Abstract
Fluorescent nanodiamonds (FNDs) are promising bio-imaging probes compared with other fluorescent nanomaterials such as quantum dots, dye-doped nanoparticles, and metallic nanoclusters, due to their remarkable optical properties and excellent biocompatibility. Nevertheless, they are prone to aggregation in physiological salt solutions, and modifying their surface to conjugate biologically active agents remains challenging. Here, inspired by the adhesive protein of marine mussels, we demonstrate encapsulation of FNDs within a polydopamine (PDA) shell. These PDA surfaces are readily modified via Michael addition or Schiff base reactions with molecules presenting thiol or nitrogen derivatives. We describe modification of PDA shells by thiol terminated poly(ethylene glycol) (PEG-SH) molecules to enhance colloidal stability and biocompatibility of FNDs. We demonstrate their use as fluorescent probes for cell imaging; we find that PEGylated FNDs are taken up by HeLa cells and mouse bone marrow-derived dendritic cells and exhibit reduced nonspecific membrane adhesion. Furthermore, we demonstrate functionalization with biotin-PEG-SH and perform long-term high-resolution single-molecule fluorescence based tracking measurements of FNDs tethered via streptavidin to individual biotinylated DNA molecules. Our robust polydopamine encapsulation and functionalization strategy presents a facile route to develop FNDs as multifunctional labels, drug delivery vehicles, and targeting agents for biomedical applications.
Keywords: Fluorescent nanodiamonds, surface modification, polydopamine, bioconjugation, single-molecule
Graphical Abstract

Fluorescent nanodiamonds are encapsulated in biocompatible shells by simple and robust self-polymerization of dopamine in mild basic aqueous solution. The polydopamine shells are modified with thiol terminated PEG molecules via Michael addition. PEGylated nanodiamonds with functionalized surfaces are used for cell imaging and monitoring DNA motion at the single-molecule level.
1. Introduction
Nano-sized materials are increasingly being developed for use in biology and medicine.[1] The recent emergence of fluorescent nanodiamonds (FNDs) has attracted significant interest for biomedical applications due to their unique optical properties.[2] The exceptional optical properties of FNDs arise from the presence of nitrogen-vacancy (NV) centers, which are localized defect sites comprising a lattice vacancy adjacent to a substitutional nitrogen in the crystalline diamond lattice.[3] NV centers show extraordinary photostability without photobleaching or photoblinking. They emit red to near infrared (NIR) fluorescence (~650 to 900 nm) and are excited broadly in the green wavelength (~530 nm) with high quantum efficiency.[4] The NIR fluorescence is distinct from cell or tissue auto-fluorescence and exhibits reduced absorption by hemoglobin, water, and proteins.[5] The large Stokes shift of FNDs improves in vivo imaging by reducing the background from biological tissues and cells.[6] FNDs also have a long fluorescence lifetime (~20 ns), which enables fluorescence imaging of single FNDs in cells and organisms by time gating.[7] In addition to their superior optical properties, multiple studies have established the biocompatibility of FNDs in numerous biological systems.[8] Due to their remarkable physicochemical properties, FNDs have been used in biomedical imaging, drug delivery, sensing, and bio-compatible composite materials.[9]
Although FNDs offer a multitude of possibilities for biomedical applications, the realization of these possibilities is hampered by limitations arising from their inherent surface properties. For example, FNDs tend to aggregate at physiological salt conditions,[10] which limits their biological applicability. To improve the colloidal stability and to facilitate conjugation of biomolecules to FNDs requires surface modification. Therefore, the reliable and efficient surface modification of FNDs is a critical prerequisite for biomedical applications.
Several covalent or noncovalent FND surface modification approaches have been reported including reduction, oxidation, hydrogenation, chlorination, amination, and electrostatic coupling of surface groups.[4a,11] Recently, encapsulation of FNDs into silica shells has been described as a promising strategy to enhance colloidal stability in biological conditions and facilitate bioconjugation on the surface of silica shells.[12] Despite these efforts, there are still concerns regarding long term stability and difficult processing steps.
Dopamine (DA), 3,4-dihydroxyphenethylamine, is a well-known molecular mimic of mussel foot protein.[13] It self-polymerizes in basic solutions and the resulting polydopamine (PDA) can deposit onto inorganic and organic surfaces including noble metals, oxides, polymers, semiconductors, and ceramics.[14] PDA, which is a major pigment in naturally occurring eumelanin, has attracted attention due to its properties including adhesion, free radical quenching, metal ion chelation, large photoacoustic cross-section, and biocompatibility.[15] In addition, the presence of catechol/quinone groups on the PDA shell permits further reactions with amine or thiol containing molecules via Schiff base formation or Michael addition, respectively. The PDA encapsulation approach therefore provides a versatile platform for surface modification with a variety of functional molecules.[14a,16]
Here we develop a simple, robust, and reproducible encapsulation process to form PDA shells on FNDs. With this simple encapsulation method, the PDA shell thickness is reliably controlled by reaction time or dopamine concentration and FNDs of various sizes are efficiently encapsulated. Furthermore, the surface of the PDA shell is readily functionalized with thiol containing moieties via Michael addition. Employing this scheme, we functionalized the PDA shell with thiol terminated poly(ethylene glycol) (PEG-SH) molecules to address two outstanding limitations of FNDs. PEG coating (i) prevents FND aggregation as well as non-specific interactions with biological molecules and (ii) provides specific conjugation sites for a molecule of interest using heterobifunctional PEG molecules as linkers. We demonstrate PEG grafted PDA shells enhance FND solubility in high salt solutions, are stable over long storage times, and improve biocompatibility of FNDs in cells while preventing nonspecific adhesion to cell membranes. Finally, we show that PEG-biotin functionalized FNDs can be used as single-molecule fluorescent probes in a DNA tethered particle motion assay.
