Abstract
Mutational analysis is an efficient approach to identifying microbial gene function. Until recently, lack of an effective tool for Anaplasmataceae yielding reproducible results has created an obstacle to functional genomics, because surrogate systems, e.g., ectopic gene expression and analysis in E. coli, may not provide accurate answers. We chose to focus on a method for high-throughput generation of mutants via random mutagenesis as opposed to targeted gene inactivation. In our search for a suitable mutagenesis tool, we considered attributes of the Himar1 transposase system, i.e., random insertion into AT dinucleotide sites, which are abundant in Anaplasmataceae, and lack of requirement for specific host factors. We chose the Anaplasma marginale tr promoter, and the clinically irrelevant antibiotic spectinomycin for selection, and in addition successfully implemented non-antibiotic selection using an herbicide resistance gene. These constructs function reasonably well in Anaplasma phagocytophilum harvested from human promyelocyte HL-60 cells or Ixodes scapularis tick cells. We describe protocols developed in our laboratory, and discuss what likely makes them successful. What makes Anaplasmataceae electroporation competent is unknown and manipulating electroporation conditions has not improved mutational efficiency. A concerted effort is needed to resolve remaining problems that are inherent to the obligate intracellular bacteria. Finally, using this approach, we describe the discovery and characterization of a putative secreted effector necessary for Ap survival in HL-60 cells.
Keywords: Anaplasma phagocytophilum, transposon mutagenesis, effector, selectable markers
1. Introduction
The family Anaplasmataceae includes important human and animal pathogens that are transmitted by ticks (Eremeeva and Dasch, 2011). The incidence of diseases caused by these bacteria has been increasing, likely due to the expanding range of tick vectors linked to global environmental changes such as warming temperatures, more humid weather patterns in endemic regions of the northern hemisphere, and human encroachment on tick host habitat. In combination, these factors support earlier and longer seasonal tick activity and greater chances of tick encounters by humans and companion animals (Robinson et al., 2015). In addition, human activities involved in animal trade and travel have resulted in the introduction of exotic tick-borne pathogens and exotic ticks into regions that were previously unaffected (Barre et al., 2011; ProMED-Mail, 2017).
Anaplasmoses and ehrlichioses are diseases that can be treated with a course of tetracycline antibiotics, but timing is important. When treatment is delayed, the illness can progress to a life-threatening pro-inflammatory syndrome in over one third of patients (Dahlgren et al., 2015; Schotthoefer et al., 2017). Immune-based prevention of these potentially life-threatening illnesses is a desirable goal, but none are currently available. Immunity against obligate intracellular bacteria involves cellular responses that are required for survival of an initial infection. In the case of Anaplasma phagocytophilum (Ap), the clearance of bacteria during primary infection in murine models is dependent on NK cells and IFN-γ and IL-12 secretion during early stages of infection followed by MHC II CD4+ T cell responses (Birkner et al., 2008). However, immune clearance of bacteria upon subsequent challenge relies on antibodies produced by memory B-cells (Feng et al., 2004; Valbuena et al., 2004).
Outer membrane fractions or killed vaccines against the bovine pathogen Anaplasma marginale have been shown to induce protection from infection with homologous but not heterologous strains (Noh et al., 2008). In Israel and Australia, cattle are routinely vaccinated with the related, mild pathogen Anaplasma centrale, which is not approved for use in the US because it results in persistent infection and can cause clinical disease in vaccinees. Nevertheless, there is evidence that transient or long-term infection with vaccine strains may be necessary to achieve protective immunity against challenge with virulent Anaplasmataceae. For instance, an avirulent strain of Ehrlichia muris that causes inapparent, chronic infections in mice efficiently protects against fatal disease from the Ixodes ovatus ehrlichia (Thirumalapura et al., 2009), and specific mutants of other intracellular bacterial pathogens with an infection-deficient phenotype have been shown to protect against virulent wild-type challenge (Bao et al., 2017). Anaplasma marginale transposon mutants expressing fluorescent markers, including one with an insertion in the upstream region of an operon encoding several outer membrane proteins thought to be required for intracellular invasion, exhibited reduced infectivity for cattle without inducing disease. These mutants protected against challenge, and might thus be vaccine candidates (Crosby et al., 2015; Felsheim et al., 2010; Hammac et al., 2013). With the recent advances in mutagenesis of Anaplasmataceae, protective, live attenuated and safe vaccines based on specific mutants with decreased infection ability can now be pursued.
There are two major approaches to mutagenesis, targeted and random, and each poses distinct advantages and challenges. Targeted mutagenesis of known genes may be a quick route to disrupt gene function and identify deficient phenotypes suitable as, e.g., vaccine candidates. On the other hand, random mutagenesis is suited for high-throughput genome-scale gene analysis. In the Rickettsiales, as many as 45% of genes encode hypothetical proteins without known function (Rikihisa, 2011). This significantly restricts the number of targets that can rationally be chosen for directed mutagenesis, though hypothetical genes may encode proteins of critical importance. Bioinformatics-based analyses may aid in the identification of desirable targets, but are often unreliable when applied to Rickettsiales. Importantly, because of their uniqueness, hypothetical proteins constitute desirable choices for development of therapeutics or preventive therapies that are predicted to have few or no off-target or side effects. Here, we summarize and discuss the current status of random mutagenesis of the Anaplasmataceae. We present the methodology that has been most successful in our experience for the generation of random mutants. We analyze the characteristics of one such mutant that suffered a disruption in the coding region for a predicted effector at the locus APH_0906, corresponding to HGE1_03857. We present the results as an example of the power of random mutagenesis for the discovery of proteins involved in infection.
2. Background/History
Mutagenesis of obligate intracellular bacteria has been a goal pursued by a number of laboratories for decades, but has resisted routine application until recently. A variety of approaches were pursued early on, including allelic exchange and transposon mutagenesis. However, reports of successful mutagenesis techniques only started to accumulate at a faster rate in 2009, although the reasons for this uptick are not entirely clear (McClure et al., 2017). A system for extracellular cultivation of Coxiella provided a breakthrough that enabled the generation of a large number of mutants, highlighting the absolute need for a host cell as a major problem hindering mutagenesis in truly obligate intracellular bacteria (Beare et al., 2012). Another impediment for development of genetic systems for Anaplasmataceae remains the fact that these organisms, unlike the Rickettsiaceae (Burkhardt et al., 2011), are unable to maintain plasmids (unpublished), and are at this time not candidates for transformation with a shuttle vector, which is a standard approach used to test bacterial gene function. Between 1994 and 2005, five publications appeared that describe successful events of transformation by non-specific DNA uptake and by transposon mutagenesis, including of typhus group rickettsiae (McClure et al., 2017). These successes were not generally adopted, in part because of the difficulties of working under BSL3 containment, and because conditions favoring transformation were not yet defined. Indeed, to date it is unknown what constitutes the state of transformation competence in Anaplasmataceae, and how to induce it. In 2006, Himar1 transposon mutagenesis was adapted to obtain stable mutants of Ap, a method which targets TA dinucleotides that are plentiful in their genomes, and does not require additional host factors for successful completion (Felsheim et al., 2006; Lampe et al., 1998). Subsequently, this work led to the generation of an Ap mutant library comprising over 1,100 mutants. This library is currently being characterized to prepare it for distribution to the research community (Herron et al., 2010).
Recently, targeted mutagenesis of Ehrlichia chaffeensis using allelic exchange has been developed, and offers a direct way to address the function of a single gene (Wang et al., 2017), including even reconstitution of function. However, here we focus on transposon mutagenesis. In combination, these approaches ring in a new era of genome analysis for the Anaplasmataceae that has application to the Rickettsiales in general.
3. Approach and Methods
a). Preparation of Anaplasmataceae for genetic transformation.
Important considerations to take into account when working with obligate intracellular bacteria are 1) how to minimize damage while purifying them from their host cells in preparation of genetic manipulation, and 2) how to maintain their viability while extracellular. The obligate intracellular Anaplasmataceae quickly lose infectivity when kept outside host cells, thus minimizing this state is important. Anaplasmataceae lack a rigid cell wall that is devoid of peptidoglycan and lipopolysaccharide (Lin and Rikihisa, 2003). In our experience, washing extracellular Anaplasmataceae bacteria in PBS compromises their integrity, possibly by attacking their cell wall, and with each washing step, investigators are left with fewer, more damaged bacteria. Instead, sucrose solutions appear to be less aggressive, and are additionally suitable as a medium for electroporation of the bacteria. Even so, washing cell-free bacteria is detrimental in our experience (based on recovery of mutants), and should be avoided to the extent possible in order to maximize the number of mutants that can be recovered, whether they are derived from mammalian or tick cells. Nevertheless, a protocol for targeted gene disruption of E. chaffeensis by allelic exchange that was recently published (Wang et al., 2017) involves washing bacteria twice in 300 mM sucrose, and gives rise to mutants within two weeks. This approach does not require maximization of mutant yield, however, and presumably a single mutant should suffice to grow into a detectable population when under selection.
An effective protocol (Table 1) then takes a minimum amount of time and requires minimal manipulation of the bacteria. Accordingly, the protocol that our lab has used most successfully involves collecting infected host cells by centrifugation at room temperature, and lysing them mechanically by vortexing in ~1.5 ml of 300 mM sucrose in the presence of ~100 μl rock tumbler grit (60/90 silicon carbide grit, Lortone, Mukilteo, WA) followed by filtration through 2 μm pore size filters (Whatman Puradisc syringe filters). Bacteria are then centrifuged at 10–11,000 × g for 10 minutes, also at room temperature, and no additional washing steps are done. Bacterial pellets are resuspended in 300 mM sucrose with 1 μg of plasmid DNA, and incubated on ice for 15–20 min prior to electroporation, as described in table 1.
Table 1.
