1 interferon production during chronic lymphocytic choriomeningitis virus infection of mice. J. Immunol. 2009;182:7178–7189. doi: 10.4049/jimmunol.0802526. [DOI] [PubMed] [Google Scholar]
- Lee J., Zhou Y.J., Ma W., Zhang W., Aljoufi A., Luh T., Lucero K., Liang D., Thomsen M., Bhagat G. Lineage specification of human dendritic cells is marked by IRF8 expression in hematopoietic stem cells and multipotent progenitors. Nat. Immunol. 2017;18:877–888. doi: 10.1038/ni.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepelley A., Louis S., Sourisseau M., Law H.K., Pothlichet J., Schilte C., Chaperot L., Plumas J., Randall R.E., Si-Tahar M. Innate sensing of HIV-infected cells. PLoS Pathog. 2011;7:e1001284. doi: 10.1371/journal.ppat.1001284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.D., Wu J., Gao D., Wang H., Sun L., Chen Z.J. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D.S., Kan A., Gao J., Crampin E.J., Hodgkin P.D., Naik S.H. DiSNE Movie Visualization and Assessment of Clonal Kinetics Reveal Multiple Trajectories of Dendritic Cell Development. Cell Rep. 2018;22:2557–2566. doi: 10.1016/j.celrep.2018.02.046. [DOI] [PubMed] [Google Scholar]
- Liu Y.J. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 2005;23:275–306. doi: 10.1146/annurev.immunol.23.021704.115633. [DOI] [PubMed] [Google Scholar]
- Liu C., Lou Y., Lizée G., Qin H., Liu S., Rabinovich B., Kim G.J., Wang Y.H., Ye Y., Sikora A.G. Plasmacytoid dendritic cells induce NK cell-dependent, tumor antigen-specific T cell cross-priming and tumor regression in mice. J. Clin. Invest. 2008;118:1165–1175. doi: 10.1172/JCI33583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lood C., Blanco L.P., Purmalek M.M., Carmona-Rivera C., De Ravin S.S., Smith C.K., Malech H.L., Ledbetter J.A., Elkon K.B., Kaplan M.J. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat. Med. 2016;22:146–153. doi: 10.1038/nm.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loschko J., Rieke G.J., Schreiber H.A., Meredith M.M., Yao K.H., Guermonprez P., Nussenzweig M.C. Inducible targeting of cDCs and their subsets in vivo. J. Immunol. Methods. 2016;434:32–38. doi: 10.1016/j.jim.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund J., Sato A., Akira S., Medzhitov R., Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J.P., Werder R.B., Loh Z., Sikder M.A.A., Curren B., Zhang V., Rogers M.J., Lane K., Simpson J., Mazzone S.B. Plasmacytoid dendritic cells protect from viral bronchiolitis and asthma through semaphorin 4a-mediated T reg expansion. J. Exp. Med. 2018;215:537–557. doi: 10.1084/jem.20170298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Wan X., Deng Z., Shi L., Hao C., Zhou Z., Zhou C., Fang Y., Liu J., Yang J. Epigenetic regulator CXXC5 recruits DNA demethylase Tet2 to regulate TLR7/9-elicited IFN response in pDCs. J. Exp. Med. 2017;214:1471–1491. doi: 10.1084/jem.20161149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macal M., Lewis G.M., Kunz S., Flavell R., Harker J.A., Zúñiga E.I. Plasmacytoid dendritic cells are productively infected and activated through TLR-7 early after arenavirus infection. Cell Host Microbe. 2012;11:617–630. doi: 10.1016/j.chom.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macal M., Tam M.A., Hesser C., Di Domizio J., Leger P., Gilliet M., Zuniga E.I. CD28 Deficiency Enhances Type I IFN Production by Murine Plasmacytoid Dendritic Cells. J. Immunol. 2016;196:1900–1909. doi: 10.4049/jimmunol.1501658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macal M., Jo Y., Dallari S., Chang A.Y., Dai J., Swaminathan S., Wehrens E.J., Fitzgerald-Bocarsly P., Zúñiga E.I. Self-Renewal and Toll-like Receptor Signaling Sustain Exhausted Plasmacytoid Dendritic Cells during Chronic Viral Infection. Immunity. 