Abstract
The GroE chaperonin system facilitates protein folding in an ATP-dependent manner. It has remained unclear why some proteins are obligate clients of the GroE system, whereas other closely related proteins are able to fold efficiently in its absence. Factors that cause folding to be slower affect kinetic partitioning between spontaneous folding and chaperone binding in favor of the latter. One such potential factor is contact order (CO), which is the average separation in sequence between residues that are in contact in the native structure. Here, we generated variants of enhanced green fluorescent protein with different COs using circular permutations. We found that GroE dependence in vitro and in vivo increases with increasing CO. Thus, our results show that CO is relevant not only for folding in vitro of relatively simple model systems but also for chaperonin dependence and folding in vivo.
Introduction
The Escherichia coli GroE chaperonin system, which comprises GroEL and its cochaperonin GroES, is a large molecular machine that assists protein folding in vitro (1) and in vivo (2) in an ATP-dependent manner (for recent reviews, see (3, 4, 5)). GroEL consists of two back-to-back stacked heptameric rings with a cavity at each end where protein folding can take place under confining and protective conditions (3, 4, 5). In vitro, the GroE chaperonin system is promiscuous and can assist in the folding of a large number of unrelated proteins (6). However, it was found that, in vivo, only ∼250 E. coli proteins interact with GroEL, of which less than 100 are obligatory clients (7, 8). It has remained unclear why some proteins require the GroE system for efficient folding and other (sometimes closely related) proteins do not (9). Features that distinguish obligatory GroE substrates from other E. coli proteins can be divided into 1) sequence motifs (10, 11) and structural properties of non-native states such as exposed hydrophobic residues that confer (or allow) binding to GroEL and 2) properties that affect the kinetic partitioning between productive folding, binding to chaperones, and misfolding and aggregation (12). Properties that affect kinetic partitioning are likely to be a major factor that distinguishes GroEL clients from nonclients, given that GroEL displays little specificity in binding unfolded and misfolded proteins (6). Kinetic partitioning is also at the heart of the iterative annealing model for GroEL action (13).
Kinetic barriers that cause slow spontaneous folding result in increased flux through pathways of chaperone binding and misfolding and aggregation. Slow folding can be due to a rugged energy landscape, which results in accumulation of folding intermediates, and/or to steps with high activation barriers such as disulfide bond formation and cis-trans proline isomerization. In the case of single-domain proteins without disulfide bonds and cis-prolines, a strong inverse correlation was found between the folding rate constant, kf, and contact order (CO) (i.e., the average separation in sequence between residues in contact in the structure) (14). This correlation is due to the higher loss of conformational entropy when nonlocal contacts are formed. However, folding rates are also affected by point mutations that alter protein stability without changing the CO (15). The relationship between folding rate and stability is reflected in Brønsted plots of logkf versus logKD-N (where KD-N is the equilibrium constant for folding) (e.g., 15). Such plots arise owing to the presence of native-like interactions in the transition state. Hence, perturbing these interactions by mutation affects the stabilities of the transition and native states (and thus folding rate and stability, respectively) in a positively correlated manner. Therefore, local stability effects and global features, such as CO, determine folding rates in combination, as shown by several studies (16, 17, 18). It follows that both types of effects are expected to affect the kinetic partitioning between spontaneous folding to the native state and chaperone binding.
Recently, we selected enhanced green fluorescent protein (eGFP) as a model system for identifying factors that determine GroEL dependence because 1) it folds in a GroEL-dependent manner (19), although it is not an E. coli protein; and 2) its folding can be monitored using fluorescence. eGFP was subjected to random mutagenesis, and screening in vivo was then carried out for variants with altered GroEL dependence (20). We found that the changes in GroEL dependence caused by the mutations could be explained using the concept of folding frustration (21). Frustration arises when different potentially stabilizing interactions in a protein are not satisfied concurrently, thereby leading to a rugged energy landscape with kinetic traps. Our results showed that mutations at frustrated positions tend to reduce GroEL dependence, whereas mutations at nonfrustrated positions tend to increase GroEL dependence. These results indicated, therefore, that local stability influences GroEL dependence. In the work described here, we generated a series of circular permutants of eGFP to test whether global topology, as reflected by CO, also affects GroEL dependence. Our results show that the GroEL dependence of eGFP folding increases with increasing CO both in vivo and in vitro. Hence, GroEL dependence is determined by a combination of local and global features that affect protein-folding rates.
Materials and Methods
Molecular biology
Circular permutants of eGFP, in which the N- and C-termini of wild-type eGFP are fused via a Gly-Ser-Gly-Gly-Thr-Gly linker, were constructed using a restriction-free cloning approach (22) and verified by DNA sequencing of their entire genes. The expression vector pET28a (Novagen/EMD Millipore Chemicals, Darmstadt, Germany) was used as a destination vector. The primers used for creating the permutations are provided in Table S1. The genes for wild-type eGFP and its circular permutants were transferred from pET28a to pMALc2 for the in vivo experiments using the forward and reverse primers 5′-TCACCAACAAGGACCATAGCATCTCGAGATGCATCACCATCACCATCACC-3′ and 5′-TATCAGGCTGAAAATCTTATCAAGCTTTTACTTATACAGCTCGTCCATGC-3′, respectively. The Ala206 → Lys mutation was introduced into all the variants to render them monomeric.