Protocol for the transformation of Anaplasma sp. bacteria.
| Step | Procedure | Volume | Centrifugation speed (xg) | Temperature | Time (min) |
|---|---|---|---|---|---|
| 1 | Collect infected cells (above 90%) | Variable | 350 | RT | 5 |
| 2 | Resuspend cells in 300 mM sucrose | 3 ml | - | RT | - |
| 3 | Add suspension into 2-ml microfuge tubes preloaded with silicone carbide tumbler grit 60/90 grade | 50–100 μl | - | RT | - |
| 4 | Vortex tubes on high to disrupt host cells | - | - | RT | 0.5 |
| 5 | Twist a 2-μm pore size filter onto the barrel of a 5-ml syringe | - | - | RT | - |
| 6 | Remove supernatant, add to syringe and filter supernatant into 2-ml microfuge tubes | Variable | - | RT | - |
| 7 | Collect bacteria | Variable | 10,000 | RT | 10 |
| 8 | Resuspend bacteria in desired volume of sucrose | Variable | - | RT | - |
| 9 | Add 1–3 μg plasmid and transfer to electroporation cuvette | Variable | - | RT | - |
| 10 | Incubate on ice | Variable | - | 4–6 °C | 15 |
| 11 | Electroporate at 400 Ohm, 25 μF, 1.8 kV (Pulse times should be 6–9 ms) | - | - | RT | - |
| 12 | Immediately recover bacteria in FBS | 0.3 ml | - | RT | - |
| 13 | Mix with host cells (HL-60 or ISE6 cells) in culture media for the desired number of microplates | 1.5 ml (or as needed) | - | RT | - |
| 14–1 | Incubate with host cells (HL-60) | 1.5 ml (or as needed) | - | 37 °C | 60 |
| 14–2 | Centrifuge suspension (ISE6) | 1.5 ml (or as needed) | 5,000 | RT | 5 |
| 14–3 | Incubate tubes with cell/bacteria pellet (ISE6) | 1.5 ml (or as needed) | - | 30 °C | 120 |
| 15 | Dilute cells in media for desired number of microplates | Variable | - | RT | - |
| 16 | Add spectinomycin and streptomycin the next day | Variable | - | RT | - |
b). Electroporation and infection of host cells.
Electroporation parameters that we have successfully used are 1.7–1.8 kV, 25 μF and 400 Ohm, yielding a pulse of 7–9 ms (Felsheim et al., 2006). Bacteria are recovered from the cuvette in fetal bovine serum (FBS), and immediately added to host cells concentrated into a minimal volume of culture medium (1–1.5 ml with 107 HL-60 or ISE6 cells). Mammalian target cells are incubated with occasional gentle agitation at 37 °C for about 90 minutes before being seeded into 96-well plates and incubated at 37 °C with 5% CO2 in humidified air. Bacteria mixed with tick cells are centrifuged at 5,000 ×g for 5 min at room temperature, and then incubated as a pellet at 30 °C for 90 min before seeding into 48 - or 96 -well plates that are incubated at 34 °C with 4% CO2 in humidified air. Selecting agents are added after 5 h, or the next day. We most commonly use constructs encoding aadA, which confers resistance to spectinomycin and streptomycin, because it is rarely used clinically. Effective concentrations for selection should be tested beforehand, but 100 μg/ml is a good starting point. During electroporation, the bacterial cell membrane is altered structurally, presumably resulting in formation of pores that facilitate uptake of DNA. When successful, electroporation causes bacteria to clump, likely due to the effect on their cell membrane. Consequently, dilution of the cells that were incubated with electroporated bacteria into multi-well plates may result in introduction of more than one mutated bacterium (if caught together in a clump) into a single well, which is what we have experienced. Using this protocol with HL-60 cells results in recovery of ~100 mutants each time, whereas with tick cells, the recovery of mutants is about 30–40 mutants from an infected ISE6 culture containing 107 infected cells, but can be much lower. In HL-60 cells, evidence of mutants expressing fluorescent markers can usually be observed within a week, whereas it takes about 2 weeks before the first mutants are detected in tick cells. Because tick cell culture plates are maintained for 4 weeks, we commonly use 48-well plates for tick cells because they accommodate a greater volume of medium per surface area than do 96 well plates, which reduces the need for feeding plate cultures. Tick cell culture plates are fed once 2 weeks after seeding.
c). Rationale of a dual host cell selection system.
Due to the obligate intracellular nature of the Anaplasmataceae, a saturating mutant library is probably not achievable. However, we have found that Ap differentially express their genome depending on whether they reside in human or tick cells, with hypothetical genes expressed in greater numbers in tick cells. There also is a core set of genes, many encoding metabolic enzymes, expressed equally in both host types (Nelson et al., 2008). Genes absolutely required for intracellular survival are essential for Ap viability, and cannot be disrupted, but those only required for growth in one of the host cell types can be knocked-out without affecting replication in the other cell line. We have previously generated an Ap mutant with a transposon insertion in an o-methyltransferase gene that is required for infection of tick cells only, and displays wild-type growth kinetics in the human promyelocyte cell line HL-60, validating this approach (Oliva Chávez et al., 2015).
4. Constructs and their features.
a). Transposon optimization.
Himar1 transposition efficiency is negatively correlated with transposon size, such that every 1-kb increase in transposon size results in a 38% decrease (Lampe et al., 1998). To keep the transposon under 2 kb in size, the fluorescent reporter and spectinomycin resistance genes (aadA) were driven by a single promoter via translational coupling. Translational coupling utilizes a small intervening sequence that ensures that both proteins are transcribed on the same RNA strand and produced with similar stoichiometry (Tian and Salis, 2015). Therefore, we originally designed two plasmids that separately encoded the transposase and the transposon (Felsheim et al., 2006), but have since then combined both functions on a single cis construct (Munderloh et al., 2012). We hypothesized that the chances of a bacterium incorporating a single plasmid were much greater than the chances of a bacterium receiving both the transposon and the transposase plasmids, offsetting the reduction of transformation efficiency due to the larger construct. The codon usage of some biomarkers and resistance proteins differ greatly from that of Ap. To optimize the expression of these proteins we routinely utilize synthetic versions of fluorescent proteins and resistance markers that employ the codon usage patterns of Bacillus subtilis (purchased from DNA2.0 Inc. http://www.dna20.com/). We included the lac operator upstream of the HIMAR1 transposase sequence to reduce expression in E. coli during plasmid production. Once within Ap, the lac operator no longer functions, and expression can proceed. This is not a crucial feature of this construct, but it improves plasmid yield and fidelity.
b). Promoters.
For our research, we have primarily used the well-characterized A. marginale tr promoter, which is homologous to the strong Ap tr1 promoter. Both function in mammalian as well as in tick cells to drive expression of genes in the msp2 cassette as a polycistronic message that is subsequently cleaved into individual components (Barbet et al., 2005). This is the site wherefrom variants of a major surface protein, MSP2, are transcribed, which is thought to underlie the organism’s capacity for immune evasion (Futse et al., 2009). There is evidence that MSP2 variants also mediate invasion of different host cells (Chavez et al., 2012), demonstrating that the mechanism is active in both mammalian and tick host environments. The tr promoter was functionally optimized (Felsheim et al., 2006) and subsequently shown to drive strong and consistent expression of fluorescent and selectable markers in Ap, Ehrlichia muris eauclairensis, and E. chaffeensis (Cheng et al., 2013; Felsheim et al., 2006; Lynn et al., 2015; Pritt et al., 2017) For reasons that remain unanswered, we have never been able to clone an optimized, functional tr1 promoter. In the future, it would be useful to develop a set of well-characterized promoters and associated regulatory elements, including those that can provide inducible or tissue-specific gene expression.
c). Selectable Markers - Antibiotics.
When working with pathogenic bacteria, the choice of a selectable marker must take into consideration which antibiotics are used for treatment of the illness they cause, and antibiotics of therapeutic value must be avoided for selection. Fortunately, antibiotics that are not effective in vivo for various reasons such as inability to penetrate into appropriate cellular or tissue compartments, may well be suitable for in vitro selection. One such antibiotic that is only rarely used, and never for treatment of any rickettsial illness, is spectinomycin. It is able to penetrate into eukaryotic cells, and thus affects bacteria residing intracellularly. The gene encoding spectinomycin resistance, aadA, also imparts resistance against streptomycin. Dual selection can alleviate problems due to single-point mutations in rpsE encoding the Sr protein of the 30s RNA, which can cause non-specific resistance to spectinomycin by changing ribosome structure.
d). Non-antibiotic Selection.
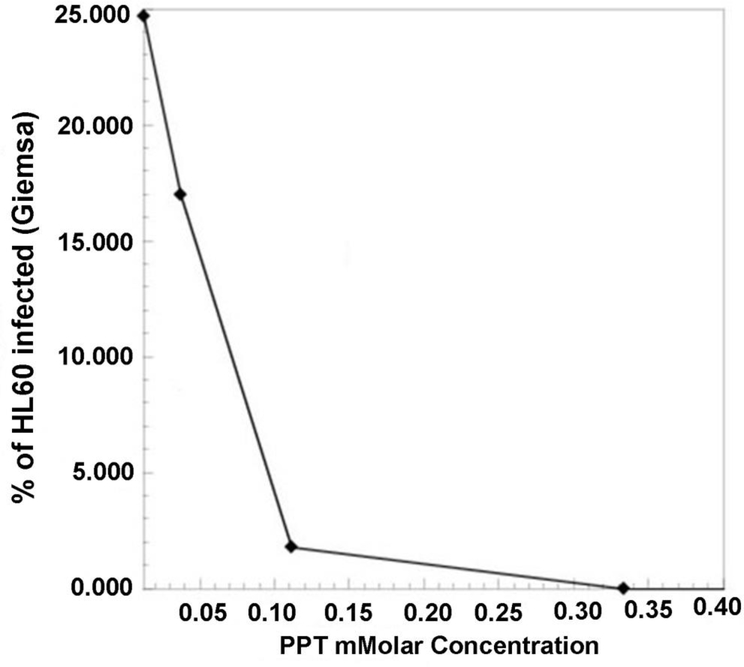
Non-antibiotic selection has been used to create field crops that can be grown in the presence of herbicides, to improve crop yields and avoid inclusion of weeds in the harvested product. Phosphinothricin (PPT), also known as Glufosinate and marketed as the herbicide Basta by Beyer, is useful for selection of transformed plants and bacteria that express resistance conferred by the bar gene (Herrero et al., 1990). Bar encodes PPT N-acetyltransferase that inactivates PPT through acetylation. It was discovered in Streptomyces hygroscopicus that also produces bialaphos (bar=bialaphos resistance), a tripeptide that when metabolized releases PPT, which acts as an analogue of glutamate, and competitively inhibits glutamine synthetase (Hoerlein, 1994; Thompson et al., 1987). Glutamine synthetase (GS) plays a central role in the regulation of eukaryotic and prokaryotic nitrogen metabolism. In susceptible organisms such as plants and bacteria, PPT causes cytosolic accumulation of ammonia and depletion of glutamine, which effectively results in killing intracellular bacteria without adversely affecting mammalian host cells. RPMI1640 cell culture medium for propagation of many human and animal cell lines does not provide sufficient glutamate or glutamine to offset the effect of PPT, so this selection system works well with them. PPT is able to reduce de number of HL-60 cells that are infected with Ap in a concentration dependent manner (Figure 1). However, tick cells are grown in medium that supplies large amounts of glutamate, rendering PPT selection inoperable. To test if PPT could be used as an effective selection marker, we transformed Ap with a plasmid containing the bar gene in replacement of the aadA gene used in (Felsheim et al., 2006). PPT-resistant Ap were generated with an efficiency equivalent to spectinomycin-resistant Ap in the presence of 100 mM PPT added the day following electroporation. Note that for transformation to generate PPT-resistant Ap, we used the original two-plasmid configuration where the transposon and the transposase are encoded on two separate plasmids that are added in equal amounts (1 μg each) to the bacteria in electroporation buffer, i.e., 300 mM sucrose (Felsheim et al., 2006). In future applications, these could also be combined into a single construct. Multiple selection mechanisms facilitate retrieval of mutants in secondary rounds of selection based on additional markers, e.g., when it is desired to recover mutant bacteria in which gene function has been restored with a functional copy of the originally disrupted gene, or when dual or triple mutants are sought.
