Abstract
Biothiols such as cysteine (Cys), homocysteine (Hcy), and glutathione (GSH) play crucial roles in maintaining redox homeostasis in bio-logical systems. This Minireview summarizes the most significant current challenges in the field of thiol-reactive probes for biomedical research and diagnostics, emphasizing the needs and opportunities that have been under-investigated by chemists in the selective probe and sensor field. Progress on multiple binding site probes to distinguish Cys, Hcy, and GSH is highlighted as a creative new direction in the field that can enable simultaneous, accurate ratiometric monitoring. New probe design strategies and researcher priorities can better help address current challenges, including the monitoring of disease states such as autism and chronic diseases involving oxidative stress that are characterized by divergent levels of GSH, Cys, and Hcy.
Keywords: biothiols, detection methods, fluorescence, imaging, multiple-binding-site probes
1. Introduction
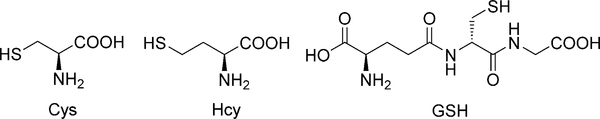
Cysteine (Cys), homocysteine (Hcy), and glutathione (GSH) are the most abundant small molecule biological thiols (Figure 1). Owing to their characteristic redox properties and nucleophilicity, they play major unique roles in human health and disease. They were initially viewedprimarily as antioxidants, protective agents serving as free radical scavengers. However, because they maintain redox homeostasis through the equilibria between their reduced free thiol and oxidized disulfide forms,[1] monitoring the relative levels of reduced and oxidized thiols in cells became a traditional means of assessing oxidative stress. More recent investigations have prompted a redefinition of oxidative stress based largely on changes in post-translational protein thiol modifications.[2] Because oxidative stress is a major aspect of the development of many chronic and degenerative illnesses including cancer, autoimmune disorders, aging, cataracts, rheumatoid arthritis, and cardiovascular and neurodegenerative diseases, monitoring the levels of biological thiols has a significant role in biomedical research and diagnostics.[3]
Figure 1.
The major amino thiols.
Analytical methods typically involving separation steps have been used previously to address the detection and determination of specific biothiols. These have included high-performance liquid chromatography (HPLC),[4] capillary electrophoresis,[5] spectrophotometry,[6] voltammetry,[7] mass spectrometry (MS),[8] and HPLC-MS/MS.[9] Fluorometry has some specific advantages over these techniques such as speed, cost, and relative operational simplicity. HPLC-MS continues to serve as arguably the most useful of these approaches for diagnostics and certain research areas; however, a relatively longer analysis time renders it sub-optimal for some researchers and, significantly, for rapid point-of-care emergency-room, bedside, or intraoperative diagnostics. It also is currently not optimal for home testing, doctor’s office testing, or for developing-nation settings that lack water and electricity. Finally, it often requires derivatization and, moreover, is relatively expensive compared to fluorescence-based sensor methods, requiring individual run charges and high instrument-maintenance costs.
A large number of thiol-reactive reagents and probes have been commercially available for many years, filling an entire chapter of the Molecular Probes Handbook.[10] However, they are non-selective among the thiols, forming covalent adducts with any sulfhydryl-containing molecules. It is important to realize that such universal thiol-reactive probes are sometimes marketed or reported in the literature as GSH (or Cys)-selective. There is an assumption that because GSH is the most abundant (1–10 mM levels) thiol in cells, any signaling from an interaction with Hcy or Cys (micromolar concentrations in cells) would be negligible. This may be true, though this assumption is typically reported without experimental evidence. However, in human blood plasma, GSH (just 3–5 pM) is less abundant than Cys (10–20 pM) as unbound thiol, and an unaware probe end-user will encounter artificially high values attributed to GSH arising from Cys and Hcy.
Chemists in the sensor and supramolecular fields have been motivated to investigate aminothiol sensors not only due to their biomedical significance but also because of the fundamental challenges inherent in creating reagents that enable the selective detection of each of the specific thiols in natural media. Prior reviews have summarized biothiol fluorescent probes from different perspectives including bioimaging applications,[11] structure and reaction classification,[12] reaction types,[13] detection methods, and reaction mechanisms.[14] There have been a large number of fluorescent probes for biothiols reported in just the past decade. Most can be divided into three categories based on function: 1) The detection of GSH/Cys/Hcy (universal thiol probes) over other amino acids,[15] 2) the selective detection one or two of GSH/Cys/Hcy (Table 1), and 2) simultaneous discrimination of Cys, Hcy, and GSH. This purpose of this Minireview is two-fold. The first is to focus on new probe design, such as multiple-binding-site fluorescent probes to concurrently distinguish Cys, Hcy, and GSH. The second is to highlight the most significant and most under-investigated challenges and applications relevant to selective thiol sensor and probe research.
Table 1:
Fluorescent probes for the selective detection of biothiols (individually or in pairs).
| Probe | Selectivity | Biological system | Probe | Selectivity | Biological system |
|---|---|---|---|---|---|
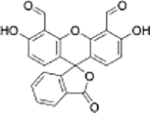 |
Cys+Hcy | Human plasma |  |
Hcy | Human plasma |
| Fluorescein derivative[19] | Fluorescein derivative[20] | ||||
 |
Hcy | Human plasma | 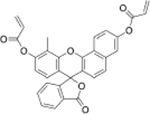 |
Cys | Human plasma |
| Fluorescein derivative[21] | SNF-probe[22] | ||||
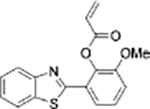 |
Cys+Hcy | Human plasma | Cys+Hcy | HepG2 cells | |
| Benzothiazole derivative[23] | Coumarin derivative[24] | ||||
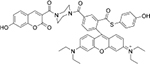
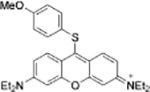
| GSH | Human plasma |  |
Cys | HepG2 cells | |
| Resorufin derivative[25] | Coumarin derivative[26] | ||||
 |
Cys+Hcy | HepG2 cells | 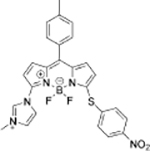 |
GSH | HeLa cells |
| FRET dyad[27] | BODIPY derivative[28] | ||||
 |
GSH | HeLa cells | 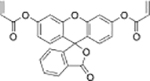 |
Cys | PC-12 cells |
| BODIPY derivative[29] | Fluorescein derivatives[30] | ||||
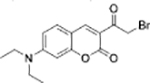
 |
GSH | B16 cells |  |
GSH | MCF-7 cells |
| Pyronin dye[31] | BODIPY dye[32] | ||||
 |
GSH | HeLa cells | 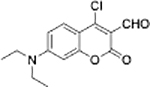 |
GSH | HeLa cells |
| KC-Br[33] | Coumarinyl aldehyde[8c] | ||||
 |
GSH | Hep3B cells |  |
GSH | Human renal cell carcinoma SiHa cells |
| Phenyl-selenium[34] | Coumarin-enone Dye[35] | ||||
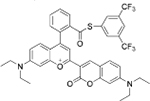 |
Cys+Hcy | HepG2 cells | 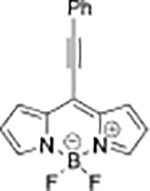 |
Cys | HeLa cells |
| NIR dye[36] | 8-alkynyl BODIPY[37] | ||||
 |
Cys | HeLa cells | 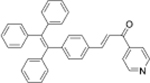 |
Cys | No study |
| DPP-AC[38] | TPE-Py[39] | ||||
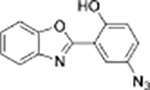 |
Cys+GSH | HeLa cells | 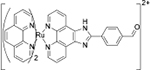 |
Cys+Hcy | No study |
| AHBO[40] | Ruthenium complex[41] | ||||
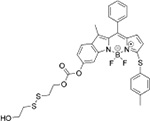 |
GSH | MKN-45 cells and HeLa cells |  |
GSH | MKN-45 cells and HeLa cells |
| S-S-BODIPY-S[42] | MeO-BODIPY-C[43] | ||||
 |
GSH+Hcy | L929 cells |  |
Cys+Hcy | HeLa cells |
| TP3[44] | L2[45] | ||||
 |
GSH | HepG2 cells |  |
Hcy | HeLa cells |
| Rhodamine derivative[46] | Pyrene derivative[47] | ||||
| Cys | HeLa cells |  |
Cys+GSH | HeLa cells | |
| Chloro-hydroxyl merocyanine derivative[48] | Chloro-dialkylamino-coumarin conjugated with malonitrile[49] |
In only a few cases of recent probe methods, investigations have led to an enhanced understanding of the fundamental redox chemistry of specific thiols.[16] Many studies to date have focused on the design of probes with optimized selectivity, signaling, and kinetics. However, most of the probes developed during the past decade have been limited to function in biological media as imaging agents in live cells, as shown in Table 1. Many of these studies involve initially describing probe selectivity for one or more of GSH/Cys/Hcy in buffer, followed by proof-of-concept imaging of function in biological media by adding excess thiol to live cells.
