Abstract
Purpose
The most common cause of acute exacerbation of chronic obstructive pulmonary disease (AECOPD) is respiratory infection. Most studies of bacterial or viral cause in AECOPD have been conducted in Western countries. We investigated bacterial and viral identification rates in AECOPD in Korea.
Materials and Methods
We reviewed and analyzed medical records of 736 cases of AECOPD at the Korea University Guro Hospital. We analyzed bacterial and viral identification rates and classified infections according to epidemiological factors, such as Global Initiative for Chronic Obstructive Lung Disease stage, mortality, and seasonal variation.
Results
The numbers of AECOPD events involving only bacterial identification, only viral identification, bacterial-viral co-identification, and no identification were 200 (27.2%), 159 (21.6%), 107 (14.5%), and 270 (36.7%), respectively. The most common infectious bacteria identified were Pseudomonas aeruginosa (13.0%), Streptococcus pneumoniae (11.4%), and Haemophilus influenzae (5.3%); the most common viruses identified were influenza virus (12.4%), rhinovirus (9.4%), parainfluenza virus (5.2%), and metapneumovirus (4.9%). The bacterial identification rate tended to be higher at more advanced stages of chronic obstructive pulmonary disease (p=0.020 overall, p=0.011 for P. aeruginosa, p=0.048 for S. pneumoniae). Staphylococcus aureus and Klebsiella pneumoniae were identified more in mortality group (p=0.003 for S. aureus, p=0.009 for K. pneumoniae). All viruses were seasonal (i.e., greater prevalence in a particular season; p<0.050). Influenza virus and rhinovirus were mainly identified in the winter, parainfluenza virus in the summer, and metapneumovirus in the spring.
Conclusion
This information on the epidemiology of respiratory infections in AECOPD will improve the management of AECOPD using antibiotics and other treatments in Korea.
Keywords: Bacteria, chronic obstructive pulmonary disease, viruses
INTRODUCTION
Chronic obstructive pulmonary disease (COPD) is an obstructive lung disease caused by smoking and air pollution.1,2 COPD patients often experience sudden aggravation of respiratory symptoms called acute exacerbation of COPD (AECOPD), and require additional treatment. AECOPD leads to deterioration in quality of life and substantial morbidity and mortality.3,4,5 A major cause of AECOPD is bacterial or viral infection.6,7,8 Bacterial and viral infections have regional variability, and epidemiological characteristics should be considered during treatment. However, most previous analyses of bacterial and viral identification rates in AECOPD have been conducted in Western countries;9,10,11,12 studies in Asia are scarce and unclear. The purpose of this study was to retrospectively analyze bacterial and viral identification rates in AECOPD in Korea based on epidemiological factors.
MATERIALS AND METHODS
Data recruitment
We reviewed and analyzed medical records of 736 cases of severe AECOPD at Korea University Guro Hospital from January 2011 to May 2017. We searched the electronic medical records database using the keywords “chronic obstructive pulmonary disease” and “acute exacerbation.” This study was approved by the Institutional Review Board of Korea University Guro Hospital (approved number: KUGH16131-002).
COPD was diagnosed based on the guidelines of the Global Initiative for Chronic Obstructive Lung Disease (GOLD). AECOPD was defined as “an acute event characterized by a worsening of the patient's respiratory symptoms that is beyond normal day-to-day variation and leads to a change in medication.”13,14 All patients had a previous pulmonary function test, and a ratio of forced expiratory volume in one second (FEV1) to forced vital capacity (FVC) of <70%. All patients were >40 years old.
Patients were excluded if 1) the cause of admission was something other than AECOPD, such as heart failure, arrhythmia, acute pulmonary edema, acute pulmonary embolism, or pneumothorax; 2) they had a comorbidity that can affect immunity, such as cancer or autoimmune disease; 3) no laboratory or culture test was conducted within 24 hr of hospitalization; or 4) they exhibited mild to moderate exacerbation of COPD.
Laboratory tests
All cultures, antigen detection assays, and polymerase chain reaction (PCR) assays of blood, sputum, urine, or naso- or oro-pharyngeal swab samples were obtained by a physician within 24 hr of admission.
