Abstract
Purpose
Many studies have proposed predictive models for type 2 diabetes mellitus (T2DM). However, these predictive models have several limitations, such as user convenience and reproducibility. The purpose of this study was to develop a T2DM predictive model using electronic medical records (EMRs) and machine learning and to compare the performance of this model with traditional statistical methods.
Materials and Methods
In this study, a total of available 8454 patients who had no history of diabetes and were treated at the cardiovascular center of Korea University Guro Hospital were enrolled. All subjects completed 5 years of follow up. The prevalence of T2DM during follow up was 4.78% (404/8454). A total of 28 variables were extracted from the EMRs. In order to verify the cross-validation test according to the prediction model, logistic regression (LR), linear discriminant analysis (LDA), quadratic discriminant analysis (QDA), and K-nearest neighbor (KNN) algorithm models were generated. The LR model was considered as the existing statistical analysis method.
Results
All predictive models maintained a change within the standard deviation of area under the curve (AUC) <0.01 in the analysis after a 10-fold cross-validation test. Among all predictive models, the LR learning model showed the highest prediction performance, with an AUC of 0.78. However, compared to the LR model, the LDA, QDA, and KNN models did not show a statistically significant difference.
Conclusion
We successfully developed and verified a T2DM prediction system using machine learning and an EMR database, and it predicted the 5-year occurrence of T2DM similarly to with a traditional prediction model. In further study, it is necessary to apply and verify the prediction model through clinical research.
Keywords: Type 2 diabetes mellitus, diabetes, machine learning, prediction, big data
INTRODUCTION
Type 2 diabetes mellitus (T2DM) is a chronic disease in which the body's blood sugar metabolism is impaired and blood sugar levels are elevated.1 It is well known to be affected by lifestyle activities, such as drinking, exercise, dietary habits, and others. T2DM along with other chronic diseases, such as hypertension, obesity, dyslipidemia, arteriosclerosis, and angina, affects the quality of life and life expectancy.2 The short-term and long-term adverse effects associated with T2DM in patients with cardiovascular risk are well known.3,4,5 Thus, both early diagnosis and prevention of T2DM are very important to preventing multiple serious and potentially life-threatening complications in patients with cardiovascular risk. Recent studies have shown that improving lifestyle and medication interventions can prevent diabetic complications, and it maybe can prevent the onset of T2DM.6,7,8,9,10,11 Therefore, it is very important to identify individuals at high risk for T2DM in order to establish prevention strategies for T2DM.
Over the last few decades, many studies have proposed models for predicting T2DM. However, predictive models that are actively used in clinical practice have not been established.12,13 The established models for predicting T2DM have typically been generated using Cox proportional or logistic regression (LR) analysis in non-diabetic patients between 5 to 15 years of follow up. These predictive models have some limitations: The performance of these predictive models have shown different results depending on the input variables, and the reproducibility of the prediction models is not guaranteed in not only established models but also other races and other populations. Also, a lot of time and resources are required to collect the data to make the model. In addition, models are just a statistical formula for multiple LR, and thus, the user accessibility of the model is not easy.
Machine learning has generally been used in the field of computer science, although it has been actively applied in the clinical medical field recently. Machine learning enables the definition of data attributes, and it allows for the prediction of various results using computational algorithms and computational power in large-scale databases with various parameters based on the available data.14,15,16,17 The purpose of this study was to develop a high-performance predictive model of T2DM using an electronic medical record (EMR) database and the machine learning method and to compare the performance of this model with predictive models developed using conventional statistical methods.
MATERIALS AND METHODS
Study population
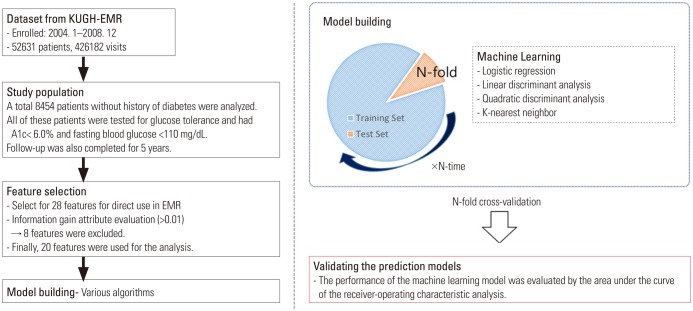
The data used in this study were obtained retrospectively from the EMR database of Korea University Guro Hospital (KUGH), and the protocol was approved by the Medical Device Institutional Review Board at KUGH (#KUGH 13017). The initially acquired subjects in the study comprised 52631 individuals (426182 visits during the study period) who visited the cardiovascular center of KUGH from January 2004 to December 2008. To clarify the results of the study, the subject with diabetes or without information on glycemic control were excluded. Finally, a total of 8454 patients who had no history of diabetes, a fasting blood glucose level of <110 mg/dL, glycated hemoglobin <6.0%, and no anti-diabetic agent treatment were enrolled in this study. All subjects completed 5 years of follow up (Fig. 1). The prevalence of T2DM was 4.78% (404/8454) during follow up.
