Abstract
Oxidation of a protein cysteinyl thiol (Cys-SH) to S-sulfenic acid (Cys-SOH) by reactive oxygen species (e.g., hydrogen peroxide), termed protein S-sulfenylation, is a reversible post-translational modification that plays a crucial role in redox regulation of protein function in various biological processes. Due to the intrinsically labile nature, protein S-sulfenylation cannot be detected or analyzed directly. Chemoselective probing has been the method of choice for analyzing S-sulfenylated proteins either in vitro or in situ, as it allows stabilization and direct detection of this transient oxidative intermediate. However, it remains challenging to globally pinpoint the specific S-sulfenylated cysteine sites onto complex proteomes and to quantify their dynamic changes upon oxidative stress. This unit describes how a benzothiazine-based chemoselective probe called BTD and mass spectrometry based chemoproteomics can be used to globally and site-specifically identify and quantify protein S-sulfenylation.
Keywords: S-sulfenylation, Chemoproteomics, Mass spectrometry, Click chemistry, Cysteine
INTRODUCTION
Cysteine, the least abundant (1–2 %) of protein-coding amino acids, is unique due to its intrinsically high reactivity and nucleophilicity (Paulsen et al., 2013). Reversible oxidation of protein cysteine thiol to sulfenic acid, termed S-sulfenylation (Cys-SOH), is a post-translational modification that has emerged as a prominent signaling pathway and shown to play critical role in regulating protein function. With an estimated half-life in minutes, Cys-SOH is considered a transient species that spontaneously convert into other, more stable thiol oxoforms (Gupta et al., 2014). High/chronic oxidative stress may cause Cys-SOH to undergo further oxidation to irreversible sulfinic (Cys-SO2H) or sulfonic acid (Cys-SO3H) forms. Due to the abundance of biological thiols (mM levels), an important and facile biological reaction of Cys-SOH is the disulfide formation. The nascent disulfide may undergo thiol-disulfide exchange to regenerate initial Cys-SH. In some rare cases (such as oxidative inactivation of PTP1B), Cys-SOH may undergo intramolecular reaction with adjacent backbone amide nitrogen resulting in the formation of a cyclic species known as cyclic S-sulfenamide that may be reduced back to thiol via disulfide formation (van Montfort et al., 2003). Due to the central role of Cys-SOH in the reversible/irreversible pathway, it serves as an important hub within the redox milieu (Reddie et al., 2008). Over the past decade, mounting evidences demonstrate the crucial role of Cys-SOH in regulation of protein activity, stability, protein-protein interactions, and many other types of biological events (Paulsen et al., 2009, Paulsen et al., 2011, Kulathu et al., 2013, Truong et al., 2013, Chouchani et al., 2016, Hourihan et al., 2016, Bersweiler et al., 2017, Qi et al., 2017, Albertolle et al., 2018). Without any doubt, systematic analysis of Cys-SOH sites and their dynamics during oxidative perturbation, can lead to the discovery of novel mechanisms of redox regulation of protein functions. However, it has proven challenging to study this transient oxidative intermediate in complex proteomes (Leonard et al., 2011, Yang et al., 2016).
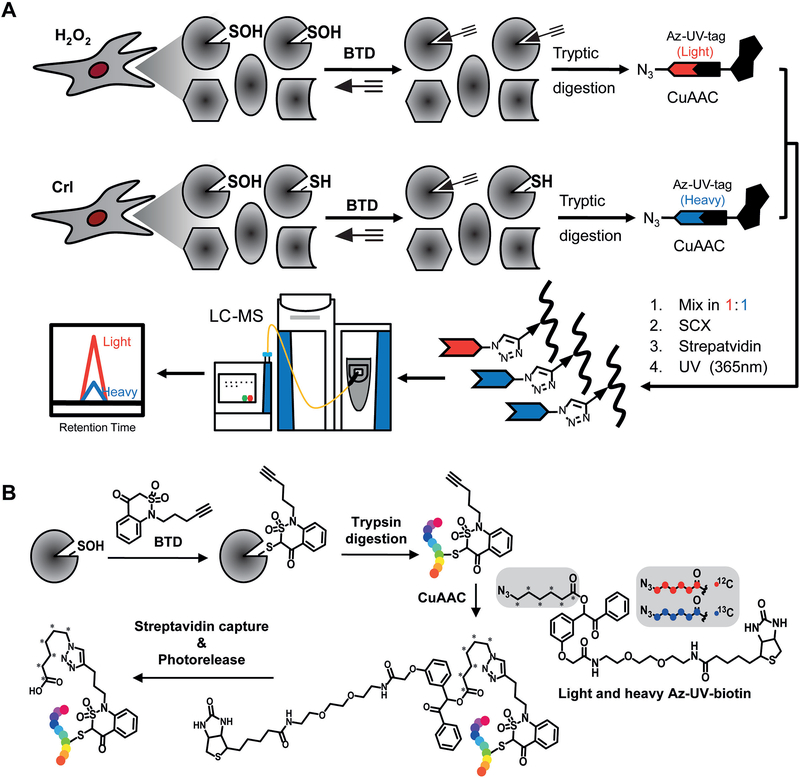
In this unit, we describe a step-by-step protocol of a chemoproteomic approach to globally, site-specifically map and quantify protein S-sulfenylation (Fig. 1A). First, cells treated with or without oxidant are incubated with a new benzothiazine-based probe (called BTD, Fig. 1B) for selective labeling of protein S-sulfenic acids in vitro (Basic Protocol 1) or in situ (Basic Protocol 2). The BTD-labeled cell samples then can be analyzed by traditional immunoblotting (Alternate Protocol 1) or state-of-the-art site-centric chemoproteomics (Basic Protocol 3). For the latter, the process can be divided into following steps: The BTD-labeled proteomes are digested into tryptic peptides, and then tagged with light and heavy azido biotin containing a photocleavable linker via copper-catalyzed azide-alkyne cycloaddition reaction (CuAAC or click chemistry). Light and heavy labeled peptides are combined, cleaned with strong cation exchange (SCX), captured with streptavidin, and eluted selectively under a certain wavelength of UV light. The photoreleased peptides are finally analyzed by mass spectrometry-based proteomics.
Figure 1. BTD-based quantitative S-sulfenylome analysis.
(A) Workflow for proteome-wide analysis of protein S-sulfenylation in RKO cells. S-sulfenylated proteins in cells treated with or without H2O2 are labeled with the BTD probe in vitro or in situ. Cell proteins then were digested with trypsin and labeled peptides are further conjugated with light (H2O2 treated) and heavy (vehicle control) azido biotin with a photocleavable linker, cleaned with SCX spin columns, captured with streptavidin beads, and photoreleased. The resulting peptides were analyzed by LC-MS/MS. (B) Chemical transformation of BTD-labeled form in the entire chemoproteomic workflow.
BASIC PROTOCOL 1 LABELING S-SULFENYLATED PROTEINS IN VITRO
The following is a standard protocol we used for in vitro labeling of S-sulfenylated proteins in cell lines and tissue samples. In this protocol, RKO cells, which are easy to subculture and manipulate, are stimulated with exogenous oxidative stress by H2O2 and labeled with the BTD probe. Labeled proteins can be detected by immunoblot or analyzed by chemoproteomics.
Materials
Reagents
RKO cell line (SGST.CN, cat. no. TCHu116) in logarithmic growth
Cell culture medium (see recipe), 4 °C
50 mM BTD (Synthesized as described previously) (Gupta et al., 2017) in DMSO (see recipe), −20 °C
Phosphate-buffered saline (PBS), 4°C
Hydrogen peroxide (H2O2), 4 °C
Catalase, −20 °C
Equipment
37 °C, humidified 5% CO2 incubator (Thermo Fisher Scientific, cat. no. 3111)
Cell scrape (Biologix, cat. no. 702180)
10-cm cell culture dishes (Corning, cat. no. 430167)
15 mL tubes (Corning, cat. no. 430790)
Eppendorf centrifuge 5417R
Microcentrifuge tube, 1.5 mL (Fisher Scientific)
Prepare cell samples
1. Grow RKO cells in 10-cm plates with MEM medium containing 10 % (vol/vol) FBS to 80–90 % confluence at 37 °C. Cell culture and treatment should be carried out under sterile conditions. If you are not working with oxidant-perturbed samples, continue to step 4.
