Abstract
Purpose
This study was undertaken to explore how miR-206 represses osteosarcoma (OS) development.
Materials and Methods
Expression levels of miR-206, PAX3, and MET mRNA were explored in paired OS and adjacent tissue specimens. A patient-derived OS cell line was established. miR-206 overexpression and knockdown were achieved by lentiviral transduction. PAX3 and MET overexpression were achieved by plasmid transfection. Treatment with hepatocyte growth factor (HGF) was utilized to activate c-Met receptor. Associations between miR-206 and PAX3 or MET mRNA in OS cells were verified by AGO2-RNA immunoprecipitation assay and miRNA pulldown assay. OS cell malignancy was evaluated in vitro by cell proliferation, metastasis, and apoptosis assays. PAX3 and MET gene expression in OS cells was assayed by RT-qPCR and Western blot. Activation of PI3K-AKT and MAPK-ERK in OS cells were assayed by evaluating Akt1 Ser473 phosphorylation and total threonine phosphorylation of Erk1/2, respectively.
Results
Expression levels of miR-206 were significantly decreased in OS tissue specimens, compared to adjacent counterparts, and were inversely correlated with expression of PAX3 and MET mRNA. miR-206 directly interacted with PAX3 and MET mRNA in OS cells. miR-206 overexpression significantly reduced PAX3 and MET gene expression in OS cells in vitro, resulting in significant decreases in Akt1 and Erk1/2 activation, cell proliferation, and metastasis, as well as increases in cell apoptosis, while miR-206 knockdown showed the opposite effects. The effects of miR-206 overexpression on OS cells were reversed by PAX3 or MET overexpression, but only partially attenuated by HGF treatment.
Conclusion
miR-206 reduces OS cell malignancy in vitro by targeting PAX3 and MET gene expression.
Keywords: Osteosarcoma, miR-206, PAX3, c-Met, PI3K-AKT, MAPK-ERK
INTRODUCTION
Osteosarcoma (OS) is the most prevalent primary bone tumor, often showing high malignancy and aggressiveness in clinic.1,2 Osteosarcoma originates from malignant osteoblasts, for which several oncogenes and tumor-promoting genes closely related to malignant transformation and OS progression have been identified: dysregulation of the expression of these genes has been found to promote OS pathogenesis and development.3,4 microRNAs (miRNAs) regulate gene expression by binding to their target mRNAs in a complementary base pairing manner, blocking mRNA translation and mediating mRNA degradation catalyzed by the RNA-induced silencing complex.5
Dysregulation in miRNA expression has been shown to be deeply involved in OS pathogenesis and development.6 Aberrant miRNA expression was found to have high prognostic value,7 and the pro- or anti-OS development mechanisms of some of these miRNAs have also been elucidated.8 Namlos, et al.9 first reported the downregulation of miR-206 in OS cells, compared to normal bone tissue, which was later verified by Bao, et al.10 who also demonstrated that decreases in miR-206 expression were significantly correlated with OS progression and that miR-206 mimic transfection could significantly reduce OS cell malignancy in vitro. The clinical significance of miR-206 in OS was further emphasized by Zhang, et al.11 who verified a significant correlation between decreased miR-206 in OS patient serum and an unfavorable prognosis. These clinical studies suggested the tumor suppressive role of miR-206 in OS progression and that the loss of miR-206 expression might be an important mechanism that promotes OS development.
The molecular mechanism of how miR-206 inhibits OS development remains largely underexplored. A recent report by Pan, et al.12 demonstrated that miR-206 mimic transfection could decrease OS cell growth and metastasis in vitro by targeting ANXA2 gene expression. miRNAs are known to regulate gene expression by binding to their target mRNAs, thus inducing mRNA degradation mediated by the RNA-induced silencing complex. MET and PAX3 mRNAs have previously been identified as direct targets of miR-206.13,14,15,16 Transcription factor Pax3 has been suggested to promote osteosarcoma and gastric cancer cell metastasis by activating MET gene expression.14,17 Meanwhile, Rees, et al.18 reported the contribution of the Pax3-c-Met axis to rhabdomyosarcoma progression. By increasing MET gene expression via lentiviral transduction in primary human osteoblasts, Patané, et al.19 and Dani, et al.20 demonstrated that MET is an oncogene in OS. Hepatocyte growth factor (HGF) is the ligand of c-Met receptor encoded by the MET gene, which activates the downstream PI3K-AKT and MAPK-ERK signaling pathway in OS cells upon HGF ligation.21 Targeting of PAX3 or MET mRNA by different miRNAs has been suggested to suppress OS cell malignancy, marked by significant decreases in cell growth and metastasis.17,22,23,24,25
In the present research, we investigated whether miR-206 inhibits OS cell growth and metastasis in vitro by targeting PAX3 and MET gene expression. Our data suggest that miR-206 overexpression in OS cells could significantly reduce cell proliferation and metastasis by reducing PAX3 and MET gene expression, thus reducing PI3K-AKT and MAPK-ERK signaling. Transiently overexpressing PAX3 or MET could reverse the anti-OS effects of miR-206 overexpression, which was only partially attenuated by HGF treatment.
MATERIALS AND METHODS
Tissue specimens
This research was approved by the ethics review committee of Chongqing Three Gorges Central Hospital. Informed consent was obtained from each patient whose tissue specimens were included in this research. OS pathologic tissue specimens and adjacent tissue specimens were sampled 3–5 cm away from the tumor edge from 25 OS patients who received radical surgery at Chongqing Three Gorges Central Hospital during 2010–2015. Tissue specimens were obtained by incisional biopsy before chemotherapy and surgery, and were stored in liquid nitrogen after resection or subjected to OS primary cell culture establishment as described below.
