Abstract
Since the most recent outbreak, the Ebola virus (EBOV) epidemic remains one of the world’s public health and safety concerns. EBOV is a negative-sense RNA virus that can infect humans and non-human primates, and causes hemorrhagic fever. It has been proposed that the T-cell immunoglobulin and mucin domain (TIM) family proteins act as cell surface receptors for EBOV, and that the interaction between TIM and phosphatidylserine (PS) on the surface of EBOV mediates the EBOV–host cell attachment. Despite these initial findings, the biophysical properties of the TIM-EBOV interaction, such as the mechanical strength of the TIM-PS bond that allows the virus-cell interaction to resist external mechanical perturbations, have not yet been characterized. This study utilizes single-molecule force spectroscopy to quantify the specific interaction forces between TIM-1 or TIM-4 and the following binding partners: PS, EBOV virus-like particle, and EBOV glycoprotein/vesicular stomatitis virus pseudovirion. Depending on the loading rates, the unbinding forces between TIM and ligands ranged from 40 to 100 pN, suggesting that TIM-EBOV interactions are mechanically comparable to previously reported adhesion molecule–ligand interactions. The TIM-4–PS interaction is more resistant to mechanical force than the TIM-1–PS interaction. We have developed a simple model for virus–host cell interaction that is driven by its adhesion to cell surface receptors and resisted by membrane bending (or tension). Our model identifies critical dimensionless parameters representing the ratio of deformation and adhesion energies, showing how single-molecule adhesion measurements relate quantitatively to the mechanics of virus adhesion to the cell.
Introduction
Ebola virus (EBOV) disease is a severe and often fatal illness in humans. First identified in 1976, and with a fatality rate of 50 to 70%, the disease has caused about 15,000 deaths1–4. EBOV is a filamentous, enveloped, non-segmented, negative-sense RNA virus that belongs to the virus family Filoviridae. Filoviruses, such as EBOV, have an extensive tissue tropism. Dendritic cells and macrophages are considered to be their first targets. Subsequent rounds of infection follow in a variety of cell types including epithelial cells such as hepatocytes, stromal cells and to a lesser degree endothelial cells5,6. There are five closely related species: Ebola virus (EBOV, formerly Zaire ebolavirus), Sudan ebolavirus (SEBOV), Taï Forest ebolavirus (TAFV), Reston ebolavirus (REBOV), and the proposed most recent addition, Bundibugyo ebolavirus (BDBV)7.
The EBOV genome encodes seven structural proteins, nucleoprotein (NP), polymerase cofactor (VP35), matrix protein (VP40), glycoprotein (GP), replication-transcription protein (VP30), minor matrix protein (VP24), RNA-dependent RNA polymerase (L) and two secreted non-structural glycoprotein (sGP and ssGP)5. In the first step of EBOV lifecycle, viral attachment through interaction between cellular molecules is followed by endocytosis, including macropinocytosis8. Subsequent trafficking of the virion through the endosomal compartment to the late endosomal/lysosomal compartment results in viral-endosomal membrane fusion and release of the viral ribonucleoprotein complex into the cytoplasm. Transcription of the negative-sense viral RNA genome by the viral polymerase complex yields mRNAs that are translated by cellular ribosomes. Upon replication, viral RNAs and structural proteins such as VP40 and GP are assembled at the plasma membrane into enveloped virus particles that bud from the host cell’s surface5,9,10, thus repeating the cycle and spreading the virus.
T-cell immunoglobulin mucin domain 1 (TIM-1) is a type 1 transmembrane glycoprotein and a member of the TIM family11. The TIM proteins are phosphatidylserine (PS) receptors, binding to PS on the surface of apoptotic bodies and clearing these dead cells from circulation. TIM-1 has also been recently recognized to enhance entry of an expansive range of viruses, including members of the picornavirus, filovirus (such as EBOV)12, flavivirus, alphavirus, arenavirus, and baculovirus families13–15. In addition to TIM-1, TIM-4, another TIM family member, has been shown to augment EBOV entry comparably to TIM-116.
For enveloped viruses, where the capsid is surrounded by a lipid bilayer that contains the viral proteins, this enhancement is believed to occur through TIM binding to PS on the viral envelope. By hijacking the cellular mechanisms utilized in the uptake of apoptotic bodies mediated by TIM, EBOV is internalized into the host cell’s endosomes. EBOV internalization by TIM-1 is found to be solely PS-dependent, and does not require the presence of the viral surface glycoprotein11,17. This mechanism, known as apoptotic mimicry was first described for the vaccinia virus18. Consequently, TIM-1, TIM-4 as well as other PS receptor complexes such as Gas6/Axl, were identified as cellular proteins engaged in this process19,20. This class of viral receptors is known as PS-mediated virus entry-enhancing receptors (PVEERs)20.