CO effects in vivo
E. coli MGM100 cells in which the chromosomal GroEL and GroES genes are under control of the arabinose promoter (23) were transformed with the pMALc2 plasmid containing the gene for wild-type eGFP or its circular permutants and then spread on Luria broth (LB) plates containing 100 μg/mL ampicillin, 50 μg/mL kanamycin, and 0.1% arabinose. Single colonies were then picked and grown for 2 h at 37°C in 50 μL of LB media containing 100 μg/mL ampicillin, 50 μg/mL kanamycin, and 0.1% arabinose. The cells were then centrifuged at 12,000 rotations per minute for 2 min, and the pellets were washed twice with 100 μL of LB media containing 100 μg/mL ampicillin and 50 μg/mL kanamycin. The cell pellets were then resuspended in 100 μL of LB media containing 100 μg/mL ampicillin and 50 μg/mL kanamycin, and 3 μL were spotted on LB plates (preincubated at 37°C for 1 h) containing 100 μg/mL ampicillin, 50 μg/mL kanamycin, 0.5 mM isopropyl β-D-1-thiogalactopyranoside, and either 0.1% arabinose or 0.1% glucose. The plates were incubated at 37°C for 6 h and then at room temperature for 12 h. The plates were then scanned using a fluorescence imager (Typhoon FLA 9500; GE Healthcare, Chicago, IL) with a laser (blue laser-diode) that has an excitation filter at 473 nm and a blue emission filter (510 nm long-pass). The respective average fluorescence intensities, <F>, of the spots corresponding to each eGFP variant were determined.
Protein purification
Purification of wild-type eGFP and its circular permutants was carried out as before (24). Purification of GroEL was carried out as described previously (20). Purification of GroES was performed as described previously (25) but with a modification. Elution from the 5-mL HisTrap HP column (Amersham Pharmacia Biotech, Uppsala, Sweden) was preceded by washing with 25 mL of 50 mM Tris-HCl buffer (pH 7.5) containing 0.5 M NaCl, 10 mM β-mercaptoethanol, and 10 mM imidazole (buffer A) and then by 100 mL of buffer A containing 10 mM KCl, 10 mM MgCl2, and 2 mM ATP. Protein concentrations were determined as described (26). The base-denatured chromophore concentrations were determined by measuring the absorbance at 447 nm as described (27). Comparison of the protein and chromophore concentrations for wild-type eGFP and the different permutants indicated that the fraction of fluorescent molecules is greater than 90% in all cases.
Folding assays
Wild-type or circularly permuted eGFP was denatured in 200 μL of 30 mM HCl at a final concentration of 12.5 μM (unless indicated otherwise) and incubated at 25°C for 1 h. The denatured protein was then diluted 100-fold in 200 μL of 50 mM (3-(N-morpholino)propanesulfonic acid) buffer (pH 7.0) containing 100 mM KCl, 10 mM Mg(CH3COO)2, 5 mM DTT, and 0.0125% Tween 20 (refolding buffer). In some experiments, the refolding buffer contained 1 mM ADP and varying amounts of GroEL from 0 to 7 μM. In other experiments, the refolding buffer contained 0.5 μM GroEL without or with 1 μM GroES, in which case folding was initiated by adding 2 mM ATP. Folding was monitored at 25°C by exciting at 465 nm and measuring the emission at 509 nm using a Fluorolog-3 spectrofluorometer (HORIBA Jobin Yvon, Bensheim, Germany) with 5 nm bandwidth. All the experiments were carried out using siliconized test tubes.