OS primary cell culture
Establishment of OS primary cell culture was performed as described by Blattmann, et al.26 with modifications. Briefly, OS tissue was resected from an OS patient and was shredded immediately after resection under sterile conditions with the protection of chilled PBS. OS tissue fragments were cultured in a petri dish in DMEM low glucose medium (Sciencell, Zhong Qiao Xin Zhou Biotechnology, Shanghai, China) supplemented with 10% FBS (Zhong Qiao Xin Zhou Biotechnology) and 1% non-essential amino acids solution (Sciencell, Zhong Qiao Xin Zhou Biotechnology) in a cell incubator with humidified atmosphere at 37℃ with 5% CO2. Cells in the petri dish were routinely sub cultured at 100% confluence. After culture for 30 days, cells in the petri dish were collected, and about 3.5×106 cells were injected subcutaneously into the left flank of two BALB/c nude mice (Vital River Laboratory Animal Technology, Beijing, China). Tumor tissues at a volume of 1000 mm3 or higher were harvested and disaggregated by mashing through a 100 µM cell strainer (Corning, Lianshuo Biological Technology, Shanghai, China). Cells were cultured in vitro for one passage, and the tumorigenicity of these cells was verified by the tumor formation assay by injecting 2×105 cells into the left flank of another two nude mice and monitoring tumor volume increases. Both mice developed xenograft tumors, which were harvested at a volume over 1500 mm3 and processed as described above to establish the OS primary cell culture. Laboratory animal use in this research was approved by the Ethical Review Committee of Chongqing Three Gorges Central Hospital.
Lentiviral transduction and plasmid transfection
miR-206 overexpression or inhibition in primary OS cells was achieved by lentiviral transduction. Pseudoviral particles for miR-206 overexpression and miR-206 knockdown (anti-miR-206) and for transduction of miR-negative control (miRNC) were purchased from Fulengen (Guangzhou, China) and were used following manufacturer's instructions. Cells with positive transduction were selected by puromycin (Solarbio, Beijing, China). PAX3 or MET gene overexpression in primary OS cells was achieved by plasmid transfection using Lip2000 transfection reagent (Solarbio). Plasmids for PAX3 or MET gene overexpression were purchased from Solarbio and were used following the manufacturer's instructions. Cells were assayed 48 hours after transfection. Positive transfection was confirmed by verifying the expression of GFP reporter gene encoded in the plasmids.
Recombinant human HGF protein was purchased from R&D system (Shanghai, China) and was used for cell treatment as described by Husmann, et al.21 with modifications. To evaluate Akt1 or Erk1/2 activation, OS cells were serum-deprived for 6 hours before HGF treatment at 100 ng/mL for 30 min. For cell viability, apoptosis, migration, and invasion assay, cells were pretreated with 100 ng/mL of HGF for 16 hours before assay with the presence of HGF at the same concentration.
Cell functional assays
Cell viability assay was performed using Cell Counting Kit-8 (Beyotime, Shanghai, China) following the manufacturer's instructions. Briefly, 2×103 cells/well were seeded on 96-well plates, and cells were incubated with CCK-8 working solution for 1 hour at 24, 48, and 72 hours post-seeding. The number of viable cells at each time point was assayed by measuring absorbance at 450 nm using a microplate reader. Cell apoptosis assay was performed by flow cytometry detecting FITC-Annexin V/IP staining using a Dead Cell Apoptosis Kit (Invitrogen, Thermo Fisher Scientific, Shanghai, China) following the manufacturer's instructions. Apoptosis was induced by serum deprivation for 48 hours. Cells after treatment were harvested and labeled with Annexin V/PI, followed by flow cytometry analyses. Cells with positive Annexin V staining were considered apoptotic cells. Cell mobility was assayed by trans-well migration assay using permeable polyester membrane inserts (8-µm pore size, 24-well format, Corning, Thermo Fisher Scientific). Serum-free medium containing 5000 cells was added into the insert, which was incubated in complete culture medium for 24 hours. Cells that migrated to the bottom of the insert membrane after incubation were stained with crystal violet (Wako, Shanghai, China) and counted under a microscope. Cell invasiveness was assayed by trans-well migration assay using a Matrigel-coated invasion chamber (8-µm pore size, 24-well format, Corning, Thermo Fisher Scientific) with similar techniques as described above.
RT-qPCR and Western blot
For RT-qPCR, total RNA was extracted from tissue homogenate or cell lysate using TRIzol reagent (Invitrogen, Thermo Fisher Scientific). To evaluate miR-206 expression levels in total RNA, an all-in-one miRNA qRT-PCR reagent kit (GeneCopoeia, Guangzhou, China) was used following the manufacturer's instructions. To evaluate PAX3 or MET mRNA levels, a BlazeTaq one-step SYBR green RT-qPCR kit (GeneCopoeia) was used following the manufacturer's instructions. Primers for qPCR were purchased from GeneCopoeia. Expression levels of miR-206 or mRNA of PAX3 or MET were semi-quantified by the 2−ΔΔCt method using snRNA U6 or GAPDH as a reference gene. Western blot was performed using homemade reducing SDS-polyacrylamide gel. Antibodies for Western blot were purchased from Abcam (Shanghai, China). Primary antibodies were used at 0.5 µg/mL, while secondary antibodies were used at 100 ng/mL. Expression levels of each protein were normalized to that of GAPDH by grayscale analysis using ImageJ software (Ver. 1.5, National Institutes of Health, Bethesda, MD, USA) before further analysis.
AGO2 RNA immunoprecipitation and miRNA pulldown
For AGO2 RNA immunoprecipitation (RIP), wild-type primary OS cells were transfected with miR-206 mimic or miRNA mimic negative control (miR-NC) for 24 hours before cell lysis. Cell lysate was then incubated with rabbit polyclonal to Argonaute-2 antibody (Abcam) and protein A-conjugated magnetic beads (Thermo Fisher Scientific) for 4 hours at 4℃ with agitation. The precipitated beads were subjected to RT-qPCR evaluating the abundance of PAX3, MET, or GAPDH mRNA associated with the beads. For miRNA pulldown assay, OS cells were transfected with biotin-labeled miR-206 mimic or miR-NC for 24 hours, followed by incubation with streptavidin-conjugated magnetic beads (Thermo Fisher Scientific) for 2 hours at 4℃ with agitation. The precipitated beads were subject to RT-qPCR evaluating the abundance of miR-206, as well as PAX3, MET, or GAPDH mRNA, associated with the beads. Biotin labeled or un-labeled miRNA mimics were purchased from Ribobio (Guangzhou, China) and were used following the manufacturer's instructions.