Bacterial cultures were performed on blood and sputum samples using standard techniques. Urinary antigen testing was performed to detect Streptococcus pneumoniae. Real-time PCR (RT-PCR) assays were performed using naso- or oro-pharyngeal swab samples to detect the following respiratory viruses: influenza virus, respiratory syncytial virus, parainfluenza virus, coronavirus, rhinovirus, enterovirus, adenovirus, bocavirus, and metapneumovirus. A rapid antigen test was used to test to detect influenza.
Pathogen identification
A bacterial pathogen was determined to be present if 1) gram-positive or gram-negative bacteria were detected in a blood or sputum sample or if 2) Streptococcus pneumoniae was detected in urine by an antigen detection assay.
A viral pathogen was determined to be present if 1) one of the viruses listed above was detected in a naso- or oro-pharyngeal swab by an RT-PCR assay or if 2) influenza virus was detected by a rapid antigen test.
Statistical analysis
Data were analyzed using SPSS 20 software (IBM Corp., Armonk, NY, USA). Data were presented as mean±standard deviation or number and percent of each group. When events were classified by GOLD stage or month, we used a one-way analysis of variance. When events were classified by in-hospital mortality, chi-square test or Fisher's test was used for discrete data, and an independent t-test or Mann-Whitney test was used for continuous data. All p-values <0.050 were considered statistically significant.
RESULTS
Study population
The number of patients involved in the 736 events was 487, and the number of patients who were hospitalized more than twice during the study period was 117. The mean patient age was 72.2±9.3 years, and the mean length of hospital stay was 10.4±9.6 days. There were 518 (70.4%) events in males and 218 (29.6%) in females. Most (434, 59.0%) of the events occurred in ex-smokers; there were 108 (14.7%) events in current smokers and 194 (26.4%) in non-smokers (Table 1).
Table 1. Characteristics of Patients with AECOPD Events.
| No. of exacerbations events/patients | 736/487 |
| Age (yr)* | 72.2±9.3 |
| Gender, no. of exacerbations events | |
| Male† | 518 (70.4) |
| Female† | 218 (29.6) |
| Smoking history, no. of exacerbations events | |
| Current-smoker† | 108 (14.7) |
| Ex-smoker† | 434 (59.0) |
| Non-smoker† | 194 (26.4) |
| Pack-year history* | 42.7±23.7 |
| Severity of COPD, no. of exacerbations events | |
| GOLD I† | 57 (7.7) |
| GOLD II† | 281 (38.2) |
| GOLD III† | 307 (41.7) |
| GOLD IV† | 91 (12.4) |
| Comorbidities, no. of exacerbations events | |
| Hypertension† | 376 (51.1) |
| Diabetes† | 172 (23.4) |
| Coronary artery disease† | 119 (16.2) |
| Cerebrovascular accident† | 34 (4.6) |
| Length of hospital stay (days)* | 10.4±9.6 |
| Mortality in hospital† | 32 (4.3) |
| Pneumococcal vaccination† | 215 (29.2) |
| Prior exacerbation history | |
| Exacerbation events in previous 1 month† | 52 (7.1) |
| Exacerbation events in previous 12 months† | 263 (35.7) |
| Baseline spirometry after bronchodilation | |
| FEV1 (liters)* | 1.2±0.5 |
| FEV1 (% of predicted value)* | 50.2±19.2 |
| Ratio of FEV1 to FVC (%)* | 48.5±12.3 |
AECOPD, acute exacerbation of chronic obstructive pulmonary disease; COPD, chronic obstructive pulmonary disease; GOLD, Global Initiative for Chronic Obstructive Lung Disease; FEV1, forced expiratory volume in one second; FVC, forced vital capacity.
*Data are presented as a mean±standard deviation, †Data are presented as a n (%).
Pathogen identification
Pathogens were identified in 466 (63.3%) events among 736 events. The numbers of patients with only bacterial identification, only viral identification, bacterial-viral co-identification, and no identification were 200 (27.2%), 159 (21.6%), 107 (14.5%), and 270 (36.7%), respectively. Among these, 113 (15.4%) events were identified as single pathogen, and 353 (48.0%) events were identified as multiple pathogens.
