Abstract
Biomarkers are needed to improve the diagnosis of neuropsychiatric disorders, which are often associated to excitatory/inhibitory imbalances in neural transmission and abnormal maturation. Here, we characterized different disease conditions by mapping changes in the expression patterns of maturation-related genes whose expression was altered by experimental neural hyperexcitation in published studies. This analysis revealed two gene expression patterns: decreases in maturity markers and increases in immaturity markers. These two groups of genes were characterized by the over-representation of genes related to synaptic function and chromosomal modification, respectively. Using these two groups in a transdiagnostic analysis of 87 disease datasets for eight neuropsychiatric disorders and 12 datasets from corresponding animal models, we found that transcriptomic pseudoimmaturity inducible by neural hyperexcitation is shared by multiple neuropsychiatric disorders, such as schizophrenia, Alzheimer disorders, and amyotrophic lateral sclerosis. Our results indicate that this endophenotype serves as a basis for the transdiagnostic characterization of these disorders.
Tomoyuki Murano et al. showed that neural hyperexcitation increases the expression of immaturity related genes. These changes in gene expression are shared among different neuropsychiatric and neurological conditions, hinting at their potential role as biomarkers.
Introduction
Neuropsychiatric disorders—such as schizophrenia, bipolar disorder, major depressive disorder, and autism spectrum disorder—are common, with over a third of the population in most countries being diagnosed with at least one such disorder at some point in their life1. Almost all neuropsychiatric disorders are currently classified mainly on the basis of clinical signs and symptoms. However, there is evidence that patients with different clinical diagnoses share similar biological features, such as genetic mutations, molecular expression, and brain activity2–6. Recently, psychiatry has undergone a tectonic shift to incorporate the concepts of modern biology. There have been recent attempts to reclassify psychiatric disorders according to biological domains (e.g., genes, neural circuits, and behavior), such as through the Research Domain Criteria (RDoC) initiative7. Therefore, identifying appropriate biomarkers that can be used for transdiagnostic assessment of neuropsychiatric disorders is essential for improving the classification of these diseases and understanding their biological basis.
Using coexpression network analysis, a recent study revealed that cross-disorder gene expression overlaps could be used to characterize five major neuropsychiatric disorders8. Some of these overlapping gene groups were well characterized biologically by Gene Ontology enrichment or cell-type specificity, but the biological properties of other gene groups were rather unclear. Thus, nonbiased coexpression network analyses do not necessarily detect modules that extract the biological features of neuropsychiatric disorders. Thus, in order to improve the characterization of neuropsychiatric disorders, it might be helpful to detect modules of coexpressed genes and conduct gene expression analysis based on the findings derived from studies on animal models of neuropsychiatric disorders.
To date, we have screened more than 180 strains of genetically engineered mice using a large-scale, and comprehensive battery of behavioral tests, and we have identified several strains with abnormal behaviors related to neuropsychiatric disorders such as schizophrenia, bipolar disorder, and intellectual disability9. We discovered common endophenotypes in the brains of multiple strains of these genetically engineered mice with behavioral abnormalities. We termed one such endophenotype in the hippocampus of adult mice the immature dentate gyrus (iDG) phenotype10–13. In this phenotype, the molecular and electrophysiological properties of adult DG neurons in the genetically engineered mice were similar to those of immature DG neurons in typically developing infants. For example, the expression of calbindin, a marker of maturity in DG neurons, was decreased, and the expression of calretinin, a marker of immaturity, was increased10–15. Molecular changes similar to some of those found in mice with iDG have been observed in the postmortem brains of patients with schizophrenia16, bipolar disorder16, and epilepsy17–19. Furthermore, there is growing evidence that changes in molecular markers of pseudoimmaturity are also present in other brain areas of patients with schizophrenia20–28, bipolar disorder26, autism26, and alcoholism29. Therefore, we proposed that pseudoimmaturity of the brain could potentially be a useful transdiagnostic biomarker9.
Pseudoimmaturity of the brain can be induced in adulthood. Previously, we found that chronic fluoxetine treatment reversed the maturation status of DG neurons in adult wild-type mice, a phenomenon that we termed dematuration30,31. Likewise, recent studies suggest that several maturation-related genes and electrophysiological properties in the DG of wild-type adult mice assume an immature-like status after treatment with pilocarpine or electroconvulsive stimulation16,32. As mentioned above, an iDG-like phenotype has been found in patients with epilepsy17–19. Therefore, we hypothesized that the neural hyperexcitation may induce pseudoimmaturity of the brain in adulthood; however, this hypothesis has not been tested in human samples.
Some studies suggest that hyperexcitation of neurons may underlie abnormalities related to certain types of neuropsychiatric disorders. Individuals with epilepsy are at increased risk of developing schizophrenia, and vice versa33,34; additionally, patients with epilepsy can display psychotic symptoms that resemble those found in patients with schizophrenia35. Excitatory/inhibitory imbalances have been proposed to be involved in the pathogenesis and pathophysiology of schizophrenia36–39. Hyperactive action-potential firing has also been observed in hippocampal granule-cell-like neurons derived from induced pluripotent stem cells (iPSCs) of patients with bipolar disorder40. Recent studies have suggested that human patients with Alzheimer’s disease and temporal lobe epilepsy may harbor common underlying disease mechanisms17,41–43. Considering these findings, we hypothesized that the immature-like gene expression patterns induced by neural hyperexcitation may overlap with the abnormal gene expression patterns in the brains of patients with neuropsychiatric disorders and the related animal models. If this is the case, we hypothesized that this overlap can be used to perform transdiagnostic characterization of neuropsychiatric disorders.
To test this hypothesis, we first performed a meta-analysis of microarray datasets, comparing the changes in gene expression in the rat DG after seizure induction with the differences in gene expression in infant mice versus adult mice. To assess consistency across species, we also conducted a similar comparison using the human fetal hippocampus. The overlap between gene sets was estimated using the Running Fisher test44, which is a nonparametric statistical method. The fold change and direction of change (up or downregulation) for each gene between the two conditions were used to define the ranked gene signatures. For comparison across different arrays, orthologs were used for each organisms. To determine the similarities between two datasets, we used a combination of rank-based enrichment statistics and ontology-based meta-analysis. This method enables us to statistically assess the pairwise correlations between any two datasets, including datasets from different species and organs28,29,45,46. The gene-expression patterns in the rat DG after seizure induction significantly overlapped with those specific to the immature mouse DG and with those specific to the early-stage human fetal hippocampus. From the set of overlapping genes, we defined two groups: maturity-marker genes and immaturity-marker genes that are inducible by neural hyperexcitation. We assessed the expression patterns of these two groups of maturation-related genes in 87 public gene-expression datasets derived from the postmortem brains of patients with various neuropsychiatric disorders and from neural cells derived from patient iPSCs. We further analyzed 12 datasets from the brains of related animal models. Through this analysis, we characterized the expression patterns of maturation-related genes that are inducible by neural hyperexcitation across different disease conditions and animal models of these diseases.
Results
Hyperexcitation induces immature-like gene expression
To examine the developmental changes in gene-expression patterns in the rodent DG, we created a microarray dataset from postnatal days 8, 11, 14, 17, 21, 25, and 29 (GSE113727) and compared it with a dataset from 33-week-old adult mice (GSE42778)12. Within the entire mouse DG dataset, the largest overlap for changes in gene expression after pilocarpine injection was for the comparison between day 8 infant and 33-week-old adult mice (Supplementary Figure 1a). We included the dataset from postnatal day 8 infant mice for subsequent analysis. The expression levels of 6552 genes were increased in the DG of infant mice compared with adult mice, whereas the expression levels of 8637 genes were decreased (absolute fold change > 1.2 and t test P < 0.05). Next, we assessed the changes in gene expression induced by neural hyperexcitation in a rodent model. We obtained publicly available microarray datasets from the DG of adult rats after seizures induced by injection of pilocarpine (GSE47752)47. The expression levels of 7073 genes were significantly changed in the DG of epileptic-seizure rats 1 day after pilocarpine injection compared with rats treated with saline (absolute fold change > 1.2, P < 0.05).