Statistical analysis
Statistical analysis was performed using GraphPad Prism software (Ver. 7.04, GraphPad Software, La Jolla, CA, USA). Each data group represents two independent replicates with three technical replicates of each. All data were normalized to the mean value of the control group and are presented as fold changes if applicable. Student's t test and one-way analysis of variance with Tukey's test as the post hoc test were performed for significance test. A p value<0.05 was considered significance.
RESULTS
miR-206 reduced PAX3 and MET gene expression via mRNA targeting
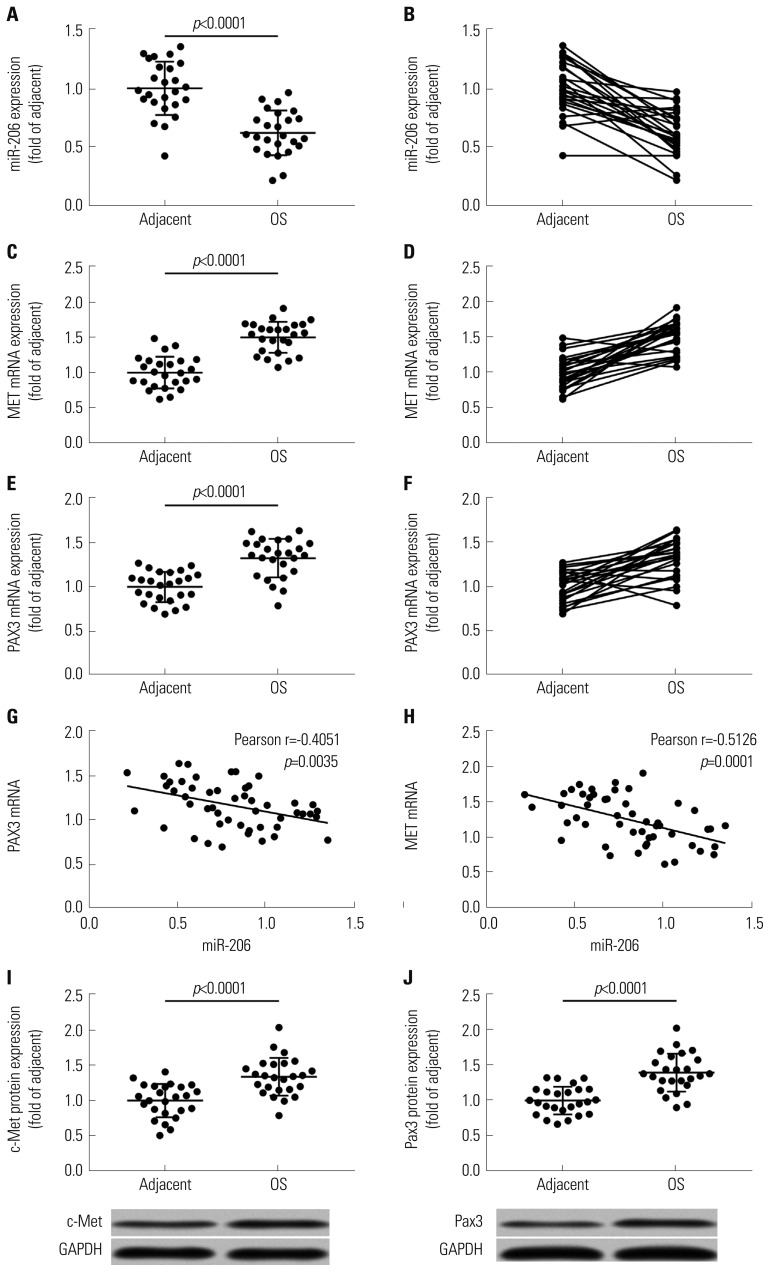
To investigate whether miR-206 would inhibit OS development by reducing PAX3 and MET gene expression, we first analyzed the expression levels of miR-206, as well as PAX3 and MET mRNA, in 25 pairs of OS and adjacent tissue specimens. RT-qPCR results clearly demonstrated a significant decrease in miR-206 expression and increases in PAX3 and MET mRNA expression in OS tissue specimens, compared to adjacent counterparts (Fig. 1A–F). Western blot analyzing protein levels of c-Met and Pax3 in these tissue specimens yielded similar results (Fig. 1I and J). Pearson correlation analysis results indicated that miR-206 expression levels in all 50 tissue specimens were inversely correlated with that of PAX3 or MET mRNA (Fig. 1G and H).
Fig. 1. miRNA-206 expression is inversely correlated with PAX3 or MET mRNA expression in osteosarcoma and adjacent tissue specimens. (A–F) mRNA expression levels of miR-206, MET mRNA, and PAX3 mRNA in 25 pairs of OS and adjacent tissue specimens. (G and H) Pearson correlation analysis determining the significance of correlation between miR-206 expression and PAX3 or MET mRNA expression. (I and J) Protein levels of c-Met and Pax3 that are encoded by MET and PAX3 gene in 25 pairs of OS and adjacent tissue specimens. OS, osteosarcoma.
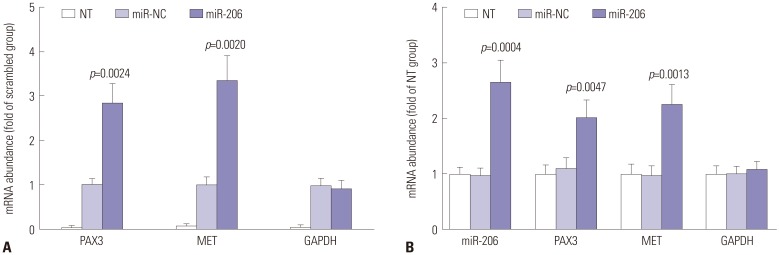
We next performed miRNA pulldown and AGO2-RIP assay to verify the direct association between miR-206 and PAX3 or MET mRNA in the primary OS cells we established. Our miRNA pulldown assay results demonstrated a potent association between PAX3 or MET mRNA and biotin-labeled miR-206 mimic (Fig. 2B), while the AGO2-RIP assay results showed a significant association between PAX3 or MET mRNA and the RISC complex after miR-206 mimic transfection, compared to miR-NC transfection (Fig. 2A). These data suggested that miR-206 could directly interact with PAX3 and MET mRNA in OS cells in vitro and mediated their gene silencing by recruiting the RISC complex.