Fig. 1. Study flow chart. KUGH: Korea University Guro Hospital, EMR: electronic medical record.
Clinical data and data collection
Personal data, such as ‘patient name’ and ‘personal identification code,’ among the data used in the analysis were provided from KUGH by generating a separate code in the dataset for privacy protection and patient identification. The predictors (or features) chosen to develop prediction of T2DM that could be extracted from the EMR were as follows: sex, age, body mass index, history of particular diseases (hypertension, coronary artery disease, myocardial infarction, coronary intervention, dyslipidemia, cerebrovascular disease, renal disease, and hyperuricemia), blood test results (fasting serum glucose, glycated hemoglobin, creatinine, total cholesterol, triglyceride, high density apolipoprotein, and low density apolipoprotein), pharmaceutical treatments for cardiovascular disease (renin-angiotensin system inhibitors, diuretics, beta blockers, calcium channel blockers, anti-anginal agent, antiplatelet agents, and statins), and pharmaceutical treatments for T2DM (meglitinides, biguanides, sulfonylureas, α-glucosidase inhibitors, thiazolidinediones, dipeptidyl peptidase-4 inhibitors, and insulin) (Table 1, Fig. 2). Among these variables, those missing information by more than a total of 30%, such as body mass index, were excluded from the model. Antidiabetic agents were used to identify the presence of diabetes in the subject at baseline or follow-up.
Table 1. Baseline Characteristics and Relative Risk Analysis for New-Onset Type 2 Diabetes Mellitus up to 5-Year Follow-up.
| Features | Total (n=8454) | T2DM (n=404) | Non-DM (n=8050) | p value | Relative risk (95% CI) |
|---|---|---|---|---|---|
| Sex, male | 3970 (46.9) | 208 (51.4) | 3762 (46.7) | 0.062 | 1.20 (0.99–1.47) |
| Age (yr) | 53.9±14.1 | 60.8±11.4 | 53.5±14.1 | <0.001 | 1.04 (1.03–1.05) |
| Hypertension | 3644 (43.1) | 242 (59.9) | 3402 (42.2) | <0.001 | 2.04 (1.66–2.50) |
| CAD | 948 (11.2) | 77 (19.0) | 871 (10.8) | <0.001 | 1.94 (1.49–2.51) |
| Prior MI | 226 (2.6) | 10 (2.4) | 216 (2.6) | 0.800 | 0.92 (0.48–1.74) |
| Prior PCI | 463 (5.4) | 44 (10.8) | 419 (5.2) | <0.001 | 2.22 (1.60–3.09) |
| Dyslipidemia | 377 (4.4) | 28 (6.9) | 349 (4.3) | 0.014 | 1.64 (1.10–2.44) |
| Stroke | 832 (9.8) | 82 (20.2) | 750 (9.3) | <0.001 | 2.47 (1.92–3.19) |
| Chronic kidney disease | 42 (0.4) | 2 (0.4) | 40 (0.4) | 0.996 | 0.99 (0.23–4.13) |
| CKD-MDRD stage | <0.001 | 1.38 (1.23–1.57) | |||
| Stage 0 | 4163 (49.2) | 161 (39.8) | 4002 (49.7) | ||
| Stage 1 | 3810 (45.0) | 199 (49.2) | 3611 (44.8) | ||