2018;48:730–744.e5. doi: 10.1016/j.immuni.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuse M.J., Briggs C.M., Parks G.D. Replication-independent activation of human plasmacytoid dendritic cells by the paramyxovirus SV5 Requires TLR7 and autophagy pathways. Virology. 2010;405:383–389. doi: 10.1016/j.virol.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Connolly J.E., Michnevitz M., Chaussabel D., Yu C.I., Glaser C., Tindle S., Pypaert M., Freitas H., Piqueras B. CD2 distinguishes two subsets of human plasmacytoid dendritic cells with distinct phenotype and functions. J. Immunol. 2009;182:6815–6823. doi: 10.4049/jimmunol.0802008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megjugorac N.J., Jacobs E.S., Izaguirre A.G., George T.C., Gupta G., Fitzgerald-Bocarsly P. Image-based study of interferongenic interactions between plasmacytoid dendritic cells and HSV-infected monocyte-derived dendritic cells. Immunol. Invest. 2007;36:739–761. doi: 10.1080/08820130701715845. [DOI] [PubMed] [Google Scholar]
- Menon M., Blair P.A., Isenberg D.A., Mauri C. A Regulatory Feedback between Plasmacytoid Dendritic Cells and Regulatory B Cells Is Aberrant in Systemic Lupus Erythematosus. Immunity. 2016;44:683–697. doi: 10.1016/j.immuni.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa M., Schmidlin H., Hazekamp M.G., Schotte R., Blom B. Development of human plasmacytoid dendritic cells depends on the combined action of the basic helix-loop-helix factor E2-2 and the Ets factor Spi-B. Eur. J. Immunol. 2008;38:2389–2400. doi: 10.1002/eji.200838470. [DOI] [PubMed] [Google Scholar]
- Naik S.H., Sathe P., Park H.Y., Metcalf D., Proietto A.I., Dakic A., Carotta S., O’Keeffe M., Bahlo M., Papenfuss A. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- Naik S.H., Perié L., Swart E., Gerlach C., van Rooij N., de Boer R.J., Schumacher T.N. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496:229–232. doi: 10.1038/nature12013. [DOI] [PubMed] [Google Scholar]
- Nehmar R., Alsaleh G., Voisin B., Flacher V., Mariotte A., Saferding V., Puchner A., Niederreiter B., Vandamme T., Schabbauer G. Therapeutic Modulation of Plasmacytoid Dendritic Cells in Experimental Arthritis. Arthritis Rheumatol. 2017;69:2124–2135. doi: 10.1002/art.40225. [DOI] [PubMed] [Google Scholar]
- O’Keeffe M., Hochrein H., Vremec D., Caminschi I., Miller J.L., Anders E.M., Wu L., Lahoud M.H., Henri S., Scott B. Mouse plasmacytoid cells: long-lived cells, heterogeneous in surface phenotype and function, that differentiate into CD8(+) dendritic cells only after microbial stimulus. J. Exp. Med. 2002;196:1307–1319. doi: 10.1084/jem.20021031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onai N., Obata-Onai A., Schmid M.A., Ohteki T., Jarrossay D., Manz M.G. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 2007;8:1207–1216. doi: 10.1038/ni1518. [DOI] [PubMed] [Google Scholar]
- Onai N., Kurabayashi K., Hosoi-Amaike M., Toyama-Sorimachi N., Matsushima K., Inaba K., Ohteki T. A clonogenic progenitor with prominent plasmacytoid dendritic cell developmental potential. Immunity. 2013;38:943–957. doi: 10.1016/j.immuni.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Oon S., Huynh H., Tai T.Y., Ng M., Monaghan K., Biondo M., Vairo G., Maraskovsky E., Nash A.D., Wicks I.P., Wilson N.J. A cytotoxic anti-IL-3Rα antibody targets key cells and cytokines implicated in systemic lupus erythematosus. JCI Insight. 2016;1:e86131. doi: 10.1172/jci.insight.86131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S.K., Kolbeck R., Sanjuan M.A. Plasmacytoid dendritic cells in autoimmunity. Curr. Opin. Immunol. 2017;44:20–25. doi: 10.1016/j.coi.2016.10.006. [DOI] [PubMed] [Google Scholar]
