Abstract
Salmonella is among the very important pathogens threating the human and animal health. Rapid and easy detection of these pathogens is crucial. In this context, antibody (Ab) based lateral flow assays (LFAs) which are simple immunochromatographic point of care test kits were developed by gold nanoparticles (GNPs) as labelling agent for Salmonella detection. For that purpose some critical parameters such as reagent concentrations on the capture zones, conjugate concentrations and ideal membrane type needed for LFAs for whole cell detection were tested for naked eye analysis. Therefore, prepared LFAs were applied to the live and heat inactivated cells when they were used alone or included in different bacterial mixtures. Among the test platforms, membrane 180 (M180) was found as an ideal membrane and 36 nm GNPs showed highly good labelling in the developed LFAs. Diluted conjugates and low concentrations of reagents affected the test signal negatively. Salmonella was detected in different bacterial mixtures, selectively in 4–5 min. The best recognized species by used Ab were S. enteritidis and S. infantis. 5 × 105S. typhimurium cells were also determined as a limit of detection of this study with mentioned parameters.
Electronic supplementary material
The online version of this article (10.1007/s13197-018-3467-5) contains supplementary material, which is available to authorized users.
Keywords: Lateral flow assay, Salmonella, Gold nanoparticles, Whole cell detection
Introduction
Food safety is the most important issue for the public health throughout the world and food quality is affected by foodborne pathogens that cause serious human and animal disease (Alves et al. 2015). Infection of humans by those pathogens occur through air, drinking water and food (Dwivedi and Jaykus 2011). Salmonella are widely distributed group of gram negative bacteria in nature and has about 2300 serotypes. The primary reservoirs of Salmonella are animals and humans. It invades the intestinal tract of hosts and causes salmonellosis leading to death. 1.4 million cases of infection are reported by Salmonella annually (Olsen et al. 2001; Voetsch et al. 2004) and almost half of the gastrointestinal infections in the US every year are accounted for Salmonella contamination in food. Among the subspecies of Salmonella enterica; S. typhi S. typhimurium, S. enteritidis, S. pullorum, S. gallinarum and S. dublin are the major agents for human and animal diseases. According to a report compiled by the European Centre for Disease Prevention and Control (ECDC) and the European Food Safety Authority (ESFA) in 2016 Salmonella infections in Europe has small increase and S. enteritidis was the most common species accounted for all salmonellosis cases (Cogan and Humphrey 2003; ECDC 2016). Additionally, S. typhimurium and S. enteritidis are also the common causes of issues in Turkey (Oral et al. 1995; Goncagül et al. 2005).
To ensure the public health, early detection of pathogens is crucial. Therefore, many kinds of identification methods or microbiological tests are in use and new detection platforms are also being tried to develop for improving the sensitivity and selectivity of detection. Selective enrichment and culture is the traditional lab method for diagnosis of Salmonella (Stone et al. 1995) before doing biochemical and serological confirmation tests. However, these steps require 5 or 7 days which is inconvenient for food sector or industrial applications. Molecular methods (D’Souza et al. 2009), immunological based methods (Wen et al. 2013), nano-biosensors (Zuo et al. 2013; Kim et al. 2015) are also available in the literature for Salmonella detection. However, many of these techniques require skilled personnel, sterile working conditions and expensive instruments to perform sensitive and specific microbial analysis and they can result in cross contamination and target losses. Besides, most of these systems can not be used in field practically (Sapparapu 2003) as they are not portable, and their application to the bacterial detection in the environmental samples or food is limited.
LFAs developed by Ab, aptamer and nucleic acids (Laderman et al. 2008; He et al. 2011; Ang et al. 2012) have been a popular platform for rapid immunoassays since their introduction in the mid-1980s. They are simple diagnostic test kits commonly based on a nitrocellulose (NC) membrane matrix and might be expressed as point of care tests. LFAs have also achieved broad penetration in a variety of markets. Compared with other analysis methods, the immunochromatography strip tests have many advantages. They are established mature technology, relative ease of manufacture, processes already developed, easily scalable to high volume production, stable shelf-lives without refrigeration, handling small volumes of multiple sample types, having high specificity, good stability, relatively low cost, and minimal education required for users and regulators (O’Farrell 2009).
Ab based immunochromatographic test strips are commonly in use since Abs can be directly immobilized onto GNPs (Shen et al. 2007) and then they are applied to the strip assays. LFAs based on GNPs/Ab conjugates have become useful innovation in nanotechnology as colloidal gold is the most widely used label today in commercial LFAs for many reasons (Chandler et al. 2000). GNPs based bioconjugates have controllable morphology, biocompatibility, high stability, ease of characterization, shortened analysis time and no development process is needed for visualization (Rosi et al. 2006; Sperling et al. 2008). Although various LFAs were reported for Salmonella, they have time consuming process such as nucleic acid isolation of Salmonella instead of cell surface recognition, further experimental steps, advanced instruments and also use the different sized GNPs (Du et al. 2017; Liu et al. 2017; Zhao et al. 2017; Shin et al. 2018) for lower detection limit. Besides, strip assays are generally tested for bacteria when they used singly (Preechakasedkit et al. 2012), and test parameters for Salmonella were not reported in detailed. Reproducibility of assay with its optimal parameters and its applicability to the live or killed target organisms when they are alone or included in bacterial mixtures are also crucial issues. Thus, here the study aimed to report ideal parameters needed for developing of GNP-based LFAs for Salmonella as a rapid and sensitive test strips for naked eye analysis when it is alone or present with multiple number of bacteria using Salmonella specific Ab.