| Stage 2 | 350 (4.1) | 28 (6.9) | 322 (4.0) | ||
| Stage 3 | 89 (1.0) | 14 (3.4) | 75 (0.9) | ||
| Stage 4 | 29 (0.3) | 2 (0.4) | 27 (0.3) | ||
| Stage 5 | 13 (0.1) | 0 (0.0) | 13 (0.1) | ||
| Hyperuricemia | 621 (7.3) | 50 (12.3) | 571 (7.0) | <0.001 | 1.85 (1.35–2.51) |
| Atrial fibrillation | 283 (3.3) | 20 (5.0) | 263 (3.3) | 0.066 | 1.54 (0.96–2.45) |
| A1c (%) | 5.51±0.30 | 5.69±0.29 | 5.50±0.30 | <0.001 | 11.5 (7.69–17.4) |
| Glucose (mL/dL) | 92.8±8.35 | 96.4±8.5 | 92.6±8.3 | <0.001 | 1.06 (1.05–1.08) |
| Medications | |||||
| ARB | 1827 (21.6) | 162 (40.0) | 1665 (20.6) | <0.001 | 2.56 (2.08–3.15) |
| ACEI | 579 (6.8) | 39 (9.6) | 540 (6.7) | 0.022 | 1.48 (1.05–2.09) |
| Diuretic | 1641 (19.4) | 164 (40.5) | 1477 (18.3) | <0.001 | 3.04 (2.47–3.73) |
| β-blockers | |||||
| Selective | 620 (7.3) | 54 (13.3) | 566 (7.0) | <0.001 | 2.04 (1.51–2.75) |
| Non-selective | 871 (10.3) | 90 (22.2) | 781 (9.7) | <0.001 | 2.66 (2.08–3.41) |
| CCB | |||||
| DHP | 1680 (19.8) | 137 (33.9) | 1543 (19.1) | <0.001 | 2.16 (1.74–2.67) |
| Non-DHP | 1023 (12.1) | 79 (19.5) | 944 (11.7) | <0.001 | 1.82 (1.41–2.36) |
| Nitrate | 1632 (19.3) | 132 (32.6) | 1500 (18.6) | <0.001 | 2.11 (1.70–2.62) |
| Aspirin | 88 (1.0) | 10 (2.4) | 78 (0.9) | 0.009 | 2.59 (1.33–5.04) |
| Clopidogrel | 814 (9.6) | 96 (23.7) | 718 (8.9) | <0.001 | 3.18 (2.49–4.05) |
| Cilostazol | 290 (3.4) | 32 (7.9) | 258 (3.2) | <0.001 | 2.59 (1.77–3.80) |
| Warfarin | 181 (2.1) | 22 (5.4) | 159 (1.9) | <0.001 | 2.85 (1.80–4.51) |
| PPI | 103 (1.2) | 14 (3.4) | 89 (1.1) | <0.001 | 3.21 (1.81–5.69) |
| Statin | 1605 (18.9) | 150 (37.1) | 1455 (18) | <0.001 | 2.67 (2.17–3.30) |
T2DM, type 2 diabetes mellitus; CI, confidence interval; CAD, coronary artery disease; MI, myocardial infarction; CKD-MDRD, chronic kidney disease–the modification of diet in renal disease; PCI, percutaneous coronary intervention; ARB, angiotensin receptor blockers; ACEI, angiotensin-converting enzyme inhibitors; CCB, calcium channel blockers; DHP, dihydropyridine; PPI, proton pump inhibitors.
Variables are expressed as mean±standard deviation or number (percentage).
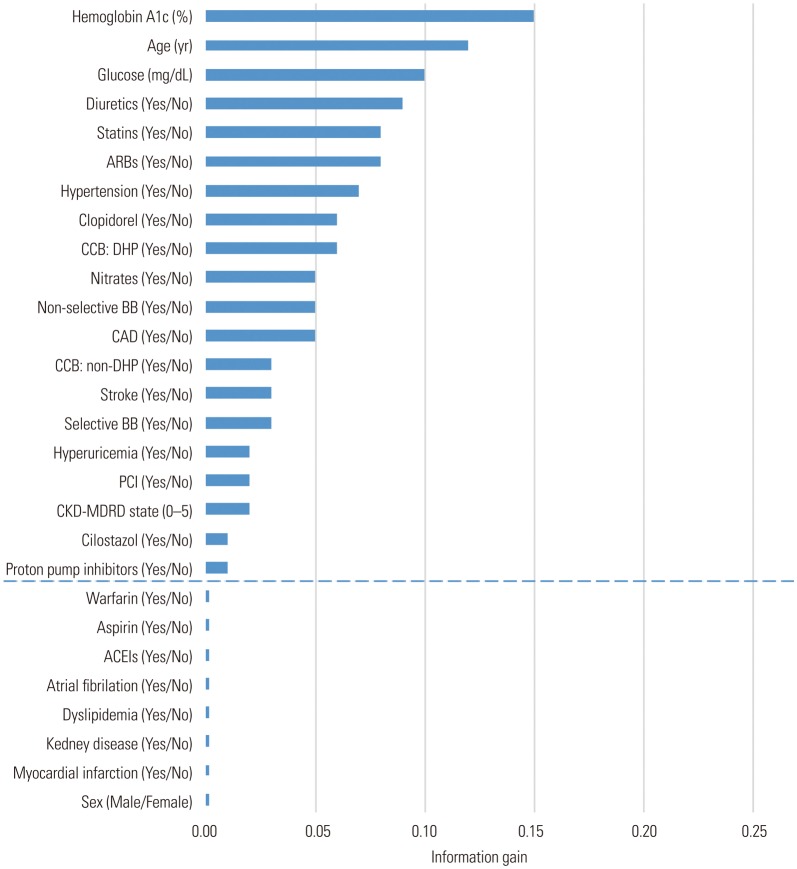
Fig. 2. Selection of features for type 2 diabetes mellitus prediction model generation using ‘Information Gain Attribute Evaluation.’ CAD, coronary artery disease; CKD-MDRD, chronic kidney disease–the modification of diet in renal disease; PCI, percutaneous coronary intervention; ARB, angiotensin receptor blockers; ACEI, angiotensin-converting enzyme inhibitors; CCB, calcium channel blockers; DHP, dihydropyridine; BB, beta blockers.