Fig. 2. miR-206 interacts with PAX3 or MET mRNA in OS cells. (A) AGO2-RIP assay determining the association between AGO2 protein and mRNA of indicated genes in OS cells after transfection with different miRNA mimics. (B) miRNA pulldown assay determining the association between miR-206 and mRNA of indicated genes in OS cells. miR-NC transfected group was used as control for significance test. OS, osteosarcoma.
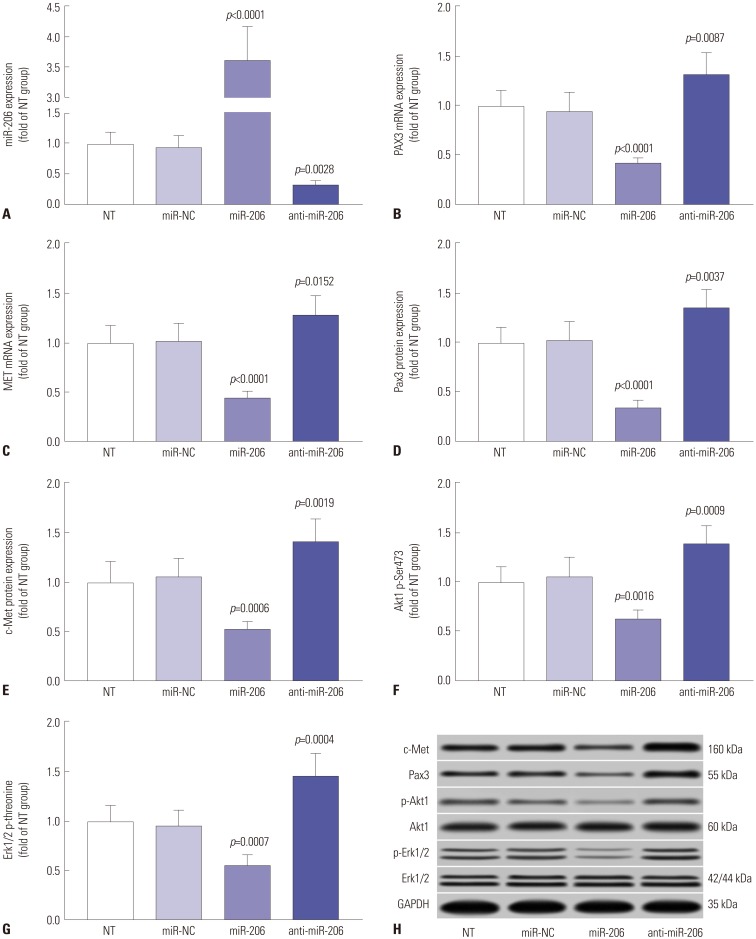
miR-206 overexpression significantly reduced PAX3 and MET gene expression in OS cells. We constructed miR-206 overexpression or knockdown OS cells by lentiviral transduction (Fig. 3A) and found that mRNA and protein expression levels of both PAX3 and MET were significantly decreased in miR-206 overexpressing OS cells and were increased in miR-206 knockdown OS cells compared to wild type cells (Fig. 3B–E, H). Activation of MAPK-ERK1/2 and Pink K-Akt signaling pathways (marked by Akt1 Ser473 phosphorylation and Erk 1/2 threonine phosphorylation) downstream of c-Met in OS cells was also significantly inhibited in miR-206 overexpressing OS cells and was increased in miR-206 knockdown OS cells, compared to their wild-type counterparts (Fig. 3F–H). These data suggested that miR-206 could reduce PAX3 and MET expression, thus inhibiting c-Met signal transduction in OS cells in vitro.
Fig. 3. miR-206 reduces PAX3 and MET expression, as well as Akt1 and Erk1/2 activation. (A–C) RT-qPCR evaluating miR-206, PAX3 mRNA and MET mRNA in OS cells after indicated lentiviral transduction. (D–H) Western blot detecting the protein expression levels of PAX3 and MET, as well as Akt1 Ser473 and Erk1/2 total threonine phosphorylation, in OS cells after indicated transduction. NT was used as control for significance test. OS, osteosarcoma; NT, non-transduced group.