Figure 1. Sensitivity of A. phagocytophilum wild-type bacteria grown in HL-60 cells to concentrations of phosphinothricin (PPT).
HL-60 cells infected with wild-type A. phagocytophilum were exposed to increasing concentrations of phosphinothricin (PPT). The number of infected cells was assessed through light microscopy after Giemsa-staining. A concentration of 100 mM PPT was subsequently chosen for selection.
Discovery of an effector.
Random mutagenesis followed by phenotype assessment, identification of insertion site, and analysis of disrupted pathways provides an efficient mean to discover the function of hypothetical genes. This is a particularly valuable approach for bacteria the genomes of which encode a large proportion of hypothetical genes. Despite continuous efforts to improve the reliability of bioinformatics software for prediction of genes encoding bacterial effectors (Lockwood et al., 2011; Meyer et al., 2013), this remains difficult. We hypothesized that himar1 transposon mutants with insertions in genes required for infection of mammalian cells could be recovered in tick cell culture. To identify candidate mutants, we screened for impaired ability to efficiently infect and replicate in human HL-60 cells. Here, we describe the generation and characterization of an Ap mutant with an insertion in a gene that encodes a putative T4SS effector previously identified in infected HL-60 cell nuclei by mass-spectrometry (Sinclair et al., 2015). We show that the protein is translocated into the cytoplasms of infected cells, where it accumulates over time. This protein is necessary for Ap survival within HL-60 cells but not for infection of ISE6 tick cells, or hamsters. Accordingly, mutation of aph_0906 using random transposition (Felsheim et al., 2006) disrupted the ability of Ap to replicate in HL-60 cells. Bioinformatic analyses also identified the presence of a putative Nuclear Localization Signal (NLS) and several protein-, DNA-and RNA-binding sites, which are conserved in APH_0906 homologs found in several Ap strains despite divergence in their protein sequence.
Materials and Methods
Culture, transformation, recovery
The human anaplasmosis isolate HGE2 was initially obtained from patient blood cultured with the human promyelocyte line, HL-60 (Goodman et al., 1996), and subsequently propagated in the I. scapularis tick cell line ISE6 (Munderloh et al., 1999). Bacteria were purified from ~107 mechanically disrupted tick cells, washed twice in 270 mM sucrose, and incubated in 50 μl sucrose with 1 μg of each transposon- (containing the gfpuv and aadA genes, pHIMAR1-UV-SS) and himar1 transposase-encoding plasmids for 15 min on ice before electroporation in a 0.2 mm gap cuvette at 1.8 kV, 400 Ohm and 25 μF using a GenePulserII (Biorad) (Felsheim et al., 2006). Bacteria were immediately recovered in 0.5 ml culture medium with 20% FBS, and spread over a confluent layer of ISE6 cells. After 30-min incubation at 34 °C, 5 ml complete medium was added, and spectinomycin plus streptomycin (100 μg/ml each) were included for selection of transformants the next day. The culture was monitored for GFP-expressing bacteria using a Nikon Diaphot inverted microscope (Nikon, New York) fitted for epifluorescence detection, and fed twice weekly with medium containing antibiotics for selection. Intracellular inclusions of transformants were first seen 4 weeks after electroporation, and transformants continued to replicate similar to wild-type.
Determination of insertion site
Southern blot analysis was used to determine the number of insertion sites. DNA from the mutant (designated ΔAPH_0906) and wild-type HGE2 bacteria grown in ISE6 cells was purified using the Puregene Core Kit A (Qiagen, Maryland) followed by phenol/chloroform extraction to remove remaining proteins. DNA (100 ηg), from each sample, was digested with BglII, HindIII and EcoRI and electrophoresed in 1% agarose gels at 30V overnight, and then blotted onto membranes and probed as described (Felsheim et al. 2006). Digoxigenin-labeled probes to detect gfpuv were generated with the PCR DIG Probe Synthesis kit (Roche, Indiana), and the pHIMAR1-UV-SS construct was used as positive control.
The location of the transposon in the APH_0906 coding region was determined by rescue cloning of BglII digested DNA into the pMOD plasmid which was electroporated into ElectroMAX DH5α cells (Invitrogen, New York). ElectroMAX DH5α cells were selected on YT plates with 50 μg/ml of spectinomycin and streptomycin. DNA from one colony was sequenced at the BioMedical Genomics Center (University of Minnesota).
Growth characteristics of ΔAPH_0906
HL-60, RF/6A and ISE6 cells were routinely maintained as described (Goodman et al., 1996; Munderloh et al., 2004, 1999). Growth of the mutant in HL-60, RF/6A and ISE6 cells was determined from two replicate cultures inoculated with bacteria purified from ISE6 cells (Oliva Chávez et al., 2015). Bacteria were resuspended in supplemented RPMI1640 (HL-60) or L15C300 medium (RF/6A and ISE6) and inoculated into 12.5-cm2 flasks containing 3.6 × 105 uninfected HL-60 cells, or confluent layers of RF/6A or ISE6 cells. Cultures were monitored weekly by epifluorescence microscopy of live cultures and by bright-field microscopy of fixed and Giemsa-stained preparations. qPCR of the single copy gene msp5 was used to determine bacterial numbers (Oliva Chávez et al., 2015).
Infection of hamsters with ΔAPH_0906
To test the ability of ΔAPH_0906 to infect mammals, nine 2-week old hamsters were injected i.p. with 500 μl of infected ISE6 cells. After 7 days, heart blood from two euthanized hamsters was cultured with HL-60 and ISE6 cells. DNA was extracted from the remaining blood. Inoculated ISE6 and HL-60 cultures were monitored bi-weekly by epifluorescence microscopy as above. Remaining hamsters were euthanized 21 days p.i., and DNA was extracted from blood and tested for presence of Ap16S rDNA using nested primer pairs EE1-EE2 and EE3-EE4 (Yang et al., 2016).
Relative gene expression of Aph_0906
Wild-type HGE2 Aph_0906 expression was compared in ISE6 and HL-60 cells incubated under standard conditions for 4, 24, 48, or 72 h. Total RNA was extracted from whole infected cells using the Absolutely RNA Miniprep Kit (Agilent, California), and remaining DNA was eliminated by two treatments of the eluate with TURBO DNAse (Ambion, New York). Gene expression was normalized against expression of RNA polymerase subunit B (rpoB) and major surface protein 5 (msp5) genes. rpoB is a housekeeping gene and part of the DNA-directed RNA polymerase pathway necessary for transcription, and msp5 is a single copy gene that is expressed in both cell lines with no significant difference in expression (Nelson et al., 2008). qRT-PCR reactions were carried out using the primers described in Table 1 with Brilliant II QRT-PCR SYBR Green Low ROX Master Mix (Agilent, California). Reaction conditions were one cycle at 50 °C for 30 min, one denaturing cycle at 95 °C for 10 min, 40 cycles that consisted of 30 sec at 95 °C, 1 min at 50 °C, and 1 min at 72 °C, and a final cycle of 1 min at 95 °C and 30 sec at 50 °C. Ct values were established during amplification and the dissociation curve was determined during the final denaturation cycle. The relative expression of aph_0906 was determined from two replicates, using the 2−ΔΔct method.
Production of recombinant APH_0906, mouse immunization
To facilitate localization of protein, we produced recombinant wt-APH_0906 protein for antibody production. Due to the size of the APH_0906 locus (4587 bp), the two halves of the ORF were cloned into the pET29a expression vector (Novagen, Germany). The portion between nucleotides 965395 – 967663 (r1APH_0906) was amplified using primers APH_0906 fw1 and APH_0906 rv1, and the second half (r2APH_0906), located between nucleotides 967516 – 969978 was amplified using primers APH_0906 fw2 and APH_0906 rv2 (Table 1). PCR conditions were one denaturing cycle at 94 °C for 3 min, 10 cycles with a denaturing step at 94 °C for 1 min, annealing at 48 °C for 1 min, and extension at 72 °C for 2 min, followed by 20 additional cycles with a denaturing step at 94 °C for 1 min, 60 °C for 1 min for annealing, and extension at 72 °C for 2 min, and a final extension step of 7 min at 72°C. Amplified products were digested with XhoI and NdeI (r1APH_0906) and XhoI and NcoI (r2APH_0906), and ligated overnight into pET29a at 15 °C. Plasmids were cloned into One Shot® TOP10 competent cells (Invitrogen, Grand Island, NY), purified using the High pure plasmid isolation kit (Roche, Indianapolis, IN), and sequenced at the Biomedical Genomics Center (University of Minnesota). Plasmids were transfected into BL21(D3) E. coli (New England Biolabs, Massachusetts) and induced with 200 μM IPTG in 100 ml of Superior Broth (AthenaES, Baltimore, Maryland) (final concentration) at 37 °C overnight with constant shaking. Recombinant r1APH_0906 and r2APH_0906 were purified using Ni-NTA Fast Start Kit columns (Qiagen, Germantown, Maryland), and dialyzed against TBS in 3 ml Slide-A-Lyzer Dialysis Cassettes with a 10-kDa molecular weight cut-off (Thermo Scientific, Illinois). Protein concentrations were measured using the BCA protein assay kit (Pierce, Illinois). The correct molecular weight of recombinant proteins was verified by electrophoresis on a 4 – 15% Mini protean TGX gel (Biorad, California) stained with Coomassie blue.
Three 6 – 8 weeks old C57BL/6J female mice (Jackson Laboratories, Maine) were injected subcutaneously at the base of the tail with 100 μg of each recombinant protein emulsified in TiterMax Research adjuvant (CytRx Co., Georgia), and boosted 14 days and 24 days later. Antiserum was collected 10 days after the third booster from heart blood after CO2 euthanasia, and serum from naive mice was used as a control.