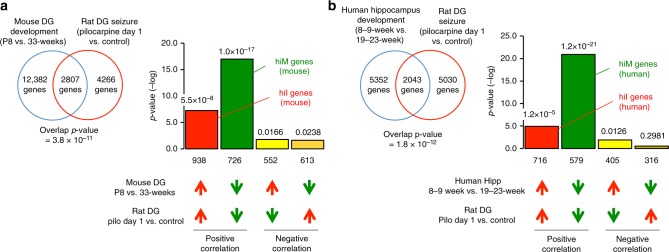
To investigate whether the neuronal hyperexcitation datasets contain immature-like gene expression patterns, we assessed the overlap between the set of genes with altered expression in immature mice and the set of genes with altered expression in adult seizure-model rats using the Running Fisher algorithm on the BaseSpace platform to determine the significance of the overlap (see Supplementary Methods for details). We found a striking degree of similarity: 2807 genes showed changes in expression in both datasets (overlap P = 3.8 × 10−11) (Fig. 1a). Among these 2807 genes, we named the 726 genes whose expression levels decreased in both datasets hyperexcitation-induced maturity-related genes (hiM genes (mouse): green bar in Fig. 1a) and the 938 genes whose expression levels increased in both datasets hyperexcitation-induced immaturity-related genes (hiI genes (mouse): red bar in Fig. 1a). Comprehensive lists of hiM and hiI genes are in Supplementary Data 1. The overlap for genes with positively correlated expression (red and green bars) was larger than the overlap for genes with negatively correlated expression (light and dark yellow bars), indicating that the directions of expressional change in the two datasets are more often similar than they are different. These results suggest that neuronal hyperexcitation induces a pattern of immature-like gene expression in the adult DG.
Fig. 1.
Neural hyperexcitation induces immature-like gene expression patterns in mouse and human. The patterns of changes in gene expression in the rat DG 1 day after pilocarpine treatment compared with developmental changes in the mouse DG and human hippocampus. Venn diagrams illustrating the overlap in genome-wide gene expression changes between the rat DG after seizure induction (GSE47752) and the DG of typically developing mouse infants (GSE113727; P8 infants compared with 33-week-old adults) (a) or the hippocampus of typically developing human fetuses (GSE25219: 19- to 23-week fetuses compared with 8- to 9-week fetuses) (b). Bar graphs illustrate the −log of the overlap P values for genes upregulated (red arrows) or downregulated (green arrows) by each condition. The Bonferroni correction was used to adjust the significance level according to the number of dataset pairs (see the Methods section and Supplementary Methods). Genes that were downregulated in both conditions were defined as mouse hiM genes (green bar in (a)), and genes that were upregulated in both conditions were defined as mouse hiI genes (red bar in (a)). Similarly, human hiM genes and human hiI genes were defined as the groups of genes with positive correlations between the conditions of development and seizures (b)
Next, we compared the changes in expression during development in the human fetal hippocampus with those in rats after seizure induction to assess consistency across species. We obtained publicly available microarray datasets for the human fetal hippocampus during development (GSE25219)48. Within the entire fetal hippocampal dataset, the largest overlap for changes in gene expression after pilocarpine injection was for the comparison between 8- to 9-week fetuses and 19- to 23-week fetuses (Supplementary Figure 1b). Again, we found a striking degree of similarity: 2043 genes showed changes in expression in both datasets (overlap P = 1.8 × 10−12) (Fig. 1b). Among these 2043 genes, we termed the 579 genes whose expression decreased in both datasets hiM genes (human) (green bar in Fig. 1b) and the 716 genes whose expression increased in both datasets hiI genes (human) (red bar in Fig. 1b). The overlaps for genes with positively correlated expression (red and green bars) were larger than the overlaps for genes with negatively correlated expression (light and dark yellow bars), suggesting that, similar to the results in mice, the gene expression changes in the rat DG after seizure induction are comparable to the reverse of the changes that occur as the human hippocampus develops.
hiM/hiI genes exhibit different biological properties
To characterize the biological features associated with the hiM and hiI gene groups in mice and humans, we conducted pathway enrichment analyses in BaseSpace. The 20 biogroups that had the largest overlap with the hiM and hiI genes are listed in Tables 1 and 2. Among mouse hiM genes, 4 out of the top 20 biogroups are associated with synapse and channel activity (e.g., transmission of nerve impulse, synapse, and synaptic transmission, Table 1); among human hiM genes, 6 out of the top 20 biogroups are also associated with synapse and channel activity (e.g., transmission of nerve impulse, synaptic transmission, axon, and synapse, Table 2). Among the mouse hiI genes, 4 out of the top 20 biogroups were associated with the nucleus (e.g., genes involved in the cell cycle and genes involved in DNA replication, Table 1). Among the human hiI genes, 15 out of the top 20 biogroups were associated with the nucleus (e.g., genes involved in the cell cycle, chromosomes, and response to DNA damage stimulus, Table 2). Notably, there is little overlap in the top 20 biogroups for the hiM and hiI genes (Table 2). Thus, the biogroups related to the hiM and hiI genes are likely to be functionally different. We further characterized the biological significances of hiM and hiI genes by comparing them with coexpression gene modules obtained from mouse DG development datasets and conducting pathway analyses of those modules. For the detail of the results, see Supplementary Figure 4.
Table 1.
Summary of results from the pathway analyses of mouse hiM and hiI genes. The 20 biogroups with the most significant similarities to mouse hiM and hiI genes
| Biogroup name | Direction | Common genes | P value |
|---|---|---|---|
| hiM genes (mouse) | |||
| Multicellular organismal signaling | Down | 56 | 4.00E−23 |
| Transmission of nerve impulse | Down | 51 | 3.10E−19 |
| Axon | Down | 45 | 7.30E−19 |
| Neuron differentiation | Down | 58 | 4.70E−17 |
| Synapse | Down | 57 | 5.70E−17 |
| Neuron development | Down | 50 | 1.50E−16 |
| Neuronal cell body | Down | 45 | 5.20E−16 |
| Cell junction | Down | 56 | 3.00E−15 |
| Cell part morphogenesis | Down | 43 | 4.70E−15 |
| Neuron projection development | Down | 42 | 6.20E−15 |
| Cell projection part | Down | 56 | 6.50E−15 |
| Cell morphogenesis involved in neuron differentiation | Down | 35 | 2.90E−14 |
| Cell morphogenesis involved in differentiation | Down | 40 | 6.70E−14 |
| Cellular chemical homeostasis | Down | 53 | 2.50E−13 |
| Cell–cell signaling | Down | 44 | 2.60E−13 |
| Synaptic transmission | Down | 37 | 3.60E−13 |
| Regulation of neurological system process | Down | 31 | 1.10E−12 |
| Regulation of transmission of nerve impulse | Down | 30 | 1.40E−12 |
| Genes involved in Neuronal System | Down | 31 | 1.70E−12 |
| Cellular ion homeostasis | Down | 49 | 1.80E−12 |
| hiI genes (mouse) | |||
| Response to wounding | Up | 60 | 1.10E−16 |
| Positive regulation of developmental process | Up | 66 | 9.30E−16 |
| Cardiovascular system development | Up | 61 | 7.70E−15 |
| Circulatory system development | Up | 61 | 7.70E−15 |
| Proteinaceous extracellular matrix | Up | 32 | 7.80E−15 |
| Genes involved in Cell Cycle | Up | 47 | 1.00E−14 |
| Extracellular matrix | Up | 37 | 1.30E−14 |
| Genes involved in Cell Cycle, Mitotic | Up | 42 | 6.30E−14 |
| Protein domain specific binding | Up | 60 | 7.00E−14 |
| Positive regulation of signal transduction | Up | 58 | 8.70E−14 |
| Genes involved in DNA replication | Up | 32 | 9.20E−14 |
| Cell cycle | Up | 67 | 3.00E−13 |
| Genes involved in adaptive immune system | Up | 56 | 3.20E−13 |
| Focal adhesion | Up | 31 | 5.60E−13 |
| Kinase binding | Up | 44 | 5.50E−12 |
| Cytoskeleton organization | Up | 54 | 5.60E−12 |
| Neuron differentiation | Up | 52 | 5.80E−12 |
| Basement membrane | Up | 19 | 7.50E−12 |
| Regulation of cell migration | Up | 36 | 1.10E−11 |
| MAPKinase Signaling Pathway | Up | 20 | 1.40E−11 |
Italic columns indicate biogroups that are related to the plasma membrane. Bold columns indicate biogroups that are related to reactions in the nucleus
Table 2.