One 10-cm plate of RKO cells (80–90 % confluence, ~1 × 107 cells) should generate ~1.5–2 mg of protein. For each sample made for S-sulfenylome analysis, 1 mg of total protein is required. We typically obtain this amount of protein from 1 dish of RKO cells. Sufficient cells for duplicates is recommended in case there is a need for a backup. It is noteworthy that appropriate cell culture density and growth conditions may vary based on cell type. To avoid cold stress, culture medium and other cell culture reagents should always be pre-warmed to 37 °C prior to use in an experiment.
2. (Optional) Before cell stimulation experiments, deprive cultured cells from serum overnight (14–16 hr) at 37 °C.
3. (Optional) After serum-deprivation, replace the medium with 15 mL fresh serum-free medium containing 0.5 mM H2O2. The control plates of cells were treated with 15 mL serum-free medium in parallel. Incubate the cells at 37 °C for 5 min.
The concentration of H2O2 and the incubation time are variable, which depend on the needs of the experiment. A control should be performed in parallel using un-stimulated cells to detect H2O2 dependent changes in protein S-sulfenylation. H2O2 can be replaced by other alternatives, such as growth factor, temperature, salt, or other stimuli, which depends on the needs of the experiment.
4. Remove the medium, wash the plates quickly three times with pre-cold PBS containing 200 unit/mL catalase. Scratch cells gently from the dishes and transfer to 15 mL tubes.
Catalase can remove H2O2 quickly to prevent artifactual oxidation during sample handling.
5. Spin down the cells by centrifugation at 100 × g for 3 min at 4 °C. Remove the supernatant after centrifugation and wash pelleted cells twice with pre-cold PBS containing 200 unit/mL catalase.
Probe labeling
6. Add 500 μL of cold lysis buffer containing 200 unit/mL catalase and 5 mM BTD probe to the pelleted cells.
Typically, one volume of cell pellet is lysed by mixing it with three volumes of lysis buffer.
7. Sonicate 10 s with 20 % output, repeat twice, to lyse the cells, and then leave them on ice for 10 min.
8. Incubate the cell lysates with rotation at 37 °C for 2 hr.
9. Analyze experimental results with one of the following methods:
a. Chemoproteomics (See Basic Protocol 3)
b. Immunoblotting (See Alternate Protocol 1)
BASIC PROTOCOL 2 LABELING S-SULFENYLATED PROTEINS IN SITU
The following protocol describes how to find an optimal condition (e.g. probe concentration and labeling time, no effects on cell viability) for in situ labeling of S-sulfenylome in intact cells.
Materials
See Basic Protocol 1
1. Follow Basic Protocol 1, steps 1 through 3.
2. Add 10 mL serum-free MEM containing 1 mM BTD (from 50 mM BTD stock solution) to each plate of cells and incubate at 37 °C for 1 hr.
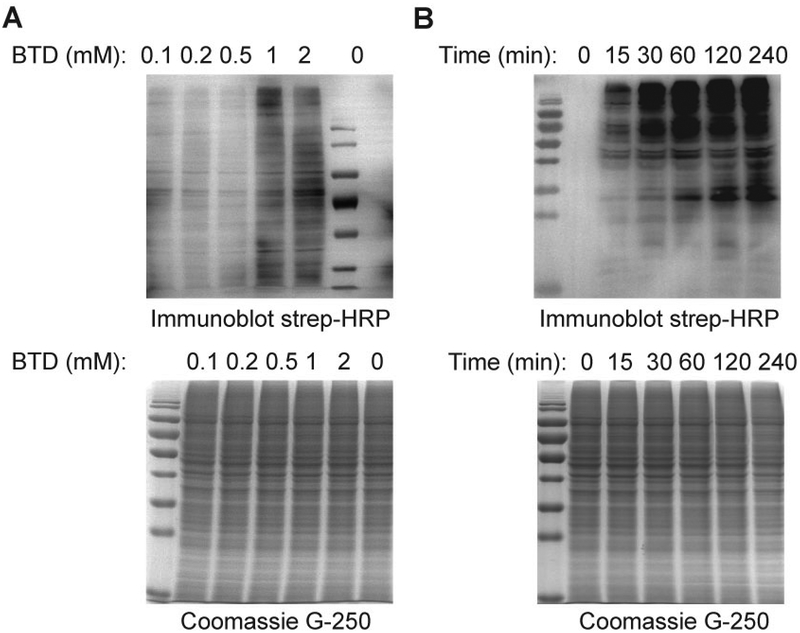
For 10 mL medium, 100 μL of 100 mM BTD is needed to achieve a final concentration of 1 mM. It should be noted that depending on the properties of specific cell lines under study, it is necessary to adjust the probe concentration if the reported procedure does not yield satisfactory results. It is recommended that the user consider optimizing the probe concentration and labeling time (see Alternate Protocol 1) to gain basic proficiency in these techniques prir to attempting the more involved system described in this protocol. Generally, we choose the concentrations of probe as follows: 0, 0.1, 0.2, 0.5, 1.0, and 2.0 mM. And we choose the following labeling time: 0, 15 min, 30 min, 1 h, 2 h, and 4h (Fig. 3). The effect of BTD on cell viability can be evaluated by MTT assay. No cytotoxicity of BTD at 1 mM or less was found to RKO cells.
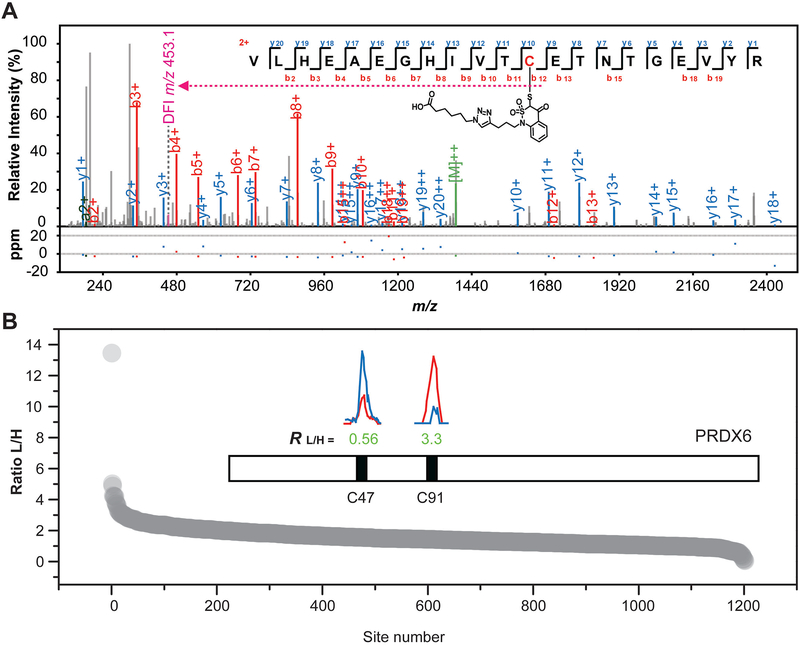
Figure 3. Global mapping and quantification of protein S-sulfenylation in RKO cells.
(A) Fully annotated HCD MS/MS spectrum of the peptide containing SNRPD3 (Small nuclear ribonucleoprotein Sm D3) C20 labeled by BTD. (B) Ranking order of the determined RL/H values of cysteines across the RKO thiol proteome. Inset: Extracted ion chromatograms are shown for changes in S-sulfenylated peptides containing PRDX6 C47 (FTPVCTTELGR) and C91 (DINAYNCEEPTEK) from H2O2 treatment (0.5 mM, 5 min) in RKO cells, with the profiles for light- and heavy-labelled peptides in red and blue, respectively. The site location and the experimental ratios calculated are displayed below the individual chromatograms, respectively.