- Panda S.K., Facchinetti V., Voynova E., Hanabuchi S., Karnell J.L., Hanna R.N., Kolbeck R., Sanjuan M.A., Ettinger R., Liu Y.J. Galectin-9 inhibits TLR7-mediated autoimmunity in murine lupus models. J. Clin. Invest. 2018;128:1873–1887. doi: 10.1172/JCI97333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin A., Otero K., Czerkowicz J.M., Kerns H.M., Shapiro R.I., Ranger A.M., Otipoby K.L., Taylor F.R., Cameron T.O., Viney J.L., Rabah D. Anti-BDCA2 monoclonal antibody inhibits plasmacytoid dendritic cell activation through Fc-dependent and Fc-independent mechanisms. EMBO Mol. Med. 2015;7:464–476. doi: 10.15252/emmm.201404719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttur F., Arnold-Schrauf C., Lahl K., Solmaz G., Lindenberg M., Mayer C.T., Gohmert M., Swallow M., van Helt C., Schmitt H. Absence of Siglec-H in MCMV infection elevates interferon alpha production but does not enhance viral clearance. PLoS Pathog. 2013;9:e1003648. doi: 10.1371/journal.ppat.1003648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttur F., Francozo M., Solmaz G., Bueno C., Lindenberg M., Gohmert M., Swallow M., Tufa D., Jacobs R., Lienenklaus S. Conventional Dendritic Cells Confer Protection against Mouse Cytomegalovirus Infection via TLR9 and MyD88 Signaling. Cell Rep. 2016;17:1113–1127. doi: 10.1016/j.celrep.2016.09.055. [DOI] [PubMed] [Google Scholar]
- Reizis B., Bunin A., Ghosh H.S., Lewis K.L., Sisirak V. Plasmacytoid dendritic cells: recent progress and open questions. Annu. Rev. Immunol. 2011;29:163–183. doi: 10.1146/annurev-immunol-031210-101345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues P.F., Alberti-Servera L., Eremin A., Grajales-Reyes G.E., Ivanek R., Tussiwand R. Distinct progenitor lineages contribute to the heterogeneity of plasmacytoid dendritic cells. Nat. Immunol. 2018;19:711–722. doi: 10.1038/s41590-018-0136-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G.L., Shirley J.L., Zolotukhin I., Kumar S.R.P., Sherman A., Perrin G.Q., Hoffman B.E., Srivastava A., Basner-Tschakarjan E., Wallet M.A. Plasmacytoid and conventional dendritic cells cooperate in crosspriming AAV capsid-specific CD8+ T cells. Blood. 2017;129:3184–3195. doi: 10.1182/blood-2016-11-751040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnblom L., Alm G.V. A pivotal role for the natural interferon alpha-producing cells (plasmacytoid dendritic cells) in the pathogenesis of lupus. J. Exp. Med. 2001;194:F59–F63. doi: 10.1084/jem.194.12.f59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland S.L., Riggs J.M., Gilfillan S., Bugatti M., Vermi W., Kolbeck R., Unanue E.R., Sanjuan M.A., Colonna M. Early, transient depletion of plasmacytoid dendritic cells ameliorates autoimmunity in a lupus model. J. Exp. Med. 2014;211:1977–1991. doi: 10.1084/jem.20132620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rua R., Lepelley A., Gessain A., Schwartz O. Innate sensing of foamy viruses by human hematopoietic cells. J. Virol. 2012;86:909–918. doi: 10.1128/JVI.06235-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage A.P., Murphy D., Maffia P., Masters L.M., Sabir S.R., Baker L.L., Cambrook H., Finigan A.J., Ait-Oufella H., Grassia G. MHC Class II-restricted antigen presentation by plasmacytoid dendritic cells drives proatherogenic T cell immunity. Circulation. 2014;130:1363–1373. doi: 10.1161/CIRCULATIONAHA.114.011090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S.I., Abe F., Kanno A., Tanimura N., Mori Saitoh Y., Fukui R., Shibata T., Sato K., Ichinohe T., Hayashi M. TLR7 mediated viral recognition results in focal type I interferon secretion by dendritic cells. Nat. Commun. 2017;8:1592. doi: 10.1038/s41467-017-01687-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvi V., Gianello V., Busatto S., Bergese P., Andreoli L., D’Oro U., Zingoni A., Tincani A., Sozzani S., Bosisio D. Exosome-delivered microRNAs promote IFN-α secretion by human plasmacytoid DCs via TLR7. JCI Insight. 