Definition and study endpoints
In this study, T2DM was defined as fasting blood glucose ≥126 mg/dL, glycated hemoglobin ≥6.5%, or the presence of a prescription for antidiabetic medication by a clinician.1 To improve the accuracy of the predictors used in the study, we cross-analyzed the records of the international conference for the ninth revision of the International Classification of Diseases (ICD-9) and clinical prescribing records recorded in the dataset. Hypertension was defined as ICD-9; 401–405 and the prescription of antihypertensive agents, myocardial infarction (ICD-9; 410–412), angina pectoris (ICD-9; 413), and cerebrovascular disease (ICD-9; 430–438). Dyslipidemia, hyperuricemia, and renal disease were defined according to relevant guidelines reflecting blood test results. Dyslipidemia was defined according to the guidelines of the National Cholesterol Education Program.18 Hyperuricemia was defined as >7.0 mg/dL for men and >6.5 mg/dL for women.19 Renal disease was assessed by the risk of an impaired glomerular filtration rate (MDRD: modification of diet in renal disease).20 The endpoint of this study was the generation of a model predicting the occurrence of T2DM within 5 years of follow-up, presenting the predictive rates of the models as the receiver operating characteristic (ROC) curve and the area under an ROC curve (AUC).
Machine learning
For this study, 28 features were available from the EMRs for model development (Table 1, Fig. 2). The use of continuous variables, such as blood test results, in machine learning model generation requires a lot of computing power and time. In this study, these continuous variables were reflected as categorical variables, such as dyslipidemia, hyperuricemia, and renal disease, for efficient allocation of resources. The T2DM prediction model was generated by LR, linear discriminant analysis (LDA), quadratic discriminant analysis (QDA), and the K-nearest neighbor (KNN) classification algorithm for machine learning. MATLAB® R2016b (MathWorks, Natick, MA, USA) was used for technical support of the machine learning techniques.
Feature selection
The selection of features for generation of the T2DM prediction model was performed by ‘information gain attribute evaluation.’21,22 This is a classifier strategy for selecting the features of the prediction model. The selected features are derived from the Ensemble classification model using a single-node decision tree and the LogitBoost algorithm.21,22,23,24
Cross-validation test
Model performance and general error estimates in the machine learning process were evaluated using stratified 10-fold cross-validation tests, which is a preferred technique in the field of data mining.25,26 This technique randomly divides the dataset into 10 equal parts, so that each part has the same number of events. A 10-fold cross-validation test is used for the validation of each part, and the remaining nine parts are used as the learning dataset, so that, ultimately, 10 LogitBoost models are generated. The performance of the entire machine learning strategy is measured by combining the validation results of the 10 generated models (Fig. 2).
Machine learning algorithms
The machine learning algorithms used in this study are summarized as follows.23,24
Logistic regression
LR is a widely used algorithm in epidemiological studies and was used as a reference for comparison with the other algorithms for analyzing data. The purpose of LR is to use the relationship between the dependent and independent variables, as detailed, for the purpose of general regression analysis for future prediction models. The LR dependent variable can be understood as a classification technique because it divides the results into specific categories for the categorical data.
Linear discriminant analysis
LDA is the most commonly used algorithm in the field of machine learning. It is a method of classifying data by learning the distribution of the data and creating a decision boundary. When classifying the given data into K classes, it is directed to find a straight line where the center (average) of each class is distant and dispersion is small.
Quadratic discriminant analysis
QDA is a more flexible classification method than LDA, which can only identify linear boundaries, because QDA can also identify secondary boundaries. Both QDA and LDA assume that the observations of each class follow a normal distribution; however, QDA assumes that the covariance matrix of each class is different from LDA. This implies that the Bayesian theorem assigns an initial estimate to the parameter. QDA assigns an observation to the class that maximizes the quantity of the parameter so that a quadratic function-type discriminant emerges.
K-nearest neighbor
The KNN algorithm is a new method to predict new data with K neighbors from the existing data when new data is provided. This is a method of classification using only the instance, without a model generation process. The hyper-parameters (detailed tuning options for efficient learning of the model) of the KNN algorithm are the number of neighbors (K) to be searched and the distance measurement method. If K is small, it overestimates the regional characteristics of the data (Overfitting), and if it is very large, the model tends to be over-normalized (Underfitting). Also, the KNN algorithm is one whose result is greatly affected by the distance measurement method chosen. In this study, we investigated the optimal K in the KNN analysis of the clinical medical data and verified the model performance according to each distance measurement method. The distance measurement method of KNN was evaluated for each city block, Euclidian, Cosine, Minkowski, Mahalanobis, Hamming, Jaccard, Correlation, Spearman, and Chebyshev models.