APH_0906 localization
Infected HL-60 cells (2.7 × 104) were inoculated into five 25-cm2 flasks containing 2 × 105 uninfected HL-60 cells in supplemented RPMI1640. Cultures were incubated at 37 °C and the contents of one flask were processed for detection of APH_0906 each day for 5 days as follows. Cells were collected at 2000 × g for 10 min, fixed in 4% paraformaldehyde in PBS overnight at 4 °C, and then washed three times with 1% BSA in PBS for 5 min. Cells were permeabilized for 10 min in 1% BSA in PBS with 0.1% saponin and 0.1% sodium azide at room temperature. To detect APH_0906 and Ap, cells were incubated with anti-r1APH0906 (1:250) and polyclonal dog anti-Ap antibodies (1:500), respectively, for 2 h at room temperature and then labeled with Cy3-labeled anti-mouse or FITC-labeled anti-dog IgG (each at 1:1,000) for 1 h at room temperature. Labeled samples were washed, centrifuged, and resuspended in 2 ml 1% BSA in PBS, and 10 μl volumes spotted onto Silane-prep™ slides (SigmaAldrich, Missouri). VectaShield mounting medium with DAPI (Vector laboratories, California) was added to label host cells nuclei. Samples were viewed on an Olympus BX61 confocal microscope (Olympus America, Pennsylvania) equipped with a DSU-D2 confocal disk-scanning unit, and images were acquired with a Photometrics QuantEM:512SC EMCCD camera (Photometrics, Arizona) or a QFire color camera (Qimaging, California). Metamorph (Molecular Devices, California) and ImageJ (US National Institutes of Health) were used to capture and compile z-projections, and Photoshop (Adobe Systems, California) was used for cropping and adjustment of brightness/contrast.
In an effort to localize APH_0906 at specific times during bacterial replication, we synchronized HL-60 cell infection with wild-type Ap and imaged samples taken 1 day, 3 days, and 5 days p.i. as described above. Bacteria were purified from a fully infected 25-cm2 flask using mechanical disruption and filtration (Oliva Chávez et al., 2015), and combined with 2.5 ×105 HL-60 cells in 200 μl of supplemented RPMI1640 medium. APH_0906 was localized within infected cells using mono-specific r2-APH0906 antibodies and imaged as above.
Bioinformatics-based comparison of APH_0906 homologs
Secondary and tertiary structures of APH_0906 were modeled using i-TASSER (Yang et al. 2015). The predicted gene ontology (GO) was inferred from the most similar protein with a crystal structure deposited into the Protein Data Bank (PDB) based on the amino acid alignment generated using LOMETS (Local Meta-Threading-Server) (Wu and Zhang, 2007). Because of restrictions on the size of the protein, two separate models for each half of the protein were generated.
The protein sequences of the APH_0906 homologs from strains Webster (APHWEB_1100; Accession # KJV60642), HGE1 (HGE1_03857; Accession # EOA61157), HGE2 (APHHGE2_1212; Accession # KJV82470), ApWI1 (APHWI1_0415; Accession # KJV85064), Annie (OTSANNIE_1184; Accession # KJV98448), CR1007 (APHCR_0401; Accession # KJZ99081), JM (WSQ_04155; Accession # AGR80798), Dog2 (YYY_04145; Accession # AGR82050), MRK (O997_04170; Accession # KDB56310), Norway variant 2 (P029_03620; ANC34432), ApMUC09 (APHMUC_0611; Accession # KJV63589), CRT35 (P030_05350; KDB57323), CRT38 (CRT38_03747; EOA62348), ApNP (APHNP_0408; Accession # KJV68008), and NCH-1 (EPHNCH_1234; Accession # KJV63015) were downloaded from GenBank (https://www.ncbi.nlm.nih.gov/protein/). Sequences were aligned using MUSCLE in Geneious R10 (Biomatters LTD, Auckland, New Zealand), generating a similarity matrix with the total number of differences in each sequence. This alignment was then used to generate phylogenetic trees using the Neighbor-Joining and UPGMA in MEGA7 (Kumar et al., 2016). The positions of protein, DNA, and RNA binding residues within the amino acid sequence of each homolog were predicted using DisoRDPbind (Peng and Kurgan, 2015), and Nuclear Localization Signals (NLSs) were predicted using NLSmapper (Kosugi et al., 2009) and NLStradamus (Nguyen Ba et al., 2009). These features were annotated for each sequence using Geneious R10. Putative Domains were investigated with SMART (Letunic et al., 2015).
Ethics statement
The use of animals in this study was approved by The Institutional Animal Care and Use Committee (IACUC) of the University of Minnesota. All animal experiments were carried out following the guidelines of the National Institutes of Health guide for the care and use of laboratory animals.
Results
Disruption of aph_0906 affects growth of Ap in HL-60 cells
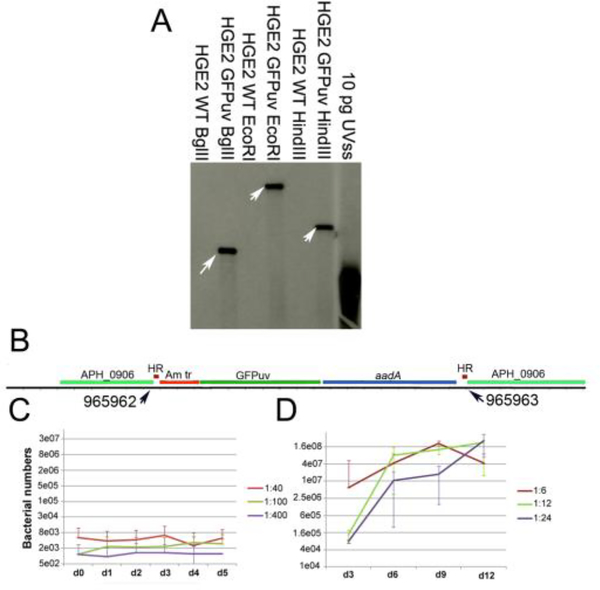
Random transposon mutagenesis of Ap (Felsheim et al., 2006) has proven valuable for analysis of genes required for mammalian pathogenesis (Chen et al., 2012) and invasion of tick vector cells (Oliva Chávez et al., 2015). Here, we describe an Ap HGE2 transposon mutant that was recovered and propagated in an I. scapularis cell line (ISE6). Southern blot analysis identified one band in BglIII, EcoRI, and HindIII digested samples (Figure 2A), suggesting that a single insertion event had occurred. Thus one E. coli clone was sequenced. The sequencing result demonstrated an insertion event into aph_0906 (Gene ID: 3930051), a gene encoding a hypothetical protein, at nucleotide positions 965962 – 965963 (Figure 2B) of the HZ genome (corresponding to positions 1161199 – 1161200 in the HGE2 genome; Locus tag LAOE01000001.1). The mutant was designated ΔAPH_0906.
Figure 2. Himar single transposition event disrupted the APH_0906 coding gene and affected the growth of A. phagocytophilum in HL-60 cells.
A) Southern blot showing a single insertion site for the himar transposition event. Genomic DNA from ΔAPH_0906 and wild-type bacteria was restriction enzyme digested and hybridized with DIG-probes from the GFPuv gene within the himar transposon. Ten pg of the plasmid containing the original UVSS gene were used as positive control. Arrows point at the single hybridization site within each digestion. B) DNA from ΔAPH_0906 was purified, digested with BglII, and electrophorated into E. coli ElectroMAX DH5α cells. Clone recue was performed and a single colony was sent for sequencing. Sequences were compared against the HZ genome and insertion sites were determined to be at positions 965962 – 965963. C) The growth of the mutant was analyzed q-PCR in HL-60. ΔAPH_0906 mutant was unable to replicate in HL-60 in all three dilutions tested. Red line represents the 1:40, the green line is the 1:100 and the purple line shows the 1:400 dilutions. D) On the other hand, growth within ISE6 cells appeared not to be affected by the mutation. Red line represents the 1:6, the green line is the 1:12 and the purple line shows the 1:24 dilutions.
Whole genome-tilling array gene expression analysis has shown that aph_0906 transcription is highly upregulated during infection of HL-60 but not ISE6 cells (Nelson et al., 2008). Indeed, microscopy of HL-60 cells inoculated with ΔAPH_0906 infected ISE6 cells or purified bacteria indicated that the bacteria were unable to form morulae and replicate in HL-60 cells, prompting more detailed analyses to determine the step at which infection by the mutant failed. ΔAPH_0906 persisted in HL-60 cells for up to 5 days post-inoculation without an increase in bacterial numbers (Figure 2C), while replication of ΔAPH_0906 in ISE6 cells was not affected (Figure 2D). These results indicate that although APH_0906 is necessary for replication within HL-60 cells, this gene is unessential for ISE6 infection. Genes that are essential for infection of the mammalian host but not for the infection of ticks have been reported in E. chaffeensis (Cheng et al., 2015). Four different mutants, generated by random mutagenesis, were impaired in their ability to infect deer and dogs, however they were detected in Amblyomma americanum ticks (Cheng et al., 2015). This indicates that the differential requirement of specific genes for infection of the tick vector and the mammalian host may be a mechanism shared between members of the Anaplasmataceae family. This could be partially explained by the differential gene expression between tick and mammalian cells (Nelson et al., 2008).
ΔAPH_0906 development in RF/6A cells is impaired, but it is able to infect hamsters
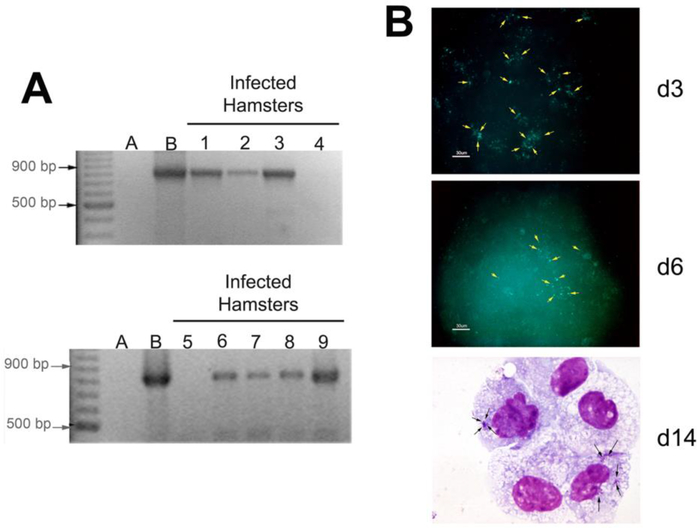
Since ΔAPH_0906 was unable to productively infect HL-60 cells, we tested its capacity to replicate in a non-human primate endothelial cell line, RF/6A, which supports the growth of wild-type bacteria, and since microvascular endothelium has been suggested to be a target of infection (Herron et al., 2005). Bacteria readily adhered to RF/6A cells, but their number appeared to decrease with time as fewer bacteria were detected by day 6, and on day 14, only a few small morulae remained visible in Giemsa-stained cells (Figure 3B).