Although TIM-1 and TIM-4 have been characterized as the PVEER for EBOV, little is known about the biomechanical properties of the TIM-1/-4 – host cell interaction that help to initiate EBOV internalization. In particular, the mechanical strength of individual interactions between TIM-1/-4 and EBOV, and how the mechanical interaction collectively drives virus adhesion, remain unclear. In this work, using atomic force microscopy (AFM)-based single-molecule force spectroscopy, a method where a single bond rupture between two molecules can be measured directly, we have quantified the mechanical strengths between TIM-1 or TIM-4 with the following interacting binding partners: PS, EBOV virus-like particle (VLP) and EBOV GP/vesicular stomatitis virus pseudovirion. As AFM can measure forces in the pN range, it is possible to investigate inter-molecular forces. This allows for even the weak interactions between tip-bound ligands and surface-bound receptor molecules to be quantified in terms of their affinities and rate constants21. Furthermore, AFM has been previously adopted to study virus-surface and virus-host cell interactions22–24. In the present study, we have demonstrated that the TIM-4–PS complex is more resistant to mechanical force than TIM-1–PS.
In order to relate the single-molecule cell receptor–virus particle binding to the behavior of the entire virus with the cell membrane, it is crucial to understand the mechanics of viral particle adhesion and engulfment. A number of approaches have been followed, including continuum, coarse-grained models, and even all atoms models25–28. Larger scale models such as triangulated membrane models and other continuum models can be used to study overall membrane adhesion without the high computational cost29,30. In this work, we wish to utilize on and off rates from single-molecule experiments to develop a continuum model of the virus-membrane adhesion process. Previously Chou et al.31 developed a model that is on the kinetics of virus binding and fusion, in particular on the competition between fusion and endocytosis. This discrete model was written in terms of on and off rates for individual processes that can be related to the adhesion energies that we will use in our model. However, the model does not explicitly include the effects of membrane stiffness and tension, which we show here to be critically important. Here, we have developed a simple biomechanical model to describe how the adhesive interaction between TIM and the viral surface drives EBOV adhesion to the host cell via a process involving membrane bending (and/or tension). A similar framework has been employed previously to study adhesion between nanoparticles and membranes32–34, and between vesicles and the plasma membrane35.
Results
In this study, we characterized the mechanical strength that enables the EBOV-host cell pair to resist the forces of microenvironment. Because EBOV is classified as a BSL-4 pathogen, we elected to use an appropriate model system: virus-like particles (VLPs), which are non-pathogenic reagents that can be studied in BSL-2 labs. VLPs are prepared by over-expression of EBOV VP40 protein in mammalian cells. Expression of solely VP40 is sufficient to assemble and form VLPs that closely resemble the size and shape of EBOV36. Co-expression of VP40 with EBOV glycoprotein (GP) enhances the production of VLPs with glycoprotein present on their surfaces, making it an ideal model system to study EBOV entry37. The VLPs used in this study consisted of green fluorescent protein (GFP)-tagged VP40 and GP, derived from EBOV15. These VLPs have been shown to interact with and be internalized by host cells in a similar fashion as the native EBOV12,15. In addition to the EBOV VLP, we used an established pseudovirus model for EBOV. This pseudovirion was derived from a recombinant vesicular stomatitis virus encoding EBOV GP (EBOV GP/rVSV), and has been shown to accurately recapitulate the entry steps of wildtype EBOV12,38. A similar EBOV GP/VSV is currently used as a Ebola vaccine and undergoing phase 1 trials in Africa and Europe39.
For the AFM studies, PS-Polyethylene Glycol (PEG), VLP, or EBOV GP/VSV was covalently attached to the AFM cantilever. Immobilization of VLP was confirmed by fluorescence microscopic images of GFP on the AFM cantilever (Fig. 1C insert). Soluble recombinant TIM-1 or TIM-4 was covalently attached to a silanized cover glass (Fig. 1A).
Figure 1.
Schematic of the experimental system. (A) The AFM cantilever is functionalized with the PS, VLP, or EBOV GP/VSV. TIM-1 or TIM-4 is immobilized on the opposing glass surface. (B) The upper panel shows three sample AFM pulling traces of the TIM-4–PS interaction. The first (upper) trace involves no interaction. The second (middle) trace shows the rupture force of the TIM-4–PS complex. The lower trace shows a typical non-specific interaction. Fu is the unbinding force. ks is the system spring constant and was derived from the slope of the force-displacement trace. The cantilever retraction rate of the measurements was 1.5 μm/s. The lower panel illustrates the four stages of stretching and rupturing a single ligand-receptor complex using AFM. (C) A schematic diagram of the live-cell AFM assay. Insert: micrograph of a VLP -functionalized cantilever showing GFP fluorescence. Bar is 20 µm.