3. Remove serum-free MEM containing BTD probe.
4. Wash plates three times with pre-cold PBS.
5. Lyse cells with 500 μL lysis buffer containing 200 unit/mL catalase.
6. Sonicate 10 s with 20 % output, repeat twice, to lyse the cells, and then leave them on ice for 10 min.
7. Analyze experimental results with one of the following methods:
a. Chemoproteomics (See Basic Protocol 3)
b. Immunoblotting (See Alternate Protocol 1)
BASIC PROTOCOL 3 ENRICHMENT AND ANALYSIS OF S-SULFENYLATED PEPTIDES LABELED WITH BTD
The following protocol describes a well-established quantitative chemoproteomic platform (Yang et al., 2015, Fu et al., 2017, Gupta et al., 2017, Sun et al., 2017, Sun et al., 2017, Tian et al., 2018) to enrich and analyze the BTD-labeled S-sulfenylated peptides. First, the BTD-labeled protein samples are digested into tryptic peptides. After CuAAC reaction, the BTD-labeled peptides will be transformed to the biotinylated peptides with a photocleavable linker. After streptavidin enrichment, the biotinylated peptides will be photoreleased as the peptides containing the BTD-triazo-hexanoic acid moiety for LC-MS/MS analysis (Fig. 1).
Materials
400 mM Dithiothreitol (DTT), prepared fresh in H2O (see recipe)
800 mM Iodoacetamide (IAM), prepared fresh in H2O (see recipe)
HPLC-grade methanol, room temperature
HPLC-grade water, room temperature
HPLC-grade acetonitrile, room temperature
Chloroform, room temperature
Trypsin digestion buffer (see recipe), room temperature
40 mM Light Azido-photocleavable-biotin (Az-UV-biotin) (see recipe), −80 °C
40 mM Heavy Azido-photocleavable-biotin (Synthesized as described previously) (Yang et al., 2015) (see recipe), −80 °C
100 mM Sodium ascorbate, prepared fresh in H2O (see recipe)
50 mM Tris ((1-benzyl-1H-1,2,3-triazol-4-yl) methyl) amine (TBTA) in DMSO and acetonitrile (see recipe), −80 °C
100 mM Copper sulfate (CuSO4), prepared fresh in water (see recipe)
HLB solvent A (see recipe), room temperature
HLB solvent B (see recipe), room temperature
SCX conditioning buffer (see recipe), room temperature
SCX loading buffer (see recipe), room temperature
SCX elution buffer (see recipe), room temperature
Streptavidin binding buffer (see recipe), room temperature
Streptavidin washing buffer (see recipe), room temperature
Photorelease buffer (see recipe), room temperature
LC-MS sampling buffer (see recipe), room temperature
Sequencing-grade trypsin, frozen (Promega, cat. no. V5113), −20 °C
Bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, cat. no. 23221), room temperature
Streptavidin-Sepharose, high performance (GE, cat. no. 17-5113-01), 4 °C
Software
pFind studio v3.1.2 (http://pfind.ict.ac.cn/software/pFind3/index.html#Downloads)
Equipment
Eppendorf centrifuge 5417R
Microcentrifuge tube, 1.5 mL (Fisher Scientific, cat. no. 509-GRD-Q)
HLB 3 cc (60 mg) SPE cartridges (Waters, cat. no. WAT094226)
HLB 1 cc (10 mg) SPE cartridges (Waters, cat. no. 186000383)
Vacuum centrifugation device (Eppendorf, cat. no.5305000)
MacroSpin SCX column (The Nest Group, cat. no. SMM HIL-SCX)
UV irradiation lamp (UVP, UVLMS-38 EL series)
Thin-walled borosilicate glass vial (VWR International, cat. no. 66011–020)
JY 92-II Sonifier (NingBo Scientz Biotechnology Co., LTD)
NanoLC system (Thermo Fisher Scientific, Easy-nLC 1200)
NanoLC precolumn bulk packing material (3 μm, 120 Å, SunChrom, USA)
NanoLC analytical column bulk packing material (1.9 μm, 120 Å, Dr. Maisch GebH, Germany)
QExactive Plus mass spectrometer (Thermo Fisher Scientific)
Reduction, alkylation, and protein precipitation
1. To 500 μL cell lysates (3~4 mg/mL) obtained from Basic Protocol 1 or 2, add 12.5 μL 400 mM DTT (Final conc. 10 mM DTT) at 37°C for 1hr with rotation and light protection.
To fully unfold the proteins, DTT is added to reduce the reversibly oxidized cysteines.
2. Add 25 μL 800 mM IAM (Final conc. 40 mM IAM) for 30 min in the dark at room temperature.
To prevent potential thiol-exchange reactions, IAM is added to block all reduced cysteines.
3. Add 500 μL of pre-cold methanol and 125 μL of chloroform to each tube to quench the alkylating reaction and to precipitate proteins (Lysate: methanol: chloroform = 4:4:1, vol/vol/vol).
4. Collect proteins at the aqueous/organic phase interface as a solid disk by centrifugation at 1,700 × g for 20 min at 4 °C.
5. Discard liquid layers and wash the pelleted protein twice in methanol/chloroform (1:1, v/v), followed by centrifugation at 16,000 × g for 10 min at 4 °C to re-pellet the proteins.
The protein pellet can be stored at −80 °C for several weeks.
Tryptic digestion
6. Dissolve the protein pellet in 0.5 mL of trypsin digestion buffer (50 mM ammonium bicarbonate) with sonication.
If precipitated protein pellet has not been thoroughly dissolved, the tryptic digestion will be incomplete. Add 0.1 M urea as a final concentration to dissolve the proteins if necessary.
7. Protein concentrations were determined with the BCA assay (Olsen et al., 2007) and adjusted to a concentration of 2 mg/mL. Typically, the final volume of protein sample made for tryptic digestion is 0.5 mL.
8. To the resuspended protein solutions, add sequencing grade trypsin at a 1:50 (enzyme/substrate) ratio and incubate overnight at 37 °C (typically 14–16 hr).
If the tryptic digestion is incomplete, the second trypsin digestion is necessary. Use an enzyme-to-substrate ratio of 1:100 (wt/wt) and incubate at 37 °C for another 4 hr.
9. Centrifuge the tube at 20,000 g for 5 min to remove any insoluble substances and save the supernatant. Reduce the volume of digests to ~0.2 mL by vacuum centrifugation.
Peptide desalting
10. Use 10-mg HLB SPE cartridges to desalt the peptide samples.
The choice of cartridge size should be based on sample input amount. The maximum mass capacities and typical sample volumes for 10-mg HLB SPE cartridges are 1–2 mg and 0.1–1 mL, respectively.
11. Condition the cartridges with 1 mL of HPLC-grade acetonitrile by gravity flow.
12. Equilibrate the cartridges with 2 mL of HPLC-grade water by gravity flow.
13. Load the sample (~200 μL) onto each cartridge by gravity flow.
14. Wash the cartridges with 1 mL of HPLC-grade water by gravity flow.
15. Elute peptides with 1 mL of HLB solvent A by gravity flow.
16. Evaporate the eluted peptide samples to dryness under pressure.
Dried peptide samples are stable, and they can be stored at −20 or −80 °C for several weeks.
CuAAC “Click chemistry”
17. To each 1 mg protein-generated peptides, add 20 μL ddH2O and 10 μL acetonitrile, check the pH (~6 typically), and then add 1 μL 40 mM light Az-UV-biotin (for H2O2 treated sample) or heavy Az-UV-biotin (for untreated sample), 4 μL 100 mM sodium ascorbate, 1 μL 50 mM TBTA, and 4 μL 100 mM CuSO4.
CuAAC: the copper-catalyzed reaction of an azide with an alkyne to form a 5-membered heteroatom ring. Before adding Az-UV-biotin, check the pH of the mixture using a pH indicator strip according to the manufactural instruction. If this is not the case, adjust the pH with formic acid (FA) or NH4OH. If the last desalting step is performed properly, the pH of the reaction mixture should be typically ~6. It is worth mentioning that this reaction formulation is typically used for peptide samples generated from 1–5 mg of input material. If different input sample materials are used and/or other click reagents are used for this reaction, the solvent type or volume, as well as ratios of these reactants, will need to be adjusted. Always perform this reaction in fresh sodium ascorbate and CuSO4 solution, because sodium ascorbate is easily oxidized.
18. Incubate the “Click” mixture at room temperature for 2 hr in the dark rotation.
After 2 hr, the color of the reaction mixture will turn into dark green because Cu(II) is reduced to Cu(I) by sodium ascorbate.
19. After reaction, add three volumes of SCX loading buffer to quench the CuAAC reaction. Centrifuge the mixture at 20,000 g for 5 min at room temperature and transfer the supernatant to a new microcentrifuge tube, which can be used immediately in the following experiments or saved at −80 °C for up to several weeks.