Figure 3. ΔAPH_0906 infects hamsters during in vivo challenge but it is not able to replicate in RF/6A cells in vitro.
A) Nine hamsters were challenge with 500 μl of ISE6 cells infected with ΔAPH_0906 bacteria by intraperitoneal (i.p.) injection. Two blood samples were taken 7 days days post-inoculation (p.i.) and the remaining 7 samples were taken at 21 days. DNA was purified from blood samples and nested PCR was used to confirm infection. The two samples taken at day 7 (Lanes 1 and 2) were positive and 5 out of 7 samples were positive at day 21 (Lanes 3–9). DNA from infected cells was used as positive control and the negative control consisted of mice injected with uninfected ISE6 cells. B) RF/6A cells were inoculated with cell free ΔAPH_0906 bacteria. Infections were observed by fluorescence microscopy at day 3 and day 6. Bacteria (pointed by yellow arrows) were observed as green fluorescence associated with cells. This fluorescence appeared to decrease as time passed. Giemsa stains from infected RF/6A at day 14 post-infection (p.i) show very small morulae, suggesting that the bacteria were not able to replicate within the infected cells.
We then tested if ΔAPH_0906 was able to infect rodents. Amplification of an 867 bp fragment of Ap 16s rDNA from blood by nested PCR demonstrated that 7 of 9 hamsters injected with ΔAPH_0906 from ISE6 cultures became infected (Figure 3A), and 5 of them (lanes 3–9) remained infected for 21 days. Blood samples from hamsters injected with uninfected ISE6 cells did not show amplification of any product (negative control, lanes A). These results suggest the ability of ΔAPH_0906 to infect mammals, although the target cell remains to be identified. We did not confirm by sequence analysis that bacteria within the hamsters had not lost their inserts although the mutants reisolated into ISE6 cells from one hamster retained green fluorescence and spectinomycin-resistance, indicating presence of the transposon. Likewise, it remains to be determined whether there is a defect in the growth of ΔAPH_090 in rodents when compared to wild-type bacteria. Overall, these results suggest that while APH_0906 expression may be essential for replication in HL-60 and RF/6A cells, the lack of this protein does not abolish infection of rodents.
aph_0906 expression in HL-60 increases over time
The change in expression of aph_0906 in HL-60 at 4, 24, 48, and 72 h was measured by qRT-PCR, using rpoB and msp5 as normalizer genes, respectively. Expression increased as infection in HL-60 cells progressed, starting at 0.005 and 0.001 at 4 h, and growing to 0.001 and 0.01 by 24 h, 0.012 and 0.03 by 48 h, and reaching 0.15 and 0.249 by 72 h (Figure 4). These numbers represent a 10-, 30-, and 249- fold upregulation of the gene by 24, 48, and 72 h, respectively, when compared to the expression at 4 h and normalized by msp5 expression. Ap has a biphasic life cycle comprising two functional forms. The invasive, or dense core form, binds to host cells and the reticulated form replicates inside a membrane bound vacuole (Troese and Carlyon, 2009). Expression of aph_0906 was lowest at 4 h pi, coinciding with internalization of Ap, and increased over the course of 72 h, when bacteria had changed into the reticulate form undergoing intracellular multiplication (Troese and Carlyon, 2009). These results suggest that the expression of aph_0906 facilitates intracellular replication but not invasion, which may explain why we were able to detect bacterial DNA in HL-60 cells for up to 5 days although bacterial numbers did not increase (Figure 2C). A similar increase in the expression of an Ap effector during intracellular infection was reported for APH_1387, which is a protein associated with the vacuolar membrane. APH_1387 expression increased from 8 h p.i. and reached its maximum by 48 h p.i. (Huang et al., 2010).
Figure 4. APH_0906 mRNA expression within ISE6 and HL-60 cells shows an increase in transcription levels in HL-60 cells.
A) The ratio of expression of the gene encoding the hypothetical protein APH_0906 during infection of HL-60 cells was measured by qRT-PCR at 4, 24, 48, and 72 h p.i. Expression of the gene increased as the infection progressed when normalized against both normalizing genes, rpoB (blue bars) and msp5 (red bars). The ratio of expression was calculated using one delta change of the Ct values (2Δtarget-normalizer). B) The fold change in mRNA transcription of APH_0906 between ISE6 cells and HL-60 cells was measured at same time points described before. The fold change was calculated using the 2−ΔΔct method. As described for the ratio of expression, the transcript levels of APH_0906 increased over time with up-regulation of the gene by 48 h and 72 h of infection when normalized by both genes, rpoB (blue bars) and msp5 (red bars)
APH_0906 is translocated into the cytoplasm of infected cells
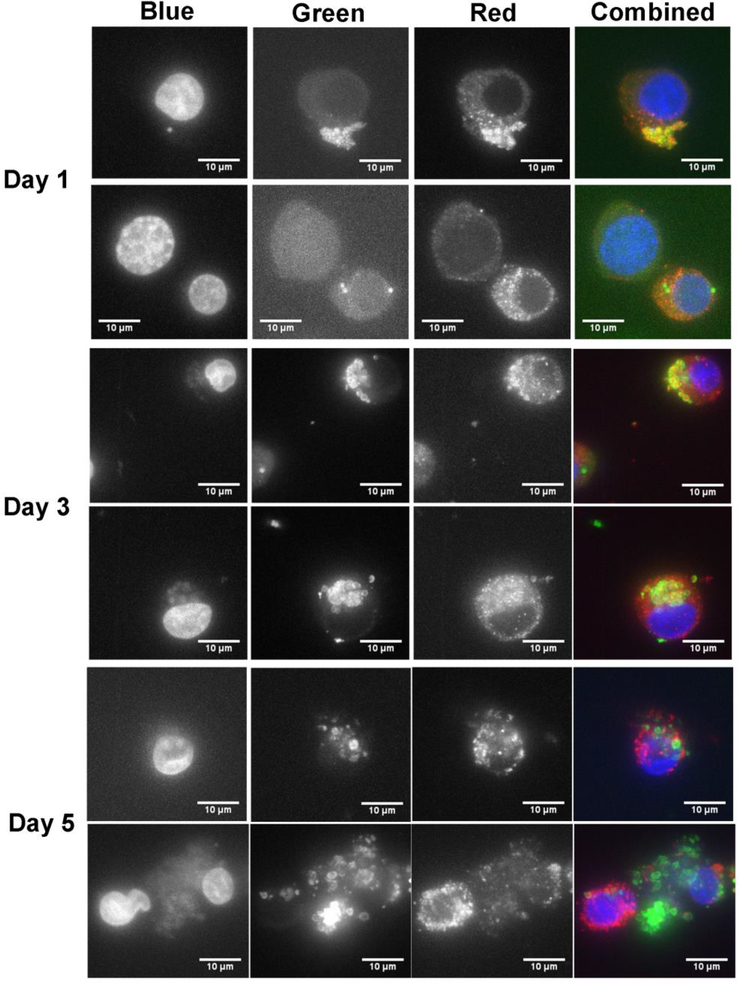
To determine the localization of APH_0906 at different time points during infection of HL-60 cells, we performed Immunofluorescence Assays (IFAs) and confocal microscopy, using antibodies against the recombinant versions of APH_0906, r1-APH0906 and r2-APH0906. Wild-type bacteria were added to uninfected HL-60 cell cultures, and samples were taken on days 1, 3, and 5 p.i. APH_0906 protein was detected using polyclonal antibodies against r1-APH_0906 or r2-APH_0906. Confocal microscopy was used to visualize the location of the protein within infected cells. On day 1 p.i., most APH_0906 co-localized with the bacteria as demonstrated by the yellow signal produced by co-localization of Cy3 (red) anti-APH_0906 and FITC (green) anti-Ap labels. A small amount of red signal was observed in the cytoplasm of the host cell in close proximity to the bacteria (Figure 5; Videos S1).
Figure 5. Subcellular localization of APH_0906 during infection of HL-60 cells.
The subcellular localization of APH_0906 was investigated with Immuno Fluorescence Assays (IFAs) of infected HL-60 cells at 1, 3, and 5 days p.i. The bacteria were labeled with dog serum against A. phagocytophilum detected with FITC anti-dog antibodies (green signal). APH_0906 was labeled using mouse serum raised against recombinant versions of APH_0906 and detected with Cy3-labeled anti-mouse antibodies (red signal). The nucleus of the cells was labeled with DAPI (blue signal). Localization of the protein at day 1 was mostly associated with the bacteria and as infection progressed the protein was translocated into the cytoplasm of the cells by day 3 and 5. A 10 μm size bar is included in the pictures for comparison.
On day 3 p.i., APH_0906 was mostly observed in the proximity of the morulae and in some cases randomly dispersed in the cytoplasm (Figure 5; Videos S2). In heavily infected cells harboring several morulae, some APH_0906 protein associated with the DAPI (blue) signal from the host cell nucleus (Figure 5; Video S3), although this association may be an artifact. By day 5 p.i., the protein appeared as aggregates that accumulated in the cytoplasm of infected cells surrounding large morula (Figure 5; Videos S3). APH_0906 became more abundant as the infection progressed confirming our expression data and the results of the tiling array (Nelson et al., 2008) (Figure 5; Video S1–3). Sinclair et al (Sinclair et al., 2015) detected APH_0906 in nuclear preparations from infected cells using proteomics. We did not conclusively observe APH_0906 in the nucleus of infected cells. These discrepancies could be due to cytoplasmic contamination in the nuclear preparations used (Sinclair et al., 2015) or that we missed the specific time point during our observations. According to our observations, APH_0906 can be observed co-localizing with morulae containing bacteria and being translocated into the cytoplasm at day 1 and day 3, but as infection progresses (day 5) the protein localizes mostly to the cytoplasm of infected cells (Figure 5). This suggests that APH_0906 might likely complete its function in the cytoplasm of infected cells.