Summary of results from the pathway analyses of human hiM and hiI genes. The 20 biogroups with the most significant similarities to human hiM and hiI genes
| Biogroup name | Direction | Common genes | P value |
|---|---|---|---|
| hiM genes (human) | |||
| Multicellular organismal signaling | Up | 87 | 4.40E−58 |
| Transmission of nerve impulse | Up | 83 | 1.40E−54 |
| Synaptic transmission | Up | 75 | 6.70E−50 |
| Neuron projection | Up | 70 | 2.60E−44 |
| Axon | Up | 42 | 2.90E−33 |
| Synapse | Up | 51 | 2.60E−32 |
| Neuron development | Up | 58 | 1.60E−30 |
| Genes involved in neuronal system | Up | 41 | 3.40E−30 |
| Genes involved in transmission across chemical synapses | Up | 32 | 1.50E−29 |
| Regulation of neurological system process | Up | 35 | 6.50E−29 |
| Regulation of transmission of nerve impulse | Up | 34 | 9.20E−29 |
| Cell projection part | Up | 53 | 1.80E−27 |
| Passive transmembrane transporter activity | Up | 44 | 1.80E−26 |
| Ion channel activity | Up | 43 | 2.20E−26 |
| Cell part morphogenesis | Up | 49 | 1.80E−24 |
| Behavior | Up | 38 | 2.90E-24 |
| Single-organism behavior | Up | 35 | 3.70E−24 |
| Cell morphogenesis involved in neuron differentiation | Up | 44 | 4.30E−24 |
| Neuron projection development | Up | 47 | 4.40E−24 |
| Dendrite | Up | 37 | 2.00E−23 |
| hiI genes (human) | |||
| Chromosome | Down | 65 | 1.30E−32 |
| Response to DNA damage stimulus | Down | 60 | 4.80E−32 |
| Genes involved in Cell Cycle | Down | 50 | 3.60E−31 |
| Genes involved in Cell Cycle, mitotic | Down | 43 | 1.20E−30 |
| Genes involved in DNA replication | Down | 34 | 2.00E−30 |
| Interphase | Down | 47 | 8.20E−30 |
| Interphase of mitotic cell cycle | Down | 46 | 5.10E−29 |
| Genes involved in mitotic M-M/G1 phases | Down | 29 | 2.60E−26 |
| Cell cycle | Down | 25 | 7.60E−23 |
| DNA repair | Down | 40 | 2.30E−22 |
| Wound healing | Down | 50 | 7.40E−22 |
| Response to ionizing radiation | Down | 21 | 1.20E−20 |
| Nuclear division | Down | 35 | 8.00E−20 |
| Mitosis | Down | 35 | 8.00E−20 |
| Cell division | Down | 38 | 2.20E−19 |
| G1/S transition of mitotic cell cycle | Down | 26 | 1.70E−18 |
| Cardiovascular system development | Down | 46 | 3.20E−18 |
| Circulatory system development | Down | 46 | 3.20E−18 |
| S phase | Down | 22 | 3.80E−18 |
| Blood coagulation | Down | 41 | 6.10E−18 |
Italic columns indicate biogroups that are related to the plasma membrane. Bold columns indicate biogroups that are related to reactions in the nucleus
We also compared datasets from the DG of typically developing infants with datasets from the rat DG at three different timepoints after seizure induction by injection of pilocarpine or kainate (day 1, day 3, and day 10) and performed principal component analysis on the changes in mouse hiM and hiI genes at different timepoints (Supplementary Figure 2a, b; Supplementary Notes). The time course of changes in the mouse hiM genes after seizure induction was different from the time course of changes in the mouse hiI genes. In addition, we conducted a spatial pattern analysis of the mouse hiM and hiI genes, which indicated that their protein products have slightly different patterns of subcellular localization (Supplementary Figure 2c; Supplementary Notes). The mouse hiM genes tend to be strongly expressed at the plasma membrane, with expression changes stabilizing by the third day after seizure induction. In contrast, the hiI genes tend to be expressed in the nucleus, and changes in expression after seizure induction are slower to stabilize. Together, these results indicate that the hiM and hiI genes have different spatiotemporal patterns of changes in expression.
Gene-expression analyses of patient samples by hiM/hiI genes
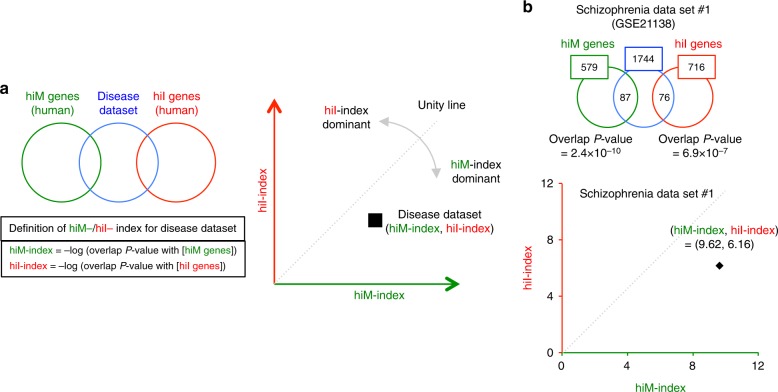
Next, we investigated whether and to what extent the expression changes in maturation-related genes induced by hyperexcitation overlap with gene expression patterns in various neuropsychiatric disorders. As above, we evaluated similarities between the changes in gene-expression patterns in different groups using overlap P values calculated by the Running Fisher algorithm (Fig. 2a). Similarity indexes for each comparison were defined as the −log of the overlap P values with hiM or hiI genes, denoted by hiM-index or hiI-index, respectively. High values of the hiM-/hiI-index indicate that there is a large overlap between the dataset analyzed and hiM/hiI genes. We obtained the hiM-/hiI-indexes for the datasets from human patients and plotted them in two-dimensional (2-D) space to show the extent of overlap between datasets and hiM/hiI genes (Fig. 2a).
Fig. 2.