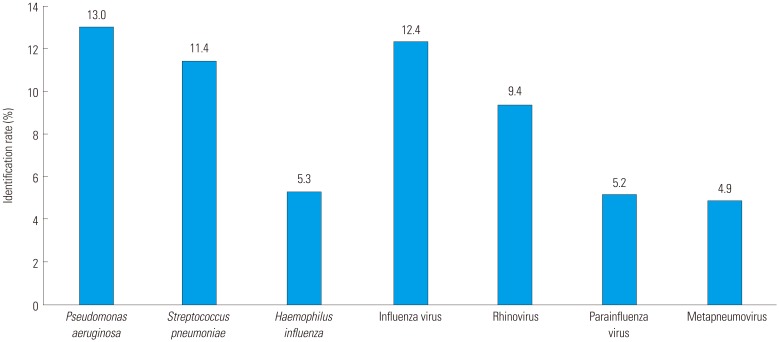
The most common bacterial pathogens were Pseudomonas aeruginosa (13.0%), Streptococcus pneumoniae (11.4%), and Haemophilus influenzae (5.3%), Staphylococcus aureus (4.8%), and Klebsiella pneumoniae (2.7%) (Fig. 1). The most common viral pathogens were influenza virus (12.4%), rhinovirus (9.4%), parainfluenza virus (5.2%), and metapneumovirus (4.9%).
Fig. 1. Pathogen identification rate.
Analysis according to GOLD stage
Of 736 AECOPD events, 91 (12.4%) were very severe COPD (stage IV, FEV1<30% of the predicted value), 307 (41.7%) were severe COPD (Stage III, 30%≤FEV1<50% of the predicted value), 281 (38.2%) were moderate COPD (Stage II, 50%≤FEV1<80% of the predicted value), and 57 (7.7%) were mild COPD (Stage I, 80%≤FEV1 of the predicted value).
The more advanced the COPD stage, the higher the bacterial identification rate tended to be (p=0.020), with P. aeruginosa and S. pneumoniae showing the strongest association (p=0.011 and p=0.048, respectively). In contrast, the viral identification rate tended to be lower at more advanced COPD stages (p=0.031). Some bacteria and viruses showed no association between identification rate and GOLD stage (Table 2).
Table 2. Bacterial and Viral Pathogen Identification according to GOLD Stage.
| All events (n=736) | GOLD I (n=57) | GOLD II (n=281) | GOLD III (n=307) | GOLD IV (n=91) | p value | |
|---|---|---|---|---|---|---|
| Only bacterial identification | 200 (27.2) | 11 (19.3) | 68 (24.2) | 91 (29.6) | 30 (33.0) | 0.020 |
| Only viral identification | 159 (21.6) | 16 (28.1) | 68 (24.2) | 60 (19.5) | 15 (16.5) | 0.031 |
| Bacterial-viral co-identification | 107 (14.5) | 4 (7.0) | 37 (13.2) | 53 (17.3) | 13 (14.3) | 0.112 |
| No identification | 270 (36.7) | 26 (45.6) | 108 (38.4) | 103 (33.6) | 33 (36.3) | 0.141 |
| Pseudomonas aeruginosa | 96 (13.0) | 4 (7.0) | 29 (10.3) | 47 (15.3) | 16 (17.6) | 0.011 |
| Streptococcus pneumoniae | 84 (11.4) | 3 (5.3) | 29 (10.3) | 38 (12.4) | 14 (15.4) | 0.048 |
| Haemophilus influenzae | 39 (5.3) | 5 (8.8) | 15 (5.3) | 17 (5.5) | 2 (2.2) | 0.158 |
| Staphylococcus aureus | 35 (4.8) | 1 (1.8) | 16 (5.7) | 10 (3.3) | 8 (8.8) | 0.333 |
| Klebsiella pneumoniae | 20 (2.7) | 1 (1.8) | 10 (3.6) | 8 (2.6)1 | 1 (1.1) | 0.509 |
| Influenza virus | 91 (12.4) | 8 (14.0) | 42 (14.9) | 32 (10.4) | 9 (9.9) | 0.112 |
| Rhinovirus | 69 (9.4) | 5 (8.8) | 21 (7.5) | 35 (11.4) | 8 (8.8) | 0.387 |
| Parainfluenza virus | 38 (5.2) | 2 (3.5) | 13 (4.6) | 15 (4.9) | 8 (8.8) | 0.165 |
| Metapneumovirus | 36 (4.9) | 3 (5.3) | 14 (5.0) | 15 (4.9) | 4 (4.4) | 0.832 |
GOLD, Global Initiative for Chronic Obstructive Lung Disease.