Materials and methods
Chemicals and reagents
Ab specific to Salmonella and immunoglobulin G (IgG) were obtained from KPL (Turkey). The sample and absorbent pads, conjugate pads and NC membranes (M240, M180, M120, M075) were supplied from Millipore (Burlington, MA, USA). HAuCl4·xH2O was purchased from Sigma (St. Louis, MO, USA). Luria broth (LB) medium was used for bacterial culture. S. typhimurium, S. enteritidis, S. infantis, B. cereus were from NanoBiz R&D lab; dry soil bacterial culture was prepared from Middle East Technical University (METU) campus; S. enteritidis (1, 2, 3) and S. infantis (1, 2, 3, 4, 5, 6, 7) isolates were given by Ankara University Faculty of Veterinary Medicine and heat treated positive control cells (E. coli, V. chloreae, S. typhimurium) were from KPL. A Nanodrop 2000 UV–Vis spectrophotometer (Thermo Scientific; Waltham, MA, USA) was used for obtaining the spectra from GNPs and conjugates. Transmission electron microscopy (TEM; 2100 F 200 kV TEM, JEOL, Peabody, MA, USA) was used to determine the size and shape of synthesized GNPs. A multiscan plate reader (Thermo Scientific) was used to obtain spectra from gold conjugates.
Synthesis of gold nanoparticles (GNPs)
GNPs were synthesized according to the citrate reduction method (Kimling et al. 2006) by making slight changes and conjugated with Salmonella Ab. The pH value of colloidal gold suspension was adjusted with K2CO3 and HCl. The evaluation of the size measurement of synthesized GNPs was performed by TEM, Malvern Zeta Potential instrument and UV–Vis spectroscopy. Zeta potentials of both naked and conjugated GNPs were also analyzed by Malvern Zeta Potential. Briefly, 500 mL of 0.01% HAuCl4·xH2O was boiled and 1% sodium tricitrate was added into the solution. After changing the color of solution from black to reddish in a 2 min, it was allowed to further boiling for about 10 min to complete the reduction before filter sterilization. After cooling, 0.05% sodium azide was added and UV–Vis spectra was measured for determining the λmax of GNPs.
Bioconjugation and characterization of GNPs
To make a gold conjugate with Salmonella Ab, the pH of GNPs was initially adjusted to 5.6–6.0–6.5–7.0–7.5–8.0–8.5–9.0 in 96-well plates. Then 40 μg/mL of Ab as final concentration was added to the each well containing GNPs and after 15 min incubation absorbance value was read at λmax. The stability and polydispersity constant of GNPs/Ab complex were evaluated by getting the ratio of absorbance at λmax:580 nm and 600 nm:λmax (Englebienne 2000), respectively after adding 10% NaCl into the solution. The optimal pH value was determined according to the graph and also by viewing the displayed color of the mixture, which had to be the same as the originally synthesized GNPs’ red color. The minimum Ab concentration desired for coating GNPs was determined in 96 well plate by adding of the Ab as 5–10–15–20–25–30–35–40–45–50 μg/mL for final concentration into each well. The absorbance value at λmax was read after incubation and 10% NaCl solution was added to each well; subsequently, the readings were retaken at λmax. To prepare a conjugate, Ab at the predetermined optimum concentration, 20 μg/mL, was added into GNPs and incubated with gentle shaking at room temperature (RT). A 10% bovine serum albumin (BSA) in sodium borate was also added into the mixture and allowed to incubate. The mixture was pelleted at 4 °C. The supernatant was discarded and pellet was washed with wash buffer (WB) containing 1% BSA for 2 times. Finally, the pellet was resuspended in 1 mL WB and called as conjugate A, and UV–Vis spectrum was recorded before storing at 4 °C. For negative controls, the same amount of GNPs was prepared without adding Ab in order to ensure that there will no non-specific capturing by other agents in conjugation on the strip assay. Table 1 contains information about the prepared GNPs with/without Ab.
Table 1.
Preparation of Salmonella Ab and GNP conjugates with negative controls and their UV–Vis spectra before and after centrifugation
| UV–Vis | 1 | 2 | 3 |
|---|---|---|---|
| Reagents in conjugate solutions | GNP/WBa | GNP/PBS/BSA/WB | GNP/Ab/BSA/WB |
| λmax before centrifuging | 526 nm | 526 nm | 526 nm |
| λmax after centrifuging | 527 nm | 528 nm | 532 nm |
aWashing buffer
Designing of lateral flow strips
The width of the strip tests was adjusted to 0.4 cm and the strip components were manually placed as specified earlier (Cam and Oktem 2017). Briefly, the sample pad was overlapped on one end of NC membrane and the absorbent pad was on the other end at a distance of 2 mm. The reagents on capture zones were manually applied with desired concentrations using a pipette tip. The treatment of samples on the sample pad was performed by either loading the 100 μL of sample or dipping the strips into the eppendorf tubes including 200 and 500 μL of the fresh cultured sample. The strips were washed with phosphate buffered saline (PBS) once if desired.
Optimization parameters of developed lateral flow strips
Stock concentrations of Salmonella Ab and IgG were prepared in PBS. Thereafter, they were spotted on the capture zones as test and control line, respectively with different concentrations. The conjugate pad was prepared by applying a mixture of 100 μL of GNP/Ab conjugate and 200 μL of conjugate pad buffer (sodium borate containing BSA, sucrose, NaCl, Tween 20, sodium azide) onto a fiberglass membrane and labeled as conjugate A. Then it was allowed to incubate for 1 h. To make a comparison of either concentrated or diluted conjugate solutions on the conjugate pad, various mixtures were prepared. Briefly, 5 and 2.5 times concentrated conjugate A was loaded on the pad and called as conjugate B and C, respectively. Conjugate D was prepared by diluting the conjugate A with a ratio of ¼. M240, M180, M120 and M075 membranes were tested to find the best membrane type for bacterial strip assay. The LOD of target bacteria was determined using heat inactivated Salmonella cells by making tenfold serial dilutions. All experiments were done in triplicate using different batches of synthesized GNPs and GNP/Ab conjugates to verify the reproducibility of the results.