Overview of the two-dimensional (2-D) analysis of disease datasets. a Genes with expression changes in the disease datasets are compared with the hiM and hiI gene groups. The hiM- and hiI-indexes were defined as the −log of the overlap P values with the hiM and hiI genes, respectively. The gene-expression patterns of the disease datasets are plotted in two-dimensional coordinates, in which the x- and y-axes are defined by the hiM- and hiI-indexes. Each dataset is characterized as hiM- or hiI-dominant by the ratio of the hiM-index to the hiI-index, and the degree of hiM- or hiI-dominance is evaluated by deviation from the unity line. The distance of each dataset from the origin shows the degree of overlap with hiM-/hiI-genes. b 2-D analysis applied to a dataset of postmortem brains (prefrontal cortex) from patients with schizophrenia (schizophrenia dataset #1). The expression levels of 1744 genes are significantly changed in this disease dataset. Of these, 87 and 76 genes overlap with the hiM/hiI genes. The overlap P values between the disease dataset and the hiM/hiI genes are 2.4 × 10−10 and 6.9 × 10−7. The hiM- and hiI-indexes for the disease dataset are therefore 9.62 and 6.16, indicating that this dataset is hiM-dominant. The results of the 2-D analysis for this dataset are plotted in the two-dimensional coordinates defined by the hiM- and hiI-indexes
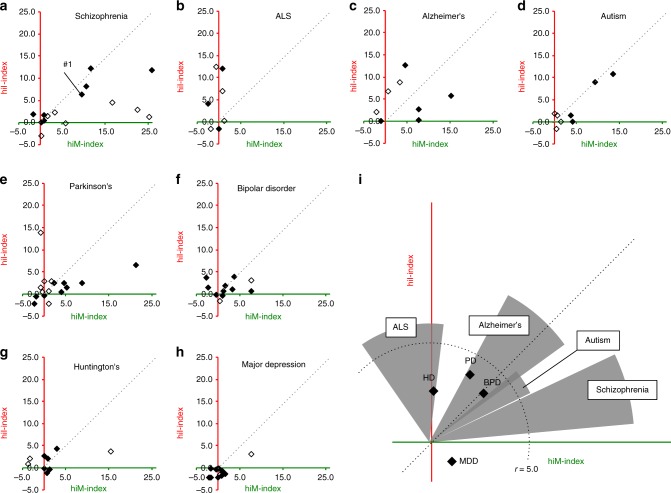
We initially performed this 2-D analysis on a dataset containing the expression profile of the prefrontal cortex in the postmortem brains of patients with schizophrenia (schizophrenia dataset #1: details in Supplementary Data 2) (Fig. 2b). The expression of 1744 genes differed between patients and healthy controls (significance level of 0.05). The numbers of hiM and hiI genes with altered expression in schizophrenia dataset #1 were 87 and 76, respectively, and the overlap P values were 2.4 × 10−10 and 6.9 × 10−7, respectively. The hiM-index and hiI-index for this dataset were 9.62 (= −log(2.4 × 10−10)) and 6.16 ( = −log(6.9 × 10−7)). This result corresponds to a point in 2-D space (Fig. 2b). Points that fall below the unity line (dashed line) indicate datasets in which changes in hiM genes are dominant, whereas points above the unity line indicate datasets with dominant changes in hiI genes. The angle from the unity line indicates the degree of hiM or hiI dominance. The same analysis was performed for other schizophrenia datasets (schizophrenia datasets #2–#16), including those obtained from different areas of the postmortem brain and from cultured neurons derived from the iPSCs of patients. Scatter plots of the results from the schizophrenia datasets are shown in Fig. 3a. Thirteen out of sixteen points were below the unity line. Most of the schizophrenia datasets exhibited hiM-index-dominant patterns, showing high hiM-index values and low hiI-index values (Fig. 3a).
Fig. 3.
Two-dimensional (2-D) analysis for disease datasets from various neuropsychiatric disorders. a–h Results of the 2-D analysis of datasets for schizophrenia (a), ALS (b), Alzheimer’s disease (c), autism (d), Parkinson’s disease (e), bipolar disorder (f), Huntington’s disease (g), and major depressive disorder (h). Each point corresponds to the results of one independent study. Filled points indicate datasets from the postmortem brain or spinal cord (ALS) of patients, and open points indicate those from cultured neural cells from patient iPSCs. i The distribution patterns of the hiM- and hiI-indexes for all diseases analyzed. The extent of the changes in hiM-/hiI-indexes is assessed by the average distance of all datasets for each disease from the origin. Four diseases whose average distances from the origin are over 5.0 are shown as circular sectors, and the others are shown as points. The radii of the circular sectors indicate the average distance of all datasets in each disease from the origin, and the central angles of the circular sectors are the average deviation ± SEM from the unity line. Each point indicates the average distance from the origin and average deviation from unity line
We extended the same analysis to 87 disease datasets from seven other neuropsychiatric diseases (amyotrophic lateral sclerosis (ALS), Alzheimer’s disease (AD), autism spectrum disorder (ASD), Parkinson’s disease (PD), bipolar disorder (BPD), Huntington’s disease (HD), and major depressive disorder (MDD); Supplementary Data 2). The results from each dataset are shown in Fig. 3b–h. The overall distribution patterns of each disease are shown in Fig. 3i. The ALS datasets tended to show higher hiI-index values than hiM-index values, indicating an hiI-index-dominant pattern (Fig. 3b). The AD datasets showed different patterns in the hiM-/hiI-index depending on the type of sample; datasets from the postmortem brains of patients with AD tended to show high values only for the hiM-index, and datasets from patient iPSCs tended to show high values only for the hiI-index (Fig. 3c). Datasets from ASD did not show any dominant patterns in either the hiM-index or the hiI-index (Fig. 3d). Most datasets from patients with PD, BPD, HD, and MDD did not show pronounced values for the hiM-index or the hiI-index (Fig. 3e–h). We applied same analyses on the microarray expression datasets from a recent report by Gandal and colleagues, in which confounding factors are controlled relatively well. Datasets from postmortem brains of patients with schizophrenia and alcohol abuse disorder showed high values for the hiM-index, and those from patients with ASD showed high values for the hiI-index (Supplementary Figure 3).
Thus, the 2-D analysis revealed that some neuropsychiatric diseases have characteristic patterns in the hiM-/hiI-indexes: for example, most datasets from patients with schizophrenia exhibited a higher hiM-index than hiI-index, whereas ALS datasets showed an hiI-index-dominant pattern (Fig. 3i). Meanwhile, different diseases sometimes shared similar changes in the hiM- or hiI-index; for example, some of the schizophrenia, ASD, and AD datasets shared a high hiM-index, and some of the ALS and AD datasets shared a high hiI-index. The other four diseases—PD, BPD, HD, and MDD—did not feature pronounced changes in the hiM-/hiI-indexes, suggesting that these diseases may not share an endophenotype of pseudoimmaturity inducible by neural hyperexcitation. These results raise the possibility that there are patterns of gene expression perturbations that are shared and distinct across these neuropsychiatric disorders.
Genetic and environmental risks induce pseudoimmaturity
Previous studies suggest that many genetic risk factors and environmental factors, such as seizure, hypoxia, and infection, contribute to the development of neuropsychiatric disorders2,49,50. We next applied the 2-D analysis technique to datasets from genetic animal models of disorders and from animals that had experienced risk events.
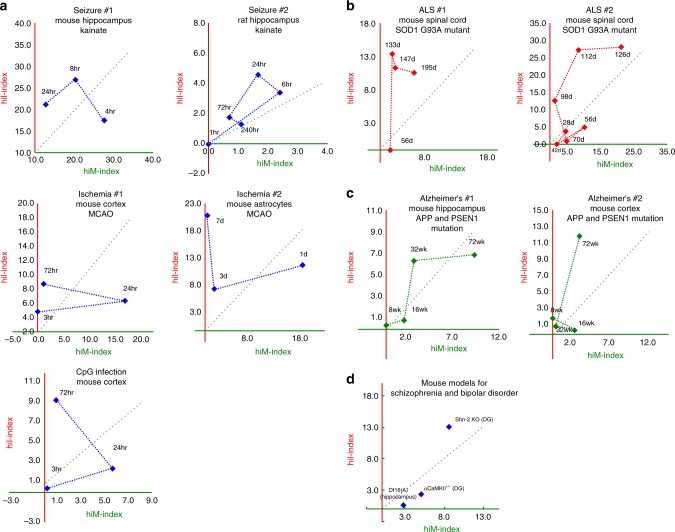
First, we obtained publicly available datasets for mice that had experienced putative risk events for schizophrenia, bipolar disorders, and Alzheimer’s disease, including seizure (#1: GSE49030, #2: GSE4236)51,52, ischemia (#1: GSE32529, #2: GSE35338)53,54, and infection (mimicked by CpG; GSE32529)53,54. All the studies used here included datasets for different timepoints after the risk event; hence, we were able to examine the time course of changes in the hiM- and hiI-indexes to reveal the short- and long-term effects of risk events on the expression patterns of hiM/hiI genes. The results showed that datasets from the hippocampus of mice treated with kainite, a seizure-inducing drug, exhibited time-course changes in the hiM- and hiI-indexes: the hiM-index tended to be dominant in the early stage after seizure induction, and the hiI-index became more dominant in the late stages (Fig. 4a: seizure #1). The results from other datasets on seizure, ischemia, and infection showed time-course pattern changes in the hiM- and hiI-indexes that roughly matched those observed in seizure dataset #1, being relatively hiM-index-dominant in the early stage and then relatively hiI-index-dominant in the later stage (Fig. 4a: seizure, ischemia, and CpG infection). These results indicate that different types of putative risk events for neuropsychiatric disorders induce roughly similar time-course changes in the expression of maturation-related genes induced by neural hyperexcitation.