Statistical analysis
In this study, the comparison of ‘continuous variables’ between the two groups was evaluated by unpaired t-test or Mann-Whitney rank test and expressed as the mean±standard deviation (SD). Comparisons of categorical variables between the two groups were assessed by χ2 or Fisher's exact test and expressed as a number and a percentage. Each parameter used to predict T2DM underwent a relative risk analysis. The performance evaluation of the learning model generated by machine learning was evaluated by the AUC of ROC analysis. The statistical significance in this study was p<0.05.27
RESULTS
In this study, a total of available 8454 patients who had no history of diabetes and were treated at the cardiovascular center of KUGH were enrolled. Also, a total of 28 features were extracted from the EMRs. Basic information for the patients at the time of registration is shown in Table 1. The prevalence of T2DM was 4.78% (404/8454) during follow up.
In order to develop a predictive model of T2DM using machine learning, the ‘information gain attribute evaluation method’ was performed (Fig. 2). Among the 28 features, parameters regarding sex, dyslipidemia, chronic renal failure, history of myocardial infarction, and cardiovascular medication (aspirin, warfarin, angiotensin-converting enzyme inhibitor) were excluded because they carried information values less than 0.01.
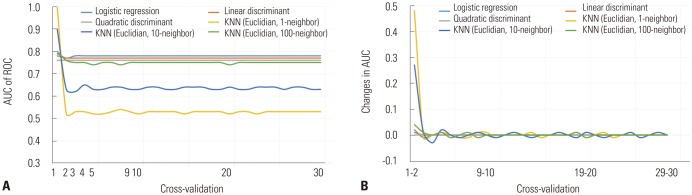
In order to verify the cross-validation test, various predictive models were generated using LR, LDA, QDA, and KNN algorithms and machine learning. KNN algorithm models were generated to Euclidean distance measurement with nearest neighbors equal to 1, 10, and 100. The change in AUC value was assessed by performing a cross-validation test from 1 to 30 fold. The SD of the AUC of the LR, LDA, QDA, and KNN (Euclidean model with the nearest neighbor 100 neighbors) models were within a range of 0.01, and the SDs of the AUCs of the Euclidean KNN model with 1 and 10 near neighbors were 0.08 and 0.05, respectively, for the cross-validation test from 1 to 30 fold. All of the predictive models maintained a change within the SD of the AUC <0.01 in the analysis after the 10-fold cross-validation test (Fig. 3).
Fig. 3. ROC analysis of the cross-validation tests ranging from 0 to 30 quartile according to the learning model. Change in AUC (A) and amount of change in AUC (B). ROC, receiver-operating characteristic; AUC, area under the curve, KNN, K-nearest neighbor.
The KNN model for the generation of the predictive model of T2DM was composed of 1, 10, 100, 200, 300, 500, and 1000 near neighbors for the optimization evaluation of the prediction model according to the hyper-parameters. Distance measurement between neighbors was Euclidian, Cosine, Minkowski, Mahalanobis, Hamming, Jaccard, Correlation, Spearman, and Chebyshev. The 10-fold cross-validation test was applied to assess the results and is shown in Table 2. In performance evaluation of the KNN model, the highest AUC was 0.77, with the near-neighbor values equal to 200 for the city block, 300 for the Euclidian, 500 for the Cosine and Minkowski, and 1000 for the Mahalanobis distance.
Table 2. Performance Evaluation of the Predictive Model for New-Onset Type 2 Diabetes Mellitus up to 5-Year Follow-up based on Hyper-Parameters (Number of Neighbors, Distance Measurement Method) in K-Nearest Neighbor Algorithm Learning Model.
| No. neighbors | Area under the curve according to the distance metrics | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| City block | Euclidian | Cosine | Minkowski | Mahalanobis | Hamming | Jaccard | Correlation | Spearman | Chebyshev | |
| 1 | 0.53 | 0.53 | 0.53 | 0.52 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 |
| 10 | 0.65 | 0.64 | 0.65 | 0.64 | 0.63 | 0.63 | 0.63 | 0.64 | 0.62 | 0.63 |
| 100 | 0.76 | 0.75 | 0.75 | 0.74 | 0.73 | 0.75 | 0.75 | 0.74 | 0.74 | 0.72 |
| 200 | 0.77 | 0.76 | 0.76 | 0.75 | 0.75 | 0.75 | 0.75 | 0.74 | 0.73 | 0.72 |
| 300 | 0.77 | 0.77 | 0.76 | 0.76 | 0.76 | 0.75 | 0.75 | 0.74 | 0.73 | 0.71 |
| 500 | 0.77 | 0.77 | 0.77 | 0.77 | 0.76 | 0.75 | 0.75 | 0.72 | 0.73 | 0.71 |
| 1000 | 0.77 | 0.77 | 0.77 | 0.77 | 0.77 | 0.75 | 0.75 | 0.70 | 0.72 | 0.71 |
The performance evaluation of the prediction model is based on the results of Fig. 3, and a 10-fold cross-validation test was applied.
Fig. 4 shows the results of the 10-fold cross-validation test regarding the T2DM predictive model using LR, LDA, QDA, and KNN with Euclidian and near-neighbor values equal to 200 or 300. Mostly, the performance of the developed T2DM prediction models converged around AUC 0.77 in this study. Compared with the LR model, in which the performance was greatest (AUC=0.78), the models of the LDA, QDA, and KNN algorithms did not show a statistically significant difference.