Data are presented as n (%).
Analysis according to in-hospital mortality
Of the 736 AECOPD events, 32 (4.3%) resulted in patient death in the hospital. Of these 32 cases, 15 (46.9%) showed only bacterial pathogen identification, a higher rate than for non-mortality events (p=0.010). In contrast, only one of the 32 cases (3.1%) was identified as viral pathogen, significantly lower than in non-mortality events (p=0.009). S. aureus and K. pneumoniae were identified more in the mortality event group (p=0.003 for S. aureus, p=0.009 for K. pneumoniae). The P. aeruginosa identification rate was 25.0% in mortality event and 12.5% in non-mortality event groups, although this difference was not statistically significant (p=0.056). Other bacteria and viruses also did not show any significant difference in identification rates between mortality and non-mortality events (Table 3).
Table 3. Bacterial and Viral Pathogen Identification according to In-hospital Mortality.
| All events (n = 736) | Mortality events in hospital (n=32) | Non-mortality events in hospital (n=704) | p value | |
|---|---|---|---|---|
| Only bacterial identification | 200 (27.2) | 15 (46.9) | 185 (26.3) | 0.010 |
| Only viral identification | 159 (21.6) | 1 (3.1) | 158 (22.4) | 0.009 |
| Bacterial-viral co-identification | 107 (14.5) | 8 (25.0) | 99 (14.1) | 0.118 |
| No identification | 270 (36.7) | 8 (25.0) | 262 (37.2) | 0.161 |
| Pseudomonas aeruginosa | 96 (13.0) | 8 (25.0) | 88 (12.5) | 0.056 |
| Streptococcus pneumoniae | 84 (11.4) | 2 (6.3) | 82 (11.6) | 0.567 |
| Haemophilus influenzae | 39 (5.3) | 2 (6.3) | 37 (5.3) | 0.684 |
| Staphylococcus aureus | 35 (4.8) | 6 (18.8) | 29 (4.12) | 0.003 |
| Klebsiella pneumoniae | 20 (2.7) | 4 (12.5) | 16 (2.27) | 0.009 |
| Influenza virus | 91 (12.4) | 3 (9.4) | 88 (12.5) | 0.787 |
| Rhinovirus | 69 (9.4) | 4 (12.5) | 65 (9.2) | 0.530 |
| Parainfluenza virus | 38 (5.2) | 1 (3.1) | 37 (5.3) | 1.000 |
| Metapneumovirus | 36 (4.9) | 0 (0.0) | 36 (5.1) | 0.396 |
Data are presented as n (%).
Analysis according to seasonal variation (monthly analysis)
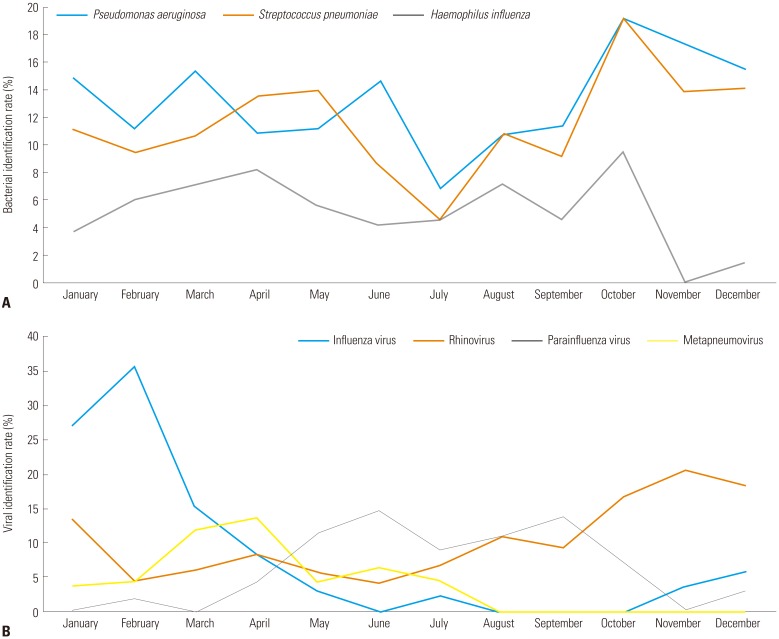
All three bacteria showed no seasonal differences in identification rates (all p-values >0.050), while all viruses did show seasonal differences (all p-values <0.050). Influenza virus and rhinovirus were mainly identified in the winter, parainfluenza virus in the summer, and metapneumovirus in the spring (Fig. 2).