Fig. 4.
Time-dependent changes in the hiM- and hiI-indexes in animals subjected to various putative risk events for neuropsychiatric disorders and in genetic mouse models of schizophrenia, bipolar disorder, ALS, and Alzheimer’s disease. a Pattern of changes in the hiM- and hiI-indexes in the mouse and rat hippocampus after treatment with kainite (seizure #1 (GSE1831) and seizure #2 (GSE4236)), in mouse cortex and astrocytes after middle cerebral artery occlusion (MCAO; ischemia #1 (GSE32529), ischemia #2 (GSE35338)), and in mouse cortex after CpG infection (GSE32529). b Pattern of changes in the hiM- and hiI-indexes of the spinal cord of an ALS mouse model with the SOD1(G93A) mutation. c Pattern of changes in the hiM- and hiI-indexes of the hippocampus and cortex of an Alzheimer’s disease mouse model with mutations in APP and PSEN1. d hiM- and hiI-indexes in mouse models of schizophrenia and bipolar disorder
Next, we obtained datasets from animal models with a genetic risk of a neurodegenerative disease: mice with transgenic expression of a G93A mutant form of human SOD1, as a model of ALS (#1: GSE46298, #2: GSE18597)55,56; transgenic mice with mutant human amyloid precursor protein (APP) and presenilin1 (PSEN1) genes, which cause familial Alzheimer’s disease (#1: GSE64398, #2: GSE64398)57; and Df16(A) heterozygous mice carrying a chromosome 16 deletion syntenic to human 22q11.2 microdeletions, as a model of schizophrenia (GSE29767)58. We also acquired datasets from Schnurri-2 (Shn-2) knockout mice as a model of schizophrenia12 and intellectual disability14,59,60 and from mice with heterozygous knockout of the α-isoform of calcium/calmodulin-dependent protein kinase II (α-CaMKII+/−)10,13 as a model of bipolar disorder61. These mice show an array of behavioral abnormalities, including locomotor hyperactivity and severe deficits in working memory. The expression profiles of several maturation marker genes in exhibit immature-like patterns in these mice10,12. The threshold current of granule cells to induce spikes was low in these mice, indicating that the granule cells of these mice are highly excitable10,12. We performed 2-D analysis on these datasets and evaluated the changes in the hiM-/hiI-indexes of these model mice. Both datasets from transgenic mice with the SOD1(G93A) mutation exhibited an hiI-index higher than the hiM-index in the later stages of disease progression (Fig. 4b). These hiI-index-dominant patterns were also observed in the results derived from human patients with ALS (Fig. 3b). In the mice with mutant human APP and PSEN1, both the hiM- and hiI-indexes increased in the dataset from the hippocampus, and only the hiI-index increased in the dataset from the cortex during the course of disease progression (Fig. 4c). These patterns are neutral or hiI-index dominant and partially mimic the results from human patients with Alzheimer’s disease (Fig. 3c). The Df16(A) heterozygous mice and α-CaMKII+/− mice showed hiM-index-dominant patterns, similar to results from human patients with schizophrenia (Fig. 4d). Shn-2 KO mice showed high values for both the hiM- and hiI-indexes (Fig. 4d). Thus, the results from the 2-D analysis of animal models are to some extent consistent with the results from human patients, indicating that these model mice have patterns of pseudoimmaturity induced by neural hyperexcitation that are similar to those in human patients. Potential confounding factors, such as types of samples, brain areas, and age, were less numerous in the animal-model analyses than in the analyses of samples from human patients, establishing the causal links between experimental manipulations corresponding to disease conditions and pseudoimmaturity inducible by hyperexcitation.
Discussion
In this study, we demonstrated that neural hyperexcitation induces changes in the pattern of gene expression in the DG that are similar to the patterns in the immature hippocampus of typically developing human fetuses. From the pool of genes, we identified two groups of genes, and found that these are shared by multiple neuropsychiatric disorders, such as schizophrenia, Alzheimer disorders, and ALS.
Many of the datasets from patients with schizophrenia and from the postmortem brains of patients with Alzheimer’s disease exhibited hiM-index-dominant pattern changes. The hiM genes, include genes encoding a GABA receptor, voltage-dependent calcium channel, glutamate receptor, and voltage-dependent sodium channel (Supplementary Data 1). These genes have been implicated in the pathological changes in the brains of patients with schizophrenia and Alzheimer’s disease62–65. Thus, many of the synaptic genes that changed in the brains of patients with schizophrenia or Alzheimer’s disease could be genes whose expression increases during maturation and decreases with neural hyperexcitation. Although reductions in the expression of some synaptic genes in these disorders are well documented, to our knowledge, our results are the first to raise the possibility that neuronal hyperexcitation may also induce reductions in such synaptic molecules. Most of the datasets from patients with ALS and Alzheimer’s disease exhibited hiI-index-dominant patterns. The hiI genes, include genes encoding DNA methyltransferase, cyclin D1, cyclin-dependent kinase 1, integrin beta 3 binding protein, and tumor protein p53 (Supplementary Data 1). These genes are known to be important in chromosomal modification and DNA repair, and abnormal functioning of these systems has been observed in patients with ALS and Alzheimer’s disease66–70. Thus, some of the genes that are considered important in the development of these disorders are immaturity-related genes, whose expression decreases during maturation and can be increased by neural hyperexcitation. As for the datasets from patients with PD, BPD, HD, and MDD, most of them did not overlap with either hiM or hiI genes, suggesting that there might not be major pathological changes in transcriptomic pseudoimmaturity inducible by neural hyperexcitation in the datasets of these four diseases. Thus, we were able to characterize the gene expression patterns in disease datasets of each disease category using the hiI- and hiM-indexes.
Our study has some limitations. First, the number of available datasets was limited. All the datasets except the one for mouse development were obtained from the BaseSpace Correlation Engine. On this platform, vast numbers (over 21,000) of complex biological and clinical datasets are available. Although we used all the gene expression dataset hits from our keyword query to avoid sampling bias, the number of datasets was still small, from 8 datasets for ALS to 16 datasets for schizophrenia. Further accumulation of the studies will improve the reliability of our results. Another limitation is that the datasets used in this study are from different types of samples, including various areas within the central nervous system, such as the hippocampus, prefrontal cortex, striatum, and spinal cord. The gene expression abnormalities in patients could differ depending on the brain area48. We also used datasets from cultured neurons differentiated from the iPSCs of human subjects, and it is controversial whether the pattern of gene expression in these neurons is comparable to that of neurons in the patients’ brains71,72. It is also possible that the altered gene expression in the postmortem brains is due to the effects of medication rather than pathological changes from the disease itself73. We compared the expression patterns of the genes derived from rodents and humans. There could be critical differences among different species, which might have caused the analysis to miss important molecular regulatory elements shared among those neuropsychiatric disorders. Other conditions that were not controlled in this study and can be potential confounding factors include the age of subjects at death, the storage conditions of the samples, the genetic background of animals, and the animal housing conditions. For these reasons, we need to be careful in interpreting the results of the analyses. It should be noted, however, that despite the variety of sample types used, we were able to identify some shared and distinct patterns of gene expression, which is mostly due to the advantages of utilizing the Running Fisher test; this test, using a rank-based nonparametric algorithm, can evaluate similarity between datasets from different species or organs44. Moreover, we applied 2-D analysis to the animal models, in which potential confounding factors such as species, sample types, and brain areas are well controlled. The results from these models were comparable to those from human samples, supporting the idea that pseudoimmaturity inducible by hyperexcitation is a feature shared by multiple neuropsychiatric disorders. It should be noted, however, that a gene expression pattern resembling those induced by neural hyperexcitation may not be induced by neural hyperexcitation per se but by other factors such as inflammation, hypoxia, and infection. Another potential advantage of using the Running Fisher test is that this method is applicable to the dataset of gene expression profiles from an individual patient or control. In the future, it might be interesting to apply this method to the characterization and classification of gene expression patterns in the brains of individual patients and in neurons derived from patients’ iPSCs.