In the case of the experiments with varying amounts of GroEL (Fig. S3), the data were analyzed assuming kinetic partitioning between spontaneous folding and GroEL binding. It is instructive to first consider simple irreversible kinetic partitioning of a species, A (e.g., unfolded protein), between two species, B (e.g., native protein) and C (misfolded or aggregated protein), described by the following scheme: . In such a case, , and one therefore obtains the following:
| (1) |
and
| (2) |
where [A]0 is the initial concentration of A. Inspection of Eq. 1 shows that the observed rate constant of folding is a function of both the folding and the misfolding or aggregation rate constants (in the latter case, there will also be a concentration dependence). It also shows that the yields depend on the relative rate constants. Slower folding will therefore result in more misfolding and/or aggregation. Here, the data are analyzed using a model of kinetic partitioning described by the following scheme: F, where EL, U, and F stand for GroEL and the unfolded and folded states of the protein substrate, respectively. In this model, binding of the substrate to GroEL is reversible, and folding can take place either spontaneously or in association with GroEL with respective rate constants ks and ka. Given this scheme for describing GroEL action, one can express the slope of the initial linear change in fluorescence, V, as a function of the total concentration of GroEL, [EL]T, as follows:
| (3) |
where Vmax is the slope in the absence of GroEL, [U]T is the total concentration of unfolded substrate (e.g., eGFP), and K is the apparent dissociation constant for the unfolded protein with GroEL. Equation 3 was derived for tight binding (i.e., without assuming that the free concentrations of unfolded protein and GroEL are equal to their total concentrations) and assuming that the concentration of the unfolded protein is initially in steady state. Inspection of Eq. 3 shows that, in the presence of excess GroEL, folding should be completely inhibited unless some folding occurs in association with GroEL. Our data show complete arrest, thereby indicating that folding in association with GroEL, which has been observed before (20), does not occur in the case of the eGFP variants studied here (Fig. S3). The data were therefore fitted to Eq. 3 with ka = 0 (using OriginPro 8).
Stability measurements
Circular dichroism (CD) spectra from 200 to 240 nm were recorded at 25°C (with steps of 1 nm and an average recording time of 3 s) to establish whether the circular permutations led to any gross structural changes. Three consecutive scans were averaged and then corrected by subtracting the spectra of buffer alone. Protein stability measurements were carried out by monitoring changes in the far ultraviolet CD signal at 218 nm as a function of increasing temperature from 50 to 94°C (steps of 2°C) with an equilibration time of 5 min at each temperature before taking a reading. All the CD measurements were performed using a Chirascan CD spectrometer (Applied Photophysics, Leatherhead, UK). The temperature was controlled using a TC125 temperature controller (Quantum Northwest, Liberty Lake, WA) and monitored with a temperature probe in the sample. All the measurements were made with a quartz suprasil cell of 1 mm pathlength (Hellma Analytics, Forest Hills, NY), and the protein concentration was 12 μM in 10 mM phosphate buffer (pH 7.0) containing 0.0125% Tween 20. Values of the apparent melting temperatures, Tm, were obtained by fitting the data as described before (24). The melts of eGFP and its circular permutants are not reversible, but the apparent Tm values obtained from irreversible melts often reflect the rank order of stabilities (e.g., (28)).
CO calculations
The relative contact orders (RCOs) of all the permutants were calculated as follows:
| (4) |
where N is the number of contacts between residues i and j with a Cα-Cα distance smaller than some cutoff and a sequence separation, ΔSi, j, of at least three residues, and L is the total number of residues in the protein. Here, a cutoff of 8 Å was used, but a very similar ranking of the RCO of the permutants was observed using cutoffs of 7, 7.5, 8.5, and 9 Å (data not shown). The Protein Data Bank structure PDB: 4EUL was used for these calculations. The effects of the linker on the sequence separation were taken into account, but it was assumed that the linker residues were not involved in any contacts. Note that CO and RCO (i.e., CO normalized by the protein length) are equivalent here because all the permutants have the same length.
Results and Discussion
Circular permutants of GFP that remain fluorescent, thus indicating that they adopt a wild-type-like native structure, were identified in previous work (27). Here, six of these nondisruptive permutations were created for eGFP (Fig. 1), and all the permutants were found to be fluorescent as expected (27). Far-ultraviolet CD spectra of wild-type eGFP and these permutants were found to be very similar, thus providing additional evidence for their structural integrity (Fig. S1). The eGFP permutants generated, therefore, have similar structural properties but different values of CO (Table 1), thereby enabling us to examine in isolation the role of topology in determining GroEL dependence in vivo and in vitro. In the absence of GroEL, the spontaneous folding of eGFP displays biphasic kinetics. We therefore tested the correlation with CO of the average of the logarithms of its two observed folding rate constants as suggested before for proteins that exhibit multistate folding kinetics (29). Such a correlation can, however, be partially masked when aggregation takes place in parallel with folding. In cases of parallel reactions, the observed rate constant of each reaction is a function of the rate constants of all the competing processes (see Materials and Methods). The observed folding rate constants are therefore expected to be sensitive to aggregation. We therefore monitored spontaneous folding at two different eGFP concentrations under which the extent of aggregation is expected to differ. An inverse correlation is found (Fig. S2) between the average of the logarithms of the two observed folding rate constants and CO, as observed for two-state folders (14). The correlation is stronger and more significant at the lower eGFP concentration (r2 values for 42.5 and 125 nM are 0.82 (p < 0.0057) and 0.58 (p < 0.047), respectively), thereby indicating that it is indeed masked, in part, by aggregation. Regardless, these correlations indicate that CO (in combination with chain length and stability) determines the folding rates even of proteins that display multistate folding kinetics (29).