20. (Optional) Combine the supernatants from “light” and “heavy” labeled sample, which can be used immediately in the following experiments.
SCX cleaning
Before the streptavidin capture step, strong cation exchange (SCX) was adopted to remove excess “Click” reagents, especially biotin-related chemicals. Peptides from CuAAC reaction mixtures are positively charged in acidified SCX loading buffer (pH 3.0), retained on the SCX columns, and then separated from the neutrally charged free biotins.
21. Use MacroSpin SCX columns with an Eppendorf 5417R centrifuge to remove excess click reagents.
22. Wash the SCX spin columns with 500 μL of HPLC-grade water by centrifugation at 104 g (= 1050 rpm in Eppendorf 5417R) for 5 min at room temperature to prevent salt precipitation in 2 mL Eppendorf microcentrifuge tubes. Repeat this step once.
23. Condition the SCX spin columns with 500 μL of SCX conditioning buffer by centrifugation at 104 g for 2 min at room temperature, and let it stand in the tube for at least 1 hr before its initial use.
24. Remove the excess SCX conditioning buffer by centrifugation at 104 g for 3 min and equilibrate the SCX spin columns with 500 μL of SCX loading buffer by centrifugation at 104 g for 5 min at room temperature. Repeat this step twice.
25. Load the samples by adding 150 μL of quenched “Click” reaction mixture onto each SCX spin columns. Centrifuge the columns at 104 g for 90 s at room temperature.
26. Wash the SCX spin columns with 150 μL of SCX loading buffer by centrifugation at 104 g for 90 s at room temperature. Repeat this step twice to wash out any trace impurities or unretained detergent.
27. Remove the collecting tubes and blot dry any moisture on the exterior of the columns. Add 150 μL of SCX elution buffers and place them in new 2-mL microcentrifuge tubes. Spin the mixture at 104 g for 90 s at room temperature and save the eluted peptide samples. Repeat this step once.
28. Transfer the eluted solutions into a 15-mL tube, and then add 10 mL of streptavidin binding buffer.
The relatively high concentration of acetonitrile in the SCX eluates is likely to affect the subsequent streptavidin capture and enrichment. Thus, they need to be diluted. We typically combine the same eluates together, transfer them to 15-mL tubes, and then add 10 mL of streptavidin binding buffer to each tube.
Streptavidin capture and photorelease
Commercially available streptavidin-Sepharose beads were used to capture biotinylated peptides. All the solutions used for streptavidin capture and enrichment are adjusted to mildly acidic pH (pH 4.5) to minimize potential hydrolysis of the photocleavable benzoin ester. Direct light exposure should also be minimized during this process.
29. Pre-wash 200 μL of the streptavidin slurry with streptavidin binding buffer twice by centrifugation at 1,700 g for 3 min at room temperature. Resuspend the streptavidin in 200 μL streptavidin binding buffer.
It is worth mentioning that the volume of streptavidin slurry is typically used for 1–2 mg of input material. If different input sample materials are used, the amount of streptavidin slurry will need to be adjusted.
30. Add 200 μL of streptavidin slurry to 15 mL tube containing the SCX eluates, and then incubate the tubes for 2 hr at room temperature with end-to-end rotation and light protection.
31. Wash the streptavidin beads with 10 mL of streptavidin binding buffer by centrifugation at 1,700 g for 3 min at room temperature.
32. Wash the streptavidin beads twice with 10 mL of streptavidin washing buffer by centrifugation at 1,700 g for 3 min at room temperature.
33. Wash the streptavidin beads twice with 10 mL of water by centrifugation at 1,700 g for 3 min at room temperature.
34. Resuspend the streptavidin beads with five volumes of photorelease buffer (25 mM NH4HCO3).
The washed streptavidin beads should be resuspended in at least five volumes of clear buffer solution to ensure the efficiency of the photorelease. Typically, we used 1.2 mL 25 mM NH4HCO3.
35. Transfer the streptavidin suspensions to thin-walled glass tubes and irradiate with 365-nm UV light for 2 hr with stirring.
The photorelease should be performed with magnetic stirring to prevent precipitation of the streptavidin beads to enhance the efficiency of the photorelease.
36. Spin down the streptavidin beads at 1,700 g for 3 min and save the supernatant.
37. Evaporate the photoreleased sample to ~100 μL by vacuum centrifugation.
Final desalting
38. Use 10 mg HLB SPE cartridges to desalt the peptide samples.
39. Condition the cartridges with 1 mL of HPLC-grade acetonitrile by gravity flow.
40. Equilibrate the cartridges with 2 mL of HPLC-grade water by gravity flow.
41. Load the sample (~100 μL) onto each cartridge by gravity flow.
42. Wash the cartridges with 1 mL of HPLC-grade water by gravity flow.
43. Elute peptides with 0.5 mL of HLB solvent A and 250 μL HLB solvent B by gravity flow.
44. Evaporate the eluted peptide samples to dryness by vacuum centrifugation.
Dried peptide samples are stable and can be stored at −20 or −80 °C for several weeks until LC-MS/MS analysis.
LC-MS/MS analysis
45. Dissolve the dried peptide samples in 12 μL of LC-MS/MS sampling buffer with vortexing.
46. Centrifuge the samples at 20,000 g for 10 min to remove any material that did not dissolve, and then place the vials in the LC autosampler, which has been cooled to 4 °C.
After centrifugation, carefully aspirate the supernatant from the sample without touching the bottom of the microcentrifuge tube.
47. For each analysis, we typically inject half of each sample onto the LC-MS/MS at a flow rate of 600 nL min-1.
If the sample amount permits, subject the sample to multiple replicate LC-MS/MS injections. Increasing the number of technical replicates usually increases the number of identified S-sulfenylation sites.
48. Analyze each sample using the LC-MS/MS parameters outlined as follows:
NanoLC column:
A 2 cm microcapillary precolumn packed with C18 (3 μm, 120 Å, SunChrom, USA). The precolumn was connected to a 12 cm 150-μm-inner diameter microcapillary analytical column packed with C18 (1.9 μm, 120 Å, Dr. Maisch GebH, Germany) and equipped with a homemade electrospray emitter tip.
NanoLC separation condition:
0 min, 7% B; 14 min, 10% B; 51 min, 20% B; 68 min, 30% B; 69–75 min, 95% B (A = water, 0.1% formic acid; B = 80 % Acetonitrile, 0.1% formic acid) at a flow rate of 600 nL/min.
Mass spectrometer:
The spray voltage is set to 2.1 kV, and the heated capillary temperature is set to 320 °C. HCD MS/MS spectra were recorded in the data-dependent mode using a Top 20 method. MS1 spectra were measured with a resolution of 70,000, an AGC target of 3e6, a max injection time of 20 ms, and a mass range from m/z 300 to 1800. HCD MS/MS spectra were acquired with a resolution of 17,500, an AGC target of 2e5, a max injection time of 50 ms, a 1.0 m/z isolation window and normalized collision energy of 28. Peptide m/z that triggered MS/MS scans were dynamically excluded from further MS/MS scans for 20 s.
There is no necessity to strictly follow step 48, since the peptide samples can be analyzed in any laboratory with access to a high-resolution LC-MS/MS.
Identification and quantification
49. We routinely use pFind studio to process the obtained shotgun MS/MS data. Major parameters used for S-sulfenylated peptide identification, quantification, and protein assembly are listed in the table(Table.1)
Table 1.