An emerging trend, and the focus of this review, is the design of multiple binding site sensors that not only enable excellent discrimination among the three main thiols but also allow for their simultaneous detection. However, there are, to date, relatively very few examples of such probes. These and related creative advances will allow researchers and clinicians to address the most significant barriers to specific thiol monitoring, thereby having a greater impact on biomedical research. Moreover, multiple binding and signaling sites in thiol probes potentially will not only promote enhanced quantitative accuracy owing to analyte-specific ratiometric signals but will also serve as a step towards more holistic single diagnostic reagents. For example, such reagents could have a role in monitoring diseases with divergent relative levels of specific thiols, such as autism, wherein children present elevated levels of Hcy[17] but depleted levels of GSH and Cys.[18]
2. Significant Challenges in the Thiol-Probe Field.
There are critical unmet needs for improved probes to address current biomedical problems, particularly in the areas of GSH and Hcy monitoring. These include specific issues and applications that have generally not been addressed by chemists in the thiol probe and sensor field either at all or at least far short in proportion to their significance to human health.
2.1. Opportunities for GSH probe and sensor development
GSH is a tripeptide possessing an internal Cys residue. It is produced in virtually all organs and is present in all tissues. It is the most abundant low-molecular weight biological thiol found in cells, appearing at millimolar levels (Cys and Hcy levels in cells are in the micromolar range). It is a major cellular antioxidant, reacting with reactive oxygen species (ROS) as well as free radicals. The exchange of its sulfhydryl proton for a free radical located on biomolecules is the basis of the biological repair process. GSH is also involved in DNA synthesis, amino acid transport, and xenobiotic detoxification. It serves as an enzyme cofactor and substrate. It regulates nitric oxide production.[50] In cells, the ratio of reduced (GSH, thiol form) to oxidized (GSSG, disulfide form) is a well-known measure of oxidative stress as well as general cellular redox status. Most GSH (> 90%) is present in the reduced (GSH, sulfhydryl) form in cells.[51]
GSH deficiency is a risk factor for major chronic diseases including heart disease, stroke, cancer, chronic respiratory diseases, and diabetes. Chronic diseases are characterized by long duration and relatively slow progression. Collectively, they cause over 60% of all deaths worldwide. GSH levels have been used to follow progression and treatment out-comes; however, the GSH levels reported suffer from poor inter-and intra-laboratory reproducibility. For example, according to one author “it is evident that the measured concentrations...are highly divergent among different research groups...findings should be reconsidered once (there is) a general agreement about the physiological levels...”)[52] This issue of inconsistency has been attributed to the variability in analytical methods, the use of different matrices (plasma vs. blood), sample oxidation, inconsistent sample preparations, and a lack of general standardized techniques)[53] Sample handling issues have been cited as main causes of the irreproducibility and variance in GSH determinations)[54] In addition, exacerbating these variance issues has been ambiguity in whether labs are reporting blood (which includes cellular millimolar levels of GSH) or plasma (micromolar ranges of GSH). There are also specific issues related to the available assays. A number of issues specific to GC- and HPLC-MS have been described. A gold standard for GSH monitoring is the glutathione-S-transferase-catalyzed reaction of monochlorobimane and GSH. However, the monochlorobimane fluorogen is unstable at ambient temperature, under ambient light, and in water. Its byproducts are fluorescent and cause elevated readings. Commercial GSH kits typically also feature relatively fragile materials and multistep procedures. Importantly, most are not selective for GSH over other thiols. Cys and DTT (dithiothreitol, the common thiol reducing agent) are common interferents. GSH-labeling kits such as ThiolTracker Violet, useful in GSH imaging, will react with other free thiols in addition to GSH.
The range of GSH depletion associated with specific chronic diseases varies from 20% to 50%, with an average of more than 30 %)[55] Researchers have discovered that blood GSH testing led to earlier detection of asymptomatic patients with structural cardiac abnormalities, as compared to an overexpression of soluble TNF receptor-1 (sTNFRl), a well-known biomarker of symptomatic heart failure severity that is also associated with oxidative stress. The patients in the study exhibited GSH decreases as high as 58%)[56]
Other examples of GSH depletion monitoring, each involving circa 20–50 % GSH decreases compared to healthy levels, include: 1) Birth defects and complications during pregnancy, 2) side-effects of post-operative therapy, 3) end-stage renal disease therapy, 4) mitochondrial diseases, 5) hospital sepsis, and 6) autism. For example, in birth defects, GSH and Hcy levels tracked pre-eclampsia and normotensive pregnancies)[57] Whole-blood GSH measurements enabled the monitoring of patient responses to glutamine (GLN) therapy following open heart surgery)[58] Mitochondrial disease may be diagnosed based on cellular GSH levels)[59] Monitoring the restoration of depleted GSH levels has also been used in end-stage renal disease)[60] In one review article it was stated that, despite the imprecision of current GSH assays, the significance of GSH monitoring in hemodialysis patients is too significant to overlook.[60a]
In summary, current GSH assays have suffered from imprecision. This has had consequence in hindering progress in a variety of serious issues. The development of simple, rapid and reliable indicator-based methods enabling selective GSH detection, with limited or no sample handling, that are practical enough to achieve broad acceptance and standardization, will have a significant, positive impact on the study and treatment of chronic disease.