All single-molecule force measurements were conducted using a custom-built AFM designed for operation in the force spectroscopy mode40–42. Using a piezoelectric translator, the functionalized cantilever was lowered onto a TIM-functionalized surface to allow binding between the TIM and PS (or other ligands) to occur. After a brief contact, the cantilever was retracted from the surface. Any binding interaction between TIM and ligand would lead to an adhesive pull-off force determined from the deflection of the cantilever via a position-sensitive two-segment photodiode. (Fig. 1A,B, lower panel).
Figure 1B shows three typical pulling traces. The first (upper) trace represents a majority (65–70%) of all the pulling curves, showing no interaction (i.e., no adhesive force) between the AFM tip and sample surface. The second (middle) trace, representing approximately 30% of the pulling curves in our single-molecule assay, shows the unbinding (i.e., pull off) force of the TIM-ligand complex (TIM-4–PS in this specific case). The unbinding force (Fu) of the TIM-ligand complex is derived from the force jump that accompanies the unbinding of the complex. ks is the system spring constant derived from the slope of the each pulling trace. The third (lower) trace, representing 5% (or less) of all pulling curves, shows a weak pull off force between AFM tip and surface. This typically occurred when one of the binding partners (TIM or ligand) is absent. We attributed these weak interactions to nonspecific interactions between the AFM tip and surface (Supplementary Figs S1 and S2). These nonspecific adhesion forces were significantly smaller than the unbinding forces of TIM-ligand interactions, averaged at approximately 18 pN and seldom exceeded 25 pN (i.e., mean + 1 SD). In addition, when the loading rate increases, the magnitudes of nonspecific interaction did not change significantly, whereas specific TIM-ligand unbinding forces increase with loading rates (Supplementary Figs S1 and S2).
To enable measurement of a single molecular interaction, the contact between the cantilever tip and the substrate was minimized by reducing both the contact duration (as low as 50 ms) and the compression force (~100 pN). The brief contact duration was chosen to ensure that, for the majority of contacts (67% or greater), no adhesion (rupture force) was observed between AFM tip and surface. Assuming the adhesion bond formation obeyed Poisson statistics, an adhesion frequency of ~33% in the force measurements implies that among the observed unbinding events, the probabilities of forming a single, double, and triple adhesion bonds between AFM tip and surface were 81%, 16%, and 2%, respectively43. Therefore, our experimental condition ensured there was a >80% probability that the adhesion event was mediated by a single bond44.
Interaction specificity was shown by the adhesion frequency measurement under the same measurement conditions. Fig. 2A shows a significant decrease in adhesion when either the TIM or its binding partner (PS, VLP or EBOV GP/VSV) was absent, confirming that the vast majority of the recorded unbinding force stemmed from specific TIM-ligand interactions.