Major parameters used for S-sulfenylated peptide identifcation and protein assembly
| Parameters | Settings |
|---|---|
| Database | Uniprot Homo Sapiens canonical database |
| Enzyme | Trypsin KR_C (Full-specific) |
| Maximum number of missed cleavages | 3 |
| Precursor tolerance | 20 ppm |
| Fragment tolerance | 20 ppm |
| Dynamic modification | Methionine oxidation (+15.9949 Da) Carbamidomethylation (+57.0214 Da) BTD (C13 12C6 H22 N4 O5 S, +418.1311 Da) |
| Quantitation type | Labeling-SILAC etc |
| Multiplicity | 2 |
| FDR | Less than 1 % at peptides level |
| Peptide mass | [600, 10000] |
| Peptide length | [6,100] |
| Number of peptides per protein | At least 1 |
| Protein FDR | 1% |
| Open Search | False |
pFind studio (http://pfind.ict.ac.cn/software/pFind/index.html) is a user-friendly informatic pipeline developed by Dr. Simin He’s Lab for shotgun proteomics, which integrates pFind, a high-performance search engine for automated peptide and protein identification from tandem mass spectra (Li et al., 2005, Wang et al., 2007, Chi et al., 2015) and pQuant, an algorithm mainly designed for MS1-based quantification (Liu et al., 2014). In this case, pQuant was used to calculate the ratios of extracted ion current (XIC) peak intensities for light and heavy isotope-labeled peptides. Notably, pQuant can automatically evaluate the accuracy of calculated isotopic ratios, which represents it main feature (Liu et al., 2014). Thus, the use of pFind studio greatly improves the throughput for processing our quantitative chemoproteomic data, whereas our previously published protocol based on BumberDash coupled with Skyline require extensive manual inspection and peak integration (Yang et al., 2015). The user manual of pFind studio (v3.1.2) can be found in the following website (http://pfind.ict.ac.cn/software/pFind3/index.html#Downloads). The time required of pFind processing depends on the complexity of MS/MS data. For a typical S-sulfenylome analysis, 15 min per RAW file is needed for both identification and quantification using a regular personal computer [Intel(R) Core(TM) i5–4460 CPU@3.2GHz; RAM: 8GB].
ALTERNATE PROTOCOL 1 IMMUNOBLOTTING DETECTION OF S-SULFENYLOME
The following is the standard protocol for detection BTD-labeled proteins by immunoblotting. This protocol can be applied to optimize the condition of probe labeling and investigate sulfenylation in cells and any other organisms of interest (Fig. 3).
Materials
40 mM Azide-biotin in DMSO (see repice), −80 °C
50 mM TBTA in DMSO (see recipe), −80 °C
Tris-Glycine mini gels (Thermo Fisher Scientific, cat. no. XP00145BOX), 4 °C
Tris-Glycine SDS running buffer (Thermo Fisher Scientific, cat. no. LC2675), room temperature
PageRuler prestained protein ladder (Thermo Fisher Scientific, cat. no. 26617), 4 °C
PageBlueTM protein staining solution (Thermo Fisher Scientific, cat. no. 24620), room temperature
Tris-Glycine transfer buffer (Thermo Fisher Scientific, cat. no. LC3675), 4 °C
PVDF membrane (0.45 μm, Merck Millipore, cat. no. IPVH00010), room temperature
20 × TBS TweenTM 20 buffer (TBST)(Thermo Fisher Scientific, cat. no. 28360), room temperature
5 % Skim milk (see recipe), 4 °C
5 % Albumin, bovine (BSA) (see recipe), 4 °C
Streptavidin-HRP antibody (Cell Signaling Technology, cat. no.3999S), −20 °C
ECL Plus western blot detection system (Tanon 4600)
For other materials, see Basic Protocol 1–3
1. Follow Basic Protocol 2, steps 1 through 5.
2. Remove the insoluble substances in the lysates of cell debris by centrifugation at 20,000 × g, 4 °C for 20 min.
3. Transfer the supernatants to new micro-centrifuge tubes and determine the protein concentrations with the BCA assay.
4. Perform a pre-cleaning step to remove endogenous biotinylated proteins prior to CuAAC reaction (see Support Protocol 1).
5. Prepare CuAAC pre-mixture (for up to 12 samples): to a 1.5 mL tube, add 2.5 μL 40 mM Azide-Biotin solution, 5.0 μL 100 mM Sodium ascorbate, 1.0 μL 50 mM TBTA ligand solution, and 5 μL 100 mM Copper Sulfate (CuSO4) solution.
Final concentration are as follows: Azido-Biotin solution: 0.2 mM, Ligand solution: 0.1 mM, Copper Sulfate (CuSO4) solution: 1.0 mM, Sodium ascorbate: 1.0 mM. It is important to note that Azido-Biotin and TBTA ligand solution
should be dissolved in DMSO in this protocol.
6. To 39 μL lysate sample (1.0–2.0 mg/mL), add 1.0 μL CuAAC pre-mixture.
7. Incubate at room temperature for 2 hr with rotation and light protection.
8. Add 10 μL of 4× Protein SDS PAGE Loading buffer to each sample and mix well.
9. Boil protein samples 5 min using a 95 °C heating block.
10. Vortex samples and centrifuge 1 min at 20,000 × g, room temperature.
11. Resolve samples by SDS-PAGE using 14% Tris-Glycine gels.
12. Transfer SDS-PAGE gel with samples onto a PVDF membrane.
13. After transfer, block the PVDF membrane with 5% milk for 1 hr at room temperature.
14. Incubate the membrane with streptavidin-HRP (1:1,000) for 1 hr at room temperature.
The antibody should be diluted in TBST.
15. Wash the membrane three times with TBST for 5 min.
16. Analyze the membrane with chemiluminescence using the ECL Plus western blot detection
system following the manufacturer’s instructions.
SUPPORT PROTOCOL 1 PRE-CLEANING ENDOGENOUS BIOTINYLATED PROTEINS
The following protocol describes a means to reduce the interference from endogenous biotinylated proteins, which, in certain instance, may result in relatively high background signal in western blotting and compromise the efficiency of enrichment experiments described in Basic Protocol 3.
Materials
See Basic Protocol 1 and 3
1. Pre-wash streptavidin sepharose with 500 μL lysis buffer three times for 3 min at 1700 × g at room temperature and resuspend them into the same buffer.
To pre-clean 100 μg cell lysate, use 10 μL of streptavidin sepharose.
2. Add 100 μg cell lysate into the resuspended streptavidin sepharose and incubate at 4 °C for 30 min.
3. Collect the supernatant by centrifugation at 1700 × g for 2 min and transfer into a clean 1.5-mL microcentrifuge tube.
REAGENTS AND SOLUTIONS
Use deionized, distilled water unless otherwise noted
Cell culture medium
50 mL of FBS solution (Thermo Fisher Scientific, cat. no. 10099141)
5 mL 100 U/mL penicillin and 100 μg/mL streptomycin (Thermo Fisher Scientific, cat. no.15070063)
450 mL of Minimum Essential Media (MEM; Thermo Fisher Scientific, cat. no.41500–034)
This medium should be stored at 4 °C and warmed to room temperature before use
BTD stock solution (50 mM)
12 mg benzothiazine-based probe (BTD)
1 mL DMSO (Sigma-Aldrich, cat. no. D8418)
Store at −80 °C for several months
Lysis buffer
50 mM HEPES, pH 7.6 (Thermo Fisher Scientific, cat. no. 15630080)
150 mM NaCl (Sigma-Aldrich, cat. no. S7653)
1% (vol/vol) Igepal (Sigma-Aldrich, cat. no. 18896)
1× protease and phosphatase inhibitor (Thermo Fisher Scientific, cat. no. A32961)
Lysis buffer without protease inhibitors can be stored at 4 °C or room temperature for 6 months. Once the protease inhibitors are added, the lysis buffer should be kept on ice or at 4 °C and used within 24 hr
DTT reduction buffer (400 mM DTT)
18 mg DTT (Sigma-Aldrich, cat. no. 10197777001)
300 μL HPLC-grade water (Fisher Scientific, cat. no. W5–4)
Prepare freshly
IAM blocking buffer (800 mM IAM)
75 mg IAM (Sigma-Aldrich, cat. no. I1149)
500 μL HPLC-grade water
Prepare freshly
Trypsin digestion buffer (50 mM NH4HCO3)
200 mg NH4HCO3 (Sigma-Aldrich, cat. no. A6141)
50 mL HPLC-grade water
Prepare freshly, and the pH of this buffer is typically ~8.0
HLB solvent A
80% acetonitrile (Fisher Scientific, cat. no. A998–4)
10% methanol (Fisher Scientific, cat. no. A452–4)
Store up to 6 months at room temperature
HLB solvent B
10% of 0.1% formic acid (FA, vol/vol) (Fisher Scientific, cat. no. A117–50)
90% HLB desalting solvent A
Store up to 6 months at room temperature
Azido-photocleavable-biotin stock solution for Basic Protocol 3 (40 mM)
15.6 mg Az-UV-biotin (KeraFast, cat. no. EVU102)
50 μL DMSO
450 μL acetonitrile
Stored at −80 °C for at least 6 months
Azide-biotin stock solution for Alternate Protocol 1 (40 mM)
10.2 mg Az-biotin (KeraFast, cat. no. EVU101)
500 μL DMSO
Stored at −80 °C for at least 6 months
TBTA Ligand stock solution used for Basic Protocol 3 (50 mM TBTA)
13 mg TBTA (Sigma-Aldrich, cat. no. 678937)
50 μL DMSO
450 μL acetonitrile
Stored at −80 °C for at least 6 months
Thawed immediately before the click chemistry reaction is performed.