Fig. 4. 10-fold cross-validation test of the predictive models of type 2 diabetes mellitus. KNN, K-nearest neighbor; AUC, area under the curve.

DISCUSSION
Recently, as medical records have become electronic databases, the utilization of big data is considered to be clinically valuable. However, if these data cannot be made clinically relevant to our real-world clinical practice, they become useless. In order to be useful and valuable, data must be analyzed, interpreted, and translated into clinical practice. Machine learning is an emerging tool for processing and utilizing big data.14,15,16,17 The development of machine learning in clinical medicine is expected to be a powerful tool for clinicians.15 Thus, studies are being actively conducted to apply machine learning to clinical medicine.22,26,28 In the present study, we generated a predictive model of T2DM using LR, LDA, QDA, and KNN algorithms and performed a cross-validation test to verify the performance of a machine learning disease prediction model. Moreover, in the prediction of T2DM, optimization of the prediction model according to the hyper-parameter settings of the KNN learning model was sought, and the performance of the optimized prediction models was compared. This approach successfully predicted the 5-year occurrence of T2DM compared with a traditional prediction model.
Machine learning develops a programmed prediction model using data, algorithms, and computing power. This process requires more computing power as the number of data variables increases. Accordingly, the efficient use of meaningful variables is important. The present study collected 28 features for analysis, including patient information, disease status, test results, and medication information, from a single-center EMR database, which was used to analyze 8454 non-diabetic subjects. In order to generate an efficient prediction model of T2DM, the information gain attribute evaluation method and a 10-fold cross-validation test were performed, and 20 out of the 28 features were selected for model generation. Generally, in the data mining and machine learning field, 10-fold cross-validation is performed to assess the validity of the generated features or models. However, use of 10-fold cross-validation in the field of clinical medical data is very limited.22,25,26 In this study, LR, LDA, and QDA learning models, as well as the KNN learning model (using 1, 10, and 100 neighbors with the Euclidian distance measurement method), were created to verify the validation test according to the learning models, and the cross-validity test was performed from 1 to 30 fold. The cut-off of the section that was most stable was searched. As shown in Fig. 3B, all of the predictive models showed a SD of the 3-fold AUC of less than 0.05 and a SD of the 10-fold AUC of less than 0.01. Our results indicated that the 10-fold cross-validation test is an effective method for verifying clinical data. Previous studies that have reported predictive models of T2DM have been developed in the form of regression formula or risk scores using regression analysis, such as Cox proportional or logistic, with predictive ranges of 0.71 to 0.91 in AUC measurement.12,13 Meanwhile, however, these predictive models were evaluated in the same cohort in which they were developed, thus allowing for overfitting.25,26 This can be a very important limitation. Thus, if the cross-validation test in the generation of predictive models is applied, it could improve problems with overfitting.
In this study, the LR model was considered as a standard regression analysis method. Thus, a LR model was created to compare the machine learning predictive models, such as LDA, QDA, and KNN model. Along with LR, the LDA, QDA, and KNN algorithms are the most common and proven machine learning algorithms in the computer science field. Since the principles of these algorithms are slightly different, it is worth exploring algorithms that exhibit optimal performance. All predictive models for performance comparisons were finally assessed with the 10-fold cross-validation test, and performance was compared by analysis of AUC. LDA and QDA algorithms are the most commonly used statistical algorithms in the field of machine learning.23,24 The KNN algorithm measures the distance of the nearest K neighbors among the given data and clustering of similar groups. This can be seen as a way of assessing risk according to the attributes of a particular group. We considered that this may be effective when the target group is local or a variable whose risk is unknown is used as a predictor in the analysis of clinical medical data. Unlike the LR, LDA, and QDA algorithms, the KNN algorithm needs to be verified for model generation because the performance of the model depends on the setting of hyper-parameters.24 The hyper-parameters of the KNN algorithm are the number of near neighbors and the distance measurement. In clinical medicine, the proper distance measurement of the KNN algorithm is unclear. As shown in Table 2 and Fig. 3, we measured the change in the predictive performance of the model according to the hyper-parameters and were able to select the best performing model. However, these methods must be re-evaluated according to the number of cohort subjects, samples, and variables that will be specific to each study.
In this study, we developed a prediction system using machine learning algorithms, including LR, LDA, QDA, and KNN models with 200 neighbors and a city block or 300 neighbors and the Euclidian. The machine learning predictive models have successfully predicted the 5-year occurrence of T2DM and showed similar prediction performance with a traditional prediction model. As shown in Fig. 4, all of the models developed in this study showed concordant discrimination, with AUCs consistently around 0.77. Our results may have been influenced by the fact that all of the predictive models for T2DM were developed using the same 20 features. This can be an important reason for the consistent performance of all the predictive models. Also, regression analysis has traditionally been a representative method for assessing the causal relationship between features and diseases in medical science. The development of predictive models using typical medical features and LR algorithms (one regression analysis) may have shown the best performance of the LR algorithm model.