Results and discussion
Synthesis of gold nanoparticles and conjugation with antibodies
The size of the synthesized GNPs was 36 nm and they were filter sterilized before use (Online Resource 1). Although it is difficult to obtain the reproducibility in regards to size distribution from batch to batch preparation of GNPs, they were homogenously distributed spherical nanoparticles. The λmax of GNPs was recorded at 526 nm wavelength as expected. The concentration of GNPs was also calculated as 1.18 × 1014 particles in 0.19 nM, theoratically (Jain et al. 2006).
The stability and polydispersity of GNPs conjugated with Ab at different pH after salt addition was evaluated (Online Resource 2a) as the pH of coupling mixture plays significant role in the labelling of GNPs process. As expected, GNP conjugate was least polydisperse and most stable at pH 9. Because to make a conjugation of GNPs in the range of 5–60 nm with Ab is generally possible at the pH values of 8–9.5 (Safenkova et al. 2010). Adding of NaCl caused nanoparticle aggregation and resulted in purplish color if the Ab concentration was low in the solution. This is possible because if the NaCl as an electrolyte is present in the solution, repulsive and attractive forces between the particles are imbalanced due to the masking of negative charges of colloidal solution (Verwey and Overbeek 1948). Increased amount of Ab enhanced the stability of GNPs, and it became constant after the concentration of 12 μg/mL Ab. However, 20 μg/mL of Ab was decided to coat the GNPs since it produces a minimal decline in absorbance at λmax and the red color intensity of conjugates was still the same as the naked GNPs after adding the salt to the solution. This amount of Ab is very less compared to the high concentrations, 150 μg/mL, (Mikawa et al. 2009) and enables making the strip cheaper. Batch to batch sample preparation including the synthesis of GNPs and conjugates were consistent and they were also found as having a good stability for long term usage which is about 10 months at 4 °C.
The UV–Vis measurements of gold conjugates were reasonable and indicated peak shifting from 526 to 532 nm after Ab coating since the absorption of GNPs is affected by the immobilized proteins on GNPs’ surface (data not shown). To result, Salmonella Abs prevented GNPs from flocculation while they were agglomerated in the absence of Abs. The size and Zeta Potential measurements were also analyzed by Malvern Zeta Sizer. Both naked and conjugated GNPs had shown perfect size distribution. It might be deduced that Abs were adsorbed onto each GNPs equally and resulted in size increment (Online Resource 3).
Zeta potential was − 53, 9 mV for naked GNPs and − 30 mV for the conjugate (Online Resource 4). It can be said that both the naked GNPs and conjugate are considerably stable. Because the value of Zeta potential which is more negative than − 30 mV or more positive than + 30 mV shows stability (Nara et al. 2010). As a result peak shifting, size increment and changed surface charges of GNPs caused by the positively charged Abs confirmed the bio-conjugation process.
Designing of lateral flow strips
The design of LFAs was performed as mentioned above. 36 nm GNPs showed a good stability and capturing efficiency in the developed strips. Buffer solutions were found to have positive effect on the flow. Increased sample volume on sample pad caused decreased color intensity on the lines. The reason may be that completion of the reaction between Ab and antigen in a very quick time suffered or high volume of sample might interrupt the interaction of conjugate and capture Abs. To add, highly concentrated sample suffered from flowing through the membranes. Although the shelf life of the strips was not evaluated in this study, the results showed that the strips prepared by 8 months of GNPs and 1 month of gold conjugate were in efficient working condition.
Antibody concentrations on the capture zones