2018;3:3. doi: 10.1172/jci.insight.98204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai M., Linehan M.M., Iwasaki A. Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science. 2010;329:1530–1534. doi: 10.1126/science.1187029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki I., Hoshino K., Sugiyama T., Yamazaki C., Yano T., Iizuka A., Hemmi H., Tanaka T., Saito M., Sugiyama M. Spi-B is critical for plasmacytoid dendritic cell function and development. Blood. 2012;120:4733–4743. doi: 10.1182/blood-2012-06-436527. [DOI] [PubMed] [Google Scholar]
- Sathe P., Vremec D., Wu L., Corcoran L., Shortman K. Convergent differentiation: myeloid and lymphoid pathways to murine plasmacytoid dendritic cells. Blood. 2013;121:11–19. doi: 10.1182/blood-2012-02-413336. [DOI] [PubMed] [Google Scholar]
- Sawai C.M., Sisirak V., Ghosh H.S., Hou E.Z., Ceribelli M., Staudt L.M., Reizis B. Transcription factor Runx2 controls the development and migration of plasmacytoid dendritic cells. J. Exp. Med. 2013;210:2151–2159. doi: 10.1084/jem.20130443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai C.M., Babovic S., Upadhaya S., Knapp D.J.H.F., Lavin Y., Lau C.M., Goloborodko A., Feng J., Fujisaki J., Ding L. Hematopoietic Stem Cells Are the Major Source of Multilineage Hematopoiesis in Adult Animals. Immunity. 2016;45:597–609. doi: 10.1016/j.immuni.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawai C.M., Serpas L., Neto A.G., Jang G., Rashidfarrokhi A., Kolbeck R., Sanjuan M.A., Reizis B., Sisirak V. Plasmacytoid dendritic cells are largely dispensable for the pathogenesis of experimental inflammatory bowel disease. Front. Immunol. 2018;9:2475. doi: 10.3389/fimmu.2018.02475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A., Loschko J., Mair K., Vogelmann R., Henkel L., Einwächter H., Schiemann M., Niess J.H., Reindl W., Krug A. Identification of CCR9- murine plasmacytoid DC precursors with plasticity to differentiate into conventional DCs. Blood. 2011;117:6562–6570. doi: 10.1182/blood-2010-12-326678. [DOI] [PubMed] [Google Scholar]
- Schmitt H., Sell S., Koch J., Seefried M., Sonnewald S., Daniel C., Winkler T.H., Nitschke L. Siglec-H protects from virus-triggered severe systemic autoimmunity. J. Exp. Med. 2016;213:1627–1644. doi: 10.1084/jem.20160189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C.L., Soen B., Martens L., Skrypek N., Saelens W., Taminau J., Blancke G., Van Isterdael G., Huylebroeck D., Haigh J. The transcription factor Zeb2 regulates development of conventional and plasmacytoid DCs by repressing Id2. J. Exp. Med. 2016;213:897–911. doi: 10.1084/jem.20151715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See P., Dutertre C.A., Chen J., Günther P., McGovern N., Irac S.E., Gunawan M., Beyer M., Händler K., Duan K. Mapping the human DC lineage through the integration of high-dimensional techniques. Science. 2017;356:356. doi: 10.1126/science.aag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigematsu H., Reizis B., Iwasaki H., Mizuno S., Hu D., Traver D., Leder P., Sakaguchi N., Akashi K. Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity. 2004;21:43–53. doi: 10.1016/j.immuni.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Sichien D., Scott C.L., Martens L., Vanderkerken M., Van Gassen S., Plantinga M., Joeris T., De Prijck S., Vanhoutte L., Vanheerswynghels M. IRF8 Transcription Factor Controls Survival and Function of Terminally Differentiated Conventional and Plasmacytoid Dendritic Cells, Respectively. Immunity. 2016;45:626–640. doi: 10.1016/j.immuni.2016.08.013. [DOI] [PubMed] [Google Scholar]