miR-206 reduced OS cell proliferation and metastasis while promoting apoptosis in vitro
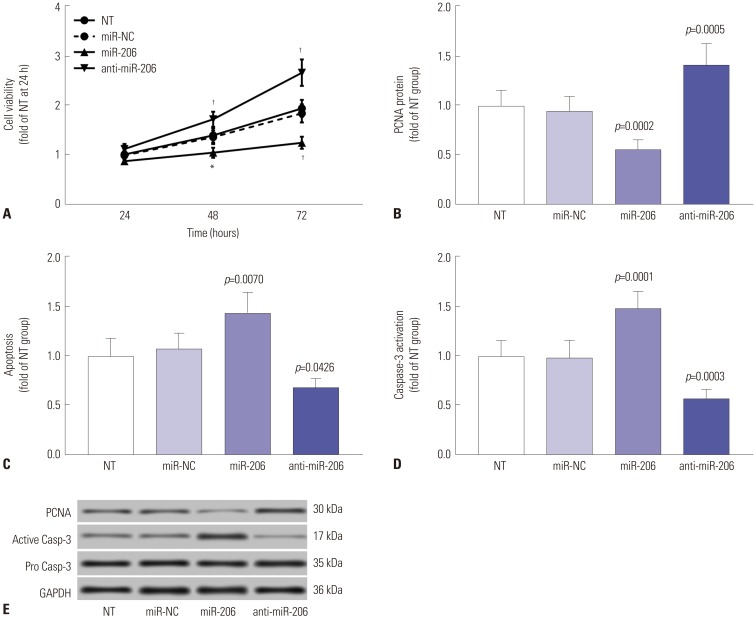
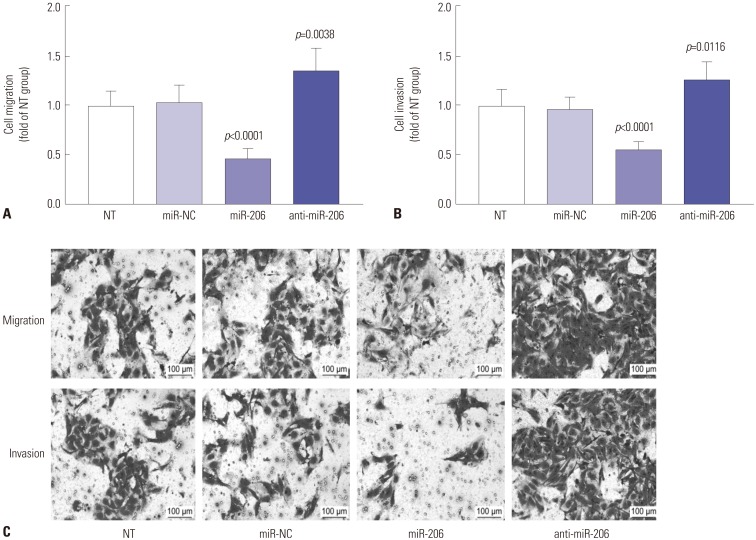
c-Met signaling plays a fundamental role in the malignant transformation of OS cells, while reducing MET gene expression or inhibiting its kinase activity has been demonstrated to inhibit cell proliferation, to induce apoptosis, and to increase therapeutic sensitivity in OS cells. We therefore investigated whether overexpressing miR-206 would achieve similar results. Cell viability assay revealed that miR-206 overexpression significantly reduces the overall cell growth in OS cells in vitro, while miR-206 knockdown showed the opposite effects (Fig. 4A). Western blot evaluating the protein expression levels of the cell proliferation marker protein PCNA revealed that OS cell proliferation was significantly reduced by miR-206 overexpression and increased by miR-206 knockdown (Fig. 4B and E). Cell apoptosis assay by flow cytometry or Western blot evaluating the activation of caspase-3 revealed that OS cell apoptosis induced by serum deprivation for 48 hours could be significantly increased by miR-206 overexpression and decreased by miR-206 knockdown (Fig. 4C–E). Considering that c-Met signaling has also been suggested to promote OS cell metastasis, we further investigated the influence of miR-206 overexpression or knockdown on OS cell migration and invasion in vitro. Our trans-well migration assay results indicated that miR-206 overexpression could significantly reduce OS cell mobility and invasiveness in vitro, which could be increased by miR-206 knockdown (Fig. 5). These data suggested that miR-206 could reduce cell proliferation and metastasis while facilitating cell apoptosis in OS cells in vitro.
Fig. 4. miR-206 reduces OS cell proliferation while promoting apoptosis in vitro. miR-206 overexpression or knockdown in OS cells in vitro was achieved by lentiviral transduction. (A) CCK-8 cell viability assay evaluating overall cell growth in vitro. (B) Western blot evaluating the protein expression levels of the cell proliferation marker PCNA. (C) Flow cytometry detection of cell apoptosis. (D) Western blot evaluating the activation of pro-apoptotic protein Caspase-3. (E) Representative results of Western blots. NT was used as a control for significance test. *p<0.01, †p<0.001. OS, osteosarcoma; NT, non-transduced group.
Fig. 5. miR-206 reduces OS cell migration (A) and invasion (B) in vitro. miR-206 overexpression or knockdown in OS cells in vitro was achieved by lentiviral transduction. (C) Representative results of cell migration and invasion assays. NT was used as a control for significance test. OS, osteosarcoma; NT, non-transduced group.
MiR-206 reduced OS cell malignancy in vitro by targeting PAX3 and MET
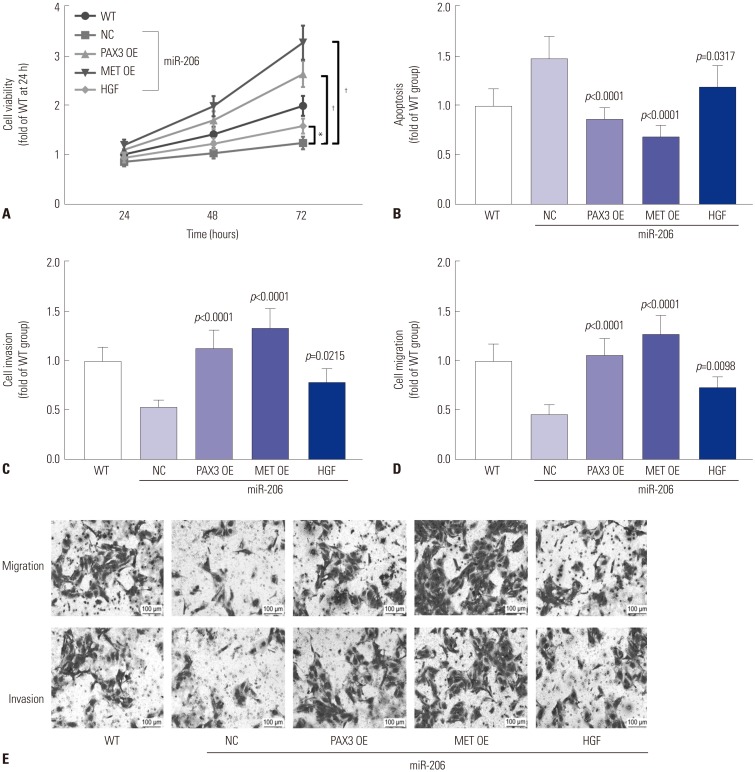
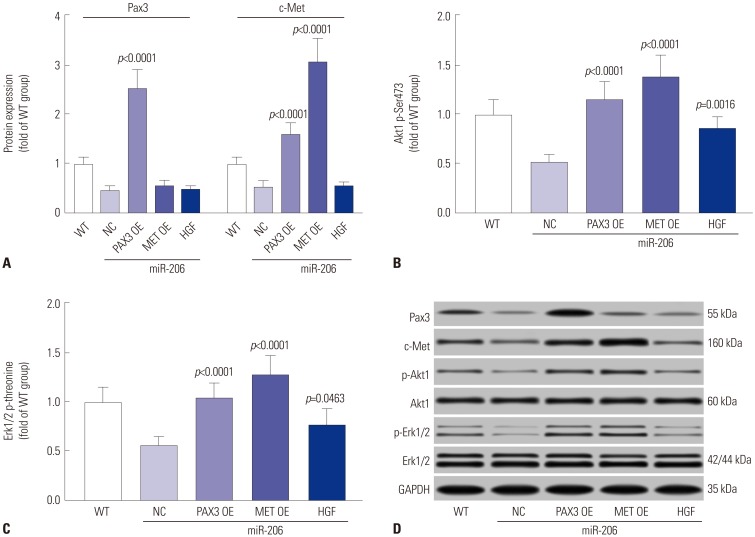
To investigate whether miR-206 reduced OS cell malignancy in vitro by targeting PAX3 and Met, we analyzed the influence of transient PAX3 or MET gene overexpression or treatment with recombinant human HGF on miR-206 overexpressing OS cells. Cell viability assay revealed that PAX3 or MET overexpression significantly reversed the observed decrease in cell growth rate of OS cells caused by miR-206 overexpression, which was only partially rescued by HGF treatment (Fig. 6A). Cell apoptosis assay results indicated that PAX3 or MET overexpression significantly