2. Results and Discussion
2.1. Synthesis and characterization of PDA-encapsulated FND nanoparticles
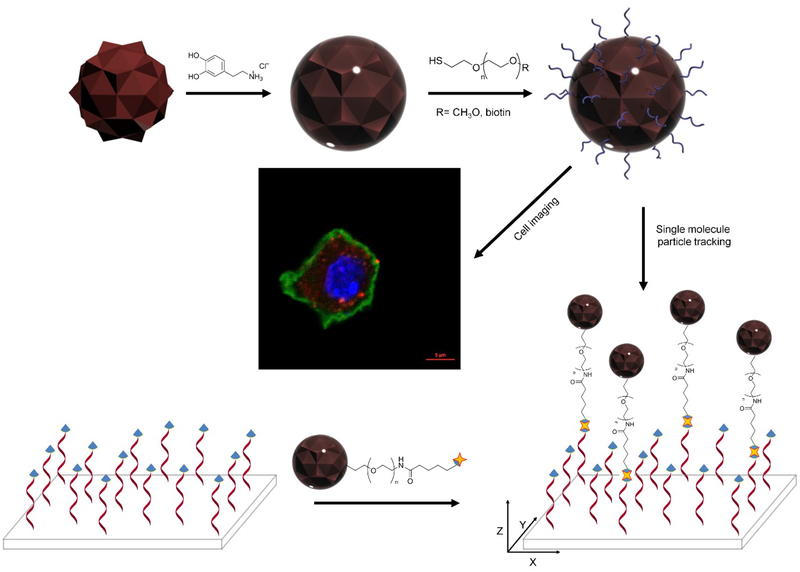
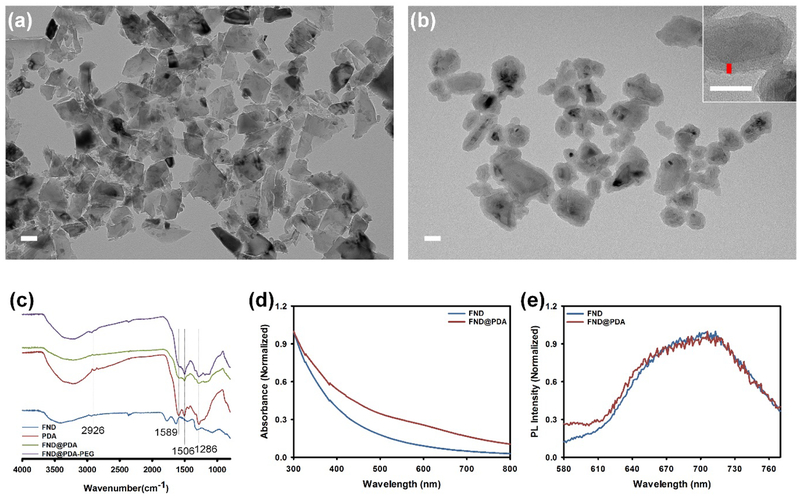
The overall scheme for the encapsulation and functionalization of FNDs with PDA shells (FND@PDA) is described in Scheme 1. FNDs were successfully incorporated into PDA shells by oxidation and self-polymerization of dopamine hydrochloride in a mild basic aqueous solution (pH 8.5).[14a,16a] First, the size, shape, and optical properties of bare 80 nm (nominal diameter) FNDs were evaluated by transmission electron microscopy (TEM), dynamic light scattering (DLS), UV-Vis, and photoluminescence (PL) spectroscopy. The results indicate FNDs have irregular shapes, sharp edges, a broad size distribution, and a mean diameter of 128 nm from DLS measurements (Figure 1 and S1). In comparison, PDA-coated FNDs (FND@PDA) have relatively smooth edges and the PDA shell could be directly observed by TEM due to differences in electron density between the FND and the PDA shell (Figure 1b). The diameter of FND@PDA measured by DLS increased to 153 nm (Figure S1). The PDA shell on FND was confirmed by Fourier transform infrared spectroscopy (FT-IR) (Figure 1c). Characteristic peaks observed in FND@PDA and PEGylated FND@PDA, but not in bare FND, at about 1506 and 1589 cm−1 correspond to the N–H shearing and bending vibrations of the amine group, respectively. The peak at around 1286 cm−1 is assigned to the phenolic C–O–H stretching vibration. The band at 2926 cm−1 is characteristic of PEG. In addition, the surface charge of FND@PDA increased to −43.6 mV (compared to −32.6 mV for bare FND), attributed to the presence of the PDA shell on the surface of the FNDs (Table S4).
Scheme 1.
Encapsulation strategy and biomedical applications of functionalized fluorescent nanodiamonds. The polydopamine encapsulated diamonds were functionalized with two different thiol terminated PEGs. mPEG-SH modified diamonds were used as fluorescent probes in cell imaging experiments and biotin-PEG-SH decorated diamonds, attached to DNA via the biotin-streptavidin interaction, were used for tracking 3-dimensional tethered particle motion.
Figure 1.
TEM images of (a) uncoated and (b) PDA-coated FND (FND@PDA). Inset: high magnification TEM image of FND@PDA with a vertical red line indicating the thickness of the PDA shell. Scale bar is 50 nm. (c) FT-IR spectra of FND, PDA, FND@PDA, and PEGylated FND@PDA. UV-Vis (d) and intensity normalized PL spectra (e) of FND and FND@PDA.))
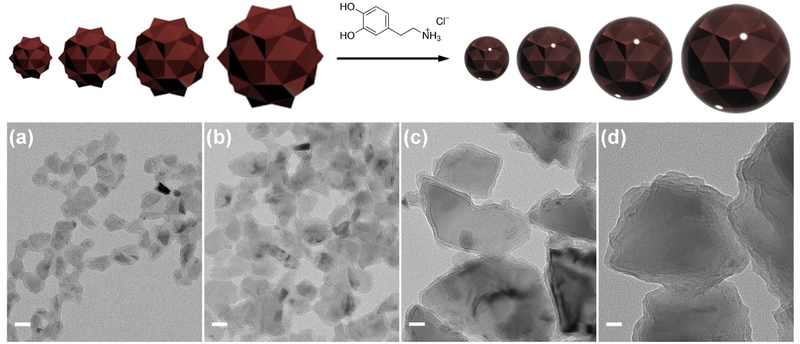
To investigate the influence of the PDA shell on the optical properties of FND, we measured the absorption and PL spectra of FND and FND@PDA (Figure 1d and 1e). Each PL spectrum was normalized by its maximum to obtain relative changes in the spectrum resulting from PDA encapsulation. The absorption of FND@PDA slightly increased compared with uncoated FND, whereas the PL spectral shape was unchanged by PDA encapsulation. Since PDA is a known fluorescence quencher,[17] we examined the PL efficiency of bare FND and FND@PDA at the single-FND level by total internal reflection fluorescence microscopy (TIRFM). FNDs with different sizes (20, 40, 80, and 150 nm nominal diameter) and processing history, were coated with PDA (Figure 2).[14b,18] The PDA shell thickness was approximately 4 nm for all FNDs under the reaction conditions (1 hr reaction time with 1.42 mM DA) except for the 150 nm FNDs, which had a PDA shell thickness of 7.5 nm, likely due to the smaller surface area to volume ratio. These reaction conditions were optimized to produce the minimum PDA shell thickness that completely encapsulates the FND. Minimizing the PDA shell thickness results in smaller final particles and improves the brightness by reducing photoluminescence losses due to quenching and direct absorption by the PDA shell. All but one of the FND samples retained greater than 75 % PL efficiency after PDA encapsulation.