Fig. 2. Seasonal variation of bacterial and viral identification rates. (A) Bacterial identification rate according to month. (B) Viral identification rate according to month.
Analysis according to bacterial-viral co-identification
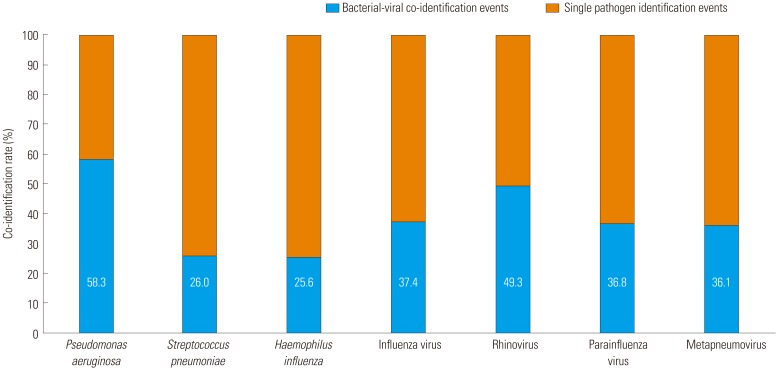
Of the 307 events involving bacterial infection, bacterial-viral co-identification was observed in 107 (34.9%). Of the 266 events involving viral infection, bacterial-viral co-identification was observed in 107 (40.2%). Of the bacteria, P. aeruginosa had the highest co-identification rate (58.3%); of the viruses, rhinovirus had the highest co-identification rate (49.3%) (Fig. 3).
Fig. 3. Bacterial-viral co-identification rate.
DISCUSSION
This was a large, retrospective, population-based, single center study of bacterial and viral pathogens associated with AECOPD in the Republic of Korea, including epidemiological factors. Bacterial and viral infections are the most common causes of AECOPD.7 The bacterial identification rate in AECOPD events has previously been reported to be 40–60% and the viral identification rate 20–40%.9,12,15,16 Our study showed similar results, with a 41.7% bacterial identification rate and 36.1% viral identification rate.
The most commonly identified bacteria in previous studies of AECOPD have been H. influenzae, Moraxella catarrhalis, and S. pneumoniae.15 However, in our study, M. catarrhalis was identified in only 8 (1.1%) of 736 AECOPD events. Along with H. influenzae and S. pneumoniae, in our study, P. aeruginosa was identified as a major AECOPD pathogen. Previous studies have found that P. aeruginosa identification rates increase with GOLD stage and that higher S. pneumoniae identification rates are associated with comorbidity.17 Our results agree with these previous studies: we found that P. aeruginosa identification increased as the GOLD stage increased. The correlation between S. pneumoniae identification rate and GOLD stage seems to be due to high comorbidity at more advanced GOLD stages. The low identification rate of M. catarrhalis may be due to regional differences. For other reasons, M. catarrhalis may not be cultured better than other bacteria, so the identification rate may be low.
New molecular diagnostic methods, such as nucleic acid amplification tests, antigen detection assays, and PCR assays, have revealed the importance of viral infection as a cause of AECOPD. Meanwhile, virus has also been identified as a major cause of exacerbation in AECOPD patients with low C-reactive protein levels.18 Respiratory viral infections affect epithelial cells in the airway and lead to microvascular dilatation, edema, inflammatory cell infiltration, and reduction of mucociliary clearance.19 In previous studies of AECOPD, rhinovirus was the most commonly identified virus.9,20 In our study, influenza and parainfluenza viruses along with rhinovirus had the highest identification rates. The high influenza identification rates in our study are likely due to influenza epidemics in Korea in 2014 and 2015; 51 of the 91 events with influenza virus identification occurred during this time. These results suggest that rhinovirus is the virus most commonly identified with AECOPD unless there is another virus outbreak.