Recent attempts such as the RDoC initiative have tried to reclassify psychiatric disorders according to biological domains (e.g., genes, neural circuits, and behavior)7. While Gandal et al.8 conducted nonbiased coexpression analyses, this study utilized gene groups derived from the findings based on the studies of animal models of neuropsychiatric disorders. Characterization by these gene groups enabled us to extract novel biological features of some neuropsychiatric disorders that are related to pseudoimmaturity inducible by neural hyperexcitation. Detecting such domains that extract the biological features of each neuropsychiatric disorder will move this diagnostic framework forward, from criteria based on signs and symptoms to those including biological dimensions.
In conclusion, the biological domain of pseudoimmaturity inducible by neural hyperexcitation is a common endophenotype among several neuropsychiatric disorders. Future studies are needed to find translational indices that correspond to these features and can be applicable to human patients for better diagnosis of these neuropsychiatric disorders. Our findings here may promote the development of biomarkers, leading to a better diagnosis of neuropsychiatric disorders.
Methods
Microarray experiments to examine mouse DG development
Wild-type mouse DGs were sampled at postnatal days 8, 11, 14, 17, 21, 25, 29 (C57BL/6J × BALB/cA background; male, n = 5)74, and microarray experiments were performed with Mouse Genome 430 2.0 Array (Affymetrix, Santa Clara, CA) as previously described10. RNA was isolated by using the TRIzol method (Invitrogen, Carlsbad, CA) from the hippocampus of mice, followed by purification, using RNeasy columns (Qiagen, Valencia, CA). Double-stranded cDNA was synthesized from the total RNA, and invitro transcription reaction was then performed on biotin-labeled RNA that was made from the cDNA. Labeled RNA was hybridized with Mouse Genome 430 2.0 Array (Affymetrix, Santa Clara, CA) containing 45,101 probe sets, and washed according to the manufacturer’s recommendations. The hybridized probe array was then stained with streptavidin-conjugated phycoerythrin, and each GeneChip was scanned by an Affymetrix GeneChip Scanner 3000 (GCS3000). GeneChip analysis was performed with Microarray Analysis Suite version 5.0. All of the gene represented on the GeneChip were globally normalized. Genes with a P value < 0.05 (without correction for multiple testing) and an absolute fold change >1.2 were included in the differentially expressed gene datasets. The microarray data, including those used in this study, were deposited in the Gene Expression Omnibus (GEO) database under accession number GSE113727. We also obtained a dataset for 33-week-old wild-type mice, which we previously reported (C57BL/6J × BALB/cA background) (GSE42778)12. We integrated these two datasets into one to construct the dataset for the development of the wild-type mouse DG used in this study (P8 versus adult, fold change >1.2, P < 0.05). All animal experiments were approved by the Institutional Animal Care and Use Committee of Fujita Health University, based on the Law for the Humane Treatment and Management of Animals and the Standards Relating to the Care and Management of Laboratory Animals and Relief of Pain. Every effort was made to minimize the number of animals used.
Data collection and processing
Except for the mouse DG developmental dataset mentioned above, the 99 gene expression datasets used in this study were obtained from publicly available databases (listed in Supplementary Data 2). We used all the hits from our keyword query for gene-expression studies to avoid sampling bias. If a single study included several different datasets, we chose the one that we considered the most comprehensive (e.g., if the study includes datasets from male, female, and all subjects, we used datasets from all subjects). All gene-expression datasets were analyzed with the BaseSpace Correlation Engine (formerly known as NextBio) (https://japan.ussc.informatics.illumina.com/c/nextbio.nb; Illumina, Cupertino, CA), a database of biomedical experiments. BaseSpace is a repository of analyzed gene-expression datasets that allows researchers to search expression profiles and other results44. The datasets registered in BaseSpace undergo several preprocessing, quality control, and organization stages. Quality control ensures the integrity of the samples and datasets and includes evaluations of prenormalization and postnormalization boxplots, missing value counts, and P value histograms (after statistical testing) with false-discovery rate analysis to establish whether the number of significantly altered genes is larger than that expected by chance.
Genes with a P value < 0.05 (without correction for multiple testing) and an absolute fold change >1.2 were included in the differentially expressed gene datasets. This sensitivity threshold is typically the lowest used with commercial microarray platforms and the default criterion in BaseSpace analyses44. Correction for multiple testing was omitted to minimize false negatives at this stage. All data from the Affymetrix GeneChip series were downloaded from the NCBI GEO database. Affymetrix Expression Console software (specifically, the robust multiarray average algorithm) was used to preprocess the data.
We used the expression values (on a log base 2 scale) to calculate the fold changes and P values between two conditions (infants–adults and patients–healthy controls). To determine the fold changes, we began with the expression values of the probes/genes in the test data sets and divided them by those of the control data sets. If the fold change was <1.0, these values were converted to the negative reciprocal, or −1/(fold change). Genes with an absolute fold change >1.2 and a t test P value < 0.05 were imported into BaseSpace Correlation Engine according to the instructions provided by the manufacturer. The rank order of these genes was determined by their absolute fold change. All statistical analyses were performed in BaseSpace, and the similarities between any two datasets were evaluated as overlap P values using the Running Fisher algorithm44. The Bonferroni correction was used to adjust the significance level according to the number of datasets pairs44.
Supplementary information
Description of Additional Supplementary Files
Acknowledgments
We thank Wakako Hasegawa, Yumiko Mobayashi, Misako Murai, Tamaki Murakami, Miwa Takeuchi, Chikako Ozeki, Yoko Kagami, Harumi Mitsuya, Aki Miyakawa, and other members of the Miyakawa laboratory for their support. This work was supported by the Grant-in-Aid for Scientific Research on Innovative Areas ‘‘Unraveling the microendophenotypes of psychiatric disorders at the molecular, cellular and circuit levels’’ (JP25116526, JP15H01297) and ‘‘Dynamic regulation of brain function by Scrap & Build system’’ (JP16H06462) from the Ministry of Education, Culture, Sports, Science and Technology; and the Strategic Research Program for Brain Sciences from Japan Agency for Medical Research and Development, AMED.
Author contribution
T. Murano contributed with the methodology, data acquisition, and data analysis. Interpretation of the data was performed by T. Murano and T. Miyakawa. The manuscript was written by T. Murano and T. Miyakawa and revised by T. Miyakawa, H.H., K.T. and M.M.
Data availability
The microarray and RNAseq data that support the findings of this study have been available in the BaseSpace Correlation Engine (https://japan.ussc.informatics.illumina.com/c/nextbio.nb; Illumina, Cupertino, CA) and Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/). All detailed information, including accession numbers, is listed in Supplementary Data 2.
Competing interests
Tsuyoshi Miyakawa received research grants from Astellas Pharma Inc. and; Katsunori Tajinda and Mitsuyuki Matsumoto are employees of Astellas Pharma Inc. The remaining authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
3/4/2019
In the original published version of this article, Supplementary Data 1 reported gene expression fold changes incorrectly. The original supplementary file reported all fold changes as raw values, whereas the main text explains that genes with fold changes <1 were converted to the negative reciprocal (−1/(fold change)). This error has been corrected in the Supplementary Data 1 file.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s42003-018-0277-2.