Figure 2.
TEM images of different sizes of FND encapsulated in PDA. The standard reaction condition was TRIS buffer (10 mM, pH=8.5) with 0.2 mg of FND, 1.42 mM of dopamine for 1 hr (Table S1 sample 6). (a) 20 nm, (b) 40 nm, (c) 80 nm, and (d) 150 nm nominal FND sizes. Scale bar is 25 nm.
One batch of 80 nm FNDs had significantly lower PL efficiency after PDA encapsulation (Table S2). To determine if this reduced PL efficiency was related to shell thickness, we controlled the PDA shell thickness by reaction time or DA concentration. The PDA shell thickness gradually increased as either the polymerization time or DA concentration were increased (Figure 3). As expected, the PL efficiency of FND decreased as the PDA shell thickness increased (Table S3 and Figure S2). However, even when the PDA shell thickness was decreased to less than 3 nm (30 min reaction), the PL efficiency of FND@PDA was only 66 % of the PL efficiency of uncoated FND (Table S3). A possible explanation for lower PL after encapsulation for this sample is quenching of fluorescence from carbon dots on the surface of the 80 nm FNDs. Raw FNDs often contain a disordered carbon and graphitic shell that reduces the PL intensity.[19] Several oxidation processes such as treatment with acid, oxygen plasma, or potassium nitrate, have been developed to remove non-diamond carbon from the surface of the FNDs.[20] The 80 nm FND had been treated with acid, however, and acid treatment of FND at temperatures exceeding 100 °C has been reported to generate carbon dots, which have broad near IR PL emission, on the surface of the nanodiamonds.[21] We suspect that the surfaces of the 80 nm FNDs were decorated with carbon dots resulting from high temperature acid treatment and that emission from these carbon dots was quenched by the PDA shell, resulting in the reduction of PL intensity proportional to PDA shell thickness.
Figure 3.
TEM images of FND@PDA with different PDA shell thicknesses (a) ~ (d) and (f) ~ (i). Scale bar is 25 nm. The PDA shell thickness was controlled by adjusting the reaction time at 1.42 mM dopamine concentration (e) or dopamine concentration for a 2 hr reaction time (j). PDA shell thickness was determined from the mean thickness measured at 10 positions of each TEM image (See Figure 1) and the error bars correspond to the standard deviation.
2.2. Synthesis and characterization of PDA-encapsulated FND nanoparticles
Both bare FND and FND@PDA are unstable and precipitate in physiological solutions, which limits their biomedical applications as imaging probes or drug delivery systems. Fortunately, the abundant catechol/quinine groups on the surface of the PDA shell can be functionalized with thiol containing groups via Michael addition under oxidizing conditions. We functionalized FND@PDA samples with thiol terminated poly(ethylene glycol) (PEG-SH) and heterobifunctional PEGs such as poly(ethylene glycol) methyl ether thiol (mPEG-SH) or alpha-biotin-omega-mercapto poly(ethylene glycol) (biotin-PEG-SH) to enhance dispersion and stability in physiological buffers, and to provide a convenient labeling scheme via the biotin-streptavidin (SA) linkage, while preserving the nontoxic nature of FND@PDA.[14a, 16c] Surface modification of FND@PDA was achieved by adding 2 kDa mPEG-SH or biotin-PEG-SH (4 mg) and 2 μL of NH4OH into a 10 mL suspension of FND@PDA (0.2 mg) in water. Functionalizing the FNDs with 2 kDa PEG prevented aggregation of particles. Based on previous reports, this molecular mass corresponds to an optimum PEG size that achieves stability while minimizing the overall particle size.[16c,22] PEG functionalized FND@PDA was purified by centrifugation.
The diameter of PEGylated FND@PDA increased from 138 to 143 nm (Figure S3). The zeta-potential decreased slightly from −43.6 to −39.7 mV due to the PEG molecules on the surface of PDA shell (Table S4). PEGylated FND@PDA remained stably dispersed for at least a month in PBS, whereas PDA-coated FNDs completely precipitated within 5 hr in PBS (Figure S4 and S5). The colloidal stability of PEGylated FND@PDA as a function of pH and ionic strength was investigated by DLS (Figure S5). The hydrodynamic diameter of PEGylated FND remained constant over the pH range of 4–10 in PBS buffer and at high ionic strength (1 M NaCl), indicating that PEGylated FND is colloidally stable over a broad range of pH and ionic strength. Introducing a second functional group into the PEG molecules allows further selective coupling reactions to attach specific functional molecules. Hence, surface modification of FND@PDA with heterobifunctional PEG (biotin-PEG-SH) facilitates not only enhancement of colloidal stability and biocompatibility, but also attachment of SA conjugated molecules via the biotin-SA interaction. To determine the number of biotin molecules available on the surface of biotinylated FND@PDA particles, we measured their size by DLS as a function of SA concentration (Scheme S1, Figure S6, and S7). Tubes containing 100 μL of biotinylated FND@PDA (0.1 mg/mL) were mixed with SA to final concentrations ranging from 0 to 45 nM. Because tetrameric SA has four high-affinity biotin binding sites, sub-saturating concentrations of SA relative to the effective biotin concentration on the FND@PDA particles induces aggregation. Conversely, saturating the surface biotins prevents aggregation of the biotinylated FND@PDA while slightly increasing the particle diameter due to conjugation of SA on the surface. The critical SA concentration at which biotinylated FND@PDA no longer aggregates is a good approximation of the concentration of surface bound biotins (Figure S6). We calculated the average number of biotins on each biotinylated FND@PDA surface to be approximately 340 based on the measured diameter as a function of SA concentration. In control experiments with FND@PDA the hydrodynamic radius remained constant as a function of SA concentration, indicating that aggregation is specific and indicative of surface bound biotins (Figure S7).