As mentioned above, P. aeruginosa is frequently identified in severe and very severe COPD patients. This is thought to be related to bronchiectasis. A high prevalence of bronchiectasis was found in patients with moderate to severe COPD.21 Chest computed tomography was performed in 526 of the 736 events, and bronchiectasis was diagnosed in 222 events. The prevalences of bronchiectasis were 10.3% for GOLD stage I, 36.9% for stage II, 49.0% for stage III, and 54.1% for stage IV. When choosing antibiotics for use in patients with severe or very severe AECOPD, antibiotics with antimicrobial activity against P. aeruginosa should be considered.
The prevalence of respiratory viruses is closely related to temperature and humidity.22,23 Korea has four distinct seasons, and worldwide, viral identification in AECOPD has shown seasonal variation. In our study, influenza virus and rhinovirus were detected in the winter, metapneumovirus in the spring and parainfluenza virus in the summer. Our results are consistent with a previous study conducted in Istanbul, in which metapneumovirus was prominent in the spring and parainfluenza virus was predominant in the summer.24 Knowledge of seasonal virus prevalence will help in prevention and treatment of respiratory viral infections in AECOPD.
The rate of mortality due to AECOPD was higher when bacteria were identified. Age, comorbidity, and home oxygen use were higher in the mortality group than in the non-mortality group. Therefore, these factors (the type of infection and the basal condition of the patient) should be taken into consideration when predicting prognosis and mortality.
When there is co-identification of bacterial and viral infection, it is not known which infection occurs first. Recent studies have shown that respiratory viral infections cause dysregulation of iron homeostasis mechanisms associated with secondary bacterial infections.25 In addition, respiratory syncytial virus promotes development of P. aeruginosa biofilms in vitro and in vivo. On the other hand, bacterial infections in the airway can promote respiratory viral infection.26 When the lungs are infected with P. aeruginosa, the tight junction between epithelium is damaged resulting in release of lactate dehydrogenase, destroying the lung barrier.27,28 Of the bacteria in our study, P. aeruginosa was most commonly co-identified with a virus. Further studies on the interactions between P. aeruginosa and viruses are needed.
Our study has some limitations. First, it is a retrospective, single center study. Therefore, many events were excluded through lack of data and exclusion criteria. For example, we evaluated pneumococcal vaccination, but did not find influenza vaccination coverage for all patients. Thus, we could not analyze the effect of influenza vaccination at the time of the influenza epidemic. Also, chest computed tomography was performed only in some cases, so it was not applicable to all events. Second, colonization could not be distinguished in the analysis of detection; in particular, it would have been advantageous if there had been previous or follow-up data from sputum culture. Third, influenza rapid antigen test was performed only in 247 (33.6%) events. Additionally, it was mainly performed only in the epidemic season. We think this would have affected the analysis of seasonal variation. Fourth, the phenotype of COPD might have a major impact on pathogen identification. However, through retrospective data, it was difficult to accurately identify COPD phenotype. Thus, we could not analyze according to COPD phenotype. However, this study has merits: it is a large study that evaluates bacterial and viral identification rates associated with AECOPD in South Korea, and it provides information to help understand the epidemiology of AECOPD.
In conclusion, bacterial and viral infections are a major cause of AECOPD. The prevalence of identified pathogens has regional variability. In our study, the most commonly identified bacterium associated with AECOPD was P. aeruginosa, and the most commonly identified virus was influenza. M. catarrhalis showed a low identification rate, which is characteristic of Korea. Viruses were shown to be prevalent in certain seasons in Korea. Understanding the epidemiology of infections in AECOPD will help in selecting antibiotics and treatment.
ACKNOWLEDGEMENTS
This study was supported by a Korea University Guro Hospital Grant (O1801541).
Footnotes
The authors have no potential conflicts of interest to disclose.