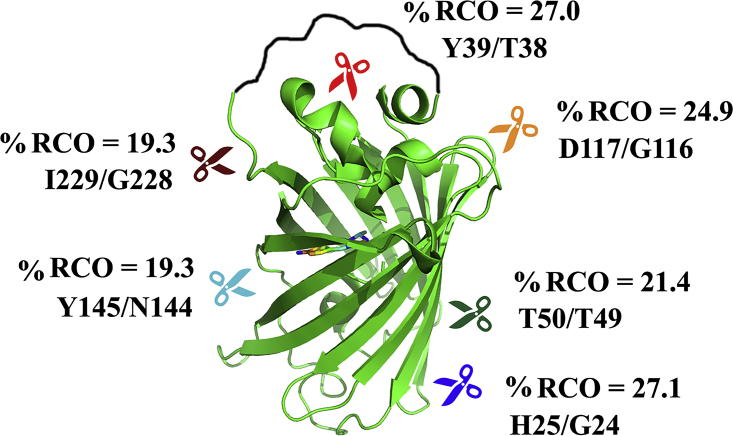
Figure 1.
Ribbon diagram of circular permutants of eGFP. The circular permutants of eGFP constructed in this study are represented using a ribbon diagram of the PDB: 4EUL structure. The linker connecting the N- and C-termini of wild-type eGFP in the permutants is shown in black and the chromophore in rainbow colors. The positions of the new termini are indicated by scissors and specified together with the resulting percent relative contact order (%RCO).
Table 1.
Properties of the eGFP Permutants Constructed in this Study
| eGFP Permutant | RCO (%) | K (nM) | Tm (K) | |
|---|---|---|---|---|
| Wild-type | 19.33 | 5.79 ± 1.72 | 0.33 ± 0.01 | 347.6 ± 0.6 |
| Y145/N144 | 19.30 | 13.00 ± 4.40 | 0.23 ± 0.02 | 335.1 ± 0.2 |
| I229/G228 | 19.34 | 7.48 ± 1.94 | 0.38 ± 0.01 | 348.3 ± 0.9 |
| T50/T49 | 21.35 | 1.96 ± 0.40 | 0.21 ± 0.01 | 349.4 ± 0.1 |
| D117/G116 | 24.88 | 1.08 ± 0.39 | 0.08 ± 0.01 | 343.3 ± 0.6 |
| Y39/T38 | 27.01 | 0.41 ± 0.18 | 0.11 ± 0.02 | 336.4 ± 1.3 |
| H25/G24 | 27.09 | 0.31 ± 0.15 | 0.04 ± 0.01 | 348.2 ± 0.1 |
The thermal melts were carried out in duplicate, and Tm values ± SDs are reported. The values of the %RCOs and dissociation constants for GroEL were determined as described under Materials and Methods. <F>glucose/<F>arabinose corresponds to the ratio of the average fluorescence intensities of a clone that expresses a particular eGFP permutant and was grown on agar plates containing glucose or arabinose, respectively.
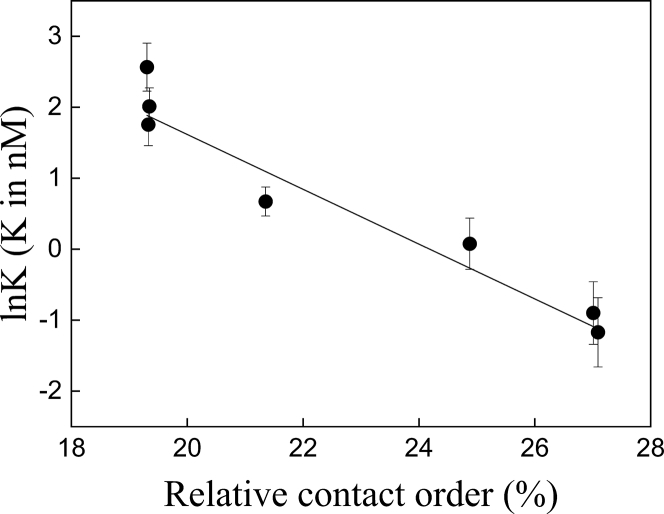
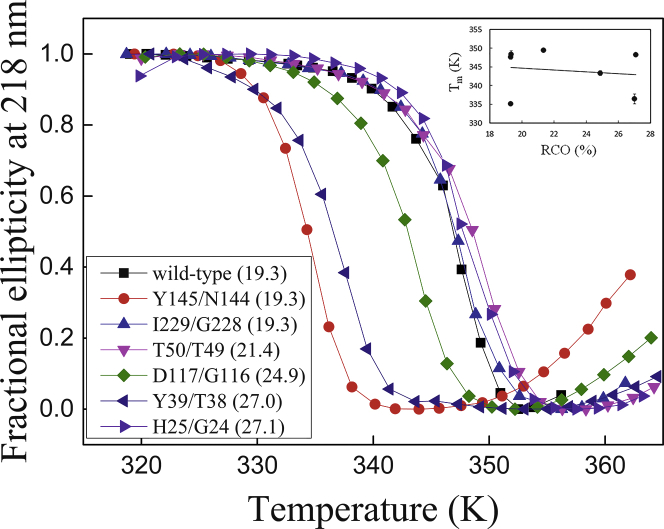
The in vitro assay for GroEL dependence was based on the recognition mentioned above that GroEL clients differ from nonclients in how they undergo kinetic partitioning between GroEL binding, spontaneous folding or misfolding, and aggregation. Given that this partitioning is affected by GroEL concentration, we denatured wild-type eGFP and the various permutants and then measured their initial rates of refolding as a function of GroEL concentration (Fig. S3). In the presence of excess GroEL, folding is retarded because the equilibrium is shifted in favor of GroEL binding. Initially, these experiments were carried out in the absence of nucleotides, but binding of some of the eGFP variants to GroEL was found to be so tight that reliable data fitting was precluded. Hence, these experiments were carried out in the presence of 1 mM ADP so that the affinity of all the eGFP variants for GroEL would be reduced. In the presence of 1 mM ADP, the affinity of denatured eGFP for GroEL is in the nM range and thus similar to that reported for Bacillus stearothermophilus lactate dehydrogenase (30). It may be seen from these experiments (Fig. 2) that the affinity of the eGFP variants for GroEL increases linearly with increasing CO (r2 = 0.94; p < 0.0003), thereby indicating that high CO shifts the kinetic partitioning between spontaneous folding and GroEL binding in favor of the latter. Importantly, the increased GroEL dependence is not due to protein destabilization (Fig. 3) because no correlation (r2 = 0.02) is observed between CO and the apparent melting temperature (Fig. 3, inset; Table 1).