2.3. Behavior of bare FND and PEGylated FND@PDA in cells
FNDs have been widely used in various biomedical applications due to their distinct physicochemical properties. However, materials intended for biomedical applications must be biocompatible. Although both FND and PDA are highly biocompatible materials,[8c,15h] we evaluated the biocompatibility of FND and PEGylated FND@PDA as a function of the concentration and incubation time using the (2-(4-Iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium sodium salt (WST-1) cell viability assay with mouse bone marrow-derived dendritic cells (BMDCs) and HeLa cells. Dendritic cells are antigen presenting cells that capture, process, and present antigens, and trigger T-cell mediated immune responses.[23] Therefore, prior to biomedical applications of functionalized FND@PDA, it is crucial to ensure that surface modified FND@PDA particles do not elicit cell toxicity or immune responses in these cells. Neither bare FND nor PEGylated FND@PDA produced significant toxicity to BMDCs or HeLa cells for any of the tested concentrations or incubation times (Figure 4). These results are consistent with direct measures of cell morphology and proliferation obtained from microscopy of treated and control cells (Figure S8).
Figure 4.
Viability of HeLa cells and mouse immature BMDC cells in the presence of FND and PEGylated FND@PDA as function of particle concentration (a) or incubation time (b). Cell viability was determined with the WST-1 method as described in the Methods. Points represent the mean of three independent measurements and the error bars correspond to the standard deviation.
We further investigated differences between bare FND and PEGylated FND@PDA interacting with mouse BMDCs and HeLa cells via cell imaging experiments. Unmodified FND and PEGylated FND@PDA were dispersed in PBS buffer and added to the cell medium (50 μg/mL final concentration) for 16 hr. After washing the cells with PBS buffer, they were co-stained with either anti-mouse CD11c-FITC (BMDCs) or Phalloidin-Alexa Fluor 488 (HeLa), and DAPI to identify the membrane, cytoskeleton, and nuclei, respectively. We observed the cellular localization of the particles using a Zeiss LSM 880 confocal microscope (Figure 5 and S9). The PEGylated FNDs were internalized into the cells (HeLa and BMDCs), whereas unmodified FNDs mostly adhered to the cell membrane. This difference in behavior is consistent with a scenario in which the low colloidal stability of unmodified FNDs results in aggregation in PBS buffer, whereas the PEGylated FNDs remain stable in PBS and individual particles can be efficiently endocytosed by the cells. Cellular uptake of nanomaterials is strongly influenced by the energy required to drive active cellular transport.[24] Aggregated particles require significantly more energy to enter the cell and consequently exhibit greater nonspecific adhesion to the cell membrane than functionalized dispersed particles.
Figure 5.
Confocal laser scanning microscopy images of bare FND and PEGylated FND@PDA stored in PBS and introduced into media of mouse immature BMDC. The displayed images are fluorescence from FNDs (red channel), CD11c-FITC, (green channel: BMDCs membrane), DAPI (blue channel: nuclei) and overlay. Scale bar is 5 μm.
2.4. DNA tethering of biotin functionalized FND@PDA for single molecule tracking
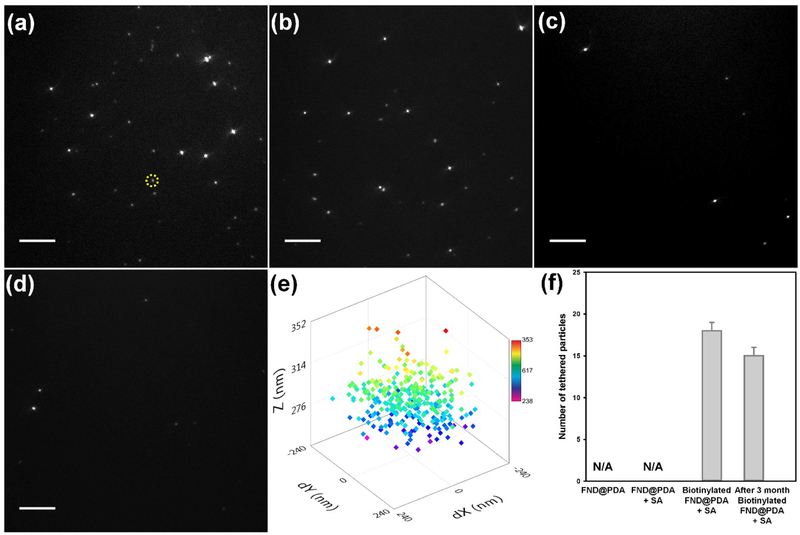
FNDs have been considered as ideal single-molecule tracking probes due to their remarkable optical properties. However, it is necessary to specifically conjugate the FND to the targeted biomolecule to employ FNDs as fluorescent probes for single-molecule tracking. Biotin-SA interactions are widely used to conjugate biomolecules to probes due to the exceptionally high affinity of biotin to SA.[1a] Hence, we used biotin-PEG-SH to functionalize the PDA shell for SA bioconjugation with biotinylated biomolecules. To confirm the applicability of biotinylated FND@PDA as a single-molecule tracking probe, the tethered particle motion (TPM) of DNA attached to FND@PDA was investigated at the single-molecule level using a total internal reflection fluorescence microscope (Figure 6). The TPM method, in which the Brownian motion of a DNA-tethered bead is monitored by tracking the bead, can report biologically relevant conformational changes of individual DNA molecules.[25] Therefore, TPM has been frequently used to study the physical and mechanical properties of biopolymers such as nucleic acids and a variety of DNA-protein interactions including DNA looping and transcription.[26] Biotinylated FND@PDA was incubated with 90 nM excess of SA. After purification of SA-coated biotinylated FND@PDA by centrifugation to remove unbound SA, the particles were introduced to the sample cell and attached to biotinylated DNA (see methods for details). After washing out the unbound particles, DNA tethered biotinylated FND@PDA nanoparticles were characterized by acquiring successive images on an EMCCD camera (20 Hz). We found on average 18 tethered FNDs per field of view with biotinylated FND@PDA, whereas no tethers were observed with either FND@PDA or SA-treated FND@PDA (Figure 6a, c, and d. Video S1, S3, and S4 in the Supporting Information). These results confirmed the existence of biotin on the surface of the PDA shell. Moreover, we could track the 3-dimensional Brownian motion of DNA tethered biotinylated FND@PDA (Figure 6e).[26a,27] To test the long-term stability of biotinylated FND@PDA, we repeated the tethering reaction after storing biotinylated FND@PDA for three months in water at room temperature (Figure 6b and Video S2). The tethering efficiency of freshly prepared and stored biotinylated FND@PDA was comparable, demonstrating long-term stability of the PDA shell and the chemical linkage between the PDA shell and biotin-PEG-SH.