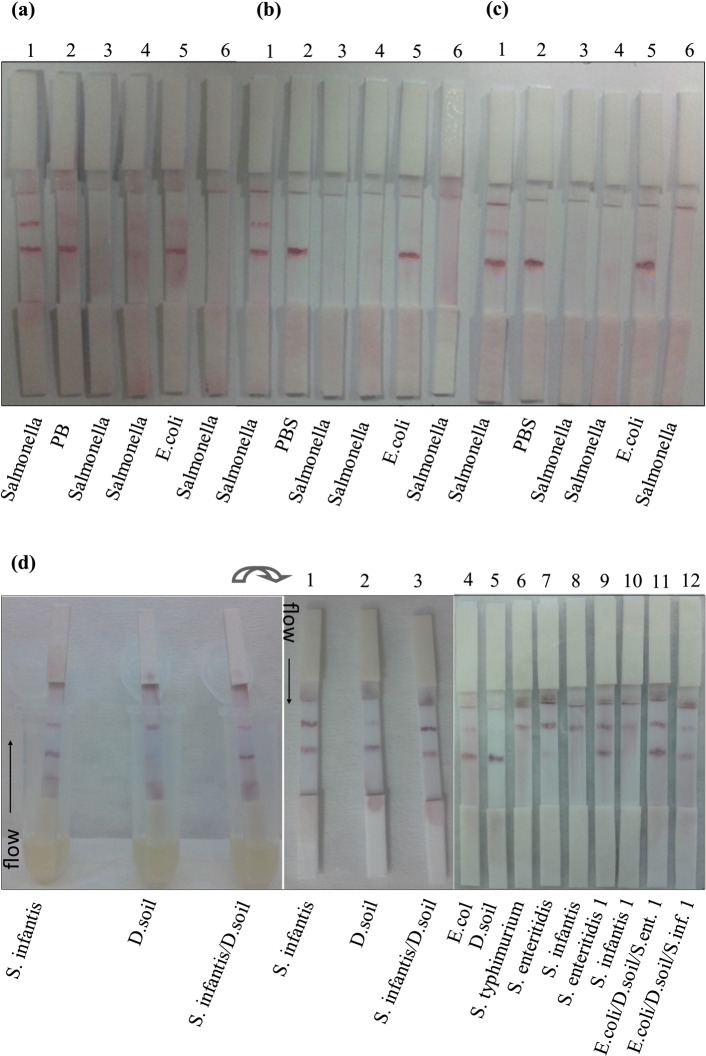
After deciding the best condition for immobilization of reagents on capture zones (data not shown), the optimal concentration of test line Ab was experienced with Salmonella positive control cells (Fig. 1a–c) and live cells (Fig. 1d) using M240 and M180 membrane, respectively. 0.3 μg/strip Ab was chosen as a suitable amount for both positive control cells and live cells and used for further experiments. This amount was found as considerably lower than others (Choi et al. 2010; Lou et al. 2012) and could allow the low cost for producers. Although high amount of Ab is seen having a high color intensity on the test line, it might cause the retention of more conjugate on the line and prevent the flow through the control line (data not shown). This means that the optimal amount of Ab is crucial for specific binding for developed dipstick assays and both the usage of excessive Ab concentration and bacterial medium might cause the accumulation. Thus, 0.3 μg Ab per strip was found in proportional with the prepared conjugate in terms of the line intensity and naked eye analysis of tested Salmonella strains. To highlight, using common structural Ab specific to Salmonella eliminated the need of extra Ab pairs for sandwich format in LFA (Singh et al. 2015) and showed high selectivity in the test. While Salmonella capturing either alone or in a bacterial mixture sample was successful, control lines of some strips were not seen (Fig. 1d6–d8, d10). This was caused by the inefficient flow of conjugate and retention of sample on conjugate pad after the interaction of high number of bacteria and media components. However, S. infantis and S. enteritidis 1 were captured sensitively and selectively compared to S. typhimurium when they were alone and in the mixture (Fig. 1 d1–d3, d9, d11). It is assumed that it is mostly related to Ab specifity to those strains. S. infantis 1 was also recognized in the mixture sensitively (Fig. 1 d12).
Fig. 1.
Different Ab concentrations on the test lines for capturing of positive control cells (a–c) and live Salmonella cells (d) using M240 and M180 membrane, respectively. Test line: 0.3 μg per strip Ab (a, d), 0.15 μg per strip Ab (b) and 0.1 μg per strip Ab (c). Control line: 5 × 106 Salmonella positive control cells. Strips 3–4 (a–c) were prepared with naked GNPs 1 come from Table 1. Strips 6 (a–c) have no capture Abs on each line. Salmonella and E.coli positive control cells were loaded on sample pad. D. soil: Dry soil bacteria sample. 500 μL of overnight cultures were transferred into the eppendorf tubes separately for dipstick assay (d). The mixture of different bacteria was prepared by adding 500 μL of each into the mix and after mixing the cultures homogenously 500 μL of mix was transferred into the tubes for dipping the strips (d)
Solution on the conjugate pad and stability of the gold conjugates
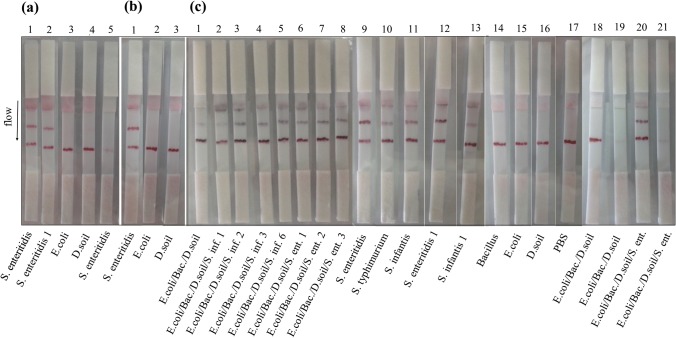
It was observed that concentrated conjugates retarded the flow on strips and resulted in weak line intensity and nonspecific adsorption of gold conjugate to the NC membrane. As expected the lower concentration of gold conjugate also showed weak binding to the target on test zones (data not shown). Therefore, conjugate A was used for further experiments as it has efficient flow and intensity on the lines. In order to ensure of the stability of GNPs and prepared strips, the conjugates were prepared by 2, 6 and 8 months of GNPs (Fig. 2). Repeating of all experiments at different dates had consistent results. According to the results, all the Salmonella strains were detected specifically and the conjugate seemed to have higher affinity to S. enteritidis strains and S. infantis compared to S. typhimurium. Thus, developed strips do not need any extra signal amplification on the capture zones (Choi et al. 2010) and still have stability for long time period. This is crucial for increasing the shelf life of the developed strips.
Fig. 2.