reduced OS cell apoptosis promoted by miR-206 overexpression, while HGF treatment showed only a moderate effect (Fig. 6B). Trans-well migration assay results also suggested that OS cell mobility and metastasis impaired by miR-206 overexpression could be significantly enhanced by PAX3 or MET overexpression and partially rescued by HGF treatment (Fig. 6C–E). Western blot analysis revealed that PAX3 or MET overexpression significantly increased Pax3 or c-Met protein expression level in OS cells, respectively, while HGF treatment showed no influence on the expression of the two proteins (Fig. 7A). Inhibition of PI3K-AKT and MAPK-ERK signaling by miR-206 were also significantly reversed by PAX3 or MET overexpression and partially attenuated by HGF treatment (Fig. 7B–D). Collectively, these results indicated that miR-206 reduces OS cell malignancy in vitro by targeting PAX3 and MET.
Fig. 6. miR-206 reduces OS cell malignancy in vitro by targeting PAX3 and MET. miR-206 overexpressing OS cells were transfected with PAX3 or MET OE plasmids or treated with HGF before assay. (A) CCK-8 cell viability assay evaluating the overall cell growth in vitro. (B) Flow cytometry detection cell apoptosis. (C and D) Cell invasion and migration assay evaluating cell metastasis in vitro. (E) Representative results of cell migration and invasion assay. WT OS cells were used as a control for significance test. *p<0.01, †p<0.001. OS, osteosarcoma; OE, overexpressing; WT, wild-type, HGF, hepatocyte growth factor; NC, negative control.
Fig. 7. miR-206 reduces PAX3 and MET gene expression and IP3K-AKT, MAPK-ERK signaling in OS cells. OS cells were treated as indicated in Fig. 6 before Western blot assay (A), Western blot detecting the protein expression levels of Pax3 or c-Met in OS cells after different treatment. (B and C) Western blot detecting Akt1 Ser473 or Erk1/2 total threonine phosphorylation in OS cells after different treatments. (D) Representative Western blot results. WT OS cells were used as a control for significance test.. OS, osteosarcoma; WT, wild-type.
DISCUSSION
Therapeutic strategies for OS treatment and OS patient outcomes have not improved for decades, and trials to introduce targeted therapy or immunotherapy for OS management have generally shown disappointing results.1,27 Identification for new therapeutic targets and development of novel treatments against OS is needed. After reviewing previous research investigating differentially expressed RNAs in OS tissue, compared to adjacent normal tissue, we noted that miR-206 is frequently downregulated in OS tissue and that its downregulation was significantly correlated with worse prognosis.10,11 However, the role of miR-206 and the mechanism of its potential anti-OS activity remain largely underexplored. In one of our preliminary studies, we investigated whether miR-206 overexpression or knockdown would influence OS cell proliferation, apoptosis, and metastasis. To do this, we established a patient-derived OS cell line and constructed miR-206 overexpressing or knockdown OS cells by lentiviral transduction. Our data clearly demonstrated that miR-206 overexpression could significantly inhibit OS cell proliferation and metastasis in vitro, while its apoptosis induced by serum deprivation was significantly increased. Meanwhile, miR-206 knockdown showed the opposite effect. These data suggested that loss of miR-206 might be an important tumorigenic mechanism in OS development.