Figure 2.
Plot of the natural logarithms of the apparent affinities of eGFP permutants for GroEL as a function of their respective relative contact orders (RCOs). The apparent affinities of eGFP permutants for GroEL, in the presence of 1 mM ADP, were determined by measuring their initial rates of refolding in the presence of different concentrations of GroEL and fitting the data (Fig. S3) to Eq. 3. The percent relative contact orders (%RCOs) were calculated using Eq. 4 as described under Materials and Methods. Error bars represent standard errors.
Figure 3.
Thermal denaturation curves of wild-type eGFP and the permutants. The CD signal at 218 nm of the different eGFP variants (with their respective %RCO indicated in parentheses) is plotted as a function of temperature. The data were fitted as before (24). The inset shows that there is no correlation between the apparent melting temperature and RCO. See Materials and Methods for further details. Error bars in the inset represent SD.
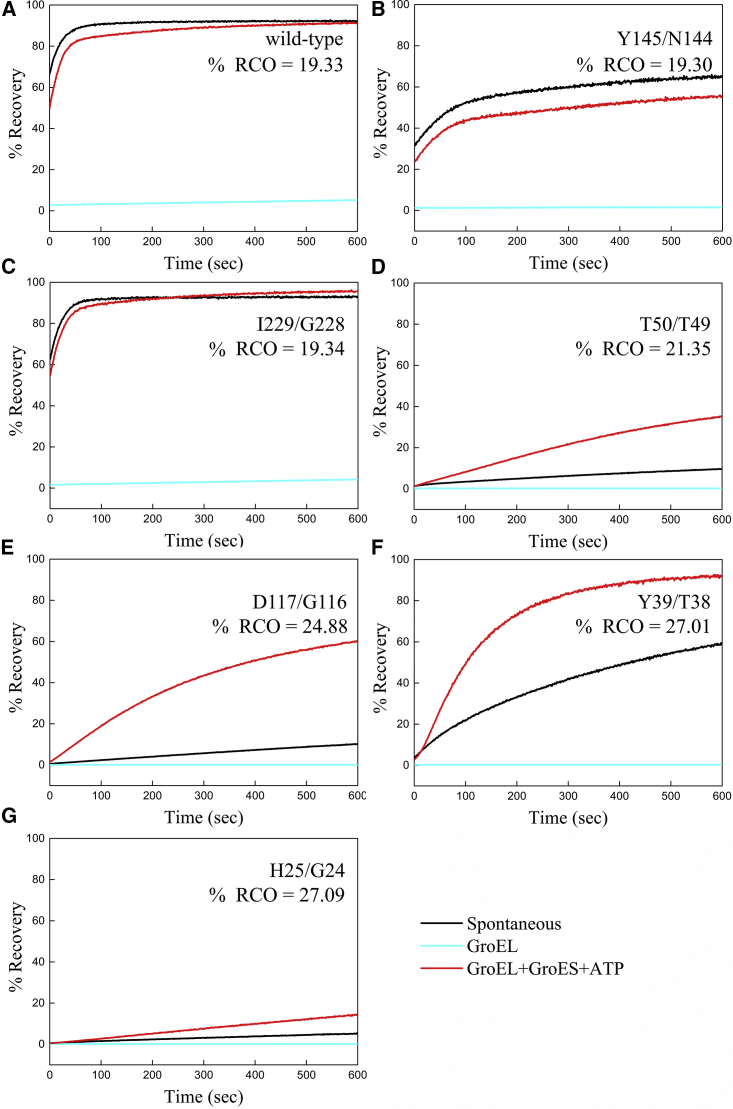
The effect of CO on GroEL dependence was also observed in reactivation experiments carried out in the presence of the full GroE system. Wild-type eGFP and its CO variants were denatured and then allowed to refold spontaneously, in the presence of GroEL alone, or in the presence of GroEL, GroES, and ATP. Remarkably, although wild-type eGFP can fold spontaneously under our experimental conditions, some of the CO variants were found to refold only in the presence of the full GroE system (Fig. 4). For example, the spontaneous folding yield of the variant with a %RCO = 24.9 is ∼10%, whereas in the presence of GroEL, GroES, and ATP, it is ∼60% (Fig. 4 E). In the case of the variant with %RCO = 27.0, the spontaneous folding yield is ∼60%, whereas in the presence of GroEL, GroES, and ATP, it is ∼100% (Fig. 4 F). Hence, increased CO can convert a nonstringent protein substrate into a stringent one or, as in the case of the variant with %RCO = 27.1, block its reactivation even in the presence of the full GroE system (Fig. 4 G). In general, the data in Fig. 4 show that the differences in yield between spontaneous folding and GroEL/ES-assisted folding are small for the variants with low CO, whereas, in the case of the variants with high CO, the yield in the presence of GroEL/ES is always higher by a significant factor. The effects of CO on a specific process such as spontaneous folding cannot be determined, however, from these data and need to be isolated from simpler experimental setups (Figs. 2 and S2).
Figure 4.