APH_0906 has no predicted domains
We used bioinformatic approaches to obtain insights into the potential function of APH_0906. BLAST analysis of the protein provided no information on potential domains and motifs present in the protein. Furthermore, no homologs of the proteins were identified outside of the Anaplasmataceae family and all hits represented proteins without known function, hence its hypothetical status. Likewise, SMART analysis of the APH_0906 protein sequence did not return any domain with significant value. The tertiary structure of the protein was inferred with i-TASSER to find proteins with known crystal structure that presented high sequence similarity to APH_0906. Comparison of the APH_0906 predicted protein sequence to proteins with known crystal structure retrieved three top models with >0.8 TM-Score. The top score corresponded to Brr2 Helicase Region (PDB ID: 4F91; TM-Score: 0.892), which is an RNA helicase that is essential for spliceosome activation (Plaschka et al., 2017) and disassembly (Santos et al., 2012). The second top-score model belonged to a homolog of Brr2 in Chaetomium thermophilum (PDB ID: 5M59; TM-Score: 0.863) (Absmeier et al. 2017) and the third top-score model corresponded to the Brr2 homolog in Saccharomyces cerevisiae (PDB ID: 4BGD; TM-Score: 0.839) (Nguyen et al., 2013), strengthening the prediction. Functional prediction by Gene Ontology of the predicted models suggested that APH_0906 may have RNA helicase (GO:0034458 and GO:0004004; GO-Score: 0.35), mRNA-binding (GO:0003729; GO-Score: 0.35) and ATP-binding (GO:005524; GO-Score: 0.41) activities. Interestingly, Ap infection of HL-60 cells differentiated into neutrophils with ATRA affects the expression of specific gene isoforms, including 9 genes encoding components of the spliceosome complex (Dumler et al., 2018). Brr2 is a key component of the spliceosome complex and is implicated in its activation, splicing catalysis and disassociation. Because Brr2 interacts with several other component of the spliceosome, authors have speculated that it may indirectly act in alternative splicing (as reviewed in (Absmeier et al., 2016)). Our bioinformatics analysis predicts similarities between APH_0906 and Brr2; however, whether or not APH_0906 binds to mRNA and can affect spliceosome assembly leading to changes in ASEs expression needs to be functionally determined.
Phylogenetic distribution and amino acid differences correlate with strain host tropism but predicted binding sites are conserved between APH_0906 homologs
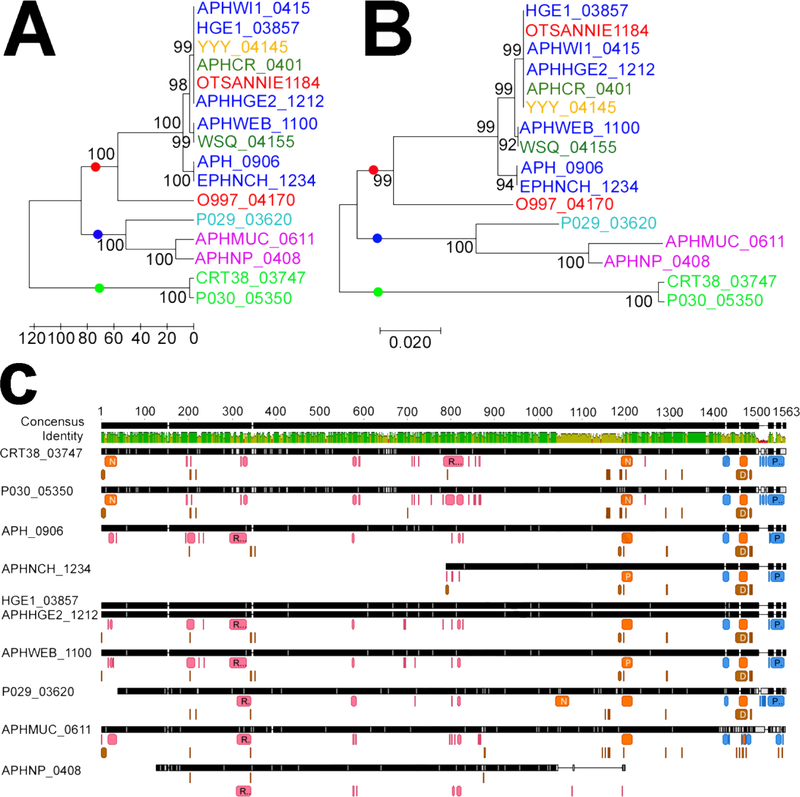
Host tropisms differences have been reported between Ap strains in the US, with a subset of strains able to infect humans, dogs, and horses (reviewed in (Dugat et al., 2015)). The phylogenetic clustering of AnkA, a known Ap effector protein, correlates with host species (Majazki et al., 2013; Scharf et al., 2011). We wanted to see if a similar phenomenon applied to other effectors. The amino acid sequence from twelve APH_0906 homologs present in different Ap strains were downloaded from NCBI, aligned, and used for phylogenetic analysis. The number of amino acid differences between homologs ranged from 0 to 337 aa (Table S1). Two trees, showing identical groupings, were constructed using UPMAG (Figure 6A) and Neighbor-Joining (Figure 6B). APH_0906 homologs appeared to cluster according to host and continent of origin. Cluster 1 contained homologs present in US strains from humans, dog, rodents, and horses, whereas Cluster 2 comprised homologs from European sheep and dog strains, and Cluster 3 included homologs from US tick isolates (Figure 6A and B). A similar grouping of human, dog, and horse strains into one cluster was reported for AnkA variants (Scharf et al., 2011). However, unlike AnkA, APH_0906 sequences of European dog strains clustered with a sheep strain of European origin and not dog strains from the US. Nevertheless, the European dog and the Norwegian sheep sequences in Cluster 2 are clearly divergent. Genomic differences between human pathogenic and non-human pathogenic strains have been reported. Human pathogenic strains presented higher homology and synteny with other human and dog isolates than with tick isolates from the US and ruminant isolates from Europe (Barbet et al., 2013). Interestingly, our phylogenetic analysis of APH_0906 showed a similar trend.
Figure 6. Bioinformatic analysis of APH_0906 and its homologs.
A) Phylogenetic relation between homologs of APH_0906 from A. phagocytophilum strains isolated from different hosts analyzed with the UPMAG and B) Neighbor-Joining methods. Strains isolated from human hosts in the US are depicted in dark blue, strains isolated from horses in the US in red, strains isolated from rodents in the US in dark green, strains isolated from ticks in the US in light green, and a strain isolated from a dog in the US is shown in yellow. Strains isolated from dogs in Europe are shown in purple and a strain isolated from a sheep in Norway is shown in light blue. The boostrap values were inferred from 1000 replicates. Three different clusters were identified with dots of different colors at the base of the branches. The red dot identifies a cluster (Cluster 1) formed by APH_0906 homologs from strains isolated from vertebrates in the US. The second cluster (Cluster 2; blue dot) is composed by homologs from strains isolated from vertebrate hosts in Europe, and the last cluster (Cluster 3; green dot) is represented by homologs from strains isolated from ticks in the US. C) MUSCLE alignment of APH_0906 homologs from different clusters showing predicted characteristics within the amino acid sequences. RNA binding sites are represented by pink boxes under the amino acid residues. DNA binding sites are shown in brown and protein binding sites are shown in blue. Predicted Nuclear Localization (NLSs) are shown in orange. Binding sites were predicted with DisoRDPbind and NLSs with NLSmapper and NLStradamus. Image was generated with Geneious R10.
Due to the differences detected in the protein sequences of the different APH_0906 homologs from other strains, we decided to compare changes in putative binding sites. APH_0906 also possesses a potential Nuclear Localization Signal (NLS). Prediction of the DNA, RNA, and protein binding sites in the different homologs showed that binding sites are highly conserved in their position (Figure 6C). Likewise, predicted NLSs were consistent in their position within most sequences with the exception of P029_03620 and the homologs from strains isolated from Camp Ripley ticks (CRT38_03747 and P030_05350) (Figure 7C and Table 2), which were derived from engorged female ticks collected from white-tailed deer. It is likely that the CRT isolates are specific to deer in this region. There was an obvious trend in the distribution of the predicted binding sites within the homologs. Predicted RNA binding sites were mostly localized within the first 900 aa from the N-terminus of the protein, whereas the protein and DNA binding sites were located within the last 600 aa of the C-terminus and corresponded to half with the predicted NLSs (Figure 6C). Only CRT38_03747 and P030_05350 possessed a predicted NLS within the N-terminus of the protein (Figure 6C and Table 2). Whether these NLSs and binding domains are functional remains to be determined, but it is interesting that differences exist between human pathogenic strains and non-human pathogenic strains. One could speculate that these differences are due to adaptations acquired by human pathogenic strains.
Table 2.
Primers used in the study.
| Primer name | Primer sequence | Target | Use | Restriction site | Piece size (bp) | Reference |
|---|---|---|---|---|---|---|
| aph_0906 fw | AGCTCATATGCACAACTATG | APH_0906 | qRT-PCR | N/A | 247 | This paper |
| aph_0906 rev | AAACGGACACTCTCTATGTT | APH_0906 | qRT-PCR | N/A | 247 | This paper |
| rpoB fw | CTTTATCCTGCTTTAGAACAACATC | rpoB | qRT-PCR | N/A | 286 | Chavez et al (2015) |
| rpoB rev | GGTCCGTATGGTCTGGTTACTC | rpoB | qRT-PCR | N/A | 286 | Chavez et al (2015) |
| msp5 F1 | TGACACTGTGGTTGAACAAGC | msp5 | qPCR and qRT-PCR | N/A | 126 | Chavez et al (2015) |
| msp5 R1 | GAAGAAAAGCCGAACATAAGC | msp5 | qPCR and qRT-PCR | N/A | 126 | Chavez et al (2015) |
| APH_0906 fw1 | ATGGTAGCATATGATGACTCTGCTGCTTAAGCCAAAC | APH_0906 | Recombinant protein | NdeI | 2268 | This paper |
| APH_0906 rv1 |
GTGATGCCTCGAGATCGATCAGAGTGTCACCGAGCAT | APH_0906 | Recombinant protein | XhoI | 2268 | This paper |
| APH_0906 fw2 | GTGCGATCCATGGAAGAAGATGGTACCGCGT | APH_0906 | Recombinant protein | NcoI | 2462 | This paper |
| APH_0906 rv2 |
ATTGCTTAGCTCGAGATGCTGCTGCTGTGATACG | APH_0906 | Recombinant protein | XhoI | 2462 | This paper |
| T7 promoter | TAATACGACTCACTATAGGG | pET28 vector | Sequencing | N/A | N/A | Novagen |
| T7 terminator | GCTAGTTATTGCTCAGCGG | pET28 vector | Sequencing | N/A | N/A | Novagen |
Conclusions
We have developed a system for the random mutagenesis of genes within members of the Anaplasmataceae family. Through our experience, we have improved this system and have developed alternatives, such as a non-antibiotic based selection system, for its use by the scientific community. This system has been extensively used to identify genes of interests in the Anaplasma spp. and Ehrlichia chaffeensis. One such mutant has an insertion within the coding region of the hypothetical protein APH_0906. This protein is important for the replication of the bacteria in vitro in HL-60 cells and RF-6A cells as the number of bacteria does not change over time. Localization of the protein in infected cells confirms its role as an effector that translocates to the cytoplasm of infected cells. Bioinformatic analysis suggests a role in mRNA binding and predicted clear differences between human pathogenic and non-human pathogenic strains.