- Siegal F.P., Kadowaki N., Shodell M., Fitzgerald-Bocarsly P.A., Shah K., Ho S., Antonenko S., Liu Y.J. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- Sisirak V., Faget J., Gobert M., Goutagny N., Vey N., Treilleux I., Renaudineau S., Poyet G., Labidi-Galy S.I., Goddard-Leon S. Impaired IFN-α production by plasmacytoid dendritic cells favors regulatory T-cell expansion that may contribute to breast cancer progression. Cancer Res. 2012;72:5188–5197. doi: 10.1158/0008-5472.CAN-11-3468. [DOI] [PubMed] [Google Scholar]
- Sisirak V., Vey N., Goutagny N., Renaudineau S., Malfroy M., Thys S., Treilleux I., Labidi-Galy S.I., Bachelot T., Dezutter-Dambuyant C. Breast cancer-derived transforming growth factor-β and tumor necrosis factor-α compromise interferon-α production by tumor-associated plasmacytoid dendritic cells. Int. J. Cancer. 2013;133:771–778. doi: 10.1002/ijc.28072. [DOI] [PubMed] [Google Scholar]
- Sisirak V., Ganguly D., Lewis K.L., Couillault C., Tanaka L., Bolland S., D’Agati V., Elkon K.B., Reizis B. Genetic evidence for the role of plasmacytoid dendritic cells in systemic lupus erythematosus. J. Exp. Med. 2014;211:1969–1976. doi: 10.1084/jem.20132522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N., Pietrancosta N., Davidson S., Dutrieux J., Chauveau L., Cutolo P., Dy M., Scott-Algara D., Manoury B., Zirafi O. Natural amines inhibit activation of human plasmacytoid dendritic cells through CXCR4 engagement. Nat. Commun. 2017;8:14253. doi: 10.1038/ncomms14253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaulding E., Fooksman D., Moore J.M., Saidi A., Feintuch C.M., Reizis B., Chorro L., Daily J., Lauvau G. STING-Licensed Macrophages Prime Type I IFN Production by Plasmacytoid Dendritic Cells in the Bone Marrow during Severe Plasmodium yoelii Malaria. PLoS Pathog. 2016;12:e1005975. doi: 10.1371/journal.ppat.1005975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout-Delgado H.W., Yang X., Walker W.E., Tesar B.M., Goldstein D.R. Aging impairs IFN regulatory factor 7 up-regulation in plasmacytoid dendritic cells during TLR9 activation. J. Immunol. 2008;181:6747–6756. doi: 10.4049/jimmunol.181.10.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M., Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat. Rev. Immunol. 2015;15:471–485. doi: 10.1038/nri3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M., Gilfillan S., Vermi W., Wang Y., Colonna M. Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8(+) T cell accrual. Immunity. 2010;33:955–966. doi: 10.1016/j.immuni.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M., Wang Y., Vermi W., Gilfillan S., Schreiber R.D., Colonna M. Type I interferon negatively controls plasmacytoid dendritic cell numbers in vivo. J. Exp. Med. 2011;208:2367–2374. doi: 10.1084/jem.20110654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M., Wang Y., Gilfillan S., Lenschow D.J., Colonna M. Cutting edge: paradoxical roles of BST2/tetherin in promoting type I IFN response and viral infection. J. Immunol. 2012;188:2488–2492. doi: 10.4049/jimmunol.1103145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M., Wang Y., Gilfillan S., Colonna M. Plasmacytoid dendritic cells contribute to systemic but not local antiviral responses to HSV infections. PLoS Pathog. 2013;9:e1003728. doi: 10.1371/journal.ppat.1003728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiecki M., Wang Y., Riboldi E., Kim A.H., Dzutsev A., Gilfillan S., Vermi W., Ruedl C., Trinchieri G., Colonna M. Cell depletion in mice that express diphtheria toxin receptor under the control of SiglecH encompasses more than plasmacytoid dendritic cells. J. Immunol. 