TBTA Ligand stock solution used for Alternate Protocol 1 (50 mM TBTA)
13 mg TBTA
500 μL DMSO
Stored at −80 °C for at least 6 months
CuSO4 solution (100 mM)
25 mg copper sulfate solid (CuSO4; Fisher Scientific, cat. no. C493–500)
1.0 mL HPLC-grade water
Prepare freshly
Sodium ascorbate solution (100 mM)
10 mg sodium ascorbate (NaOAc; Sigma-Aldrich, cat. no. 236400)
0.5 mL HPLC-grade water
Prepare freshly
SCX conditioning buffer (0.2 M monosodium phosphate, 0.3 M sodium acetate)
6.9 g monosodium phosphate (NaH2PO4; Sigma-Aldrich, cat. no. S9638)
10.2 g sodium acetate (NaOAc; Sigma-Aldrich, cat. no. 236400)
250 mL HPLC-grade water
The pH of this solution is ~5.5
Store at least 2 months at room temperature
SCX loading buffer (5 mM NaH2PO4, 25% (vol/vol) acetonitrile, pH 3.0)
103.5 mg monosodium phosphate
150 mL HPLC-grade water
adjust the pH to 3.0 with HCl
finally add 50 ml of acetonitrile
Store at least 2 months at room temperature
SCX elution buffer, 0.4 M (5 mM NaH2PO4, 0.4 M NaCl, 25% (vol/vol) acetonitrile, pH 3.0)
4.6 g NaCl
200 mL SCX loading buffer
Store at least 2 months at room temperature
Streptavidin binding buffer (50 mM NaOAc, pH 4.5)
6.8 g sodium acetate
1 L HPLC-grade water
adjust the pH to 4.5 using acetic acid
Store at least 1 months at room temperature
Streptavidin washing buffer (50 mM NaOAc and 2 M NaCl, pH 4.5)
29 g NaCl250 mL streptavidin binding buffer
Store at least 1 months at room temperature
Photorelease buffer (25 mM NH4HCO3)
100 mg NH4HCO3
50 mL HPLC-grade water
Prepare freshly
LC-MS sampling buffer
0.1 % FA solution (Fisher Scientific, cat. no. A117–50)
5 % (vol/vol) acetonitrile
Store at least 1 months at room temperature
4× Protein SDS PAGE Loading buffer
4 % SDS (Sigma-Aldrich, cat. no.74255)
40 mM Tris-HCl pH8.0
40% Glycerol (Sigma-Aldrich, cat. no.G5516–500ML)
200 mM DTT
0.05 % Coomassie® Brilliant blue G 250 (Sigma-Aldrich, cat. no.1154440025)
Store at least 6 months at −20 °C
5 % Skim milk
5 g Skim milk (BD, cat. no.232100)
100 mL 1×TBST
Prepare freshly
5% Albumin, bovine (BSA)
5 g BSA (Amresco, cat. no.0332–110g)
100 mL 1×TBST
Prepare freshly
COMMENTARY
Background Information
Due to the intrinsically labile nature, protein S-sulfenic acid cannot be detected or analyzed directly with only very few exceptions (Choi et al., 1998). In the last several decades, a wide-choice of dimedone (β-dicarbonyl) and its analogs is available for selective labeling and detection of cysteine sulfenic acids (Furdui et al., 2014). For instance, the dimedone-labeled cysteine S-sulfenylation can be detected by western blotting using its corresponding antibody (Seo et al., 2009). Alternatively, S-sulfenylated proteins can be labeled by dimedone analogs with report groups like fluorescent group or biotin, which allow visualization and/or enrichment of this modification (Poole et al., 2005, Charles et al., 2007, Poole et al., 2007). However, these dimedone-based probes cannot be used to label S-sulfenylated proteins in living cells, since they usually cannot penetrate the cell membrane. To address this issue, Carroll Lab introduced the first in situ sulfenic acid probe called DAz-1, which is a dimedone tagged with an azide chemical handle that can be selectively detected with phosphine reagents via the Staudinger ligation for identification, enrichment and visualization of modified proteins (Reddie et al., 2008). In 2012, Carroll lab developed another cell-permeable alkyne-tagged dimedone analog, DYn-2, which exhibited superior stability and higher efficiency for in situ labeling of Cys-SOH as compared to other dimedone-based probes (Paulsen et al., 2011). In combination with mass spectrometry based proteomics, we and others have successfully use the DYn-2 probe for global profiling of protein S-sulfenylation in plants and human cells (Yang et al., 2014, Akter et al., 2015). Without any doubt, the dimedone-based probes have greatly expanded the inventory of S-sulfenylated proteins and/or sites. However, the moderate reactivity of dimedone warhead towards cysteine sulfenic acid prevent its utility in proteomic application in two-fold: 1) an extremely large amount of input (20–30 mg) and experimental materials (e.g. sequence-grade trypsin, desalting columns, SCX spin columns) is required, 2) labeling reaction mediated by the dimedone-based probe cannot over compete with the rapid turnover of many cysteine sulfenic acids during in situ and in vitro labeling of cell or tissue samples. In addition to dimedone, many other types of chemistry have been explored to selectively label S-sulfenic acids (Qian et al., 2012, Poole et al., 2014, McGarry et al., 2016, Alcock et al., 2018). However, none of them has been applied for global and site-specific analysis of this modification. To develop the next generation of chemoproteomic probe for global analysis of S-sulfenylome, Carroll Lab first developed a novel library containing ~100 cyclic carbon-nucleophiles that react selectively with Cys-SOH (Gupta et al., 2016). Building upon on this resource, they further developed four new alkyne-tagged probes for selective labeling of protein S-sulfenic acids (Gupta et al., 2017). Among of them, a benzothiazine-based probe, called BTD exhibits the highest level of S-sulfenylome reactivity (kobs=1700 M-1s-1), which represents a rate enhancement of more than 2 orders of magnitude as compared to the dimedone-based probe, DYn-2 (kobs=10 M-1s-1) (Gupta et al., 2017). More importantly, BTD has been demonstrated to be highly compatible with our site-centric chemoproteomic platform (Yang et al., 2015, Fu et al., 2017, Sun et al., 2017, Tian et al., 2018). Hence, the new BTD probe can be used to globally, site-specifically map and quantify cysteine S-sulfenylation in complex proteomes in a much more efficient fashion.
Critical Parameters
pH
pH values need to be carefully inspected and/or adjusted in several key steps of this protocol. First, if the secondary tryptic digestion need to be performed, one should check the pH value and make sure it around 8.0 before adding additional enzymes. Second, SCX loading buffer should be kept at pH 3.0 to enable fully protonate peptides in the “Click” mixture. Third, streptavidin capture should be performed in mildly acidic pH (pH 4.5) to minimize potential hydrolysis of the photocleavable benzoin ester.
HLB desalting
If HLB cartridges are overloaded or the flow rates used for the sample loading are too high, it will lead to low yield of eluted peptides. We typically calculate the proper loading amount according to the Manufactures’ instruction and use gravity to load sample solution onto the cartridges.
SCX
As the same as HLB cartridges, SCX spin columns also cannot be overloaded. For a sample generated from 1 mg of starting material, we typically use one MacroSpin SCX spin column. In addition, we typically handle SCX columns at the indicated speed, angle, and duration in the Eppendorf 5417R. If one uses the centrifuge from other vendors, those parameters need to be carefully adjusted to make sure the bedding material is fully wet during the spin-based cleaning step.