Reactivation of wild-type eGFP and the permutants by the GroE system. Wild-type eGFP (A) and the permutants (B–G) were acid denatured and then allowed to refold either spontaneously or in the presence of 0.5 μM GroEL or 0.5 μM GroEL, 1 μM GroES (i.e., GroEL and GroES each at 7 μM protomer concentration), and 2 mM ATP. Folding was monitored by measuring the regain of eGFP fluorescence, and the data for each permutant, indicated by its N- and C-termini and RCO, were normalized by the fluorescence of a sample at the same concentration before denaturation. See Materials and Methods for further details.
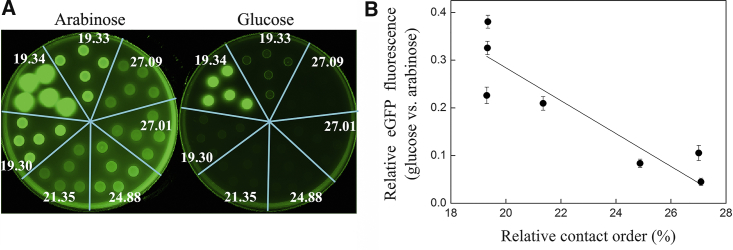
Testing for the effect of CO on GroEL dependence in vivo was achieved using the E. coli MGM100 strain in which the chromosomal GroEL and GroES genes are under control of the arabinose promoter (23). In these cells, arabinose induces GroE expression, and glucose suppresses it. Hence, clones expressing eGFP permutants that are less GroEL dependent were expected, in the presence of glucose, to have higher eGFP folding yields (and thus be more fluorescent) than clones expressing wild-type eGFP. Conversely, clones expressing eGFP permutants that are more GroEL dependent were expected, in the presence of glucose, to contain less folded eGFP and thus be less fluorescent than clones expressing wild-type eGFP. It may be seen that all the clones are more fluorescent in the presence of arabinose than glucose, thus reflecting eGFP’s GroEL dependence (Fig. 5 A). In addition, clones expressing permutants with a higher CO are less fluorescent than the other clones in the presence of either arabinose or glucose (Fig. 5 A), thus reflecting the effect of CO on folding. The fluorescence in the presence of glucose, normalized by the fluorescence in the presence of arabinose, provides a measure of the effect of CO on GroEL dependence in vivo, in which other possible (e.g., photophysical) effects of the permutations cancel out. A higher value for this measure indicates decreased GroEL dependence in vivo. It may be seen (Fig. 5 B) that GroEL dependence in vivo increases linearly with increasing CO (r2 = 0.82; p < 0.005). Hence, CO is shown to affect GroEL dependence both in vitro and in vivo.
Figure 5.
Effect of contact order (CO) on GroEL dependence in vivo. (A) E. coli MGM100 cells expressing wild-type eGFP or one of its permutants were spotted on LB plates containing 100 μg/mL ampicillin, 50 μg/mL kanamycin, 0.5 mM isopropyl β-D-1-thiogalactopyranoside, and either 0.1% arabinose or 0.1% glucose, which induce or block GroE expression, respectively. Following incubation, the plates were scanned using a fluorescence imager as described in Materials and Methods. The strains were spotted in order of increasing CO of the eGFP variant they express, starting with the strain expressing wild-type eGFP at the top and continuing anticlockwise. (B) A plot of the ratio between the average fluorescence intensities on glucose- versus arabinose-containing plates of the clones expressing the various eGFP permutants, , as a function of their respective RCOs. This correlation was reproduced in four other independent experiments. Error bars represent standard errors.