2014;192:4409–4416. doi: 10.4049/jimmunol.1303135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H., Fukaya T., Eizumi K., Sato Y., Sato K., Shibazaki A., Otsuka H., Hijikata A., Watanabe T., Ohara O. Plasmacytoid dendritic cells are crucial for the initiation of inflammation and T cell immunity in vivo. Immunity. 2011;35:958–971. doi: 10.1016/j.immuni.2011.10.014. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Asabe S., Wieland S., Garaigorta U., Gastaminza P., Isogawa M., Chisari F.V. Plasmacytoid dendritic cells sense hepatitis C virus-infected cells, produce interferon, and inhibit infection. Proc. Natl. Acad. Sci. USA. 2010;107:7431–7436. doi: 10.1073/pnas.1002301107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tel J., Aarntzen E.H., Baba T., Schreibelt G., Schulte B.M., Benitez-Ribas D., Boerman O.C., Croockewit S., Oyen W.J., van Rossum M. Natural human plasmacytoid dendritic cells induce antigen-specific T-cell responses in melanoma patients. Cancer Res. 2013;73:1063–1075. doi: 10.1158/0008-5472.CAN-12-2583. [DOI] [PubMed] [Google Scholar]
- Terra M., Oberkampf M., Fayolle C., Rosenbaum P., Guillerey C., Dadaglio G., Leclerc C. Tumor-Derived TGFβ Alters the Ability of Plasmacytoid Dendritic Cells to Respond to Innate Immune Signaling. Cancer Res. 2018;78:3014–3026. doi: 10.1158/0008-5472.CAN-17-2719. [DOI] [PubMed] [Google Scholar]
- Tomasello E., Naciri K., Chelbi R., Bessou G., Fries A., Gressier E., Abbas A., Pollet E., Pierre P., Lawrence T. Molecular dissection of plasmacytoid dendritic cell activation in vivo during a viral infection. EMBO J. 2018;37:e98836. doi: 10.15252/embj.201798836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhaya S., Sawai C.M., Papalexi E., Rashidfarrokhi A., Jang G., Chattopadhyay P., Satija R., Reizis B. Kinetics of adult hematopoietic stem cell differentiation in vivo. J. Exp. Med. 2018;215:2815–2832. doi: 10.1084/jem.20180136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bon L., Affandi A.J., Broen J., Christmann R.B., Marijnissen R.J., Stawski L., Farina G.A., Stifano G., Mathes A.L., Cossu M. Proteome-wide analysis and CXCL4 as a biomarker in systemic sclerosis. N. Engl. J. Med. 2014;370:433–443. doi: 10.1056/NEJMoa1114576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villadangos J.A., Young L. Antigen-presentation properties of plasmacytoid dendritic cells. Immunity. 2008;29:352–361. doi: 10.1016/j.immuni.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Villani A.C., Satija R., Reynolds G., Sarkizova S., Shekhar K., Fletcher J., Griesbeck M., Butler A., Zheng S., Lazo S. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:356. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster B., Werneke S.W., Zafirova B., This S., Coléon S., Décembre E., Paidassi H., Bouvier I., Joubert P.E., Duffy D. Plasmacytoid dendritic cells control dengue and Chikungunya virus infections via IRF7-regulated interferon responses. eLife. 2018;7:7. doi: 10.7554/eLife.34273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland S.F., Takahashi K., Boyd B., Whitten-Bauer C., Ngo N., de la Torre J.C., Chisari F.V. Human plasmacytoid dendritic cells sense lymphocytic choriomeningitis virus-infected cells in vitro. J. Virol. 2014;88:752–757. doi: 10.1128/JVI.01714-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmers F., Subedi N., van Buuringen N., Heister D., Vivié J., Beeren-Reinieren I., Woestenenk R., Dolstra H., Piruska A., Jacobs J.F.M. Single-cell analysis reveals that stochasticity and paracrine signaling control interferon-alpha production by plasmacytoid dendritic cells. Nat. Commun. 2018;9:3317. doi: 10.1038/s41467-018-05784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohn C., Ober-Blöbaum J.L., Haak S., Pantelyushin S., Cheong C., Zahner S.P., Onderwater S., Kant M., Weighardt H., Holzmann B. Langerin(neg) conventional dendritic cells produce IL-23 to drive psoriatic plaque formation in mice. Proc. Natl. Acad. Sci. USA. 2013;110:10723–10728. doi: 10.1073/pnas.1307569110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf A.I., Buehler D., Hensley S.E., Cavanagh L.L., Wherry E.J., Kastner P., Chan S., Weninger W. Plasmacytoid dendritic cells are dispensable during primary influenza virus infection. J. Immunol. 2009;182:871–879. doi: 10.4049/jimmunol.182.2.871. [DOI] [PubMed] [Google Scholar]