Figure 6.
DNA tethering experiments under four different conditions. TIRFM images for DNA tethering with (a) biotinylated FND@PDA treated with SA (FND@PDA-PEG-biotin+SA), (b) FND@PDA-PEG-biotin stored for three months at RT in water, treated with SA, (c) PDA-coated FND (FND@PDA), and (d) FND@PDA treated with SA (FND@PDA+SA). Scale bar in panels a-d is 10 μm. (e) An example 3-D spatial distribution of a DNA tethered FND@PDA-PEG-biotin+SA obtained by tracking the Brownian motion of the particle (indicated by the yellow circle in panel a). Color scale bar provides a measure of the z position (in nm) of the tethered FND. (f) The average number of DNA molecules tethered by FND per a field of view for four different conditions. No DNA tethered FND particles were observed for either FND@PDA or FND@PDA+SA. A few nonspecifically stuck immobile particles are distinguished from tethered particles which undergo Brownian motion, (please see the video S3 and S4). The average number of FNDs tethered per field of view (73 μm × 73 μm) with the day-old and 3-month old FND@PDA-PEG-biotin+SA are 18 ± 1 [N (number of different field of views) = 6)] and 15 ± 1 [N = 8)] respectively. The reported error is the standard error of the mean.
3. Conclusion
We present a simple and robust method to encapsulate fluorescent nanodiamonds within a PDA shell that can be readily covalently functionalized to provide specific binding groups. We produced methoxy-PEG or biotin-PEG functionalized FNDs and demonstrated their use in two example applications that demonstrate the merits of this strategy; single-molecule tracking of DNA tethered motion and imaging within cells. We found this PDA encapsulation method is robust and reproducible across FNDs with various sizes and surface properties. Unlike other functionalization schemes, the method is simple and easily completed on the benchtop in less than one day using only mild aqueous buffers. This permits facile PEGylation of nanodiamonds, enhancing their colloidal stability in physiological buffers over long storage periods and facilitating broad use of fluorescent nanodiamonds in biomedical applications.
4. Experimental Section
Chemicals and Materials:
Dopamine hydrochloride (DA), tris(hydroxymethyl)aminomethane (TRIS), Streptavidin (SA), WST-1 Cell Proliferation Reagent, ammonium hydroxide solution (NH4OH, 25 %), gentamicin, Poly-L-lysine solution (0.1 % w/v in H2O) and 2-mercaptoethanol were purchased from Sigma-Aldrich. mPEG-SH (2 kDa) and biotin-PEG-SH (2 kDa,) were purchased from NANOCS. RPMI 1640 was purchased from Lonza. L-glutamine was purchased from Gibco. 10 % heat-inactivated fetal calf serum was purchased from BioScience. GM-CSF was purchased from Peprotech. Anti-mouse CD11c-FITC was purchased from eBioscience. Poly-L-lysine was purchased from Invitrogen. Fluorescent nanodiamonds were supplied by Adamas Nanotechnologies and Columbus NanoWorks. Deionized (DI) water with a resistivity of 18.2 MΩ·cm was from Milli-Q Water Purification System.
General method for synthesizing FND@PDA:
DA was dissolved in Tris-HCl (pH=8.5, 10 mM), FNDs in suspension were added, and the suspension was incubated at room temperature (25 °C) for a determined time. Solution compositions and incubation times are listed in Table S1. PDA shell thickness was controlled by varying the reaction time or dopamine concentration. The reaction mixture was then centrifuged for 15 min at 41000 g (20,000 rpm), the precipitate was re-dispersed in 10 mL of DI water, and the obtained FND@PDA solution was filtered through a 0.2-μm syringe filter to remove aggregates.
Surface Modification of PDA-coated FND with mPEG-SH (PEGylated FND@PDA) or biotin PEG-SH (biotinylated FND@PDA):
Surface modification of FND@PDA was performed by adding 4 mg of 2 kDa mPEG-SH (or biotin-PEG-SH) and 2 μL of NH4OH into a suspension of 0.02 mg/mL FND@PDA in 10 mL of DI water, followed by stirring for 16 hr at room temperature (25 °C). The PEG modified FND@PDA was isolated by centrifugation for 20 min at 41000 g (20,000 rpm) and the supernatant was removed. The isolated PEGylated FND@PDA (or biotinylated FND@PDA) was then re-dispersed in water. This washing process of centrifugation followed by re-dispersing the PEGylated FND@PDA (or biotinylated FND@PDA) in water was repeated 2 times.
BMDCs and HeLa cell cultures:
Mice were bred and maintained in-house at the NCI-Frederick animal facility. All mice were cared for in accordance with National Institutes of Health guidelines with the approval of the National Cancer Institute Animal Care and Use Committee. 4 × 106 bone marrow cells per well were cultured in tissue-culture-treated 6-well plates in 4 mL of complete medium, 10 % heat-inactivated fetal calf serum, and GM-CSF (20 ng/mL) following a standard protocol.[28] The culture medium was replaced by fresh warmed medium with GM-CSF (20 ng/mL) every 2 days. On day 7, non-adherent loosely adherent DCs were harvested by gentle washing with PBS, and used as the starting source of material for most experiments.
Human cervical adenocarcinoma (HeLa) cells were maintained in Dulbecco’s modified eagle medium (DMEM; Gibco) supplemented with 10 % fetal bovine serum, penicillin and streptomycin, under a humidified atmosphere containing 5 % CO2.