- Study design and concept: Juwhan Choi, Kyung Hoon Min.
- Acquisition of data: Juwhan Choi, Jee Youn Oh, Young Seok Lee, Gyu Young Hur.
- Analysis and interpretation of data: Juwhan Choi, Sung Yong Lee, Jae Jeong Shim, Kyung Ho Kang, Kyung Hoon Min.
- Drafting of the manuscript: Juwhan Choi, Kyung Hoon Min.
- Critical revision of the manuscript for important intellectual content: Juwhan Choi, Kyung Hoon Min.
- Statistical analysis: Juwhan Choi.
- Study supervision: Juwhan Choi, Kyung Hoon Min.
References
- 1.Martin P, Glasgow H, Patterson J. Chronic obstructive pulmonary disease (COPD): smoking remains the most important cause. N Z Med J. 2005;118:U1409. [PubMed] [Google Scholar]
- 2.Bloemsma LD, Hoek G, Smit LAM. Panel studies of air pollution in patients with COPD: systematic review and meta-analysis. Environ Res. 2016;151:458–468. doi: 10.1016/j.envres.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 3.Wedzicha JA, Seemungal TA. COPD exacerbations: defining their cause and prevention. Lancet. 2007;370:786–796. doi: 10.1016/S0140-6736(07)61382-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seemungal TA, Hurst JR, Wedzicha JA. Exacerbation rate, health status and mortality in COPD--a review of potential interventions. Int J Chron Obstruct Pulmon Dis. 2009;4:203–223. doi: 10.2147/copd.s3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spencer S, Calverley PM, Burge PS, Jones PW. Impact of preventing exacerbations on deterioration of health status in COPD. Eur Respir J. 2004;23:698–702. doi: 10.1183/09031936.04.00121404. [DOI] [PubMed] [Google Scholar]
- 6.McManus TE, Marley AM, Baxter N, Christie SN, O'Neill HJ, Elborn JS, et al. Respiratory viral infection in exacerbations of COPD. Respir Med. 2008;102:1575–1580. doi: 10.1016/j.rmed.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papi A, Bellettato CM, Braccioni F, Romagnoli M, Casolari P, Caramori G, et al. Infections and airway inflammation in chronic obstructive pulmonary disease severe exacerbations. Am J Respir Crit Care Med. 2006;173:1114–1121. doi: 10.1164/rccm.200506-859OC. [DOI] [PubMed] [Google Scholar]
- 8.De Serres G, Lampron N, La Forge J, Rouleau I, Bourbeau J, Weiss K, et al. Importance of viral and bacterial infections in chronic obstructive pulmonary disease exacerbations. J Clin Virol. 2009;46:129–133. doi: 10.1016/j.jcv.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohan A, Chandra S, Agarwal D, Guleria R, Broor S, Gaur B, et al. Prevalence of viral infection detected by PCR and RT-PCR in patients with acute exacerbation of COPD: a systematic review. Respirology. 2010;15:536–542. doi: 10.1111/j.1440-1843.2010.01722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin SH, Kuo PH, Hsueh PR, Yang PC, Kuo SH. Sputum bacteriology in hospitalized patients with acute exacerbation of chronic obstructive pulmonary disease in Taiwan with an emphasis on Klebsiella pneumoniae and Pseudomonas aeruginosa. Respirology. 2007;12:81–87. doi: 10.1111/j.1440-1843.2006.00999.x. [DOI] [PubMed] [Google Scholar]
- 11.Li XJ, Li Q, Si LY, Yuan QY. Bacteriological differences between COPD exacerbation and community-acquired pneumonia. Respir Care. 2011;56:1818–1824. doi: 10.4187/respcare.00915. [DOI] [PubMed] [Google Scholar]
- 12.Chang CH, Tsao KC, Hu HC, Huang CC, Kao KC, Chen NH, et al. Procalcitonin and C-reactive protein cannot differentiate bacterial or viral infection in COPD exacerbation requiring emergency department visits. Int J Chron Obstruct Pulmon Dis. 2015;10:767–774. doi: 10.2147/COPD.S76740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rabe KF, Hurd S, Anzueto A, Barnes PJ, Buist SA, Calverley P, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176:532–555. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez-Roisin R. Toward a consensus definition for COPD exacerbations. Chest. 2000;117(5 Suppl 2):398S–401S. doi: 10.1378/chest.117.5_suppl_2.398s. [DOI] [PubMed] [Google Scholar]