2.2. Opportunities for Hoy probe and sensor development
Hyperhomocysteinemia (HHcy) is defined as total plasma homocysteine (tHcy, the sum of reduced free sulfhydryl and oxidized disulfide forms of Hcy) above circa 12–15 pM. Despite decades of research, Hcy remains highly controversial. There is no general agreement on Hcy’s association with disease)[61] Extensive large scale clinical trials, involving tens of thousands of patients, have failed to establish any definitive relationship between Hcy lowering and reduced cardiovascular disease (CVD) risk. This stunning dichotomy with the observational evidence that Hcy is linked to CVD and several other major disorders has rendered Hcy’s role in disease a highly contentious topic.[61]
Hcy is the one-carbon homologue of Cys. However, unlike Cys, Hcy is typically incorporated into proteins post-translationally, either through disulfide bonds to Cys residues or amide bonds to Lys residues. Hyperhomocysteinemia has been linked to numerous major pathologies including CVD, birth defects, osteoporosis, Alzheimer’s disease, and renal failure, among many others.
The term total homocysteine (tHcy) is a misnomer. In the clinical protocols used to measure tHcy levels, at least half of the Hcy forms (as its metabolites) are not accounted for. For example, Hcy-thiolactone (HTL) is found in all human cells, is more toxic than Hcy to human endothelial cells, and reacts readily with protein Lys residues affording N-Hcy-protein, a new protein thiol (Figure 2). N-Hcy-protein levels were not accounted for in the clinical trials involving Hcy lowering. There is strong evidence that N-Hcy-protein is associated with coronary artery disease (CAD), induces cell death, causes autoimmune responses, and interferes with blood clotting)[62] The proteolytic degradation of N-Hcy-protein produces the isopeptide Nε-Hcy-Lys. Levels of N-Hcy-protein and Nε-Hcy-Lys are in the micromolar range in circulation, within the range of what has been termed tHcy. Nε-Hcy-Lys is also (in a limited number of patients, however) a potential independent predictor of heart attack within 12 h of chest pain symptoms)[63] We have reported that N-Hcy-protein formation promotes free radical damage to proteins and protein carbonyl formation)[16]
Figure 2.
Significant derivatives of Hey include its thiolactone form (HTL), which reacts with Lys residues to form post-translational modifications. The Hcy-Lys isopeptide results from protein degradation. HTL, N-Hcy-protein, and Nε-Hcy-Lys each have shown evidence of significant health impact. However, there is a lack of selective abiotic optical indicators dyes and probes that have targeted these analytes. Their physiological and structural relationships render them intriguing targets for multiple binding site probes.
HPLC can enable the monitoring of Hcy levels along with other structurally related analytes, such as Cys. HPLC methods have also been reported for determining levels of HTL[64] and N-Hcy-protein[65] as well as Nε-Hcy-Lys.[66] Hcy has been monitored by clinicians mainly by HPLC; however, it has not incorporated the simultaneous monitoring of the structurally related Hcy forms such as HTL, N-Hcy-pro-tein,[65] and Ne-Hcy-Lys. Clinical monitoring is also relatively low throughput and involves a laborious sample pretreatment. HPLC is run sequentially, with each sample taking approximately 10–30 min. Immuno- and enzymatic assays can only monitor one analyte at a time. The Axis-Shield immunoassay method requires instruments that can handle multiple reagents per test and the relatively specialized fluorescent polarization (FP) detection. The time required is approx-imately 30–45 min per test. The precision of the immunoassay is often lower than that of HPLC or enzymatic assays. Diazyme has created a promising commercial enzyme-cycling test for Hcy that is relatively faster, less expensive, and less labor intensive than HPLC and immunoassays. However, Hcy metabolites such as HTL (after ring opening), N-Hcy-protein, and Nε-Hcy-Lys could, in principle, be monitored by fluorescent probe methods analogous to those used for Hcy because they contain similar aminothiol structural elements.[16]
Reagents that afford a selective optical response corresponding to Hcy as well as its metabolites, as a more global measure of Hcy, could facilitate basic research, clinical studies, and diagnostics in this field. It has been shown that selective-probe approaches can be rapid and can eliminate the need for sample preparation for the analysis of Hcy in human samples.[67] Moreover, these methods have already shown promise for N-Hcy-protein determination.[16] The fact that Hcy, Cys, and GSH can be discriminated by multiple-binding-site probes encourages optimism that analogous designs can enable simultaneous detection of Hcy and its metabolites to facilitate elucidation of their interdependent roles in disease. On the other hand, there have been fewer reports of molecular sensors that are selective for Hcy, as compared to those enabling Cys or GSH selectivity. Cys levels in plasma are typically 15–20-fold higher than those of Hcy. Therefore, the selective detection of Hcy by abiotic chemo-sensors or dosimeters has been challenging, with some of the reports describing a lack of sensitivity at physiological thiol levels, fluorescence at only short wavelength excitation, or optimal function limited to non-aqueous solvents.
In summary, there is still a need for Hcy-selective probes to facilitate the understanding of its role in disease as either a causative agent or as a spectator biomarker. The HPLC methods are laborious. In any case, practical Hcy probes, especially those enabling simultaneous or individual monitoring of HTL, N-Hcy-protein, and Nε-Hcy-Lys, are more challenging and currently lacking compared to Cys and GSH probes.
3. Multiple-Binding-Site Probes for Simultaneous GSH/Cys/Hcy Monitoring
3.1. Nucleophilic aromatic substitution (SnAr) reactions for the discrimination of Cys, Hcy, and GSH
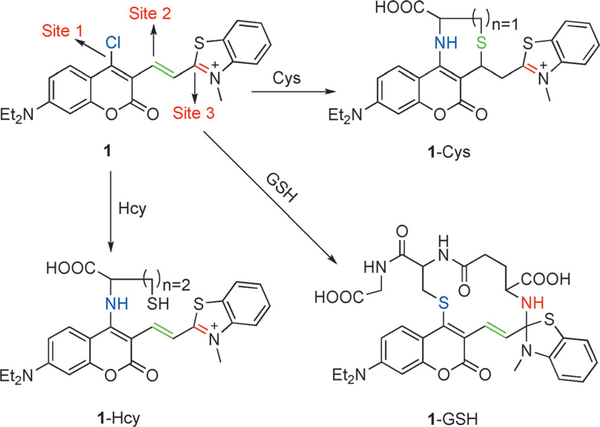
A fluorophore bearing a labile substituent can react with thiols to yield a thioether by nucleophilic aromatic substitution (SNAr). This has been used mainly for the selective detection of Cys and/or Hcy over GSH. The amino group of Cys and Hcy (but not GSH) displaces sulfur through a 5- or 6-membered ring transition state. The distinct photophysical properties of thioether- and amino-substituted dyes can enable the selective detection of GSH, Cys, and/or Hcy. For example, Liu et al. reported chlorinated coumarin-hemicyanine fluorescence probe 1 with three potential reaction sites (Figure 3). Site 1 occupies the 4-position of the coumarinmoiety of 1 and undergoes nucleophilic substitution with thiols. Site 2 is a Michael acceptor with differential reactivity for Cys and Hcy. Site 3 is a benzothiazolium group promoting discrimination between GSH and Cys/Hcy. Cys and Hcy can substitute for the chloride at site 1 to afford the corresponding thio-coumarins, followed by rearrangement to the N-substituted aryl ring. The thiol derived from Cys undergoes rapid conjugate addition yielding highly fluorescent amino-cou-marin 1-Cys. The reaction is slower in the case of the Hcy derivative, possibly owing to a less energetically-favorable 8-membered ring transition state. GSH forms 1-GSH, involving thiocoumarin formation and macrocyclization.
Figure 3.
The reactions of 1 with Cys, Hcy, and GSH.