CuAAC
To enable efficient “Click” derivatization of BTD-labeled peptides, CuAAC reaction should be performed in ~30% (vol/vol) acetonitrile with pH ~6. The addition of organic phase is used for thorough dissolving the hydrophobic ligand, TBTA. If the pH in the reaction mixture is too low, this “Click” reaction can be inhibited. On the other hand, if the “Click” solution is neutral or basic, the photocleavable benzoin ester linker tend to be hydrolyzed, thereby also resulting low yield of biotinylated peptides.
Troubleshooting
Difficulty in dissolving the precipitated proteins
Indeed, the method/chloroform precipitated protein pellets are not easy to dissolve. However, one can use the commercially available ultrasonic cell disruptor or homogenizer with microtips to handle samples in microcentrifuge tubes. To prevent the protein solutions from overheat during sonication, one can consider performing this step on ice. It should be note that the commonly used ultrasonic cleaners won’t work for such purpose.
Low number of identification
Many issues can lead to low number of identified -SOH sites. If there was almost nothing (none of modified and unmodified peptides) in the pFind report, one could open the RAW MS/MS file and inspect the MS signals. The most common issue might come from the SCX cleaning step. Additionally, sometimes, if LC column clogged during sample running, it would lead to a failure of LC-MS/MS analysis and left only background signals. If one could identify many unmodified peptides but not BTD-labeled ones, it might suggest the low yield of biotinylated peptides formed in CuAAC or a failure of streptavidin capture. In addition to pH value of CuAAC reaction, it should also be noted that the click reagents need to be added into the digested peptide samples in the same order as indicated and in a timely manner. For streptavidin capture, one need to pre-wash the agarose beads and keep the organic content, which come from SCX eluting buffer, as low as possible (Typically < 5%).
Understanding Results
Using the protocol described in this unit and only 2 mg of protein lysates as input materials, in total, we identified 1,867 S-sulfenylated cysteine sites on 1202 proteins in RKO cells upon hydrogen peroxide (H2O2) treatment in a single LC-MS/MS run (Supplemental Table 1). In contrast, in order to achieve a similar coverage (1,105 Cys-SOHs on 778 proteins) of RKO S-sulfenylome using the DYn-2 probe, we previously acquired 30 mg input materials and performed many replicates and LC-MS/MS runs (Yang et al., 2014, Yang et al., 2015). A typical BTD-based identification is demonstrated by a fully annotated HCD MS/MS spectrum with a diagnostic fragment ion (DFI, m/z 453.1) generated by the well-known C-S fragmentation pathway (Fig. 3A).
Furthermore, 64.4% (1202) of the identified Cys-SOHs in RKO cells could be quantified with pQuant interference score less than 0.5 (Fig. 3B and Supplemental Table 1). However, in our previously published work using the light and heavy DYn-2 probes, only 360 Cys-SOHs could be quantified in RKO cells in response to the same stimulus (0.5 mM H2O2, 5min), while 7.5-fold input materials were needed. Interestingly, although the RKO cells used in these two studies were cultured under different conditions, two independent analyses detected the similar dynamic Cys-SOH changes. For example, both analyses detected the decrease of S-sulfenylation level in the catalytic cysteine C47 of PRDX6 after H2O2 treatment, whereas an opposite behavior was found for the PRDX6 C91 (Fig. 3B) (Yang et al., 2014).
Time Considerations
Cells should be cultured 2 or 4 days before the experiment, so they will be sub-confluent on the day of the experiment. Cell lysis, probe labeling, reduction, alkylation, precipitation and BCA quantitation are typically finished in one day, although probe-labeled cell samples can be saved in −80°C until further use. Trypsin digestion can be performed overnight. The 1st peptide desalting step (including the concentration step) typically takes 4–5 hr. After 2 hr of CuAAC reaction, the reaction mixture is typically saved in −80°C. SCX cleaning (~3 hr), streptavidin capture and photorelease (5–6 hr), and final desalting (3–4 hr) can take another one or two days. The time needed for LC-MS/MS and data analysis depends on the number of samples to be processed. For one sample, this entire protocol will take 3–4 days.
Supplementary Material
Figure 2. Western blotting detection of protein S-sulfenylation in RKO cells.
Intact RKO cells were labeled with the BTD probe for the indicated concentration (A) and time (B). BTD-labeled S-sulfenylated proteins can be conjugated to azide-biotin via CuAAC and detected by streptavidin-HRP immunoblot. Equal protein loading is demonstrated by Coomassie G-250 dye staining of the SDS-PAGE gel.
Significance Statement.
Cysteine S-sulfenylation, the reversible oxidation of protein cysteinyl thiol to sulfenic acid, has emerged as a potential mechanism to regulate protein functions and redox signaling. Proteome-wide analysis of this modification will allow comprehensive assessment of its role in redox control and signaling pathways, although it has proven challenging. In this unit, we describe how a benzothiazine-based chemoselective probe called BTD and mass spectrometry based chemoproteomics can be used to globally and site-specifically identify and quantify protein S-sulfenylation. It represents the best possible practice to date and state-of-the-art for quantitative S-sulfenylome analysis.
ACKNOWLEGEMENT
Part of this work was supported by the National Key R&D Program of China (no. 2016YFA0501303), the National Natural Science Foundation of China (nos. 31770885, 31500666 and 81573395) and Beijing Nova Program (no. 171100001117014) to J.Y. and the National Institutes of Health (R01 GM102187 and R01 CA174864 to K.S.C).
LIETERATURE CITED
- Akter S, Huang J, Bodra N, De Smet B, Wahni K, Rombaut D, Pauwels J, Gevaert K, Carroll K, Van Breusegem F & Messens J (2015). DYn-2 Based Identification of Arabidopsis Sulfenomes. Mol Cell Proteomics, 14, 1183–1200. doi: 10.1074/mcp.M114.046896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertolle ME, Phan TTN, Pozzi A & Guengerich FP (2018). Sulfenylation of Human Liver and Kidney Microsomal Cytochromes P450 and Other Drug-Metabolizing Enzymes as a Response to Redox Alteration. Mol Cell Proteomics, 17, 889–900. doi: 10.1074/mcp.RA117.000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcock LJ, Perkins MV & Chalker JM (2018). Chemical methods for mapping cysteine oxidation. Chem Soc Rev, 47, 231–268. doi: 10.1039/c7cs00607a. [DOI] [PubMed] [Google Scholar]
- Bersweiler A, D’Autreaux B, Mazon H, Kriznik A, Belli G, Delaunay-Moisan A, Toledano MB & Rahuel-Clermont S (2017). A scaffold protein that chaperones a cysteine-sulfenic acid in H2O2 signaling. Nat Chem Biol, 13, 909–915. doi: 10.1038/nchembio.2412. [DOI] [PubMed] [Google Scholar]
- Charles RL, Schroder E, May G, Free P, Gaffney PR, Wait R, Begum S, Heads RJ & Eaton P (2007). Protein sulfenation as a redox sensor: proteomics studies using a novel biotinylated dimedone analogue. Mol Cell Proteomics, 6, 1473–1484. doi: 10.1074/mcp.M700065-MCP200. [DOI] [PubMed] [Google Scholar]
- Chi H, He K, Yang B, Chen Z, Sun RX, Fan SB, Zhang K, Liu C, Yuan ZF, Wang QH, Liu SQ, Dong MQ & He SM (2015). pFind-Alioth: A novel unrestricted database search algorithm to improve the interpretation of high-resolution MS/MS data. J Proteomics, 125, 89–97. doi: 10.1016/j.jprot.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Choi HJ, Kang SW, Yang CH, Rhee SG & Ryu SE (1998). Crystal structure of a novel human peroxidase enzyme at 2.0 A resolution. Nat Struct Biol, 5, 400–406. [DOI] [PubMed] [Google Scholar]