- Workman C.J., Wang Y., El Kasmi K.C., Pardoll D.M., Murray P.J., Drake C.G., Vignali D.A. LAG-3 regulates plasmacytoid dendritic cell homeostasis. J. Immunol. 2009;182:1885–1891. doi: 10.4049/jimmunol.0800185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Satpathy A.T., Kc W., Liu P., Murphy T.L., Murphy K.M. Bcl11a controls Flt3 expression in early hematopoietic progenitors and is required for pDC development in vivo. PLoS ONE. 2013;8:e64800. doi: 10.1371/journal.pone.0064800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Briseño C.G., Grajales-Reyes G.E., Haldar M., Iwata A., Kretzer N.M., Kc W., Tussiwand R., Higashi Y., Murphy T.L., Murphy K.M. Transcription factor Zeb2 regulates commitment to plasmacytoid dendritic cell and monocyte fate. Proc. Natl. Acad. Sci. USA. 2016;113:14775–14780. doi: 10.1073/pnas.1611408114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K., Nündel K., Watkins A.A., Dhawan T., Bonegio R.G., Ubellacker J.M., Marshak-Rothstein A., Rifkin I.R. Phenotype and function of B cells and dendritic cells from interferon regulatory factor 5-deficient mice with and without a mutation in DOCK2. Int. Immunol. 2013;25:295–306. doi: 10.1093/intimm/dxs114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Cai B., Wang M., Tan P., Ding X., Wu J., Li J., Li Q., Liu P., Xing C. Cross-Regulation of Two Type I Interferon Signaling Pathways in Plasmacytoid Dendritic Cells Controls Anti-malaria Immunity and Host Mortality. Immunity. 2016;45:1093–1107. doi: 10.1016/j.immuni.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun T.J., Lee J.S., Machmach K., Shim D., Choi J., Wi Y.J., Jang H.S., Jung I.H., Kim K., Yoon W.K. Indoleamine 2,3-Dioxygenase-Expressing Aortic Plasmacytoid Dendritic Cells Protect against Atherosclerosis by Induction of Regulatory T Cells. Cell Metab. 2016;23:852–866. doi: 10.1016/j.cmet.2016.04.010. [DOI] [PubMed] [Google Scholar]
- Zhan Y., Chow K.V., Soo P., Xu Z., Brady J.L., Lawlor K.E., Masters S.L., O’keeffe M., Shortman K., Zhang J.G., Lew A.M. Plasmacytoid dendritic cells are short-lived: reappraising the influence of migration, genetic factors and activation on estimation of lifespan. Sci. Rep. 2016;6:25060. doi: 10.1038/srep25060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Casartelli N., Lemoine S., Mozeleski B., Azria E., Le Ray C., Schwartz O., Launay O., Leclerc C., Lo-Man R. Plasmacytoid dendritic cells engagement by influenza vaccine as a surrogate strategy for driving T-helper type 1 responses in human neonatal settings. J. Infect. Dis. 2014;210:424–434. doi: 10.1093/infdis/jiu103. [DOI] [PubMed] [Google Scholar]
- Zhang H., Gregorio J.D., Iwahori T., Zhang X., Choi O., Tolentino L.L., Prestwood T., Carmi Y., Engleman E.G. A distinct subset of plasmacytoid dendritic cells induces activation and differentiation of B and T lymphocytes. Proc. Natl. Acad. Sci. USA. 2017;114:1988–1993. doi: 10.1073/pnas.1610630114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga E.I., Liou L.Y., Mack L., Mendoza M., Oldstone M.B. Persistent virus infection inhibits type I interferon production by plasmacytoid dendritic cells to facilitate opportunistic infections. Cell Host Microbe. 2008;4:374–386. doi: 10.1016/j.chom.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]