The comparison of GNPs/Ab conjugates prepared at different dates using M180 membrane. Test line: 0.3 μg per strip Salmonella Ab. Control line: 0.6 μg per strip anti goat IgG. The conjugates were prepared by 2 (a), 6 (b) and 8 (c) months old GNPs. Strips A5, C19 and C21 were prepared by naked 1 GNPs coming from Table 1. Sample preparation was performed by mixing 100 μL of overnight culture and 100 μL of PBS (a, b) and the sample was loaded by adding 33 and 25 μL of each overnight growth bacteria into 100 μL PBS as total volume separately for the mixture containing three and four different bacteria samples, respectively (c). Bacteria alone was loaded as 100 μL from overnight culture
Comparison of the membrane types
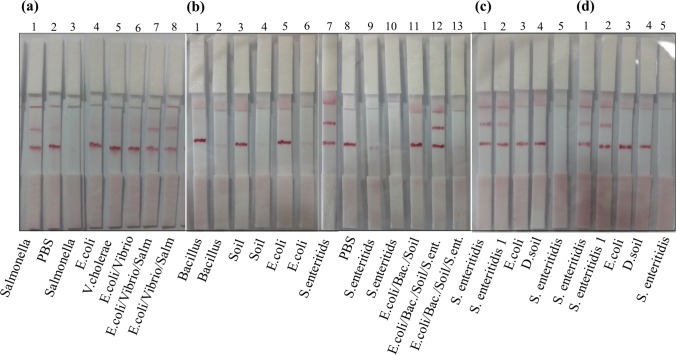
Since the detection time for end users of strips is crucial, four types of Millipore membranes having different flow rate were compared using both heat treated cells and live cells (Fig. 3). The most appropriate NC membrane was determined based on color intensity, stability, and reaction time. The targets were recognized selectively by all membranes. However, the test line intensity and releasing of conjugates from the conjugate pads were not efficient for M120 and M075 (c, d) even if they had rapid flow rate. Weak line intensity might be related to inefficient Ab immobilization on the test lines of those membranes. M240 has also weak color intensity on the test zone and flow take a long time, 7 min. However, M180 showed the best results in terms of those factors. Therefore, it can be decided as the best membrane type for whole cell detection compared to the other membranes used for the recognition of nucleic acids of Salmonella (Du et al. 2017; Liu et al. 2013).
Fig. 3.
The comparison of membrane types for Salmonella test strips. a M240, b M180, c M120, d M075. Strips a3, b2, b4, b6, b9, b10, b13, c5, d5 were prepared with naked 1 GNPs. 5 × 106, 3 × 106 and 3 × 106 positive control cells were loaded in 100 μL PBS for Salmonella, E. coli and V. cholerae, respectively for single and mixture format (a). Mixtures were prepared by adding of 33 and 25 µL of each live bacteria into 100 µL PBS for three and four types of bacteria, separately for dipstick assay (b). 100 µL of live bacteria was added into 100 µL PBS for single bacteria dipstick assay (b–d). Salm: Salmonella positive control cells. D.soil: Dry soil bacteria sample
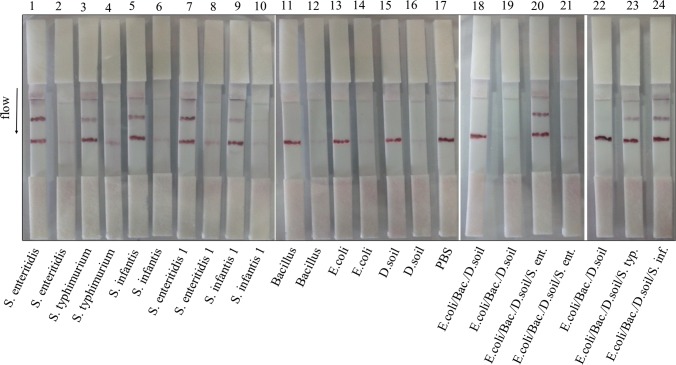
After deciding the membrane type, its specificity in mixture containing other live bacteria was further experimented using M180 membrane (Fig. 4). S. enteritidis strains, S. typhimurium and S. infantis were detected either alone or in the mixture specifically. However, the recognition of S. enteritidis strains (strips 1, 7, 20) was better than others under both conditions (strips 3, 5, 23, 24). This result is important with respect to the specific detection of pathogens in their environment as they are living with various kinds of microorganisms in real life. Thus, M180 might be an ideal test membrane for multiple detection. To say that, dipping the strips into high volume of bacteria in medium without PBS affected the flow time and releasing of conjugate from the conjugate pad. Thus, PBS was added into the samples and prevented the agglomeration.
Fig. 4.
Dipstick assay for live Salmonella strains alone and in the mixture using M180 membrane. Test line: 0.3 μg per strip Salmonella Ab. Control line: 0.6 μg per strip antigoat IgG. Overnight cultures were applied to the strips. The strips were dipped into 200 μL of total bacteria with PBS. The bacteria were used in 100 μL volume when they were used singly. The strips 2–4–6–8–10–12–14–16–19–21 were prepared by naked 1 GNPs as shown in Table 1. D. soil: Dry soil bacteria, Bac: B. cereus, S. ent: S. enteritidis, S. typ: S. typhimurium
Determination of limit of detection (LOD)
LOD of captured Salmonella cells was determined as 5 × 105 cells in 100 µL (Fig. 5), which are known as enough cell number for gastroenteritis in humans caused by S. typhimurium (BVL 1996), in a 3 min with naked eye. The strips developed in this study had a lower LOD than the 106 CFU/mL detection limit of S. enteritidis recognized by gold based immunochromatographic assay using commercial Abs (Moongkarndi et al. 2011). Although lower detection limits were reported (Zhang et al. 2006; Moongkarndi et al. 2011; Yuan et al. 2014) they need to biosensor development, silver enhacement, and nucleic acid isolation of Salmonella for detection by LFA (Liu et al. 2013) instead of cell surface recognition. Different membrane types and using increased Ab concentrations on the test line did not affect the sensitivity and specifity of detection limit, significantly (data not shown).
Fig. 5.