Considering that miRNAs play their tumor promoting or suppressive roles primarily by reducing gene expression via mRNA targeting, we attempted to explore oncogenes and tumor-promoting genes that have been shown to be regulated by miR-206. PAX3 and MET were two such genes that we found.15,16 MET is known to play a pivotal role in OS oncogenesis and development,28 while PAX3 has been found to activate MET transcription.29 Inhibition of MET or PAX3 gene expression has been shown to reduce OS development:17, 28 notably, Zhang, et al.14 demonstrated that loss of miR-206 might facilitate gastric cancer metastasis due to restoration of PAX3 expression and c-Met signaling. We therefore hypothesized that miR-206 might reduce OS cell malignancy, possibly by targeting PAX3 and MET expression. In this study we revealed an inverse correlation in expression levels between miR-206 and PAX3 or MET mRNA in OS patient tissue specimens, and our miRNA pulldown assay, as well as AGO2-RIP assay, confirmed a direct interaction between miR-206 and PAX3 or MET mRNA, suggesting that miR-206 could reduce PAX3 and MET gene expression in OS cells by directly targeting their mRNA. We also found significant correlation between miR-206 expression levels and OS progression using OS patient clinical records, which was not presented in this manuscript because similar results have been repeatedly reported.10,11
The concept that miR-206 could reduce OS cell malignancy was further emphasized by our Western blot assay and cell functional assay results. Downregulation in cell viability and metastasis potential, as well as upregulation in cell apoptosis, in OS cells induced by miR-206 overexpression was associated with reduced Pax3 and c-Met protein expression levels and with decreased PI3K-AKT and MAPK-ERK signaling, while miR-206 knockdown showed the opposite effect. Our data seem to suggest that miR-206 overexpression exerts more potent effects in altering cell function and gene expression, compared to miR-206 knockdown, possibly because these patient-derived OS cells already had decreased miR-206 expression. In this study, we also found that PAX3 or MET overexpression could overwhelm the influence of miR-206 overexpression on OS cells, which was only partially attenuated by HGF treatment. Our Western blot data suggested that PAX3 overexpression could rescue c-Met protein expression level downregulated by miR-206 overexpression in OS cells. Since HGF is the ligand of c-Met receptor, we speculated that miR-206 overexpression in OS cells did not completely abolish c-Met protein expression (as indicated in Fig. 1E and H), which could still be activated by HGF ligation and mediate PI3K-AKT and MAPK-ERK activation. Thus, our data suggested that c-Met activated by HGF can promote OS cell malignancy by activating PI3K-AKT and MAPK-ERK signaling and that miR-206 can reduce OS cell malignancy by targeting PAX3 and MET expression.
Notably, Western blot assay results presented in Pan, et al.12 publication suggested that pan-Akt protein expression level in MG-63 human OS cells would be significantly decreased by miR-206 mimic transfection and increased by miR-206 inhibitor transfection. However, our data revealed no significant change in Akt1 protein expression levels in OS cells with different miR-206 expression levels, and similar results were also achieved in Western blot detecting the protein expression levels of Akt2 (data not shown). We currently found no bioinformatic or experimental evidence suggesting that miR-206 would target AKT1/2 gene expression. It is generally believed that gene expression in cells is regulated by multiple different mechanisms. We hypothesized that a sharp, short-term increase or decrease in miR-206 expression in OS cells, as achieved by Pan, et al.12 utilizing miRNA mimic or inhibitor transfection, might interfere with certain mechanisms regulating AKT1/2 gene expression; however, these interferences might be compensated by other mechanisms and restore AKT1/2 gene expression during long-term miR-206 overexpression, resulting in what we have observed. Also, use of different cells might also interfere with the results. Nevertheless, this potential influence of miR-206 on AKT1/2 gene expression should be further investigated.
It should also be pointed that the present research only investigated the role of miR-206 in OS cells in vitro, and experiments should be performed to verify the anti-OS effect of miR-206 in vivo. Loss of miR-206 might be corrected by application of miR-206 agomirs by intra-tumoral or tail vein injection. miRNA agomirs function similarly to their miRNA counterparts; however, they diffuse through cell membranes more easily than miRNA mimics and often show higher resistance to RNA degradation due to hypermethylation, phosphorothioation, and cholesterol modification. Application of miRNA agomirs in vivo is a possible technique to treat miRNA dysregulation-related disease. Based on our results and previous publications, we proposed that use of miR-206 agomir might aid in OS treatment, which should be investigated in future research.
Overall, our data demonstrated for the first time that miR-206 reduces OS cell malignancy in vitro by targeting PAX3 and MET expression, resulting in significant decreases in PI3K-AKT and MAPK-ERK signaling. Restoring miR-206 expression might be a possible approach to improve OS treatment outcomes and patient prognosis.
Footnotes
The authors have no potential conflicts of interest to disclose.
- Conceptualization: Fang-Biao Zhan.
- Data curation: Xian-Wei Zhang, Shi-Long Feng.
- Formal analysis: Fang-Biao Zhan, Xian-Wei Zhang.
- Funding acquisition: Xian-Wei Zhang.
- Investigation: Fang-Biao Zhan, Shi-Long Feng.
- Methodology: Xian-Wei Zhang, Shi-Long Feng, Jun Cheng.
- Project administration: Fang-Biao Zhan.
- Resources: Jun Cheng, Bo Li.
- Software: Jun Cheng, You Zhang.
- Validation: Li-Zhong Xie, Qian-Rong Deng.
- Writing—original draft: You Zhang.
- Writing—review & editing: Bo Li, Li-Zhong Xie, Qian-Rong Deng.