Conclusions
In previous work, we showed that GroEL dependence both in vitro and in vivo is affected by local frustration (20). Here, we show that GroEL dependence in vitro and in vivo is also affected by topology (i.e., it increases with increasing CO). Our results, therefore, indicate that CO is relevant not only for folding in vitro of relatively simple model systems but also for chaperonin dependence and folding in vivo of more complex systems. Taken together with our earlier work (20), our findings show that increased GroEL dependence can result from both local and global properties that slow folding and thus shift kinetic partitioning between spontaneous folding and GroEL binding toward the latter. Different mechanisms of action have been proposed for GroEL in which its cavity is either 1) just a “passive cage” in which aggregation is prevented (31) or 2) a chamber in which the folding process is optimized because of confinement and, perhaps, other factors (32). In addition, it has been reported that encapsulated misfolded substrates can undergo forced unfolding (33). These mechanisms, all of which involve initial kinetic partitioning between spontaneous folding, misfolding, and GroEL binding, are not necessarily mutually exclusive because GroEL has evolved to assist a range of substrates with potentially different needs. Nevertheless, our previous work (20) and the results reported here suggest that rescuing misfolded species is a key aspect of GroEL function. Future structure-based predictions of GroEL dependence will require identifying and determining the weights of additional factors that can affect kinetic partitioning.
Author Contributions
B.B. and T.M. performed research. R.U. and A.H. designed the research. B.B., T.M., R.U., and A.H. analyzed the data, and A.H. wrote the manuscript.
Acknowledgments
A.H. is an incumbent of the Carl and Dorothy Bennett Professorial Chair in Biochemistry.
This work was supported by the Minerva Foundation with funding from the Federal German Ministry for Education and Research (to A.H.), grant 2015170 of the US-Israel Binational Science Foundation (to A.H.), and grant 772/13 of the Israel Science Foundation (to R.U.).
Editor: James Shorter.
Footnotes
Supporting Materials and Methods, three figures, and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)31284-0.
Contributor Information
Ron Unger, Email: ron@biomodel.os.biu.ac.il.
Amnon Horovitz, Email: amnon.horovitz@weizmann.ac.il.
Supporting Material
References
- 1.Goloubinoff P., Christeller J.T., Lorimer G.H. Reconstitution of active dimeric ribulose bisphosphate carboxylase from an unfoleded state depends on two chaperonin proteins and Mg-ATP. Nature. 1989;342:884–889. doi: 10.1038/342884a0. [DOI] [PubMed] [Google Scholar]
- 2.Ostermann J., Horwich A.L., Hartl F.U. Protein folding in mitochondria requires complex formation with hsp60 and ATP hydrolysis. Nature. 1989;341:125–130. doi: 10.1038/341125a0. [DOI] [PubMed] [Google Scholar]
- 3.Saibil H.R., Fenton W.A., Horwich A.L. Structure and allostery of the chaperonin GroEL. J. Mol. Biol. 2013;425:1476–1487. doi: 10.1016/j.jmb.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 4.Hayer-Hartl M., Bracher A., Hartl F.U. The GroEL-GroES chaperonin machine: a nano-cage for protein folding. Trends Biochem. Sci. 2016;41:62–76. doi: 10.1016/j.tibs.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Gruber R., Horovitz A. Allosteric mechanisms in chaperonin machines. Chem. Rev. 2016;116:6588–6606. doi: 10.1021/acs.chemrev.5b00556. [DOI] [PubMed] [Google Scholar]
- 6.Viitanen P.V., Gatenby A.A., Lorimer G.H. Purified chaperonin 60 (groEL) interacts with the nonnative states of a multitude of Escherichia coli proteins. Protein Sci. 1992;1:363–369. doi: 10.1002/pro.5560010308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerner M.J., Naylor D.J., Hartl F.U. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 8.Fujiwara K., Ishihama Y., Taguchi H. A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 2010;29:1552–1564. doi: 10.1038/emboj.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azia A., Unger R., Horovitz A. What distinguishes GroEL substrates from other Escherichia coli proteins? FEBS J. 2012;279:543–550. doi: 10.1111/j.1742-4658.2011.08458.x. [DOI] [PubMed] [Google Scholar]