In our study, the source data of the predictive model were readily obtained from an EMR database. We suspect that prediction models could be programmed into EMR databases, facilitating race or locality-optimized diagnosis or prediction models of a disease. Furthermore, when patient information is updated and unknown parameters are discovered and applied, the performance of these models may be improved. Further, this may help reduce the development costs of prediction models. With this expectation, many researchers are working on applying machine learning or deep learning to medicine. However, at the moment, the performance improvements with machine learning do not yet expand beyond the abilities of humans. In our study, the maximum performance among all of the developed models showed an AUC of 0.78, which was not significantly different from that of a previous study. Similarly, Gulshan, et al.28 verified the deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. In the evaluation of retinal fundus photographs of diabetic patients, the deep running algorithm showed high sensitivity and specificity for detecting diabetic retinopathy; however, there was no statistical difference with current ophthalmologic assessment. As such, while machine learning methods using computer power, mathematical algorithms, and EMR data can provide convenience in model development and use, it seems that this does not yet show the performance level to replace humans. Further research is needed to determine the feasibility of applying machine learning in a clinical setting and to determine whether machine learning can lead to improved outcomes in comparison to clinical assessments.
In this study, there were several limitations. First, the source data of this study included subjects with cardiovascular risks, so the results of this study cannot be generalized to everyone. Thus, in further study, it is necessary to collect cases and improve performance based on the model from this study. Second, ICD codes were used to diagnose disease. The use of ICD codes indicates the presence or absence of disease, but does not reflect the progression of the disease. To overcome these drawbacks, the results of blood tests and medication information were used for analysis, although this may not be enough. Third, the study used an EMR database, and missing information from the EMR was not reflected. This can influence the outcomes of the study. Fourth, unlike previous traditional studies, our study applied 10-fold validation in the development of the models. However, throughout the study, model development and validation was conducted with only one database. Thus, it is necessary to collect additional cases and verify the model derived in this study using other data sources. Finally, the LR model was considered as a standard regression analysis method in this study. Nevertheless, the LR model is derived from machine learning and may show the different performance than regression formulas and risk scores based on regression analysis.
ACKNOWLEDGEMENTS
This work was supported by Korea Institute for Advancement of Technology (KIAT) grant funded by the Korea government (MOTIE) (P0006675, Development of blood glucose measurement system for self-monitoring of blood glucose).
Footnotes
The authors have no potential conflicts of interest to disclose.
- Conceptualization: Byoung Geol Choi, Seung-Woon Rha.
- Data curation: Byoung Geol Choi.
- Formal analysis: Byoung Geol Choi, Jun Hyuk Kang.
- Funding acquisition: Suhng Wook Kim.
- Investigation: Byoung Geol Choi, Seung-Woon Rha.
- Methodology: Byoung Geol Choi, Jun Hyuk Kang.
- Project administration: Seung-Woon Rha.
- Software: Byoung Geol Choi, Jun Hyuk Kang.
- Supervision: Jun Hyuk Kang, Seung-Woon Rha, Yung-Kyun Noh.
- Validation: Byoung Geol Choi.
- Writing—original draft: Byoung Geol Choi.
- Writing—review & editing: Byoung Geol Choi, Seung-Woon Rha, Suhng Wook Kim, Ji Young Park.