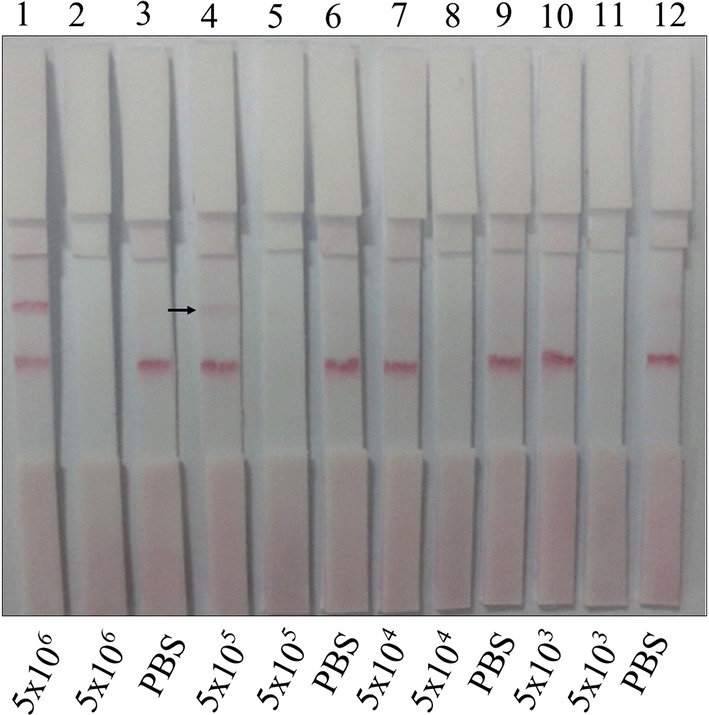
LOD of Salmonella positive control cells by developed LF strips using M180 membrane. Test line: 0.6 μg per strip Salmonella Ab. Control line: 3 × 106 Salmonella positive control cells. Strips 2, 5, 8 and 11 were prepared by naked 1 GNPs come from Table 1. From 5 × 106 to 5 × 103 Salmonella positive control cells were loaded
Conclusion
Immunochromatographic test strips were prepared by using colloidal GNPs with optimized parameters needed for the development of Salmonella LFAs. Dipstick assays successfully recognized the Salmonella cells with a desired number that is enough for triggering the illness without advanced instruments and experiments. S. enteritidis and S. infantis were the best recognized bacteria among the experienced strains. Strip assays could detect the targets when they were in the medium and buffer. M180 membrane was decided as the best membrane type for developing the assay for whole cell detection. 36 nm sized GNPs can be estimated as a perfect size and good label for Ab based LFA for bacterial detection. Because both the nanoparticles and their conjugates with Abs were found as highly stable after about 8 months at 4 °C. This is crucial for increasing the shelf life of the developed strips. Since the dipstick format of assays are generally attractive for the end users, recognition of Salmonella by developed strips might be applied to the real samples as it is shown in this study. Thus, those findings can make these strips a potential candidate for capturing the pathogenic Salmonella in water or food samples and will be helpful for various test developers in terms of the LFA parameters. The strips are currently being planned to be integrated in nationwide screening programs by our team.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1 Synthesized GNPs. (a) Colloidal naked GNPs, (b) UV–Vis spectrum of naked GNPs, (c, d) TEM images of naked GNPs. Scale bar: 50 nm. (TIFF 2492 kb)
Online Resource 2 Optimal pH value (a) and Ab concentration (b) for coating GNPs. (TIFF 1783 kb)
Online Resource 3 Size distributions of naked GNPs (a) and Ab/GNPs conjugate (conjugate A) (b). (TIFF 2089 kb)
Online Resource 4 Zeta potential measurements of naked (a) and GNPs/Ab conjugate (conjugate A) (b). (TIFF 1735 kb)
Acknowledgements
This research was supported by the Faculty Development Program (ÖYP-BAP-08-11-DPT-2011K121010), and Scientific and Technological Research Council of Turkey (TÜBİTAK-TEYDEB) projects funded to Nanobiz Nanobiotechnological Systems R&D Limited. The authors acknowledge to Ankara University Faculty of Veterinary Medicine for bacterial samples and Dr. Ceren Berkman for help.