- 10.Chaudhuri T.K., Gupta P. Factors governing the substrate recognition by GroEL chaperone: a sequence correlation approach. Cell Stress Chaperones. 2005;10:24–36. doi: 10.1379/CSC-64R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stan G., Brooks B.R., Thirumalai D. Identifying natural substrates for chaperonins using a sequence-based approach. Protein Sci. 2005;14:193–201. doi: 10.1110/ps.04933205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balcin D., Hayer-Hartl M., Hartl F.U. In vivo aspects of protein folding and quality control. Science. 2016;353:aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- 13.Todd M.J., Lorimer G.H., Thirumalai D. Chaperonin-facilitated protein folding: optimization of rate and yield by an iterative annealing mechanism. Proc. Natl. Acad. Sci. USA. 1996;93:4030–4035. doi: 10.1073/pnas.93.9.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plaxco K.W., Simons K.T., Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. J. Mol. Biol. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 15.Fersht A.R., Itzhaki L.S., Otzen D.E. Single versus parallel pathways of protein folding and fractional formation of structure in the transition state. Proc. Natl. Acad. Sci. USA. 1994;91:10426–10429. doi: 10.1073/pnas.91.22.10426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fersht A.R. Transition-state structure as a unifying basis in protein-folding mechanisms: contact order, chain topology, stability, and the extended nucleus mechanism. Proc. Natl. Acad. Sci. USA. 2000;97:1525–1529. doi: 10.1073/pnas.97.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinner A.R., Karplus M. The roles of stability and contact order in determining protein folding rates. Nat. Struct. Biol. 2001;8:21–22. doi: 10.1038/83003. [DOI] [PubMed] [Google Scholar]
- 18.Lindberg M.O., Tångrot J., Oliveberg M. Folding of circular permutants with decreased contact order: general trend balanced by protein stability. J. Mol. Biol. 2001;314:891–900. doi: 10.1006/jmbi.2001.5186. [DOI] [PubMed] [Google Scholar]
- 19.Makino Y., Amada K., Yoshida M. Chaperonin-mediated folding of green fluorescent protein. J. Biol. Chem. 1997;272:12468–12474. doi: 10.1074/jbc.272.19.12468. [DOI] [PubMed] [Google Scholar]
- 20.Bandyopadhyay B., Goldenzweig A., Horovitz A. Local energetic frustration affects the dependence of green fluorescent protein folding on the chaperonin GroEL. J. Biol. Chem. 2017;292:20583–20591. doi: 10.1074/jbc.M117.808576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreiro D.U., Komives E.A., Wolynes P.G. Frustration in biomolecules. Q. Rev. Biophys. 2014;47:285–363. doi: 10.1017/S0033583514000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bandyopadhyay B., Peleg Y. Facilitating circular permutation using Restriction Free (RF) cloning. Protein Eng. Des. Sel. 2018;31:65–68. doi: 10.1093/protein/gzx061. [DOI] [PubMed] [Google Scholar]
- 23.McLennan N., Masters M. GroE is vital for cell-wall synthesis. Nature. 1998;392:139. doi: 10.1038/32317. [DOI] [PubMed] [Google Scholar]
- 24.Sokolovski M., Bhattacherjee A., Horovitz A. Thermodynamic protein destabilization by GFP tagging: a case of interdomain allostery. Biophys. J. 2015;109:1157–1162. doi: 10.1016/j.bpj.2015.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franck J.M., Sokolovski M., Horovitz A. Probing water density and dynamics in the chaperonin GroEL cavity. J. Am. Chem. Soc. 2014;136:9396–9403. doi: 10.1021/ja503501x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gill S.C., von Hippel P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 27.Topell S., Hennecke J., Glockshuber R. Circularly permuted variants of the green fluorescent protein. FEBS Lett. 1999;457:283–289. doi: 10.1016/s0014-5793(99)01044-3. [DOI] [PubMed] [Google Scholar]
- 28.Lavinder J.J., Hari S.B., Magliery T.J. High-throughput thermal scanning: a general, rapid dye-binding thermal shift screen for protein engineering. J. Am. Chem. Soc. 2009;131:3794–3795. doi: 10.1021/ja8049063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivankov D.N., Garbuzynskiy S.O., Finkelstein A.V. Contact order revisited: influence of protein size on the folding rate. Protein Sci. 2003;12:2057–2062. doi: 10.1110/ps.0302503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staniforth R.A., Burston S.G., Clarke A.R. Affinity of chaperonin-60 for a protein substrate and its modulation by nucleotides and chaperonin-10. Biochem. J. 1994;300:651–658. doi: 10.1042/bj3000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tyagi N.K., Fenton W.A., Horwich A.L. Double mutant MBP refolds at same rate in free solution as inside the GroEL/GroES chaperonin chamber when aggregation in free solution is prevented. FEBS Lett. 2011;585:1969–1972. doi: 10.1016/j.febslet.2011.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakraborty K., Chatila M., Hayer-Hartl M. Chaperonin-catalyzed rescue of kinetically trapped states in protein folding. Cell. 2010;142:112–122. doi: 10.1016/j.cell.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 33.Lin Z., Madan D., Rye H.S. GroEL stimulates protein folding through forced unfolding. Nat. Struct. Mol. Biol. 2008;15:303–311. doi: 10.1038/nsmb.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.