References
- 1.American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2013;36(Suppl 1):S67–S74. doi: 10.2337/dc13-S067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wexler DJ, Grant RW, Wittenberg E, Bosch JL, Cagliero E, Delahanty L, et al. Correlates of health-related quality of life in type 2 diabetes. Diabetologia. 2006;49:1489–1497. doi: 10.1007/s00125-006-0249-9. [DOI] [PubMed] [Google Scholar]
- 3.Laakso M. Hyperglycemia and cardiovascular disease in type 2 diabetes. Diabetes. 1999;48:937–942. doi: 10.2337/diabetes.48.5.937. [DOI] [PubMed] [Google Scholar]
- 4.Romero SP, Garcia-Egido A, Escobar MA, Andrey JL, Corzo R, Perez V, et al. Impact of new-onset diabetes mellitus and glycemic control on the prognosis of heart failure patients: a propensity-matched study in the community. Int J Cardiol. 2013;167:1206–1216. doi: 10.1016/j.ijcard.2012.03.134. [DOI] [PubMed] [Google Scholar]
- 5.Twito O, Ahron E, Jaffe A, Afek S, Cohen E, Granek-Catarivas M, et al. New-onset diabetes in elderly subjects: association between HbA1c levels, mortality, and coronary revascularization. Diabetes Care. 2013;36:3425–3429. doi: 10.2337/dc12-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tuomilehto J, Lindström J, Eriksson JG, Valle TT, Hämäläinen H, Ilanne-Parikka P, et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med. 2001;344:1343–1350. doi: 10.1056/NEJM200105033441801. [DOI] [PubMed] [Google Scholar]
- 7.Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park JY, Rha SW, Choi B, Choi JW, Ryu SK, Kim S, et al. Impact of low dose atorvastatin on development of new-onset diabetes mellitus in Asian population: three-year clinical outcomes. Int J Cardiol. 2015;184:502–506. doi: 10.1016/j.ijcard.2015.03.047. [DOI] [PubMed] [Google Scholar]
- 9.Rha SW, Choi BG, Seo HS, Park SH, Park JY, Chen KY, et al. Impact of statin use on development of new-onset diabetes mellitus in Asian population. Am J Cardiol. 2016;117:382–387. doi: 10.1016/j.amjcard.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 10.Almdal T, Scharling H, Jensen JS, Vestergaard H. The independent effect of type 2 diabetes mellitus on ischemic heart disease, stroke, and death: a population-based study of 13,000 men and women with 20 years of follow-up. Arch Intern Med. 2004;164:1422–1426. doi: 10.1001/archinte.164.13.1422. [DOI] [PubMed] [Google Scholar]
- 11.Wilson PW, D'Agostino RB, Parise H, Sullivan L, Meigs JB. Metabolic syndrome as a precursor of cardiovascular disease and type 2 diabetes mellitus. Circulation. 2005;112:3066–3072. doi: 10.1161/CIRCULATIONAHA.105.539528. [DOI] [PubMed] [Google Scholar]
- 12.Abbasi A, Peelen LM, Corpeleijn E, van der Schouw YT, Stolk RP, Spijkerman AM, et al. Prediction models for risk of developing type 2 diabetes: systematic literature search and independent external validation study. BMJ. 2012;345:e5900. doi: 10.1136/bmj.e5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt MI, Duncan BB, Bang H, Pankow JS, Ballantyne CM, Golden SH, et al. Identifying individuals at high risk for diabetes: The Atherosclerosis Risk in Communities study. Diabetes Care. 2005;28:2013–2018. doi: 10.2337/diacare.28.8.2013. [DOI] [PubMed] [Google Scholar]
- 14.Waljee AK, Higgins PD. Machine learning in medicine: a primer for physicians. Am J Gastroenterol. 2010;105:1224–1226. doi: 10.1038/ajg.2010.173. [DOI] [PubMed] [Google Scholar]
- 15.Obermeyer Z, Emanuel EJ. Predicting the future - big data, machine learning, and clinical medicine. N Engl J Med. 2016;375:1216–1219. doi: 10.1056/NEJMp1606181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deo RC. Machine learning in medicine. Circulation. 2015;132:1920–1930. doi: 10.1161/CIRCULATIONAHA.115.001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darcy AM, Louie AK, Roberts LW. Machine learning and the profession of medicine. JAMA. 2016;315:551–552. doi: 10.1001/jama.2015.18421. [DOI] [PubMed] [Google Scholar]
- 18.Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults. Executive summary of the Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult Treatment Panel III) JAMA. 2001;285:2486–2497. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 19.Lai SW, Tan CK, Ng KC. Epidemiology of fatty liver in a hospital-based study in Taiwan. South Med J. 2002;95:1288–1292. [PubMed] [Google Scholar]
- 20.Poggio ED, Wang X, Greene T, Van Lente F, Hall PM. Performance of the modification of diet in renal disease and Cockcroft-Gault equations in the estimation of GFR in health and in chronic kidney disease. J Am Soc Nephrol. 2005;16:459–466. doi: 10.1681/ASN.2004060447. [DOI] [PubMed] [Google Scholar]
- 21.Hall MA, Holmes G. Benchmarking attribute selection techniques for discrete class data mining. IEEE Trans Knowl Data Eng. 2003;15:1437–1447. [Google Scholar]
- 22.Motwani M, Dey D, Berman DS, Germano G, Achenbach S, Al-Mallah MH, et al. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis. Eur Heart J. 2017;38:500–507. doi: 10.1093/eurheartj/ehw188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotsiantis SB. Supervised machine learning: a review of classification techniques. Informatica. 2007;31:249–268. [Google Scholar]
- 24.Aha DW, Kibler D, Albert MK. Instance-based learning algorithms. Machine Learning. 1991;6:37–66. [Google Scholar]
- 25.Molinaro AM, Simon R, Pfeiffer RM. Prediction error estimation: a comparison of resampling methods. Bioinformatics. 2005;21:3301–3307. doi: 10.1093/bioinformatics/bti499. [DOI] [PubMed] [Google Scholar]
- 26.Witten IH, Frank E, Hall MA, Pal CJ. Data mining: practical machine learning tools and techniques. 4th ed. Cambridge (MA): Morgan Kaufmann; 2016. [Google Scholar]
- 27.Bradley AP. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recogn. 1997;30:1145–1159. [Google Scholar]
- 28.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]