References
- Alves J, Niguma NH, Oliveira TC. Detection of Salmonella spp. in eight complex food matrices using polymerase chain reaction assay. J Food Saf. 2015;35:453–457. doi: 10.1111/jfs.12194. [DOI] [Google Scholar]
- Ang GY, Yu CY, Yean CY. Ambient temperature detection of PCR amplicons with a novel sequence-specific nucleic acid lateral flow biosensor. Biosens Bioelectron. 2012;38:151–156. doi: 10.1016/j.bios.2012.05.019. [DOI] [PubMed] [Google Scholar]
- BVL (Bundesamt für Verbraucherschutz und Lebensmittelsicherheit) (1996) Position statement of the ZKBS on classifying Salmonella typhimurium LT2 strains and Salmonella typhimurium strains with stable mutations in aroA, galE or cya and crp genes as recipient organisms in genetic engineering operations. Ref. No: 6790-10-42
- Cam D, Oktem HA. Optimizations needed for lateral flow assay for rapid detection of pathogenic E. coli. Turk J Biol. 2017;41:954–968. doi: 10.3906/biy-1705-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler J, Gurmin T, Robinson N. The place of gold in rapid tests. IVD Technol. 2000;6:37–49. [Google Scholar]
- Choi DH, Lee SK, Oh YK, Bae BW, Lee SD, Kim S, Shin YB, Kim MG. A dual gold nanoparticle conjugate-based lateral flow assay (LFA) method for the analysis of troponin I. Biosens Bioelectron. 2010;25:1999–2002. doi: 10.1016/j.bios.2010.01.019. [DOI] [PubMed] [Google Scholar]
- Cogan TA, Humphrey TJ. The rise and fall of Salmonella enteritidis in the UK. J Appl Microbiol. 2003;94:114–119. doi: 10.1046/j.1365-2672.94.s1.13.x. [DOI] [PubMed] [Google Scholar]
- D’Souza DH, Critzer FJ, Golden DA. Real-time reverse-transcriptase polymerase chain reaction for the rapid detection of Salmonella using inv A primers. Foodborne Pathog Dis. 2009;6:1097–1106. doi: 10.1089/fpd.2009.0322. [DOI] [PubMed] [Google Scholar]
- Du XJ, Zhou TJ, Li P, Wang S. A rapid Salmonella detection method involving thermophilic helicase-dependent amplification and a lateral flow assay. Mol Cell Probes. 2017;34:37–44. doi: 10.1016/j.mcp.2017.05.004. [DOI] [PubMed] [Google Scholar]
- Dwivedi HP, Jaykus LA. Detection of pathogens in foods: the current state-of-the-art and future directions. Crit Rev Microbiol. 2011;37:40–63. doi: 10.3109/1040841X.2010.506430. [DOI] [PubMed] [Google Scholar]
- ECDC (2016). Salmonellosis—annual epidemiological report 2016 (2014 data). Annual epidemiological report on communicable diseases in Europe
- Englebienne P. Immune and receptor assays in theory and practice. New York: CRC Press; 2000. pp. 233–234. [Google Scholar]
- Goncagül G, Günaydin E, Carli KT. Prevalence of Salmonella serogroups in chicken meat. Turk J Vet Anim Sci. 2005;29:103–106. [Google Scholar]
- He Y, Zhang S, Zhang X, Baloda M, Gurung AS, Xu H, Zhang X, Liu G. Ultrasensitive nucleic acid biosensor based on enzyme–gold nanoparticle dual label and lateral flow strip biosensor. Biosens Bioelectron. 2011;26:2018–2024. doi: 10.1016/j.bios.2010.08.079. [DOI] [PubMed] [Google Scholar]
- Jain PK, Lee KS, El-Sayed IH, El-Sayed MA. Calculated absorption and scattering properties of gold nanoparticles of different size, shape, and composition: applications in biological imaging and biomedicine. Phys Chem B. 2006;110:7238–7248. doi: 10.1021/jp057170o. [DOI] [PubMed] [Google Scholar]
- Kim G, Moon JH, Moh CY, Lim JG. A microfluidic nano-biosensor for the detection of pathogenic Salmonella. Biosens Bioelectron. 2015;67:243–247. doi: 10.1016/j.bios.2014.08.023. [DOI] [PubMed] [Google Scholar]
- Kimling J, Maier M, Okenve B, Kotaidis V, Ballot H, Plech A. Turkevich method for gold nanoparticle synthesis revisited. J Phys Chem B. 2006;110:15700–15707. doi: 10.1021/jp061667w. [DOI] [PubMed] [Google Scholar]
- Laderman EI, Whitworth E, Dumaual E, Jones M, Hudak A, Hogrefe W, Carney J, Groen J. Rapid, sensitive, and specific lateral-flow immunochromatographic point-of-care device for detection of herpes simplex virus type 2-specific immunoglobulin G antibodies in serum and whole blood. Clin Vaccine Immunol. 2008;15:159–163. doi: 10.1128/CVI.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Yeung CY, Chen PH, Yeh MK, Hou SY. Salmonella detection using 16S ribosomal DNA/RNA probe-gold nanoparticles and lateral flow immunoassay. Food Chem. 2013;141:2526–2532. doi: 10.1016/j.foodchem.2013.05.089. [DOI] [PubMed] [Google Scholar]
- Liu HB, Zang YX, Du XJ, Li P, Wang S. Development of an isothermal amplification-based assay for the rapid visual detection of Salmonella bacteria. J Dairy Sci. 2017;100:7016–7025. doi: 10.3168/jds.2017-12566. [DOI] [PubMed] [Google Scholar]
- Lou S, Ye JY, Li KQ, Wu A. A gold nanoparticle-based immunochromatographic assay: the influence of nanoparticulate size. Analyst. 2012;137:1174–1181. doi: 10.1039/C2AN15844B. [DOI] [PubMed] [Google Scholar]
- Mikawa AY, Santos SAT, Kenfe FR, da Silva FH, da Costa PI. Development of a rapid one-step immunochromatographic assay for HCV core antigen detection. J Virol Methods. 2009;158:160–164. doi: 10.1016/j.jviromet.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Moongkarndi P, Rodpai E, Kanarat S. Evaluation of an immunochromatographic assay for rapid detection of Salmonella enterica serovars Typhimurium and Enteritidis. J Vet Diagn Invest. 2011;23:797–801. doi: 10.1177/1040638711408063. [DOI] [PubMed] [Google Scholar]