Cytotoxicity assay (WST-1):
BMDCs and HeLa cells were transferred to a 96-well tissue culture plate in a final volume of 100 μL/well culture medium. Cells were treated with various amounts of PEGylated FND@PDA or FND in triplicate and incubated overnight. 10 μL cell proliferation reagent WST-1 was added to the wells and mixed gently followed by a 4 hr incubation. Cell viability was determined by absorbance at 450 nm for WST-1 as described in the manual provided by the kit manufacturer.
Immunofluorescence microscopy:
BMDCs (or HeLa cells) were incubated with 50 μg/mL PEGylated FND@PDA or FND for 16 hr at 37 °C and washed with PBS. Cells were fixed with 4 % paraformaldehyde in PBS for 30 min at room temperature, and washed twice with PBS. BMDCs were stained with anti-mouse CD11c-FITC and HeLa cells were stained with Alexa Fluor 488 Phalloidin on ice for 30 min, and washed. Cells were plated on poly-L-lysine-coated glass coverslips for 30 min at room temperature for analysis by Zeiss LSM 880 confocal laser scanning microscopy. Samples were mounted in ProLong Gold antifade reagent with DAPI (Invitrogen).
Preparation of SA-conjugated FND:
SA-conjugated biotinylated FND@PDA was prepared by binding SA in excess to biotinylated FND@PDA (Figure S6). 100 μL (90 nM) of SA in Wash Buffer (WB; 1x PBS, 0.03 % w/v BSA, and 0.01 % v/v Tween-20) was added to a suspension of biotinylated FND@PDA (0.1 mg/mL) in water (100 μL). The mixture was agitated overnight. The resulting dispersion was centrifuged for 10 min at 13400 g (12,000 rpm), and the precipitate was re-dispersed in 100 μL of WB.
DNA tethering experiment:
The detailed DNA tethering procedure and instrumentation for single molecule fluorescence assay were previously published.[26a] In brief, 2.5 kb DNA labelled with biotin and digoxigenin was generated by PCR of pET28b DNA plasmid with 5´ biotin- and 5´ digoxin-primers respectively (Operon). 2.5 kb DNA molecules (25 pM) were incubated with anti-digoxigenin (25 nM) in 1x PBS for 15 min to allow anti-digoxigenin-digoxigenin binding. The DNA-anti-digoxigenin mixture was then added into a pre-assembled sample cell and incubated at 4 °C overnight to permit non-specific absorption of anti-digoxigenin to the sample cell surface. The sample cell was then washed with 200 μl WB to remove unbound DNA molecules. To tether SA-conjugated biotinylated FND@PDA to DNA, 40 μl of SA-biotin FND@PDA (0.1 mg/mL) was introduced into the sample cell and incubated overnight at 4 °C or 1 hr at RT. After washing with 600 μl of WB, FNDs tethered by individual surface immobilized DNA molecules were visualized on an EMCCD (Andor iXon+ DU-897, 512X512 Array with 16 μm pixels) using a custom-built micro-mirror TIRF instrument (ex: 560 nm and em: 640 nm).[26a] For 3-D tracking of DNA tethered FND, we used a custom written tracking program in Labview which is based on the algorithm utilized in Mosaic Particle tracker 2-D.[27] Using this program, we identified the x and y positions of the FND in each image and also obtained the sum of the pixel intensities Isum within a cut-off radius (90 % percentile in brightness) of the FND. The height of the FND above the surface of the flow-cell (nm) was determined from the height calibration curve relating Isum to the height of the particle.[26a] The x and y displacements were calculated by converting the x and y positions in pixels to positions in nm via measured calibrations of the x and y pixel dimensions.
FND Characterization:
UV–vis absorption (UV–vis) spectra for bare and PDA coated FNDs were measured with a HP/Agilent 8453 Spectrophotometer, and the corresponding PL spectra were recorded in a Tecan Spark 10M Multi-Mode Microplate Reader. PL spectra were collected using 530 nm excitation wavelength. FT-IR measurements were carried out in a Varian 1000, and the hydrodynamic radius was measured in a DynoPro Nanostar (Waytt) and then evaluated in Dynamic software with regularization method. Surface charge of particles was measured with a Zetasizer Nano ZS (Malvern Instruments). A JEOL 1400 transmission electron microscope operating at an accelerating voltage of 120 kV was used to characterize the size, PDA shell thickness, and structure of FND and FND@PDA. To measure the PL intensity of bare or PDA-coated FND, 100 μL of each particle (in deionized water) was added to 100 μL poly-L-lysine solution. The poly-L-lysine coated particles were deposited onto cover glass by spin coating 100 μL of the mixture. The coated cover glass was then attached to a slide with double sticky tape forming a chamber in which water was added. Individual PL intensity at the single-molecule level of FND and FND@PDA were measured using a Nikon Eclipse Ti-E microscope equipped with an H-TIRF module attachment, a CFI60 Apochromat TIRF 100X Oil Immersion Objective Lens (N.A. 1.49, W.D. 0.12mm, F.O.V 22mm) and an EMCCD camera (Andor iXon Ultra 888 EMCCD, 1024X1024 array with13 μm pixel). The 561 nm laser line was used for excitation and emission was collected through a 700 nm (CWL) wavelength filter with a 75 nm (FWHM) bandwidth. All data were acquired using a 100 ms exposure time for bare and PDA-coated FND images. HeLa cell morphology and proliferation were observed under a light microscope (EVOS FL Cell Imaging System) and photographs were taken using a Sony ICX445 monochrome CCD camera. Cell imaging experiments were carried out in a Zeiss LSM 880 META confocal microscope system using a 63× oil immersion objective lens. The fluorescence images were collected at room temperature at 532 nm excitation wavelength using LSM 880 software, in scanning range 612–729 nm, pinhole at diameter 57.1 μm (1 Airy unit) and a laser dwell time of 1.03 μsec per pixel.
Supplementary Material
Acknowledgements
H.-S. Jung and K.-J. Cho contributed equally to this work. We thank Dr. Martin Brechbiel, Dr. Rolf Swenson and Dr. Jonathan Silver for helpful discussions, the NHLBI Electron Microscopy and Biophysical Core Facility for help with transmission electron microscopy and scattering measurements, Dr. Marie-Paule Strub for providing access to ultra-centrifuge system, and Dr. James R. Sellers for use of the Nikon H-TIRF microscope. This research was supported by the NHLBI and NCI DIR programs of the National Institutes of Health.
H.-S. Jung was supported by a grant of the Korea Health Technology R&D project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI15C0979).