Supplementary Material
Video S1. Video constructed with confocal photographs composed of the z-stack of an A. phagocytophilum infected HL-60 cell at day 1 showing the localization of APH_0906.
Red signal represents the localization of APH_0906 labeled Cy3 anti-mouse antibodies reaction against anti-rAPH_0906 mouse serum. The green signal shows the localization of A. phagocytophilum labeled by FITC anti-dog antibodies recognizing anti-A. phagocytophilum dog serum. The nucleus of the cell is labeled with DAPI (blue). Yellow signal represents colocalization of APH_0906 with A. phagocytophilum bacteria. Video presents 1 fps and each frame represents a z-section of the cell.
Video S2. Video constructed with confocal photographs composed of the z-stack of an A. phagocytophilum infected HL-60 cell at day 3 showing the localization of APH_0906.
Video S3. Video constructed with confocal photographs composed of the z-stack of an A. phagocytophilum infected HL-60 cell at day 5 showing the localization of APH_0906.
Table 3.
Nuclear Localization Signals (NLSs) predicted for the homologs of APH_0906.
| Protein | Host | NLS Mapper | NLStradamus | ||
|---|---|---|---|---|---|
| NLS Sequence | Score | NLS Sequence | Cutoff | ||
| APH_0906 | Human | KRHGRVLPTPMDPKVLQDLRSSNLLAA | 4.8 | TDGKKPSTTVPKKPPRPARGAK | 0.6 |
| EPHNCH_1234 | Human | KRHGRVLPTPMDPKVLQDLRSSNLLAA | 4.8 | TDGKKPSTTVPKKPPRPARGAK | 0.6 |
| APHWEB_1100 | Human | KRHGRVLPTPMDPKVLQDLRSSNLLAA | 4.8 | TDGKKPSTTVPKKPPRPARGAK | 0.6 |
| HGE1_03857 | Human | KRHGRVLPTPMDPKVLQDLRSSNLLAA | 4.8 | TDGKKPSTTVPKKPPRPARGAK | 0.6 |
| OTSANNIE_1184 | Human | KRHGRVLPTPMDPKVLQDLRSSNLLAA | 4.8 | TDGKKPSTTVPKKPPRPARGAK | 0.6 |
| O997_04170 | Horse | KRHGRVLPTSMDPKVLQDLRSYNLLAA | 4.8 | KGTDGKKPSTTAPKKPPRSARGAK | 0.6 |
| RGGYRNSKDVVEVIASGPDGLFTARPYLVKLR | |||||
| KGS | 6.1 | ||||
| P029_03620 | Sheep | RKRHGRVLPTPTDPEVLQDLRSSNLLAA | 5.8 | DGKKSSTTAPKKPPRTARGAK | 0.6 |
| PIKRHGSVLPTPMDPKVLQDLRSSNLLAA | 5.2 | ||||
| CRT38_03747 | Tick | PSKRSTALLGSAIDFLLCRDQNSNPARQRI | 4.9 | GKKPSTTVHKKPPRPARGAK | 0.6 |
| PIKRHGSVLPTPMDPKVLQDLRSSNLLAA | 5.2 | ||||
| P030_05350 | Tick | PSKRSTALLGSAIDFLLCRDQNSNPARQRI | 4.9 | GKKPSTTVHKKPPRPARGAK | 0.6 |
| APHNP_0408 | Dog | None | N/A | None | N/A |
| APHMUC_0611 | Dog | LKRHGRVLPTPMDPKALQDLISSNLLAA | 4.8 | PPKKPPRHA | 0.6 |
Acknowledgments
Funding: This research was supported by a generous grant from NIH to UGM (R01 AI042792).
Abbreviations
- Ap
Anaplasma phagocytophilum
- BSA
Bovine Serum Albumin
- Himar
Hematobia irritans mariner element
- h
hours
- i.p.
intraperitoneally
- min
minutes
- msp5
single-copy gene encoding Major Surface Protein 5 (MSP5)
- PBS
phosphate buffered saline
- p.i.
post infection
- rpoB
RNA polymerase subunit B
- T4SS
type 4 secretion system
- tr
transcriptional regulator
- TBS
tris-buffered saline
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Absmeier E, Santos KF, Wahl MC, 2016. Functions and regulation of the Brr2 RNA helicase during splicing. Cell Cycle 15, 3362–3377. 10.1080/15384101.2016.1249549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Tian M, Li P, Liu J, Ding C, Yu S, 2017. Characterization of Brucella abortus mutant strain Delta22915, a potential vaccine candidate. Vet. Res 48, 17 10.1186/s13567-017-0422-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet AF, Agnes JT, Moreland AL, Lundgren AM, Alleman AR, Noh SM, Brayton KA, Munderloh UG, Palmer GH, 2005. Identification of functional promoters in the msp2 expression loci of Anaplasma marginale and Anaplasma phagocytophilum. Gene 353, 89–97. 10.1016/j.gene.2005.03.036 [DOI] [PubMed] [Google Scholar]
- Barbet AF, Al-Khedery B, Stuen S, Granquist EG, Felsheim RF, Munderloh UG, 2013. An emerging tick-borne disease of humans is caused by a subset of strains with conserved genome structure. Pathogens 2, 544–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barre N, Happold J, Delathiere J-M, Desoutter D, Salery M, de Vos A, Marchal C, Perrot R, Grailles M, Mortelecque A, 2011. A campaign to eradicate bovine babesiosis from New Caledonia. Ticks Tick. Borne. Dis 2, 55–61. 10.1016/j.ttbdis.2010.11.001 [DOI] [PubMed] [Google Scholar]
- Beare PA, Larson CL, Gilk SD, Heinzen RA, 2012. Two systems for targeted gene deletion in Coxiella burnetii. Appl. Environ. Microbiol 78, 4580–4589. 10.1128/AEM.00881-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkner K, Steiner B, Rinkler C, Kern Y, Aichele P, Bogdan C, von Loewenich FD, 2008. The elimination of Anaplasma phagocytophilum requires CD4+ T cells, but is independent of Th1 cytokines and a wide spectrum of effector mechanisms. Eur. J. Immunol 38, 3395–3410. 10.1002/eji.200838615 [DOI] [PubMed] [Google Scholar]
- Burkhardt NY, Baldridge GD, Williamson PC, Billingsley PM, Heu CC, Felsheim RF, Kurtti TJ, Munderloh UG, 2011. Development of shuttle vectors for transformation of diverse Rickettsia species. PLoS One 6, e29511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez ASO, Felsheim RF, Kurtti TJ, Ku P-S, Brayton KA, Munderloh UG, 2012. Expression patterns of Anaplasma marginale Msp2 variants change in response to growth in cattle, and tick cells versus mammalian cells. PLoS One 7, e36012 10.1371/journal.pone.0036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Severo MS, Sakhon OS, Choy A, Herron MJ, Felsheim RF, Wiryawan H, Liao J, Johns JL, Munderloh UG, Sutterwala FS, Kotsyfakis M, Pedra JH, 2012. Anaplasma phagocytophilum dihydrolipoamide dehydrogenase 1 affects host-derived immunopathology during microbial colonization. Infect Immun 80, 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Nair ADS, Indukuri VV, Gong S, Felsheim RF, Jaworski D, Munderloh UG, Ganta RR, 2013. Targeted and random mutagenesis of Ehrlichia chaffeensis for the identification of genes required for in vivo infection. PLoS Pathog. 9, e1003171 10.1371/journal.ppat.1003171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Nair ADS, Jaworski DC, Ganta RR, 2015. Mutations in Ehrlichia chaffeensis Causing Polar Effects in Gene Expression and Differential Host Specificities. PLoS One 10, e0132657 10.1371/journal.pone.0132657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby FL, Brayton KA, Magunda F, Munderloh UG, Kelley KL, Barbet AF, 2015. Reduced Infectivity in cattle for an outer membrane protein mutant of Anaplasma marginale. Appl. Environ. Microbiol 81, 2206–2214. 10.1128/AEM.03241-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren FS, Heitman KN, Drexler NA, Massung RF, Behravesh CB, 2015. Human granulocytic anaplasmosis in the United States from 2008 to 2012: a summary of national surveillance data. Am J Trop Med Hyg 93, 66–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugat T, Lagree AC, Maillard R, Boulouis HJ, Haddad N, 2015. Opening the black box of Anaplasma phagocytophilum diversity: current situation and future perspectives. Front Cell Infect Microbiol 5, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumler JS, Sinclair SH, Shetty AC, 2018. Alternative Splicing of Differentiated Myeloid Cell Transcripts after Infection by Anaplasma phagocytophilum Impacts a Selective Group of Cellular Programs. Front. Cell. Infect. Microbiol 8, 14 10.3389/fcimb.2018.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremeeva ME, Dasch GA, 2011. Anaplasmataceae as Human Pathogens : Biology, Ecology and Epidemiology, in: Revue Tunisienne d’Infectiologie, Intracellular Bacteria: From Biology to Clinic. pp. S7–S14.