- Nara S, Tripathi V, Singh H, Shrivastav TG. Colloidal gold probe based rapid immunochromatographic strip assay for cortisol. Anal Chim Acta. 2010;682:66–71. doi: 10.1016/j.aca.2010.09.041. [DOI] [PubMed] [Google Scholar]
- O’Farrell B (2009) Evolution in lateral flow—based immunoassay systems. In: Lateral flow immunoassay. pp 1–33, Humana Press, New York
- Olsen SJ, Bishop R, Brenner FW, Roels TH, Bean N, Tauxe RV, Slutsker L. The changing epidemiology of Salmonella: trends in serotypes isolated from humans in the United States, 1987–1997. J Infect Dis. 2001;183:753–761. doi: 10.1086/318832. [DOI] [PubMed] [Google Scholar]
- Oral R, Sümer S, Can D, Tiraş Ü, Yener H. Yenidoğan Ünitesinde Salmonella Typhimurium Salgını. Turkiye Klinikleri J Pediatr. 1995;4:50–55. [Google Scholar]
- Preechakasedkit P, Pinwattana K, Dungchai W, Siangproh W, Chaicumpa W, Tongtawe P, Chailapakul O. Development of a one-step immunochromatographic strip test using gold nanoparticles for the rapid detection of Salmonella typhi in human serum. Biosens Bioelectron. 2012;31:562–566. doi: 10.1016/j.bios.2011.10.031. [DOI] [PubMed] [Google Scholar]
- Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. doi: 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- Safenkova IV, Zherdev AV, Dzantiev BB. Correlation between the composition of multivalent antibody conjugates with colloidal gold nanoparticles and their affinity. J Immunol Methods. 2010;357:17–25. doi: 10.1016/j.jim.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Sapparapu G (2003) Development of immunological reagents for detecting Salmonella Enterica serovar Typhimurium (Master of Science dissertation, University of Florida)
- Shen G, Shen G, Yu R. Au nanoparticle network-type thin films formed via mixed assembling and cross-linking route for biosensor application: quartz crystal microbalance study. Anal Biochem. 2007;365:1–6. doi: 10.1016/j.ab.2006.11.038. [DOI] [PubMed] [Google Scholar]
- Shin JH, Hong J, Go H, Park J, Kong M, Ryu S, Kim KP, Roh E, Park JK. Multiplexed detection of Foodborne pathogens from contaminated lettuces using a handheld multistep lateral flow assay device. J Agric Food Chem. 2018;66:290–297. doi: 10.1021/acs.jafc.7b03582. [DOI] [PubMed] [Google Scholar]
- Singh J, Sharma S, Nara S. Evaluation of gold nanoparticle based lateral flow assays for diagnosis of enterobacteriaceae members in food and water. Food Chem. 2015;170:470–483. doi: 10.1016/j.foodchem.2014.08.092. [DOI] [PubMed] [Google Scholar]
- Sperling RA, Gil PR, Zhang F, Zanella M, Parak WJ. Biological applications of gold nanoparticles. Chem Soc Rev. 2008;37:1896–1908. doi: 10.1039/b712170a. [DOI] [PubMed] [Google Scholar]
- Stone GG, Oberst RD, Hays MP, McVey S, Galland JC, Curtiss R, Kelly SM, Chengappa MM. Detection of Salmonella typhimurium from rectal swabs of experimentally infected beagles by short cultivation and PCR-hybridization. J Clin Microbiol. 1995;33:1292–1295. doi: 10.1128/jcm.33.5.1292-1295.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwey EJW, Overbeek JTG. The free energy of a double layer system. Theory of the stability of lyophobic colloids. New York: Elsevier; 1948. p. 51. [Google Scholar]
- Voetsch AC, Van Gilder TJ, Angulo FJ, Farley MM, Shallow S, Marcus R, Tauxe RV. Emerging infections program FoodNet working group. FoodNet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin Infect Dis. 2004;38:127–134. doi: 10.1086/381578. [DOI] [PubMed] [Google Scholar]
- Wen CY, Hu J, Zhang ZL, Tian ZQ, Ou GP, Liao YL, Li Y, Xie M, Sun ZY, Pang DW. One-step sensitive detection of Salmonella typhimurium by coupling magnetic capture and fluorescence identification with functional nanospheres. Anal Chem. 2013;85:1223–1230. doi: 10.1021/ac303204q. [DOI] [PubMed] [Google Scholar]
- Yuan J, Tao Z, Yu Y, Ma X, Xia Y, Wang L, Wang Z. A visual detection method for Salmonella typhimurium based on aptamer recognition and nanogold labeling. Food Control. 2014;37:188–192. doi: 10.1016/j.foodcont.2013.09.046. [DOI] [Google Scholar]
- Zhang S, Huang TS, Bridgman R, Weese J. Development and characterization of monoclonal and polyclonal antibodies against Salmonella enterica Typhimurium for biosensor detection. J Food Sci. 2006;71:M100–M104. doi: 10.1111/j.1365-2621.2006.tb15631.x. [DOI] [Google Scholar]
- Zhao Y, Jiang X, Qu Y, Pan R, Pang X, Jiang Y, Man C. Salmonella detection in powdered dairy products using a novel molecular tool. J Dairy Sci. 2017;100:3480–3496. doi: 10.3168/jds.2016-12535. [DOI] [PubMed] [Google Scholar]
- Zuo P, Li X, Dominguez DC, Ye BC. A PDMS/paper/glass hybrid microfluidic biochip integrated with aptamer-functionalized graphene oxide nano-biosensors for one-step multiplexed pathogen detection. Lab Chip. 2013;13:3921–3928. doi: 10.1039/c3lc50654a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online Resource 1 Synthesized GNPs. (a) Colloidal naked GNPs, (b) UV–Vis spectrum of naked GNPs, (c, d) TEM images of naked GNPs. Scale bar: 50 nm. (TIFF 2492 kb)
Online Resource 2 Optimal pH value (a) and Ab concentration (b) for coating GNPs. (TIFF 1783 kb)
Online Resource 3 Size distributions of naked GNPs (a) and Ab/GNPs conjugate (conjugate A) (b). (TIFF 2089 kb)
Online Resource 4 Zeta potential measurements of naked (a) and GNPs/Ab conjugate (conjugate A) (b). (TIFF 1735 kb)