References
- 1.Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma - connecting aetiology, biology and therapy. Nat Rev Endocrinol. 2017;13:480–491. doi: 10.1038/nrendo.2017.16. [DOI] [PubMed] [Google Scholar]
- 2.Gill J, Ahluwalia MK, Geller D, Gorlick R. New targets and approaches in osteosarcoma. Pharmacol Ther. 2013;137:89–99. doi: 10.1016/j.pharmthera.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14:722–735. doi: 10.1038/nrc3838. [DOI] [PubMed] [Google Scholar]
- 4.Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):vii320–vii325. doi: 10.1093/annonc/mdq276. [DOI] [PubMed] [Google Scholar]
- 5.Iwakawa HO, Tomari Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015;25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Ram Kumar RM, Boro A, Fuchs B. Involvement and clinical aspects of microRNA in osteosarcoma. Int J Mol Sci. 2016;17:E877. doi: 10.3390/ijms17060877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim YH, Goh TS, Lee CS, Oh SO, Kim JI, Jeung SH, et al. Prognostic value of microRNAs in osteosarcoma: a meta-analysis. Oncotarget. 2017;8:8726–8737. doi: 10.18632/oncotarget.14429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmini G, Marini F, Brandi ML. What is new in the miRNA world regarding osteosarcoma and chondrosarcoma? Molecules. 2017;22:E417. doi: 10.3390/molecules22030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Namløs HM, Meza-Zepeda LA, Barøy T, Østensen IH, Kresse SH, Kuijjer ML, et al. Modulation of the osteosarcoma expression phenotype by microRNAs. PLoS One. 2012;7:e48086. doi: 10.1371/journal.pone.0048086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao YP, Yi Y, Peng LL, Fang J, Liu KB, Li WZ, et al. Roles of microRNA-206 in osteosarcoma pathogenesis and progression. Asian Pac J Cancer Prev. 2013;14:3751–3755. doi: 10.7314/apjcp.2013.14.6.3751. [DOI] [PubMed] [Google Scholar]
- 11.Zhang C, Yao C, Li H, Wang G, He X. Serum levels of microRNA-133b and microRNA-206 expression predict prognosis in patients with osteosarcoma. Int J Clin Exp Pathol. 2014;7:4194–4203. [PMC free article] [PubMed] [Google Scholar]
- 12.Pan BL, Tong ZW, Wu L, Pan L, Li JE, Huang YG, et al. Effects of microRNA-206 on osteosarcoma cell proliferation, apoptosis, migration and invasion by targeting ANXA2 through the AKT signaling pathway. Cell Physiol Biochem. 2018;45:1410–1422. doi: 10.1159/000487567. [DOI] [PubMed] [Google Scholar]
- 13.Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, et al. MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst. 2010;102:706–721. doi: 10.1093/jnci/djq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Xia L, Zhao L, Chen Z, Shang X, Xin J, et al. Activation of PAX3-MET pathways due to miR-206 loss promotes gastric cancer metastasis. Carcinogenesis. 2015;36:390–399. doi: 10.1093/carcin/bgv009. [DOI] [PubMed] [Google Scholar]
- 15.Yan D, Dong Xda E, Chen X, Wang L, Lu C, Wang J, et al. MicroRNA-1/206 targets c-Met and inhibits rhabdomyosarcoma development. J Biol Chem. 2009;284:29596–29604. doi: 10.1074/jbc.M109.020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirai H, Verma M, Watanabe S, Tastad C, Asakura Y, Asakura A. MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. J Cell Biol. 2010;191:347–365. doi: 10.1083/jcb.201006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q, Yang G, Qian Y. Loss of MicroRNA-489-3p promotes osteosarcoma metastasis by activating PAX3-MET pathway. Mol Carcinog. 2017;56:1312–1321. doi: 10.1002/mc.22593. [DOI] [PubMed] [Google Scholar]
- 18.Rees H, Williamson D, Papanastasiou A, Jina N, Nabarro S, Shipley J, et al. The MET receptor tyrosine kinase contributes to invasive tumour growth in rhabdomyosarcomas. Growth Factors. 2006;24:197–208. doi: 10.1080/08977190600759923. [DOI] [PubMed] [Google Scholar]
- 19.Patané S, Avnet S, Coltella N, Costa B, Sponza S, Olivero M, et al. MET overexpression turns human primary osteoblasts into osteosarcomas. Cancer Res. 2006;66:4750–4757. doi: 10.1158/0008-5472.CAN-05-4422. [DOI] [PubMed] [Google Scholar]
- 20.Dani N, Olivero M, Mareschi K, van Duist MM, Miretti S, Cuvertino S, et al. The MET oncogene transforms human primary bone-derived cells into osteosarcomas by targeting committed osteoprogenitors. J Bone Miner Res. 2012;27:1322–1334. doi: 10.1002/jbmr.1578. [DOI] [PubMed] [Google Scholar]
- 21.Husmann K, Ducommun P, Sabile AA, Pedersen EM, Born W, Fuchs B. Signal transduction and downregulation of C-MET in HGF stimulated low and highly metastatic human osteosarcoma cells. Biochem Biophys Res Commun. 2015;464:1222–1227. doi: 10.1016/j.bbrc.2015.07.108. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Sun X, Wu J, Li Z. MicroRNA-613 suppresses proliferation, migration and invasion of osteosarcoma by targeting c-MET. Am J Cancer Res. 2016;6:2869–2879. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Niu G, Li B, Sun J, Sun L. miR-454 is down-regulated in osteosarcomas and suppresses cell proliferation and invasion by directly targeting c-Met. Cell Prolif. 2015;48:348–355. doi: 10.1111/cpr.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao H, Li M, Li L, Yang X, Lan G, Zhang Y. MiR-133b is down-regulated in human osteosarcoma and inhibits osteosarcoma cells proliferation, migration and invasion, and promotes apoptosis. PLoS One. 2013;8:e83571. doi: 10.1371/journal.pone.0083571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan K, Gao J, Yang T, Ma Q, Qiu X, Fan Q, et al. MicroRNA-34a inhibits the proliferation and metastasis of osteosarcoma cells both in vitro and in vivo. PLoS One. 2012;7:e33778. doi: 10.1371/journal.pone.0033778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blattmann C, Thiemann M, Stenzinger A, Roth EK, Dittmar A, Witt H, et al. Establishment of a patient-derived orthotopic osteosarcoma mouse model. J Transl Med. 2015;13:136. doi: 10.1186/s12967-015-0497-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isakoff MS, Bielack SS, Meltzer P, Gorlick R. Osteosarcoma: current treatment and a collaborative pathway to success. J Clin Oncol. 2015;33:3029–3035. doi: 10.1200/JCO.2014.59.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scagliotti GV, Novello S, von Pawel J. The emerging role of MET/HGF inhibitors in oncology. Cancer Treat Rev. 2013;39:793–801. doi: 10.1016/j.ctrv.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Kubic JD, Little EC, Lui JW, Iizuka T, Lang D. PAX3 and ETS1 synergistically activate MET expression in melanoma cells. Oncogene. 2015;34:4964–4974. doi: 10.1038/onc.2014.420. [DOI] [PMC free article] [PubMed] [Google Scholar]