- 15.Sethi S. Infectious etiology of acute exacerbations of chronic bronchitis. Chest. 2000;117(5 Suppl 2):380S–385S. doi: 10.1378/chest.117.5_suppl_2.380s. [DOI] [PubMed] [Google Scholar]
- 16.Sethi S, Murphy TF. Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin Microbiol Rev. 2001;14:336–363. doi: 10.1128/CMR.14.2.336-363.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boixeda R, Almagro P, Díez-Manglano J, Cabrera FJ, Recio J, Martin-Garrido I, et al. Bacterial flora in the sputum and comorbidity in patients with acute exacerbations of COPD. Int J Chron Obstruct Pulmon Dis. 2015;10:2581–2591. doi: 10.2147/COPD.S88702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biancardi E, Fennell M, Rawlinson W, Thomas PS. Viruses are frequently present as the infecting agent in acute exacerbations of chronic obstructive pulmonary disease in patients presenting to hospital. Intern Med J. 2016;46:1160–1165. doi: 10.1111/imj.13213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frickmann H, Jungblut S, Hirche TO, Groß U, Kuhns M, Zautner AE. The influence of virus infections on the course of COPD. Eur J Microbiol Immunol (Bp) 2012;2:176–185. doi: 10.1556/EuJMI.2.2012.3.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwak HJ, Park DW, Kim JE, Park MK, Koo GW, Park TS, et al. Prevalence and risk factors of respiratory viral infections in exacerbations of chronic obstructive pulmonary disease. Tohoku J Exp Med. 2016;240:131–139. doi: 10.1620/tjem.240.131. [DOI] [PubMed] [Google Scholar]
- 21.Jin J, Yu W, Li S, Lu L, Liu X, Sun Y. Factors associated with bronchiectasis in patients with moderate-severe chronic obstructive pulmonary disease. Medicine (Baltimore) 2016;95:e4219. doi: 10.1097/MD.0000000000004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gamba-Sanchez N, Rodriguez-Martinez CE, Sossa-Briceño MP. Epidemic activity of respiratory syncytial virus is related to temperature and rainfall in equatorial tropical countries. Epidemiol Infect. 2016;144:2057–2063. doi: 10.1017/S0950268816000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paynter S. Humidity and respiratory virus transmission in tropical and temperate settings. Epidemiol Infect. 2015;143:1110–1118. doi: 10.1017/S0950268814002702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goktas S, Sirin MC. Prevalence and seasonal distribution of respiratory viruses during the 2014 - 2015 season in Istanbul. Jundishapur J Microbiol. 2016;9:e39132. doi: 10.5812/jjm.39132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendricks MR, Lashua LP, Fischer DK, Flitter BA, Eichinger KM, Durbin JE, et al. Respiratory syncytial virus infection enhances Pseudomonas aeruginosa biofilm growth through dysregulation of nutritional immunity. Proc Natl Acad Sci U S A. 2016;113:1642–1647. doi: 10.1073/pnas.1516979113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bellinghausen C, Gulraiz F, Heinzmann AC, Dentener MA, Savelkoul PH, Wouters EF, et al. Exposure to common respiratory bacteria alters the airway epithelial response to subsequent viral infection. Respir Res. 2016;17:68. doi: 10.1186/s12931-016-0382-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rejman J, Di Gioia S, Bragonzi A, Conese M. Pseudomonas aeruginosa infection destroys the barrier function of lung epithelium and enhances polyplex-mediated transfection. Hum Gene Ther. 2007;18:642–652. doi: 10.1089/hum.2006.192. [DOI] [PubMed] [Google Scholar]
- 28.Dunn MM, Dunne M, Kamp DW. Polymorphonuclear leukocyte- and Pseudomonas aeruginosa-induced damage to a human pulmonary epithelial cell line. J Infect Dis. 1990;162:172–177. doi: 10.1093/infdis/162.1.172. [DOI] [PubMed] [Google Scholar]