Probe 1 is virtually non-emissive at the excitation wavelength of 360 nm. Upon addition of Cys, a new emission peak appeared at 420 nm, taking 60 min to reach saturation. A 53-fold increase in the fluorescence signal was observed. By comparison, at 420 nm Hcy and GSH only promoted 7- and 4-fold emission enhancements, respectively, indicating the ability of probe 1 to discriminate Cys from Hcy/GSH. When 500 nm was selected as the excitation wavelength for detecting Hcy, only a weak emission at 625 nm was observed that diminished upon Hcy addition. When λex = 450 nm, upon addition of GSH a new emission peak appeared at 512 nm. GSH produced a 67-fold increase in fluorescence signal at the concentrations studied. When Cys, Hcy, and N-acetylcysteine were added no change in the peak at 512 Nm was observed. Overall, these results demonstrated that probe 1 enabled the highly selective detection of Cys and GSH from different emission channels (Figure 3).[68] Moreover, probe 1 was shown to enable the simultaneously imaging of Cys and GSH in COS-7 cells using different emission channels. Overall, this is a highly promising result and a very interesting probe design. Much of the understanding of the probe properties and selective signaling was also derived from a careful evaluation of the absorption spectra along with an analysis of intermediate reaction structures. Although the 60 min equilibration kinetics for Cys detection could be improved, and the signaling promoted by Hcy was suboptimal, this work embodies an excellent blueprint for the development of sensors for simultaneous GSH/Cys/Hcy detection.
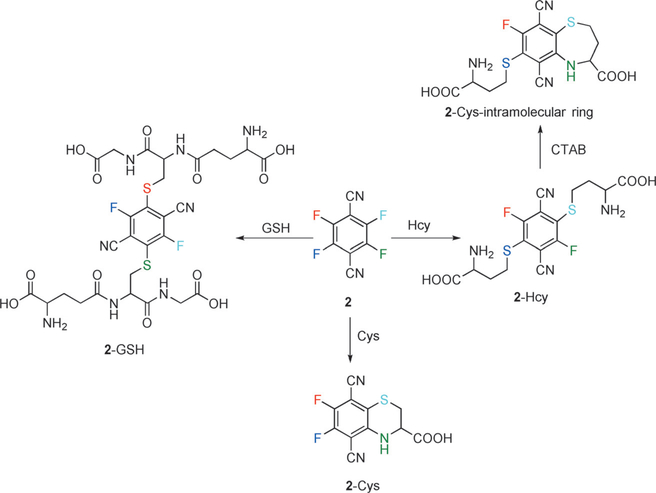
Zhang et al. reported a commercial compound 2 (4F-2CN) as an efficient fluorescent probe with four potential reaction sites for detecting biological thiols (Figure 4).[69] They hypothesized that the formation of 2-Cys was initiated by thiol substitution of a fluoro group, followed by cyclization to displace a second fluoro group. Conversely, the formation of 2-Hcy (or 2-GSH) entailed substitution of two fluorine atoms by two equivalents of thiol. Probe 2 responded to Cys and emitted strong green fluorescence, whereas it reacted with Hcy/GSH and generated blue fluorescence. Thus, probe was capable of simultaneously detecting Cys and Hcy/GSH using dual emission channels. Addition of CTAB (cetyl trimethylammonium bromide) was observed to alter the color of the reaction product of 2 and Hcy (from blue to green), but no alteration of the fluorescence color occurred for Cys and GSH. Furthermore, probe 2 was used to image Cys/Hcy/GSH in HeLa cells. Potential drawbacks could bethe long time (up to 2 h) to reach an optimal signal and a relatively short wavelength excitation used in some experiments. However, the researchers discovered that selective two-photon detection for Cys could be achieved upon excitation at 860 nm. This was ascribed to a combination of molecular rigidity and favorable electronic properties in the case of the Cys-probe adduct. The selective two-photon properties, combined with the clever use of CTAB, the relative simplicity of the commercial probe, and the interplay between the thiol adduct structures and characteristic electronic properties, should enable analogous design strategies for optimal simultaneous analyte detection and imaging.
Figure 4.
The reactions of 2 with Cys, Hcy, and GSH.
3.2. Condensation and reduction reactions for the discrimination of Cys, Hcy, and GSH
Miao et al. reported sulfonamide-based self-quenched fluorescent probe 3 possessing two potential reaction sites for simultaneously sensing biothiols in physiological media with high sensitivity (Figure 5).[70] The probe reacted with GSH through sulfonamide cleavage to yield 3-GSH (CBT). When the probe reacted with Cys or Hcy, the corresponding heterocycles formed. Each product exhibited different emission maxima (3-GSH, λem = 490 nm; 3-Hcy, λem = 517 nm; 3-Cys, λem = 522 nm). Furthermore, 3 was also successfully used for discriminating between Cys and GSH in HepG2 cells byratiometric quantification.
Figure 5.
The reactions of 3 with Cys, Hey, and GSH.
Although fluorescence spectra needed to be acquired in buffer after 2 h at 37 °C using short wavelength (350 nm) excitation, the reactions of Hcy and Cys with this probe are noteworthy alternatives to the formation of thiazinane and thiazolidine heterocycles that arise from aldehyde-functionalized fluorophores (examples shown in Table 1). The different emission signals produced as a function of the 5- and 6-membered heterocyclic rings can serve as a useful starting point for the development of highly selective, longer wave-length probes with enhanced kinetics.
3.3. Native chemical ligation approach value for discrimination of Cys, Hcy, and GSH
Native chemical ligation (NCL) is a significant tool used in the synthesis of proteins. Classical NCL involves a reaction cascade beginning with transthioesterification of a thioester at a C-terminus of a peptide through the reaction of an N-terminal Cys residue from another peptide. The thioester-linked intermediate undergoes intramolecular amide bond formation. The sequence is relatively rapid, involving a five-membered intramolecular transition state.
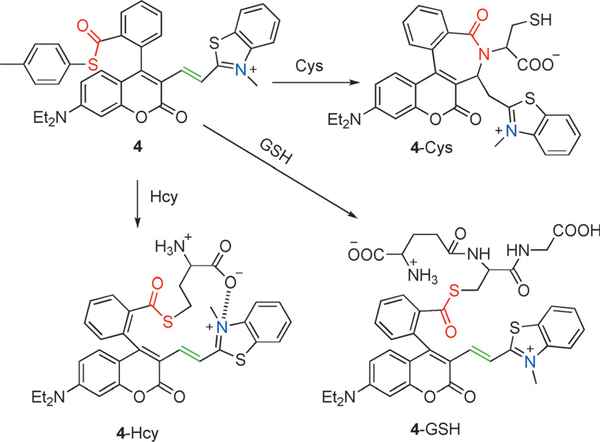
Liu et al. designed a carboxylic acid-functionalized cou-marin-hemicyanine fluorescent probe 4 with three potential reaction sites, a thioester group, a coumarin-hemicyanine unit, and a benzothiazolium N atom (Figure 6).[71] Cys under-went a native chemical ligation (NCL) reaction with 4 followed by cyclization to produce 4-Cys. However, the reaction of 4 and Hcy resulted in thioester 4-Hcy, which was stabilized by the electrostatic attraction between the carboxylate and heterocyclic benzothiazolium nitrogen. This stabilization was shown by molecular simulations to be more favorable for Hcy than Cys, and thereby inhibited amide formation and subsequent cyclization. Similarly, in the case of GSH, thioester probe 4-GSH was produced without amide formation owing to a ten-membered ring transition state. They examined the emission behavior of 4 upon the addition of Cys, Hcy, and GSH, by using two different excitations at 405 nm and 545 nm. Probe 4 displayed a weak emission at 494 nm (λex = 405 nm) and a strong emission at 644 nm (λex = 545 nm). Upon the addition of Cys, the emission intensity at 494 nm gradually increased with the simultaneous decrease in the emission at 644 nm. In this case, an approximate 96-fold enhancement of the ratiometric value (I494/I644) was observed.However, the addition of Hcy (or GSH) induced only a slight change in the ratiometric value (1.6 and 0.78-fold for Hcy and GSH, respectively). Although, probe 4 could not achieve different signal responses, it could engender distinctive reactivity among the biothiols. Furthermore, probe 4 could be used to image Cys in the human renal cell carcinoma 786–0 cells.