- Chouchani ET, Kazak L, Jedrychowski MP, Lu GZ, Erickson BK, Szpyt J, Pierce KA, Laznik-Bogoslavski D, Vetrivelan R, Clish CB, Robinson AJ, Gygi SP & Spiegelman BM (2016). Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1. Nature, 532, 112–116. doi: 10.1038/nature17399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Liu KK, Sun MA, Tian CP, Sun R, Morales Betanzos C, Tallman KA, Porter NA, Yang Y, Guo DJ, Liebler DC & Yang J (2017). Systematic and quantitative assessment of hydrogen peroxide reactivity with cysteines across human proteomes. Mol Cell Proteomics, 16, 1815–1828. doi: 10.1074/mcp.RA117.000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdui CM & Poole LB (2014). Chemical approaches to detect and analyze protein sulfenic acids. Mass Spectrom Rev, 33, 126–146. doi: 10.1002/mas.21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V & Carroll KS (2014). Sulfenic acid chemistry, detection and cellular lifetime. Biochim Biophys Acta, 1840, 847–875. doi: 10.1016/j.bbagen.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V & Carroll KS (2016). Profiling the Reactivity of Cyclic C-Nucleophiles towards Electrophilic Sulfur in Cysteine Sulfenic Acid. Chem Sci, 7, 400–415. doi: 10.1039/C5SC02569A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V, Yang J, Liebler DC & Carroll KS (2017). Diverse Redoxome Reactivity Profiles of Carbon Nucleophiles. J Am Chem Soc, 139, 5588–5595. doi: 10.1021/jacs.7b01791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hourihan JM, Moronetti Mazzeo LE, Fernandez-Cardenas LP & Blackwell TK (2016). Cysteine Sulfenylation Directs IRE-1 to Activate the SKN-1/Nrf2 Antioxidant Response. Mol Cell, 63, 553–566. doi: 10.1016/j.molcel.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulathu Y, Garcia FJ, Mevissen TE, Busch M, Arnaudo N, Carroll KS, Barford D & Komander D (2013). Regulation of A20 and other OTU deubiquitinases by reversible oxidation. Nat Commun, 4, 1569. doi: 10.1038/ncomms2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard SE & Carroll KS (2011). Chemical ‘omics’ approaches for understanding protein cysteine oxidation in biology. Curr Opin Chem Biol, 15, 88–102. doi: 10.1016/j.cbpa.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Li D, Fu Y, Sun R, Ling CX, Wei Y, Zhou H, Zeng R, Yang Q, He S & Gao W (2005). pFind: a novel database-searching software system for automated peptide and protein identification via tandem mass spectrometry. Bioinformatics, 21, 3049–3050. doi: 10.1093/bioinformatics/bti439. [DOI] [PubMed] [Google Scholar]
- Liu C, Song CQ, Yuan ZF, Fu Y, Chi H, Wang LH, Fan SB, Zhang K, Zeng WF, He SM, Dong MQ & Sun RX (2014). pQuant improves quantitation by keeping out interfering signals and evaluating the accuracy of calculated ratios. Anal Chem, 86, 5286–5294. doi: 10.1021/ac404246w. [DOI] [PubMed] [Google Scholar]
- McGarry DJ, Shchepinova MM, Lilla S, Hartley RC & Olson MF (2016). A Cell-Permeable Biscyclooctyne As a Novel Probe for the Identification of Protein Sulfenic Acids. ACS Chem Biol, 11, 261 3300–3304. doi: 10.1021/acschembio.6b00742. [DOI] [PubMed] [Google Scholar]
- Paulsen CE & Carroll KS (2009). Chemical dissection of an essential redox switch in yeast. Chem Biol, 16, 217–225. doi: 10.1016/j.chembiol.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Paulsen CE & Carroll KS (2013). Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem Rev, 113, 4633–4679. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE & Carroll KS (2011). Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat Chem Biol, 8, 57–64. doi: 10.1038/nchembio.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole LB, Klomsiri C, Knaggs SA, Furdui CM, Nelson KJ, Thomas MJ, Fetrow JS, Daniel LW & King SB (2007). Fluorescent and affinity-based tools to detect cysteine sulfenic acid formation in proteins. Bioconjug Chem, 18, 2004–2017. doi: 10.1021/bc700257a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole LB, Zeng BB, Knaggs SA, Yakubu M & King SB (2005). Synthesis of chemical probes to map sulfenic acid modifications on proteins. Bioconjug Chem, 16, 1624–1628. doi: 10.1021/bc050257s. [DOI] [PubMed] [Google Scholar]
- Poole TH, Reisz JA, Zhao W, Poole LB, Furdui CM & King SB (2014). Strained cycloalkynes as new protein sulfenic acid traps. J Am Chem Soc, 136, 6167–6170. doi: 10.1021/ja500364r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Keenan HA, Li Q, Ishikado A, Kannt A, Sadowski T, Yorek MA, Wu IH, Lockhart S, Coppey LJ, Pfenninger A, Liew CW, Qiang G, Burkart AM, Hastings S, Pober D, Cahill C, Niewczas MA, Israelsen WJ, Tinsley L, Stillman IE, Amenta PS, Feener EP, Vander Heiden MG, Stanton RC & King GL (2017). Pyruvate kinase M2 activation may protect against the progression of diabetic glomerular pathology and mitochondrial dysfunction. Nat Med, 23, 753–762. doi: 10.1038/nm.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Wani R, Klomsiri C, Poole LB, Tsang AW & Furdui CM (2012). A simple and effective strategy for labeling cysteine sulfenic acid in proteins by utilization of beta-ketoesters as cleavable probes. Chem Commun (Camb), 48, 4091–4093. doi: 10.1039/c2cc17868k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddie KG & Carroll KS (2008). Expanding the functional diversity of proteins through cysteine oxidation. Curr Opin Chem Biol, 12, 746–754. doi: 10.1016/j.cbpa.2008.07.028. [DOI] [PubMed] [Google Scholar]
- Reddie KG, Seo YH, Muse Iii WB, Leonard SE & Carroll KS (2008). A chemical approach for detecting sulfenic acid-modified proteins in living cells. Mol Biosyst, 4, 521–531. doi: 10.1039/b719986d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YH & Carroll KS (2009). Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proc Natl Acad Sci U S A, 106, 16163–16168. doi: 10.1073/pnas.0903015106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Fu L, Liu KK, Tian CP, Yang Y, Tallman KA, Porter NA, Liebler DC & Yang J (2017). Chemoproteomics Reveals Chemical Diversity and Dynamics of 4-Oxo-2-nonenal Modifications in Cells. Mol Cell Proteomics, 16, 1789–1800. doi: 10.1074/mcp.RA117.000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Shi F, Liu K, Fu L, Tian C, Yang Y, Tallman KA, Porter NA & Yang J (2017). A Chemoproteomic Platform To Assess Bioactivation Potential of Drugs. Chem Res Toxicol, 30, 1797–1803. doi: 10.1021/acs.chemrestox.7b00183. [DOI] [PubMed] [Google Scholar]
- Tian C, Liu K, Sun R, Fu L & Yang J (2018). Chemoproteomics Reveals Unexpected Lysine/Arginine-Specific Cleavage of Peptide Chains as a Potential Protein Degradation Machinery. Anal Chem, 90, 794–800. doi: 10.1021/acs.analchem.7b03237. [DOI] [PubMed] [Google Scholar]
- Truong TH & Carroll KS (2013). Redox regulation of protein kinases. Crit Rev Biochem Mol Biol, 48, 332–356. doi: 10.3109/10409238.2013.790873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Montfort RL, Congreve M, Tisi D, Carr R & Jhoti H (2003). Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature, 423, 773–777. doi: 10.1038/nature01681. [DOI] [PubMed] [Google Scholar]
- Wang LH, Li DQ, Fu Y, Wang HP, Zhang JF, Yuan ZF, Sun RX, Zeng R, He SM & Gao W (2007). pFind 2.0: a software package for peptide and protein identification via tandem mass spectrometry. Rapid Commun Mass Spectrom, 21, 2985–2991. doi: 10.1002/rcm.3173. [DOI] [PubMed] [Google Scholar]
- Yang J, Carroll KS & Liebler DC (2016). The Expanding Landscape of the Thiol Redox Proteome. Mol Cell Proteomics, 15, 1–11. doi: 10.1074/mcp.O115.056051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Gupta V, Carroll KS & Liebler DC (2014). Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat Commun, 5, 4776. doi: 10.1038/ncomms5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Gupta V, Tallman KA, Porter NA, Carroll KS & Liebler DC (2015). Global, in situ, site-specific analysis of protein S-sulfenylation. Nat Protoc, 10, 1022–1037. doi: 10.1038/nprot.2015.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Tallman KA, Porter NA & Liebler DC (2015). Quantitative chemoproteomics for site-specific analysis of protein alkylation by 4-hydroxy-2-nonenal in cells. Anal Chem, 87, 2535–2541. doi: 10.1021/ac504685y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.