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Hak-Sung Jung, Laboratory of Single Molecule Biophysics, NHLBI, NIH, Bethesda, MD 20892, USA.
Kyung-Jin Cho, Experimental Immunology Branch, NCI, NIH, Bethesda, MD 20892, USA.
Yeonee Seol, Laboratory of Single Molecule Biophysics, NHLBI, NIH, Bethesda, MD 20892, USA.
Yasuharu Takagi, Laboratory of Single Molecule Biophysics, NHLBI, NIH, Bethesda, MD 20892, USA; Laboratory of Molecular Physiology, NHLBI, NIH, Bethesda, MD 20892, USA.
Andrew Dittmore, Laboratory of Single Molecule Biophysics, NHLBI, NIH, Bethesda, MD 20892, USA.
Paul A. Roche, Experimental Immunology Branch, NCI, NIH, Bethesda, MD 20892, USA
Keir C. Neuman, Laboratory of Single Molecule Biophysics, NHLBI, NIH, Bethesda, MD 20892, USA
References
- [1].a) Niemeyer CM, Angew. Chem. Int. Ed 2001, 40, 4128; [DOI] [PubMed] [Google Scholar]; b) Alivisatos P, Nat. Biotechnol 2003, 22, 47; [DOI] [PubMed] [Google Scholar]; c) Ferrari M, Nat. Rev. Cancer 2005, 5, 161. [DOI] [PubMed] [Google Scholar]
- [2].a) Mochalin VN, Shenderova O, Ho D, Gogotsi Y, Nat. Nanotechnol 2012, 7, 11; [DOI] [PubMed] [Google Scholar]; b) Montalti M, Cantelli A, Battistelli G, Chem. Soc. Rev 2015, 44, 4853; [DOI] [PubMed] [Google Scholar]; c) Hsiao WW-W, Hui YY, Tsai P-C, Chang H-C, Acc. Chem. Res 2016, 49, 400. [DOI] [PubMed] [Google Scholar]
- [3].Kurtsiefer C, Mayer S, Zarda P, Weinfurter H, Phys. Rev. Lett 2000, 85, 290. [DOI] [PubMed] [Google Scholar]
- [4].a) Fu C-C, Lee H-Y, Chen K, Lim T-S, Wu H-Y, Lin P-K, Wei P-K, Tsao P-H, Chang H-C, Fann W, Proc. Natl. Acad. Sci. U. S. A 2007, 104, 727; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Abbas M, Koenderink AF, New J. Phys 2013, 15, 043017. [Google Scholar]
- [5].a) Grosenick D, Wabnitz H, Rinneberg HH, Moesta KT, Schlag PM, Appl. Opt 1999, 38, 2927; [DOI] [PubMed] [Google Scholar]; b) Weissleder R, Ntziachristos V, Nat. Med 2003, 9, 123. [DOI] [PubMed] [Google Scholar]
- [6].a) Peng X, Song F, Lu E, Wang Y, Zhou W, Fan J, Gao Y, J. Am. Chem. Soc 2005, 127, 4170; [DOI] [PubMed] [Google Scholar]; b) Yu J, Diao X, Zhang X, Chen X, Hao X, Li W, Zhang X, Lee C-S, Small 2014, 10, 1125. [DOI] [PubMed] [Google Scholar]
- [7].a) Collins AT, Thomaz MF, Jorge MIB, J. Phys. C-Solid State. Phys 1983, 16, 2177; [Google Scholar]; b) Wu T-J, Tzeng Y-K, Chang W-W, Cheng C-A, Kuo Y, Chien C-H, Chang H-C, Yu J, Nat. Nanotechnol 2013, 8, 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a) Yu S-J, Kang M-W, Chang H-C, Chen K-M, Yu Y-C, J. Am. Chem. Soc 2005, 127, 17604; [DOI] [PubMed] [Google Scholar]; b) Kuang-Kai L, Chia-Liang C, Chia-Ching C, Jui IC, Nanotechnology 2007, 18, 325102; [Google Scholar]; c) Schrand AM, Dai L, Schlager JJ, Hussain SM, Osawa E, Diamond Relat. Mater 2007, 16, 2118. [Google Scholar]
- [9].a) Chang Y-R, Lee H-Y, Chen K, Chang C-C, Tsai D-S, Fu C-C, Lim T-S, Tzeng Y-K, Fang C-Y, Han C-C, Chang H-C, Fann W, Nat. Nanotechnol 2008, 3, 284; [DOI] [PubMed] [Google Scholar]; b) Sarkar SK, Bumb A, Wu X, Sochacki KA, Kellman P, Brechbiel MW, Neuman KC, Biomed. Opt. Express 2014, 5, 1190; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhao L, Xu Y-H, Qin H, Abe S, Akasaka T, Chano T, Watari F, Kimura T, Komatsu N, Chen X, Adv. Funct. Mater 2014, 24, 5348; [Google Scholar]; d) Maze JR, Stanwix PL, Hodges JS, Hong S, Taylor JM, Cappellaro P, Jiang L, Dutt MVG, Togan E, Zibrov AS, Yacoby A, Walsworth RL, Lukin MD, Nature 2008, 455, 644; [DOI] [PubMed] [Google Scholar]; e) Zhang Q, Mochalin VN, Neitzel I, Knoke IY, Han J, Klug CA, Zhou JG, Lelkes PI, Gogotsi Y, Biomaterials 2011, 32, 87. [DOI] [PubMed] [Google Scholar]
- [10].a) Dahoumane SA, Nguyen MN, Thorel A, Boudou J-P, Chehimi MM, Mangeney C, Langmuir 2009, 25, 9633; [DOI] [PubMed] [Google Scholar]; b) Sreenivasan VKA, Ivukina EA, Deng W, Kelf TA, Zdobnova TA, Lukash SV, Veryugin BV, Stremovskiy OA, Zvyagin AV, Deyev SM, J. Mater. Chem 2011, 21, 65. [Google Scholar]
- [11].a) Wolcott A, Schiros T, Trusheim ME, Chen EH, Nordlund, Diaz RE, Gaathon O, Englund D, Owen JS, J. Phys. Chem. C 2014, 118, 26695; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Girard HA, Petit T, Perruchas S, Gacoin T, Gesset C, Arnault JC, Bergonzo P, Phys. Chem. Chem. Phys 2011, 13, 11517; [DOI] [PubMed] [Google Scholar]; c) Miller JB, Brown DW, Langmuir 1996, 12, 5809; [Google Scholar]; Zhang d T., Neumann A, Lindlau J, Wu Y, Pramanik G, Naydenov B, Jelezko F, Schüder F, Huber S, Huber M, Stehr F, Högele A, Weil T, Liedl T, J. Am. Chem. Soc 2015, 137, 9776. [DOI] [PubMed] [Google Scholar]