Figure 6.
The reactions of 4 with Cys, Hcy, and GSH.
Additionally, Liu et al. reported fluorescent probe 5 with two potential reaction sites (Figure 7) to detect biothiols in pure PBS buffer.[72] The probe itself was non-fluorescent owing to the donor-excited photoinduced electron transfer (d-PET) process. The Cys-induced Michael addition-rearrangement cascade reaction led to an amino-substituted product 5 c with strong fluorescence. This was ascribed to inhibiting C=C isomerization-induced fluorescence quenching by an intramolecular N—H—O hydrogen bond. The Hcy (or GSH)-induced Michael addition reaction led to a thiol-substituted product 5b (or 5-GSH), which lacked a similar intramolecular hydrogen-bonding interaction and thus displayed poor fluorescence due to efficient C=C isomerization-induced fluorescence quenching. Even in the presence of Hcy (or GSH), the probe could detect Cys with obvious fluorescence enhancement. Although probe 5 failed to distinguish between all three of Cys, Hcy, and GSH, the but-3-yn-2-one group and the isomerization effects was effective for distinguishing between Cys and Hcy and embodies a potentially useful foundation for probes with even greater selectivity among the thiols. Using a laser scanning confocal microscope, the authors demonstrated that probe 5 could selectively image Cys in the human renal cell carcinoma 786–0 cells.
Figure 7.
The reactions of 5 with Cys, Hcy, and GSH.
4. Nanoprobes as Materials for the Potential Discrimination of Cys, Hcy, and GSH
Although multiple-binding-site fluorescent probes can effectively distinguish Cys, Hcy, and GSH, these fluorescent probes may suffer from complex synthesis routes. Additionally, chemical and photostability may be problematic and hinder practical applications. Nanoparticle probes possess excellent photostability, good water solubility, chemical inertness, low toxicity, favorable biocompatibility, and relatively low cost.[73]
Targeted research on the use of quantum dots (QDs)[74] for bioanalytes of small molecular weight has been successful in the selective detection of biothiols over other amino acids without thiol groups. Guo et al. used N,S-carbon dots (N,S-CDs) complexed with CuII, in which the metal acts as the quencher, and the N,S-CDs fluorescence was reactivated by biothiols. The N,S-CDs probe was successful in the selective detection of biothiols in human serum but so far lacks selectivity for each of GSH/Cys/Hcy.[15t]
The number of nanoparticle probes that can selectively detect one or two of GSH/Cys/Hcy has been increasing during the past decade.[75] However, nanoparticle probes that can simultaneously distinguish Cys, Hcy, and GSH have not yet been reported. This is a fertile area for further research since nanoparticles are amenable to functionalization with divergent receptors.[76]
5. Summary and Outlook
In conclusion, multiple binding site probes can be used to not only distinguish the structurally related biothiols Cys, Hcy, and GSH but also enable their simultaneous monitoring, provided probe reactivity and specificity is optimal. This is significant because levels of each of the thiols are related to redox events associated with major diseases. The use of multiple-binding-site probes is a new and unique approach but is not the only strategy to address the issues facing modern thiol research. Future studies in this area can address critical issues in GSH and Hcy monitoring such as inter-laboratory inconsistencies and facilitate clinical trials. The design principles for simultaneous thiol monitoring can also be applied to other significant targets such as the Hcy metabolites, the perthiols, H2S, thiophenols, and many others. Moreover, there are many opportunities to create optimized probes based on the foundational principles described in the examples from the literature presented herein. Probes should be evaluated not only for their function in biological media such as cell culture and plasma for detecting one specific thiol but also should demonstrate clear selectivity in mixtures of thiols with dynamically changing levels of each analyte.[23] Future progress can enable the monitoring of disease progression and treatment by using multiple biomarkers in a simple, rapid and reliable manner. This will lead to more efficient clinical trials, disease diagnostics and staging, and tracking of therapeutic outcomes and will help enhance the understanding of the chemical and biological basis of disease.
Acknowledgements
The work was supported by the National Natural Science Foundation of China (21672131), Talents Support Program of Shanxi Province (2014401), Scientific Instrument Center of Shanxi University and the National Institutes of Health (R15EB016870).
Biography

Caixia Yin joined the group of Shie Ming, Peng at Taiwan University as a postdoctoral, fellow (2006–2007). She visited University, of Missouri, USA, in 2014–2016. Now she, is a Professor of Inorganic Chemistry at the Institute of Molecular Science at Shanxi University. Her current research interests are molecular recognition and sensor chemistry.

Kangming Xiong obtained his B.Sc. in July 2014 from Shanxi University in China. In September 2014, he joined Prof. Caixia Yin’s group at the Institute of Molecular Science, Shanxi University, to pursue his M.Sc. degree. His current research is in molecular chemistry.

Fangjun Huo visited the University of Mis-souri, USA, in 2014–2016. Now he is an Associate Professor of Organic Chemistry in the Research Institute of Applied Chemistry at Shanxi University. His current research interests are sensor chemistry and supramolecular chemistry.

James C. Salamanca holds a B.A. from Portland State University. He joined the Strongin Croup and was the 2016 recipient of the Dawn Dressier Health Science Studies Endowed Award. His current research interests include biomedical applications of chemosensors for metabolites associated - with disease pathogenicity in humans.