References
- 1.Cross-national comparisons of the prevalences and correlates of mental disorders. WHO International Consortium in Psychiatric Epidemiology. Bull. World Health Organ. 2000;78:413–426. [PMC free article] [PubMed] [Google Scholar]
- 2.Cardno AG, Owen MJ. Genetic relationships between schizophrenia, bipolar disorder, and schizoaffective disorder. Schizophr. Bull. 2014;40:504–515. doi: 10.1093/schbul/sbu016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall J, Trent S, Thomas KL, O’Donovan MC, Owen MJ. Genetic risk for schizophrenia: convergence on synaptic pathways involved in plasticity. Biol. Psychiatry. 2015;77:52–58. doi: 10.1016/j.biopsych.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Forero DA, et al. A network of synaptic genes associated with schizophrenia and bipolar disorder. Schizophr. Res. 2016;172:68–74. doi: 10.1016/j.schres.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 5.Douaud G, et al. A common brain network links development, aging, and vulnerability to disease. PNAS. 2014;111:17648–17653. doi: 10.1073/pnas.1410378111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Argyelan M, et al. Resting-state fMRI connectivity impairment in schizophrenia and bipolar disorder. Schizophr. Bull. 2014;40:100–110. doi: 10.1093/schbul/sbt092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Insel TR, Cuthbert BN. Brain disorders? Precisely. Science. 2015;348:499–500. doi: 10.1126/science.aab2358. [DOI] [PubMed] [Google Scholar]
- 8.Gandal MJ, et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018;359:693–697. doi: 10.1126/science.aad6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagihara, H., Takao, K., Walton, N. M., Matsumoto, M. & Miyakawa, T. Immature dentate gyrus: an endophenotype of neuropsychiatric disorders. Neural Plast.2013, Article ID 318596 (2013). [DOI] [PMC free article] [PubMed]
- 10.Yamasaki N, et al. Alpha-CaMKII deficiency causes immature dentate gyrus, a novel candidate endophenotype of psychiatric disorders. Mol. Brain. 2008;1:6. doi: 10.1186/1756-6606-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohira K, et al. Synaptosomal-associated protein 25 mutation induces immaturity of the dentate granule cells of adult mice. Mol. Brain. 2013;6:12. doi: 10.1186/1756-6606-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takao K, et al. Deficiency of Schnurri-2, an MHC enhancer binding protein, induces mild chronic inflammation in the brain and confers molecular, neuronal, and behavioral phenotypes related to schizophrenia. Neuropsychopharmacology. 2013;38:1409–1425. doi: 10.1038/npp.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagihara H, et al. Circadian gene circuitry predicts hyperactive behavior in a mood disorder mouse model. Cell Rep. 2016;14:2784–2796. doi: 10.1016/j.celrep.2016.02.067. [DOI] [PubMed] [Google Scholar]
- 14.Nakao A, et al. Immature morphological properties in subcellular-scale structures in the dentate gyrus of Schnurri-2 knockout mice: a model for schizophrenia and intellectual disability. Mol. Brain. 2017;10:60. doi: 10.1186/s13041-017-0339-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagihara H, Fujita M, Umemori J, Hashimoto M, Miyakawa T. Immature-like molecular expression patterns in the hippocampus of a mouse model of dementia with Lewy body-linked mutant β-synuclein. Mol. Brain. 2018;11:38. doi: 10.1186/s13041-018-0378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin R, et al. The immature dentate gyrus represents a shared phenotype of mouse models of epilepsy and psychiatric disease. Bipolar Disord. 2013;15:405–421. doi: 10.1111/bdi.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.You JC, et al. Epigenetic suppression of hippocampal calbindin-D28k by ΔFosB drives seizure-related cognitive deficits. Nat. Med. 2017;23:1377. doi: 10.1038/nm.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maglóczky Z, Halász P, Vajda J, Czirják S, Freund TF. Loss of calbindin-D28K immunoreactivity from dentate granule cells in human temporal lobe epilepsy. Neuroscience. 1997;76:377–385. doi: 10.1016/S0306-4522(96)00440-X. [DOI] [PubMed] [Google Scholar]
- 19.Karádi K, et al. Correlation between calbindin expression in granule cells of the resected hippocampal dentate gyrus and verbal memory in temporal lobe epilepsy. Epilepsy Behav. 2012;25:110–119. doi: 10.1016/j.yebeh.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Zhang ZJ, Reynolds GP. A selective decrease in the relative density of parvalbumin-immunoreactive neurons in the hippocampus in schizophrenia. Schizophr. Res. 2002;55:1–10. doi: 10.1016/S0920-9964(01)00188-8. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto T, et al. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J. Neurosci. 2003;23:6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fung SJ, et al. Expression of interneuron markers in the dorsolateral prefrontal cortex of the developing human and in schizophrenia. Am. J. Psychiatry. 2010;167:1479–1488. doi: 10.1176/appi.ajp.2010.09060784. [DOI] [PubMed] [Google Scholar]
- 23.Pantazopoulos H, Woo TUW, Lim MP, Lange N, Berretta S. Extracellular matrix-glial abnormalities in the amygdala and entorhinal cortex of subjects diagnosed with schizophrenia. Arch. Gen. Psychiatry. 2010;67:155–166. doi: 10.1001/archgenpsychiatry.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berretta S. Extracellular matrix abnormalities in schizophrenia. Neuropharmacology. 2012;62:1584–1597. doi: 10.1016/j.neuropharm.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagihara H, Ohira K, Takao K, Miyakawa T. Transcriptomic evidence for immaturity of the prefrontal cortex in patients with schizophrenia. Mol. Brain. 2014;7:41. doi: 10.1186/1756-6606-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gandal MJ, Nesbitt AM, McCurdy RM, Alter MD. Measuring the maturity of the fast-spiking interneuron transcriptional program in autism, schizophrenia, and bipolar disorder. PLoS ONE. 2012;7:e41215. doi: 10.1371/journal.pone.0041215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mellios N, et al. Molecular determinants of dysregulated GABAergic gene expression in the prefrontal cortex of subjects with schizophrenia. Biol. Psychiatry. 2009;65:1006–1014. doi: 10.1016/j.biopsych.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Hashimoto T, et al. Alterations in GABA-related transcriptome in the dorsolateral prefrontal cortex of subjects with schizophrenia. Mol. Psychiatry. 2008;13:147–161. doi: 10.1038/sj.mp.4002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murano T, Koshimizu H, Hagihara H, Miyakawa T. Transcriptomic immaturity of the hippocampus and prefrontal cortex in patients with alcoholism. Sci. Rep. 2017;7:srep44531. doi: 10.1038/srep44531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kobayashi K, et al. Reversal of hippocampal neuronal maturation by serotonergic antidepressants. PNAS. 2010;107:8434–8439. doi: 10.1073/pnas.0912690107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohira K, Takeuchi R, Iwanaga T, Miyakawa T. Chronic fluoxetine treatment reduces parvalbumin expression and perineuronal netsin gamma-aminobutyric acidergic interneurons of the frontal cortex in adultmice. Mol. Brain. 2013;6:43. doi: 10.1186/1756-6606-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imoto Y, Segi-Nishida E, Suzuki H, Kobayashi K. Rapid and stable changes in maturation-related phenotypes of the adult hippocampal neurons by electroconvulsive treatment. Mol. Brain. 2017;10:8. doi: 10.1186/s13041-017-0288-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang YT, et al. Bidirectional relation between schizophrenia and epilepsy: a population-based retrospective cohort study. Epilepsia. 2011;52:2036–2042. doi: 10.1111/j.1528-1167.2011.03268.x. [DOI] [PubMed] [Google Scholar]
- 34.Hyde TM, Weinberger DR. Seizures and schizophrenia. Schizophr. Bull. 1997;23:611–622. doi: 10.1093/schbul/23.4.611. [DOI] [PubMed] [Google Scholar]
- 35.Cascella NG, Schretlen DJ, Sawa A. Schizophrenia and epilepsy: is there a shared susceptibility? Neurosci. Res. 2009;63:227. doi: 10.1016/j.neures.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coyle JT. Glutamate and schizophrenia: beyond the dopamine hypothesis. Cell. Mol. Neurobiol. 2006;26:363–382. doi: 10.1007/s10571-006-9062-8. [DOI] [PubMed] [Google Scholar]