- [12].a) Bumb A, Sarkar SK, Billington N, Brechbiel MW, Neuman KC, J. Am. Chem. Soc 2013, 135, 7815; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rehor I, Slegerova J, Kucka J, Proks V, Petrakova V, Adam M-P, Treussart F, Turner S, Bals S, Sacha P, Ledvina M, Wen AM, Steinmetz NF, Cigler P, Small 2014, 10, 1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Waite, Herbert J, Taner ML, Science 1981, 212, 1038. [DOI] [PubMed] [Google Scholar]
- [14].a) Lee, Dellatore SM, Miller WM, Messersmith PB, Science 2007, 318, 426; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ye Q, Zhou F, Liu W, Chem. Soc. Rev 2011, 40, 4244. [DOI] [PubMed] [Google Scholar]
- [15].a) d’Ischia M, Napolitano A, Ball V, Chen C-T, Buehler MJ, Acc. Chem. Res 2014, 47, 3541; [DOI] [PubMed] [Google Scholar]; b) Liu Y, Ai K, Lu L, Chem. Rev 2014, 114, 5057; [DOI] [PubMed] [Google Scholar]; c) Lee H, Scherer NF, Messersmith PB, Proc. Natl. Acad. Sci. U. S. A 2006, 103, 12999; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Meredith P, Sarna T, Pigm. Cell Res 2006, 19, 572; [DOI] [PubMed] [Google Scholar]; e) Meredith P, Powell BJ, Riesz J, Nighswander-Rempel SP, Pederson MR, Moore EG, Soft Matter 2006, 2, 37; [DOI] [PubMed] [Google Scholar]; f) Repenko T, Fokong S, De Laporte L, Go D, Kiessling F, Lammers T, Kuehne AJC, Chem. Commun 2015, 51, 6084; [DOI] [PubMed] [Google Scholar]; g) Hong S, Kim KY, Wook HJ, Park SY, Lee KD, Lee DY, Lee H, Nanomedicine 2011, 6, 793; [DOI] [PubMed] [Google Scholar]; h) Liu X, Cao J, Li H, Li J, Jin Q, Ren K, Ji J, ACS Nano 2013, 7, 9384. [DOI] [PubMed] [Google Scholar]
- [16].a) Lee H, Rho J, Messersmith PB, Adv. Mater 2009, 21, 431; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Xu LQ, Yang WJ, Neoh K-G, Kang E-T, Fu GD, Macromolecules 2010, 43, 8336; [Google Scholar]; c) Ju K-Y, Lee Y, Lee S, Park SB, Lee J-K, Biomacromolecules 2011, 12, 625. [DOI] [PubMed] [Google Scholar]
- [17].Qiang W, Li W, Li X, Chen X, Xu D, Chem. Sci 2014, 5, 3018. [Google Scholar]
- [18].Black KC, Rivera JG, Zelasko-Leon DC, Messersmith PB, Nanomedicine 2013, 8, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].a) Fang X, Mao J, Levin EM, Schmidt-Rohr K, J. Am. Chem. Soc 2009, 131, 1426; [DOI] [PubMed] [Google Scholar]; b) Havlik J, Petrakova V, Rehor I, Petrak V, Gulka M, Stursa J, Kucka J, Ralis J, Rendler T, Lee S-Y, Reuter R, Wrachtrup J, Ledvina M, Nesladek M, Cigler P, Nanoscale 2013, 5, 3208. [DOI] [PubMed] [Google Scholar]
- [20].Stehlik S, Varga M, Ledinsky M, Jirasek V, Artemenko A, Kozak H, Ondic L, Skakalova V, Argentero G, Pennycook T, Meyer JC, Fejfar A, Kromka A, Rezek B, J. Phys. Chem. C 2015, 119, 27708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shenderova O, Hens S, Vlasov I, Turner S, Lu Y-G, Van Tendeloo G, Schrand A, Burikov SA, Dolenko TA, Part. Part. Syst. Charact 2014, 31, 580. [Google Scholar]
- [22].a) Liu Z, Dong K, Liu J, Han X, Ren J, Qu X, Small 2014, 10, 2429; [DOI] [PubMed] [Google Scholar]; b) Ju KY, Lee S, Pyo J, Choo J, Lee JK, Small 2015, 11, 84. [DOI] [PubMed] [Google Scholar]
- [23].Guermonprez P, Valladeau J, Zitvogel L, Théry C, Amigorena S, Annu. Rev. Immunol 2002, 20, 621. [DOI] [PubMed] [Google Scholar]
- [24].Salvati A, Åberg C, dos Santos T, Varela J, Pinto P, Lynch I, Dawson KA, Nanomedicine 2011, 7, 818. [DOI] [PubMed] [Google Scholar]
- [25].a) Schafer DA, Gelles J, Sheetz MP, Landick R, Nature 1991, 352, 444; [DOI] [PubMed] [Google Scholar]; b) Kovari DT, Yan Y, Finzi L, Dunlap D, in Single Molecule Analysis: Methods and Protocols, (Ed: Peterman EJG), Springer; New York, New York, NY: 2018, 317. [Google Scholar]
- [26].a) Seol Y, Neuman KC, in Single Molecule Analysis: Methods and Protocols, (Ed: Peterman EJG), Springer; New York, New York, NY: 2018, 297; [Google Scholar]; b) Dunlap D, Zurla C, Manzo C, Finzi L, in Single Molecule Analysis: Methods and Protocols, (Eds: Peterman EJG, Wuite GJL), Humana Press, Totowa, NJ: 2011, 295. [Google Scholar]
- [27].Sbalzarini IF, Koumoutsakos P, J. Struct. Biol 2005, 151, 182. [DOI] [PubMed] [Google Scholar]
- [28].Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman RM, J. Exp. Med 1992, 176, 1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.