Professor Robert M. Strongin is a Professor of Organic Chemistry at Portland State University. His major interests are in physical organic and synthetic organic chemistry. He specializes in redox chemistry as applied j to probe and pharmaceutical design, as well as to understanding the molecular basis of disease and tobacco-related toxin formation.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Cai-Xia Yin, Key Laboratory of Chemical Biology and Molecular Engineering of Ministry of Education Institute of Molecular Science,Key Laboratory of Materials for Energy Conversion and Storage of Shanxi Province, Shanxi University, Taiyuan 030006 (China)
Kang-Ming Xiong, Key Laboratory of Chemical Biology and Molecular Engineering of Ministry of Education Institute of Molecular Science,Key Laboratory of Materials for Energy Conversion and Storage of Shanxi Province, Shanxi University, Taiyuan 030006 (China)
Fang-Jun Huo, Research Institute of Applied Chemistry, Shanxi University, Taiyuan 030006 (China)
James C. Salamanca, Department of Chemistry, Portland state University, Portland, OR, 97201 (USA)
Robert M. Strongin, Department of Chemistry, Portland state University, Portland, OR, 97201 (USA)
References
- [1].Wood ZA, Schroder E, Harris JR, Poole LB, Trends Biochem. Sci 2003, 28, 32–40. [DOI] [PubMed] [Google Scholar]
- [2].Harris C, Hansen JM, Methods Mol. Biol 2012, 889, 325–346. [DOI] [PubMed] [Google Scholar]
- [3].Pham-Huy LA, He H, Pham-Huy C, Int. J. Biomed. Sci 2008, 4, 89–96. [PMC free article] [PubMed] [Google Scholar]
- [4].Vacek J, Klejdus B, Petrlova J, Lojkova L, Kuban V, Analyst 2006, 131, 1167–1174. [DOI] [PubMed] [Google Scholar]
- [5].a) Stamler JS, Loscalzo J, Anal. Chem 1992, 64, 779–785; [DOI] [PubMed] [Google Scholar]; b)Inoue T, Kirchhoff JR, Anal. Chem 2002, 74, 1349–1354. [DOI] [PubMed] [Google Scholar]
- [6].a) Rahman I, Kode A, Biswas SK, Nat. Protoc 2007,1, 3159–3165; [DOI] [PubMed] [Google Scholar]; b) Liang G, Ronald J, Chen Y, Ye D, Pandit P, Ma ML, Rutt B, Rao J, Angew. Chem. Int. Ed 2011, 50, 6283–6286; Angew. Chem 2011,123, 6407–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kaczynska A, Pelsers MMAL, Bochowicz A, Kostru-biec M, Glatz JFC, Pruszczyk P, Clin. Chim. Acta 2006, 371, 117–123. [DOI] [PubMed] [Google Scholar]
- [8].a) MacCoss MJ, Fukagawa NK, Matthews DE, Anal Chem. 1999, 71, 4527–4533; [DOI] [PubMed] [Google Scholar]; b) Vellasco AP, Haddad R, Eberlin MN, Hoehr NF, Analyst 2002, 127, 1050–1053; [DOI] [PubMed] [Google Scholar]; c) Xu C, Li H, Yin B, Biosens. Bioelectron 2015, 72, 275–281. [DOI] [PubMed] [Google Scholar]
- [9].Seiwert B, Karst U, Anal. Chem 2007, 79, 7131–7138. [DOI] [PubMed] [Google Scholar]
- [10].Spence MT, Johnson ID, The Molecular Probes Handbook: a Guide to Fluorescent Probes and Labeling Technologies, Live Technologies Corporation, Carlsbad, 2010. [Google Scholar]
- [11].Chen H, Tang Y, Lin W, TrAC Trends Anal Chem. 2016, 76, 166–181. [Google Scholar]
- [12].Zhou Y, Yoon J, Chem. Soc. Rev 2012, 41, 52–67. [DOI] [PubMed] [Google Scholar]
- [13].a) Yin L-L, Chen Z-Z, Tong L-L, Xu K-H, Tang B, Chin. J. Anal. Chem 2009,37,1073 −1081; [Google Scholar]; b) Yin C, Huo F, Zhang J, Martinez-Manez R, Yang Y, Lv H, Li S, Chem. Soc. Rev 2013, 42, 6032–6059; [DOI] [PubMed] [Google Scholar]; c) Niu L-Y, Chen Y-Z, Zheng H-R, Wu L-Z, Tung C-H, Yang Q-Z, Chem. Soc. Rev 2015,44, 6143–6160; [DOI] [PubMed] [Google Scholar]; d) Chen X, Zhou Y, Peng X, Yoon J, Chem. Soc. Rev 2010, 39, 2120–2135; [DOI] [PubMed] [Google Scholar]; e) Hoyle CE, Bowman CN, Angew. Chem. Int. Ed 2010, 49, 1540–1573; Angew. Chem 2010,122, 1584–1617; [DOI] [PubMed] [Google Scholar]; f) Wang K, Peng H, Wang B, J. Cell. Biochem 2014,115, 1007–1022. [DOI] [PubMed] [Google Scholar]
- [14].a) Jung HS, Chen X, Kim JS, Yoon J, Chem. Soc. Rev 2013, 42, 6019–6031; [DOI] [PubMed] [Google Scholar]; b) Wang J, Liu H-B, Tong Z, Ha C-S, Coord. Chem. Rev 2015, 303, 139–184. [Google Scholar]
- [15].a) Zhang Q, Yu D, Ding S, Feng G, Chem. Commun 2014, 50, 14002–14005; [DOI] [PubMed] [Google Scholar]; b) Zhang H, Zhang C, Liu R, Yi L, Sun H, Chem. Commun 2015, 51, 2029–2032; [DOI] [PubMed] [Google Scholar]; c) Sun J, Zhang L, Zhang X, Hu Y, Ge C, Fang J, Analyst 2016, 141, 2009–2015; [DOI] [PubMed] [Google Scholar]; d) Zhu X, Li Y, Zan W, Zhang J, Chen Z, Liu X, Qi F, Yao X, Zhang X, Zhang H, Photochem. Photobiol. Sci 2016, 15, 412–419; [DOI] [PubMed] [Google Scholar]; e) Dong C, Zhou C-Q, Yang J-W, Liao T-C, Chen J-X, Yin C-X, Chen W-H, RSC Adv. 2015, 5, 32990–32993; [Google Scholar]; f) Zhang Q, Ding S, Zhai Q, Feng G, RSC Adv. 2015, 5, 62325–62330; [Google Scholar]; g) Cao X, Lin W, Yu Q, J. Org. Chem 2011, 76, 7423–7430; [DOI] [PubMed] [Google Scholar]; h) Zhu X, Gao H, Zan W, Li Y, Zhang J, Liu X, Wei X, Qi F, Yao X, Zhang H, Tetrahedron 2016, 72, 2048–2056; [Google Scholar]; i) Men Y, Li Z, Zhang J, Tong Z, Xi Z, Qiu X, Yi L, Tetrahedron Lett. 2015, 56, 5781–5786; [Google Scholar]; j) Chen W, Luo H, Liu X, Qi F, Yang D, Song X, Tetrahedron Lett. 2015, 56, 7176–7179; [Google Scholar]; k) Wang S-P, Deng W-J, Sun D, Yan M, Zheng H, Xu J-G, Org. Biomol. Chem 2009, 7, 4017–4020; [DOI] [PubMed] [Google Scholar]; l) Liu X-D, Sun R, Xu Y, Xu Y-J, Ge J-F, Lu J-M, Sens. Actuators B 2013, 178, 525–531; [Google Scholar]; m) Shin I-S, Gwon S-Y, Kim S-H, Spectrochim. Acta Part A 2014, 120, 642–645; [DOI] [PubMed] [Google Scholar]; n) Kand D, Kalle AM, Varma SJ, Talukdar P, Chem. Commun. 2012, 48, 2722 −2724; [DOI] [PubMed] [Google Scholar]; o) Wang J, Zhou C, Zhang J, Zhu X, Liu X, Wang Q, Zhang H, Spectrochim. Acta Part A 2016,166, 31–37; [DOI] [PubMed] [Google Scholar]; p) Liu Y, Xiang K, Tian B, Zhang J, Tetrahedron Lett. 2016, 57, 2478–2483; [Google Scholar]; q) Zhang Y, Huo F, Yin C, Yue Y, Hao J, Chao J, Liu D, Sens. Actuators B 2015, 207, 59–65; [Google Scholar]; r) Qu L, Yin C, Huo F, Li J, Chao J, Zhang Y, Sens. Actuators B 2014, 195, 246–251; [Google Scholar]; s) Liu T, Huo F, Li J, Chao J, Zhang Y, Yin C, Sens. Actuators B 2016, 232, 619–624; [Google Scholar]; t) Guo Y, Yang L, Li W, Wang X, Shang Y, Li B, Microchim. Acta 2016, 183, 1409–1416. [Google Scholar]