- 37.Mouri A, Noda Y, Enomoto T, Nabeshima T. Phencyclidine animal models of schizophrenia: Approaches from abnormality of glutamatergic neurotransmission and neurodevelopment. Neurochem. Int. 2007;51:173–184. doi: 10.1016/j.neuint.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 38.Kehrer, C., Maziashvili, N., Dugladze, T. & Gloveli, T. Altered excitatory-inhibitory balance in the NMDA-hypofunction model of schizophrenia. Front Mol Neurosci1, 6 (2008). [DOI] [PMC free article] [PubMed]
- 39.O’Donnell C, Gonçalves JT, Portera-Cailliau C, Sejnowski TJ. Beyond excitation/inhibition imbalance in multidimensional models of neural circuit changes in brain disorders. eLife. 2017;6:e26724. doi: 10.7554/eLife.26724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mertens J, et al. Differential responses to lithium in hyperexcitable neurons from patients with bipolar disorder. Nature. 2015;527:95–99. doi: 10.1038/nature15526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vossel KA, et al. Incidence and impact of subclinical epileptiform activity in Alzheimer’s disease. Ann. Neurol. 2016;80:858–870. doi: 10.1002/ana.24794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lam AD, et al. Silent hippocampal seizures and spikes identified by foramen ovale electrodes in Alzheimer’s disease. Nat. Med. 2017;23:678–680. doi: 10.1038/nm.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vossel KA, Tartaglia MC, Nygaard HB, Zeman AZ, Miller BL. Epileptic activity in Alzheimer’s disease: causes and clinical relevance. Lancet Neurol. 2017;16:311–322. doi: 10.1016/S1474-4422(17)30044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kupershmidt I, et al. Ontology-based meta-analysis of global collections of high-throughput public data. PLoS ONE. 2010;5:e13066. doi: 10.1371/journal.pone.0013066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takao K, Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. PNAS. 2015;112:1167–1172. doi: 10.1073/pnas.1401965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryan SD, et al. Isogenic human iPSC Parkinson’s model shows nitrosative stress-induced dysfunction in MEF2-PGC1α transcription. Cell. 2013;155:1351–1364. doi: 10.1016/j.cell.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dingledine, R. et al. Transcriptional profile of hippocampal dentate granule cells in four rat epilepsy models. Sci. Data.4, 170061 (2017). [DOI] [PMC free article] [PubMed]
- 48.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Os J, Kenis G, Rutten BPF. The environment and schizophrenia. Nature. 2010;468:203–212. doi: 10.1038/nature09563. [DOI] [PubMed] [Google Scholar]
- 50.DeMichele-Sweet MaA, et al. Genetic risk for schizophrenia and psychosis in Alzheimer disease. Mol. Psychiatry. 2017;23:963–972. doi: 10.1038/mp.2017.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson DN, et al. Microarray analysis of postictal transcriptional regulation of neuropeptides. J. Mol. Neurosci. 2005;25:285. doi: 10.1385/JMN:25:3:285. [DOI] [PubMed] [Google Scholar]
- 52.Hermey G, et al. Genome-wide profiling of the activity-dependent hippocampal transcriptome. PLoS ONE. 2013;8:e76903. doi: 10.1371/journal.pone.0076903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vartanian KB, et al. LPS preconditioning redirects TLR signaling following stroke: TRIF-IRF3 plays a seminal role in mediating tolerance to ischemic injury. J. Neuroinflamm. 2011;8:140. doi: 10.1186/1742-2094-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stevens SL, et al. Multiple preconditioning paradigms converge on interferon regulatory factor-dependent signaling to promote tolerance to ischemic brain injury. J. Neurosci. 2011;31:8456–8463. doi: 10.1523/JNEUROSCI.0821-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nardo G, et al. Transcriptomic indices of fast and slow disease progression in two mouse models of amyotrophic lateral sclerosis. Brain. 2013;136:3305–3332. doi: 10.1093/brain/awt250. [DOI] [PubMed] [Google Scholar]
- 56.Lerman BJ, et al. Deletion of galectin-3 exacerbates microglial activation and accelerates disease progression and demise in a SOD1 G93A mouse model of amyotrophic lateral sclerosis. Brain Behav. 2012;2:563–575. doi: 10.1002/brb3.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matarin M, et al. A genome-wide gene-expression analysis and database in transgenic mice during development of amyloid or taupathology. Cell Rep. 2015;10:633–644. doi: 10.1016/j.celrep.2014.12.041. [DOI] [PubMed] [Google Scholar]
- 58.Xu B, Hsu PK, Stark KL, Karayiorgou M, Gogos JA. Derepression of a neuronal inhibitor due to miRNA dysregulation in a schizophrenia-related microdeletion. Cell. 2013;152:262–275. doi: 10.1016/j.cell.2012.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Srivastava S, et al. Loss-of-function variants in HIVEP2 are a cause of intellectual disability. Eur. J. Hum. Genet. 2016;24:556–561. doi: 10.1038/ejhg.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steinfeld H, et al. Mutations in HIVEP2 are associated with developmental delay, intellectual disability, and dysmorphic features. Neurogenetics. 2016;17:159–164. doi: 10.1007/s10048-016-0479-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Butler G, et al. Limbic epilepsy in transgenic mice carrying a Ca2+/calmodulin-dependent kinase II alpha-subunit mutation. PNAS. 1995;92:6852–6855. doi: 10.1073/pnas.92.15.6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harrison PJ. The hippocampus in schizophrenia: a review of the neuropathological evidence and its pathophysiological implications. Psychopharmacology. 2004;174:151–162. doi: 10.1007/s00213-003-1761-y. [DOI] [PubMed] [Google Scholar]
- 63.Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience. 2013;251:90–107. doi: 10.1016/j.neuroscience.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fatemi, S. H., Folsom, T. D. & Thuras, P. D. GABAA and GABAB receptor dysregulation in superior frontal cortex of subjects with schizophrenia and bipolar disorder. Synapse71 (2017). [DOI] [PubMed]
- 65.Lisman JE, et al. Circuit-based framework for understanding neurotransmitter and risk gene interactions in schizophrenia. Trends Neurosci. 2008;31:234–242. doi: 10.1016/j.tins.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chestnut BA, et al. Epigenetic regulation of motor neuron cell death through DNA methylation. J. Neurosci. 2011;31:16619–16636. doi: 10.1523/JNEUROSCI.1639-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martin LJ, Wong M. Aberrant regulation of DNA methylation in amyotrophic lateral sclerosis: a new target of disease mechanisms. Neurotherapeutics. 2013;10:722–733. doi: 10.1007/s13311-013-0205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen MD, et al. Cell cycle regulators in the neuronal death pathway of amyotrophic lateral sclerosis caused by mutant superoxide dismutase 1. J. Neurosci. 2003;23:2131–2140. doi: 10.1523/JNEUROSCI.23-06-02131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dorszewska J, et al. Mutations of TP53 gene and oxidative stress in Alzheimer’s disease patients. Adv. Alzheimer’s Dis. 2014;3:24. doi: 10.4236/aad.2014.31004. [DOI] [Google Scholar]
- 70.Moskalev AA, et al. Gadd45 proteins: relevance to aging, longevity and age-related pathologies. Ageing Res. Rev. 2012;11:51–66. doi: 10.1016/j.arr.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brennand KJ, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wen Z, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515:414–418. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chan MK, et al. Evidence for disease and antipsychotic medication effects in post-mortem brain from schizophrenia patients. Mol. Psychiatry. 2011;16:1189–1202. doi: 10.1038/mp.2010.100. [DOI] [PubMed] [Google Scholar]
- 74.Hagihara H, Toyama K, Yamasaki N, Miyakawa T. Dissection of hippocampal dentate gyrus from adult mouse. J. Vis. Exp. 2009;33:1543. doi: 10.3791/1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The microarray and RNAseq data that support the findings of this study have been available in the BaseSpace Correlation Engine (https://japan.ussc.informatics.illumina.com/c/nextbio.nb; Illumina, Cupertino, CA) and Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/). All detailed information, including accession numbers, is listed in Supplementary Data 2.