- Felsheim RF, Chávez ASO, Palmer GH, Crosby L, Barbet AF, Kurtti TJ, Munderloh UG, 2010. Transformation of Anaplasma marginale. Vet. Parasitol 167 10.1016/j.vetpar.2009.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsheim RF, Herron MJ, Nelson CM, Burkhardt NY, Barbet AF, Kurtti TJ, Munderloh UG, 2006. Transformation of Anaplasma phagocytophilum. BMC Biotechnol 6, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H-M, Whitworth T, Olano JP, Popov VL, Walker DH, 2004. Fc-dependent polyclonal antibodies and antibodies to outer membrane proteins A and B, but not to lipopolysaccharide, protect SCID mice against fatal Rickettsia conorii infection. Infect. Immun 72, 2222–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futse JE, Brayton KA, Nydam SD, Palmer GH, 2009. Generation of antigenic variants via gene conversion: Evidence for recombination fitness selection at the locus level in Anaplasma marginale. Infect. Immun 77, 3181–3187. 10.1128/IAI.00348-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman JL, Nelson C, Vitale B, Madigan JE, Dumler JS, Kurtti TJ, Munderloh UG, 1996. Direct cultivation of the causative agent of human granulocytic ehrlichiosis. N Engl J Med 334, 209–215. [DOI] [PubMed] [Google Scholar]
- Hammac GK, Ku P-S, Galletti MF, Noh SM, Scoles GA, Palmer GH, Brayton KA, 2013. Protective immunity induced by immunization with a live, cultured Anaplasma marginale strain. Vaccine 31, 3617–3622. 10.1016/j.vaccine.2013.04.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero M, de Lorenzo V, Timmis KN, 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol 172, 6557–6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron MJ, Ericson ME, Kurtti TJ, Munderloh UG, 2005. The interactions of Anaplasma phagocytophilum, endothelial cells, and human neutrophils. Ann N Y Acad Sci 1063, 374–382. [DOI] [PubMed] [Google Scholar]
- Herron MJ, Felsheim RF, Nelson CM, Oliva Chávez AS, Munderloh UG, 2010. Construction and Screening of an Anaplasma phagocytophilum Mutant Library., in: 24th Meeting of the American Society for Rickettsiology Stevenson, WA. [Google Scholar]
- Hoerlein G, 1994. Glufosinate (phosphinothricin), a natural amino acid with unexpected herbicidal properties. Rev. Environ. Contam. Toxicol 138, 73–145. [DOI] [PubMed] [Google Scholar]
- Huang B, Troese MJ, Ye S, Sims JT, Galloway NL, Borjesson DL, Carlyon JA, 2010. Anaplasma phagocytophilum APH_1387 is expressed throughout bacterial intracellular development and localizes to the pathogen-occupied vacuolar membrane. Infect Immun 78, 1864–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H, 2009. Systematic identification of cell cycledependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A 106, 10171–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K, 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe DJ, Grant TE, Robertson HM, 1998. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics 149, 179–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P, 2015. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43, D257–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Rikihisa Y, 2003. Ehrlichia chaffeensis and Anaplasma phagocytophilum lack genes for lipid A biosynthesis and incorporate cholesterol for their survival. Infect. Immun 71, 5324–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockwood S, Voth DE, Brayton KA, Beare PA, Brown WC, Heinzen RA, Broschat SL, 2011. Identification of Anaplasma marginale type IV secretion system effector proteins. PLoS One 6, e27724 10.1371/journal.pone.0027724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn GE, Oliver JD, Nelson CM, Felsheim RF, Kurtti TJ, Munderloh UG, 2015. Tissue distribution of the Ehrlichia muris-like agent in a tick vector. PLoS One 10, e0122007 10.1371/journal.pone.0122007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majazki J, Wuppenhorst N, Hartelt K, Birtles R, von Loewenich FD, 2013. Anaplasma phagocytophilum strains from voles and shrews exhibit specific ankA gene sequences. BMC Vet Res 9, 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure EE, Chávez ASO, Shaw DK, Carlyon JA, Ganta RR, Noh SM, Wood DO, Bavoil PM, Brayton KA, Martinez JJ, McBride JW, Valdivia RH, Munderloh UG, Pedra JHF, 2017. Engineering of obligate intracellular bacteria: progress, challenges and paradigms. Nat. Rev. Microbiol 15, 544–558. 10.1038/nrmicro.2017.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DF, Noroy C, Moumene A, Raffaele S, Albina E, Vachiery N, 2013. Searching algorithm for type IV secretion system effectors 1.0: a tool for predicting type IV effectors and exploring their genomic context. Nucleic Acids Res 41, 9218–9229. 10.1093/nar/gkt718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munderloh UG, Felsheim RF, Burkhardt NY, Herron MJ, Oliva Chávez AS, Nelson CM, Kurtti TJ, 2012. The Way Forward: Improving Genetic Systems., in: Intracellular Pathogens II: Rickettsiales. p. Vol. 89, No. 3. [Google Scholar]
- Munderloh UG, Jauron SD, Fingerle V, Leitritz L, Hayes SF, Hautman JM, Nelson CM, Huberty BW, Kurtti TJ, Ahlstrand GG, Greig B, Mellencamp MA, Goodman JL, 1999. Invasion and intracellular development of the human granulocytic ehrlichiosis agent in tick cell culture. J Clin Microbiol 37, 2518–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munderloh UG, Lynch MJ, Herron MJ, Palmer AT, Kurtti TJ, Nelson RD, Goodman JL, 2004. Infection of endothelial cells with Anaplasma marginale and A. phagocytophilum. Vet Microbiol 101, 53–64. [DOI] [PubMed] [Google Scholar]
- Nelson CM, Herron MJ, Felsheim RF, Schloeder BR, Grindle SM, Chavez AO, Kurtti TJ, Munderloh UG, 2008. Whole genome transcription profiling of Anaplasma phagocytophilum in human and tick host cells by tiling array analysis. BMC Genomics 9 10.1186/1471-2164-9-364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Ba AN, Pogoutse A, Provart N, Moses AM, 2009. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinformatics 10, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TH, Li J, Galej WP, Oshikane H, Newman AJ, Nagai K, 2013. Structural basis of Brr2-Prp8 interactions and implications for U5 snRNP biogenesis and the spliceosome active site. Structure 21, 910–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh SM, Brayton KA, Brown WC, Norimine J, Munske GR, Davitt CM, Palmer GH, 2008. Composition of the surface proteome of Anaplasma marginale and its role in protective immunity induced by outer membrane immunization. Infect. Immun 76, 2219–2226. 10.1128/IAI.00008-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva Chávez AS, Fairman JW, Felsheim RF, Nelson CM, Herron MJ, Higgins L, Burkhardt NY, Oliver JD, Markowski TW, Kurtti TJ, Edwards TE, Munderloh UG, Oliva Chavez AS, Fairman JW, Felsheim RF, Nelson CM, Herron MJ, Higgins L, Burkhardt NY, Oliver JD, Markowski TW, Kurtti TJ, Edwards TE, Munderloh UG, 2015. An O-Methyltransferase Is Required for Infection of Tick Cells by Anaplasma phagocytophilum. PLoS Pathog 11, e1005248 10.1371/journal.ppat.1005248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Kurgan L, 2015. High-throughput prediction of RNA, DNA and protein binding regions mediated by intrinsic disorder. Nucleic Acids Res 43, e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C, Lin P-C, Nagai K, 2017. Structure of a pre-catalytic spliceosome. Nature 546, 617–621. 10.1038/nature22799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritt BS, Allerdice MEJ, Sloan LM, Paddock CD, Munderloh UG, Rikihisa Y, Tajima T, Paskewitz SM, Neitzel DF, Hoang Johnson DK, Schiffman E, Davis JP, Goldsmith CS, Nelson CM, Karpathy SE, 2017. Proposal to reclassify Ehrlichia muris as Ehrlichia muris subsp. muris subsp. nov. and description of Ehrlichia muris subsp. eauclairensis subsp. nov., a newly recognized tick-borne pathogen of humans. Int. J. Syst. Evol. Microbiol 67, 2121–2126. 10.1099/ijsem.0.001896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ProMED-Mail, 2017. PRO/AH/EDR> Invasive tick – USA: (NJ) [WWW Document]. Arch. Number 20171123.5462146. URL http://www.promedmail.org/post/5462146 (accessed 11.23.17).
- Rikihisa Y, 2011. Mechanisms of obligatory intracellular infection with Anaplasma phagocytophilum. Clin. Microbiol. Rev 24, 469–489. 10.1128/CMR.00064-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SJ, Neitzel DF, Moen RA, Craft ME, Hamilton KE, Johnson LB, Mulla DJ, Munderloh UG, Redig PT, Smith KE, Turner CL, Umber JK, Pelican KM, 2015. Disease risk in a dynamic environment: the spread of tick-borne pathogens in Minnesota, USA. Ecohealth 12, 152–163. 10.1007/s10393-014-0979-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos KF, Jovin SM, Weber G, Pena V, Luhrmann R, Wahl MC, 2012. Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome. Proc Natl Acad Sci U S A 109, 17418–17423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf W, Schauer S, Freyburger F, Petrovec M, Schaarschmidt-Kiener D, Liebisch G, Runge M, Ganter M, Kehl A, Dumler JS, Garcia-Perez AL, Jensen J, Fingerle V, Meli ML, Ensser A, Stuen S, von Loewenich FD, 2011. Distinct host species correlate with Anaplasma phagocytophilum ankA gene clusters. J Clin Microbiol 49, 790–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotthoefer AM, Schrodi SJ, Meece JK, Fritsche TR, Shukla SK, 2017. Pro-inflammatory immune responses are associated with clinical signs and symptoms of human anaplasmosis. PLoS One 12, e0179655 10.1371/journal.pone.0179655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair SH, Garcia-Garcia JC, Dumler JS, 2015. Bioinformatic and mass spectrometry identification of Anaplasma phagocytophilum proteins translocated into host cell nuclei. Front Microbiol 6, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumalapura NR, Crossley EC, Walker DH, Ismail N, 2009. Persistent infection contributes to heterologous protective immunity against fatal ehrlichiosis. Infect. Immun. 77, 5682–5689. 10.1128/IAI.00720-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CJ, Movva NR, Tizard R, Crameri R, Davies JE, Lauwereys M, Botterman J, 1987. Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J. 6, 2519–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Salis HM, 2015. A predictive biophysical model of translational coupling to coordinate and control protein expression in bacterial operons. Nucleic Acids Res. 43, 7137–7151. 10.1093/nar/gkv635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troese MJ, Carlyon JA, 2009. Anaplasma phagocytophilum dense-cored organisms mediate cellular adherence through recognition of human P-selectin glycoprotein ligand 1. Infect Immun 77, 4018–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valbuena G, Jordan JM, Walker DH, 2004. T cells mediate cross-protective immunity between spotted fever group rickettsiae and typhus group rickettsiae. J. Infect. Dis 190, 1221–1227. 10.1086/423819 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wei L, Liu H, Cheng C, Ganta RR, 2017. A genetic system for targeted mutations to disrupt and restore genes in the obligate bacterium, Ehrlichia chaffeensis. Sci. Rep 7, 15801 10.1038/s41598-017-16023-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zhang Y, 2007. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res 35, 3375–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Liu Z, Niu Q, Liu J, Xie J, Chen Q, Chen Z, Guan G, Liu G, Luo J, Yin H, 2016. Evaluation of different nested PCRs for detection of Anaplasma phagocytophilum in ruminants and ticks. BMC Vet. Res 12, 35 10.1186/s12917-016-0663-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Video constructed with confocal photographs composed of the z-stack of an A. phagocytophilum infected HL-60 cell at day 1 showing the localization of APH_0906.
Red signal represents the localization of APH_0906 labeled Cy3 anti-mouse antibodies reaction against anti-rAPH_0906 mouse serum. The green signal shows the localization of A. phagocytophilum labeled by FITC anti-dog antibodies recognizing anti-A. phagocytophilum dog serum. The nucleus of the cell is labeled with DAPI (blue). Yellow signal represents colocalization of APH_0906 with A. phagocytophilum bacteria. Video presents 1 fps and each frame represents a z-section of the cell.
Video S2. Video constructed with confocal photographs composed of the z-stack of an A. phagocytophilum infected HL-60 cell at day 3 showing the localization of APH_0906.
Video S3. Video constructed with confocal photographs composed of the z-stack of an A. phagocytophilum infected HL-60 cell at day 5 showing the localization of APH_0906.