- [16].Sibrian-Vazquez M, Escobedo JO, Lim S, Samoei GK, Strongin RM, Proc. Natl Acad. Sci. USA 2010, 107, 551–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mohammad NS, Shruti PS, Bharathi V, Prasad CK, Hussain T, Alrokayan SA, Naik U, Devi ARR, Psychiatr. Genet 2016, 26, 281–286. [DOI] [PubMed] [Google Scholar]
- [18].James SJ, Melnyk S, Jernigan S, Cleves MA, Halsted CH, Wong DH, Cutler P, Bock K, Boris M, Bradstreet JJ, Am. J. Med. Genet. Part B 2006, 141, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rusin O, St Luce NN, Agbaria RA, Escobedo JO, Jiang S, Warner IM, Dawan FB, Lian K, Strongin RM, J. Am. Chem. Soc 2004, 126, 438–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Barve A, Lowry M, Escobedo JO, Thainashmuthu J, Strongin RM, Fluoresc J. 2016, 26, 731–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Barve A, Lowry M, Escobedo JO, Huynh K, Hakuna L, Strongin R, Chem. Commun. 2014, 50, 8219–8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yang X, Guo Y, Strongin RM, Org. Biomol Chem 2012, 10, 2739–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yang X, Guo Y, Strongin RM, Angew. Chem. Int. Ed 2011, 50, 10690–10693; Angew. Chem 2011, 123, 10878–10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yue Y, Huo F, Li X, Wen Y, Yi T, Salamanca J, Escobedo JO, Strongin RM, Yin C, Org. Lett 2017, 19, 82–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].a) Guo Y, Yang X, Hakuna L, Barve A, Escobedo JO, Lowry M, Strongin RM, Sensors 2012, 12, 5940–5950; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hakuna L, Doughan B, Escobedo JO, Strongin RM, Analyst 2015, 140, 3339–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yue Y, Yin C, Huo F, Chao J, Zhang Y, Sens. Actuators B 2016, 223, 496–500. [Google Scholar]
- [27].Yuan L, Lin W, Xie Y, Zhu S, Zhao S, Chem. Eur J 2012, 18, 14520–14526. [DOI] [PubMed] [Google Scholar]
- [28].Niu L-Y, Yang Q-Q, Zheng H-R, Chen Y-Z, Wu L-Z, Tung C-H, Yang Q-Z, RSC Adv. 2015, 5, 3959–3964. [Google Scholar]
- [29].Niu L-Y, Guan Y-S, Chen Y-Z, Wu L-Z, Tung C-H, Yang Q-Z, J. Am. Chem. Soc. 2012, 134, 18928–18931. [DOI] [PubMed] [Google Scholar]
- [30].Wang H, Zhou G, Gai H, Chen X, Chem. Commun 2012, 48, 8341–8343. [DOI] [PubMed] [Google Scholar]
- [31].Liu J, Sun Y-Q, Zhang H, Huo Y, Shi Y, Guo W, Chem. Sci 2014, 5, 3183–3188. [Google Scholar]
- [32].Isik M, Guliyev R, Kolemen S, Altay Y, Senturk B, Tekinay T, Akkaya EU, Org. Lett 2014, 16, 3260–3263. [DOI] [PubMed] [Google Scholar]
- [33].He L, Xu Q, Liu Y, Wei H, Tang Y, Lin W, ACS Appl. Mater. Interfaces 2015, 7, 12809–12813. [DOI] [PubMed] [Google Scholar]
- [34].Kim Y, Mulay SV, Choi M, Yu SB, Jon S, Churchill DG, Chem. Sci 2015, 6, 5435–5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Liu Y, Lv X, Liu J, Sun Y-Q, Guo W, Chem. Eur J 2015, 21, 4747–4754. [DOI] [PubMed] [Google Scholar]
- [36].Lv H, Yang X-F, Zhong Y, Guo Y, Li Z, Li H, Anal Chem. 2014, 86, 1800–1807. [DOI] [PubMed] [Google Scholar]
- [37].Liu Y, Lv X, Hou M, Shi Y, Guo W, Anal. Chem 2015, 87, 11475–11483. [DOI] [PubMed] [Google Scholar]
- [38].Zhang X, Hang Y, Qu W, Yan Y, Zhao P, Hua J, RSC Adv. 2016, 6, 20014–20020. [Google Scholar]
- [39].Zhao N, Gong Q, Zhang RX, Yang J, Huang ZY, Li N, Tang BZ, J. Mater. Chem. C 2015, 3, 8397–8402. [Google Scholar]
- [40].Zhang D, Yang Z, Li H, Pei Z, Sun S, Xu Y, Chem. Commun 2016, 52, 749–752. [DOI] [PubMed] [Google Scholar]
- [41].Chen H, Li X, Wu Y, Gao W, Bai R, Dalton Trans. 2012, 41, 13292–13297. [DOI] [PubMed] [Google Scholar]
- [42].Wang F, Zhou L, Zhao C, Wang R, Fei Q, Luo S, Guo Z, Tian H, Zhu W-H, Chem. Sci 2015, 6, 2584–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wang F, Guo Z, Li X, Li X, Zhao C, Chem. Eur. J 2014, 20, 11471–11478. [DOI] [PubMed] [Google Scholar]
- [44].Song L, Jia T, Lu W, Jia N, Zhang W, Qian J, Org. Biomol Chem 2014, 12, 8422–8427. [DOI] [PubMed] [Google Scholar]
- [45].Das P, Mandal AK, Chandar NB, Baidya M, Bhatt HB, Ganguly B, Ghosh SK, Das A, Chem. Eur. J 2012, 18, 15382–15393. [DOI] [PubMed] [Google Scholar]
- [46].Hou X, Guo X, Chen B, Liu C, Gao F, Zhao J, Wang J, Sens. Actuators B 2015, 209, 838–845. [Google Scholar]
- [47].Lee HY, Choi YP, Kim S, Yoon T, Guo Z, Lee S, Swamy KMK, Kim G, Lee JY, Shin I, Yoon J, Chem. Commun 2014, 50, 6967–6969. [DOI] [PubMed] [Google Scholar]
- [48].Chen H, Tang Y, Ren M, Lin W, Chem. Sci 2016, 7, 1896–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Li Y, Liu W, Zhang P, Zhang H, Wu J, Ge J, Wang P, Biosens. Bioelectron 2017, 90, 117–124. [DOI] [PubMed] [Google Scholar]
- [50].a) Meister A, Anderson ME, Annu. Rev. Biochem 1983, 52, 711–760; [DOI] [PubMed] [Google Scholar]; b) Perricone C, De Carolis C, Perricone R, Auto-immun. Rev 2009, 8, 697–701. [DOI] [PubMed] [Google Scholar]
- [51].Franco R, Schoneveld O, Pappa A, Panayiotidis M, Arch. Physiol Biochem 2007, 113, 234–258. [DOI] [PubMed] [Google Scholar]
- [52].Iwasaki Y, Saito Y, Nakano Y, Mochizuki K, Sakata O, Ito R, Saito K, Nakazawa H, Chromatogr J. B 2009, 877, 3309–3317. [DOI] [PubMed] [Google Scholar]
- [53].Fahrenholz T, Wolle MM, Kingston HS, Faber S, Kern JC, Pamuku M, Miller L, Chatragadda H, Kogelnik A, Anal. Chem. 2015, 87, 1232–1240. [DOI] [PubMed] [Google Scholar]
- [54].Rossi R, Milzani A, Dalle-Donne I, Giustarini D, Lusini L, Colombo R, DiSimplicio P, Clin. Chem 2002, 48, 742–753. [PubMed] [Google Scholar]
- [55].Lang CA, Mills BJ, Mastropaolo W, Liu MC, J. Lab. Clin. Med 2000,135, 402–405. [DOI] [PubMed] [Google Scholar]
- [56].Damy T, Kirsch M, Khouzami L, Caramelle P, Le Corvoisier P, Roudot-Thoraval F, Dubois-Randé J-L, Hittinger L, Pavoine C, Pecker F, PLoS One 2009, 4, e4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Raijmakers MT, Roes EM, Poston L, Steegers EA, Peters WH, Eur. J. Obstet. Gynecol. Reprod. Biol 2008,138, 39–44. [DOI] [PubMed] [Google Scholar]
- [58].Engel J, Mühling J, Kwapisz M, Heidt M, Acta Anaesthesiol. Scand 2009, 53, 1317–1323. [DOI] [PubMed] [Google Scholar]
- [59].Atkuri KR, Cowan TM, Kwan T, Ng A, Herzenberg LA, Herzenberg LA, Enns GM, Proc. Natl. Acad. Sci. USA 2009, 106, 3941–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].a) Santangelo F, Witko-Sarsat V, Drüeke T, Descamps-Latscha B, Nephrol Dial Transplant. 2004, 19, 1951–1955; [DOI] [PubMed] [Google Scholar]; b) Moberly JB, Logan J, Borum PR, Story KO, Webb LE, Jassal SV, Mupas L, Rodela H, Alghamdi GA, Moran JE, J. Am. Soc. Nephrol 1998, 9, 1093–1099. [DOI] [PubMed] [Google Scholar]
- [61].a) Smulders YM, Blom HJ, J. Inherited Metab. Dis 2011, 34, 93–99; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Debreceni B, Debreceni L, Cardiovasc. Ther 2014, 32, 130–138; [DOI] [PubMed] [Google Scholar]; c) Joseph J, Loscalzo J, Nutrients 2013, 5, 3235–3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jakubowski H, Nutr J. 2006, 136, 1741S–1749S. [DOI] [PubMed] [Google Scholar]
- [63].Zaabczyk M, Glowacki R, Machnik A, Herod P, Kazek G, Jakubowski H, Undas A, Clin. Chem. Lab. Med 2011, 49, 729–735. [DOI] [PubMed] [Google Scholar]
- [64].Chwatko G, Jakubowski H, Anal Biochem. 2005, 337, 271–277. [DOI] [PubMed] [Google Scholar]
- [65].a) Jakubowski H, Anal. Biochem. 2008, 380, 257–261; [DOI] [PubMed] [Google Scholar]; b) Glowacki R, Bald E, Jakubowski H, Amino Acids 2011, 41, 187–194. [DOI] [PubMed] [Google Scholar]
- [66].Mazur P, Kozynacka A, Glowacki R, Pfitzner R, Fijorek K, Sadowski J, Undas A, Eur. J. Vasc. Endovasc. Surg 2012, 43, 588–593. [DOI] [PubMed] [Google Scholar]
- [67].a) Hakuna L, Escobedo JO, Lowry M, Barve A, McCallum N, Strongin RM, Chem. Commun 2014, 50, 3071–3073; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Escobedo JO, Wang W, Strongin RM, Nat. Protoc 2006, 1, 2759–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Liu J, Sun Y-Q, Huo Y, Zhang H, Wang L, Zhang P, Song D, Shi Y, Guo W, J. Am. Chem. Soc 2014, 136, 574–577. [DOI] [PubMed] [Google Scholar]
- [69].Zhang H, Liu R, Liu J, Li L, Wang P, Yao SQ, Xu Z, Sun H, Chem. Sci 2016, 7, 256–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Miao Q, Li Q, Yuan Q, Li L, Hai Z, Liu S, Liang G, Anal Chem. 2015, 87, 3460–3466. [DOI] [PubMed] [Google Scholar]
- [71].Liu J, Sun Y-Q, Zhang H, Huo Y, Shi Y, Shi H, Guo W, RSC Adv. 2014, 4, 64542–64550. [Google Scholar]
- [72].Liu Y, Zhang S, Lv X, Sun Y-Q, Liu J, Guo W, Analyst 2014, 139, 4081–4087. [DOI] [PubMed] [Google Scholar]
- [73].a) Baker SN, Baker GA, Angew. Chem. Int. Ed 2010, 49, 6726–6744; Angew. Chem 2010, 122, 6876–6896; [DOI] [PubMed] [Google Scholar]; b) Qu S, Wang X, Lu Q, Liu X, Wang L, Angew. Chem. Int. Ed 2012, 51, 12215–12218; Angew. Chem 2012, 124, 12381–12384; [DOI] [PubMed] [Google Scholar]; c) Wu C, Wang C, Han T, Zhou X, Guo S, Zhang J, Adv. Healthcare Mater 2013, 2, 1613–1619; [DOI] [PubMed] [Google Scholar]; d) Ye R, Xiang C, Lin J, Peng Z, Huang K, Yan Z, Cook NP, Samuel ELG, Hwang C-C, Ruan G, Ceriotti G, Raji A-RO, Marti AA, Tour JM, Nat. Commun 2013, 4, 2943. [DOI] [PubMed] [Google Scholar]
- [74].Xiangzhao A, Qiang M, Xingguang S, Microchim. Acta 2013, 180, 269–277. [Google Scholar]
- [75].a) Deng J, Lu Q, Hou Y, Liu M, Li H, Zhang Y, Yao S, Anal Chem. 2015, 87, 2195–2203; [DOI] [PubMed] [Google Scholar]; b) Lin J-H, Chang C-W, Tseng W-L, Analyst 2010, 135, 104–110; [DOI] [PubMed] [Google Scholar]; c) Lin L, Song X, Chen Y, Rong M, Wang Y, Zhao L, Zhao T, Chen X, Anal Chim. Acta 2015, 891, 261–268; [DOI] [PubMed] [Google Scholar]; d) Stobiecka M, Deeb J, Hepel M, Biophys. Chem 2010, 146, 98–107. [DOI] [PubMed] [Google Scholar]
- [76].a) Huo F, Kang J, Yin C, Zhang Y, Chao J, Sens. Actuators B 2015, 207, 139–143; [Google Scholar]; b) Yao Z, Feng X, Li C, Shi G, Chem. Commun 2009, 5886–5888; [DOI] [PubMed] [Google Scholar]; c) Zhang M, Yu M, Li F, Zhu M, Li M, Gao Y, Li L, Liu Z, Zhang J, Zhang D, Yi T, Huang C, J. Am. Chem. Soc 2007,129,10322 −10323; [DOI] [PubMed] [Google Scholar]; d) Zhang X, Ren X, Xu Q-H, Loh KP, Chen Z-K, Org. Lett 2009, 11, s1257–1260. [DOI] [PubMed] [Google Scholar]