Figure 2.
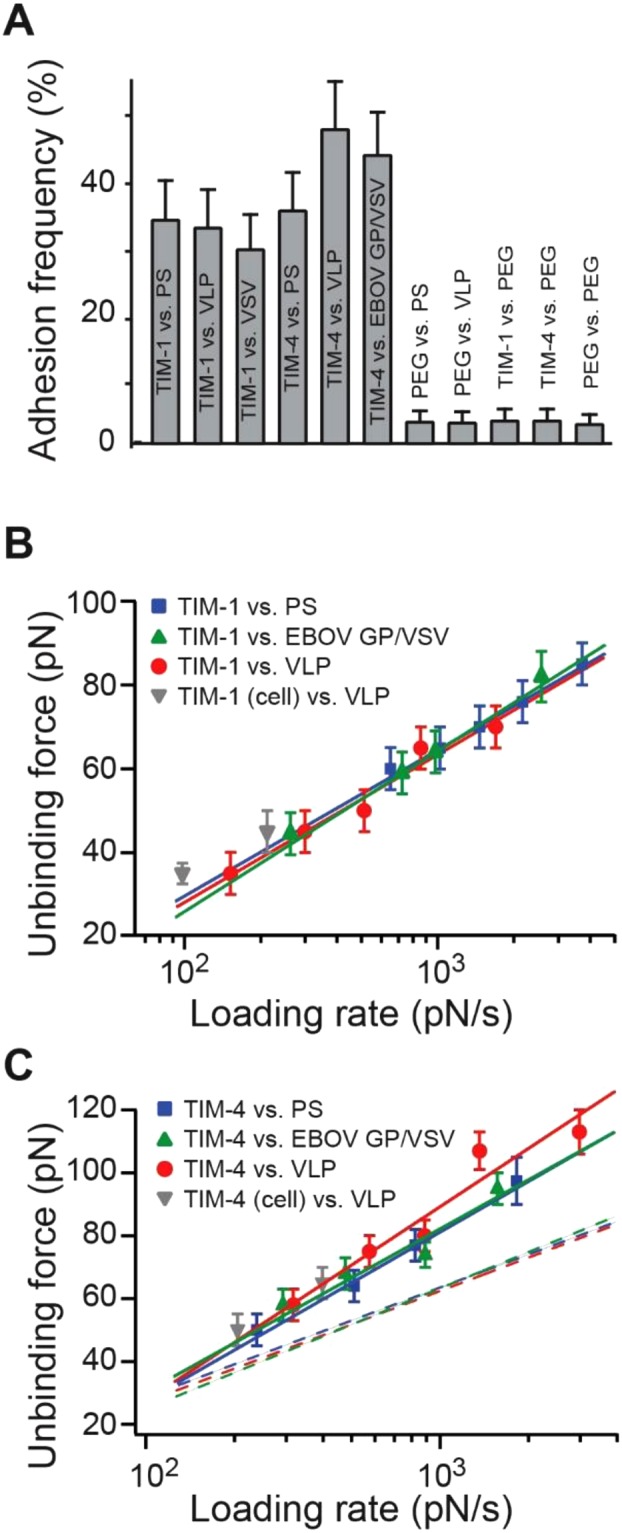
AFM measurement of TIM-ligand interactions. (A) The adhesion frequency of the AFM measurements for different interacting pairs. Contact force, contact time and retraction speed for all the interacting AFM tip and surfaces were set at 200 pN, 0.43 s and 1.5 µm/s, respectively. Error bars are Poisson errors (i.e., the square root of the adhesion number). (B) The dynamic force spectrum (i.e., the plot of the most probable unbinding force, Fu, as a function of the loading rate, rF) of the TIM-1–PS, TIM-1–VLP, TIM-1 (cell bound)–VLP and TIM-1–EBOV GP/VSV interactions. Unbinding forces at different loading rates were plotted as histograms (Fig. S1). Loading rates were determined directly from the force-extension data by multiplying the system spring constant (Fig. 1B) of the unbinding pulling trace and the retraction speed of the cantilever. The peak of each histogram (i.e., the most probable unbinding force) was plotted against the loading rate; uncertainty in the peak forces is shown as half of the bin width. Solid lines are linear fits to equation (2) for TIM-1–ligand interactions. EBOV GP/VSV is an abbreviation for the EBOV glycoprotein/vesicular stomatitis virus pseudovirions. (C) The dynamic force spectra of the TIM-4–PS, TIM-4–VLP, TIM-4 (cell bound)–VLP, and TIM-4–EBOV GP/VSV interactions. Solid lines are linear fits to equation [2] for TIM-4–ligand interactions. Dashed line is the linear fits for TIM-1–ligand interactions taken from (B).
The biophysical properties of TIM-ligand interactions were studied by the means of a dynamic force spectrum (DFS) (Fig. 2B). The DFS is the plot of most probable unbinding force as a function of the loading rate. The loading rate is obtained by multiplying the system’s spring constant (Fig. 1B) and the pulling speed of each force curve. The unbinding forces of each TIM-ligand interactions were first grouped into 4 to 5 groups by their loading rates. The distribution of forces within the same group was analyzed by histograms (Supplementary Figs S1 and S2). The most probable unbinding forces were then determined from the modes of each histograms. Fig. 2B shows that the unbinding force of the TIM-1–PS, TIM-1–VLP, and TIM-1–EBOV GP/VSV bonds increased linearly with the logarithm of the loading rate, ranging similarly from 40 to 80 pN over a loading rate of 200 to 4,000 pN/s, respectively.
In order to confirm our findings on a cell surface, HEK293T cells were transfected with TIM-1. Under two similar loading rates, the interaction of cell-bound TIM-1 with VLP was mechanically comparable to that when purified soluble TIM-1 was covalently coupled to glass cover slips (Fig. 2B).
A more detailed analysis of the biophysical properties of TIM-1–ligand interactions was conducted by fitting the acquired DFS data to the Bell-Evans model. The model describes the influence of an external force on the rate of bond dissociation45. According to this model, a pulling force, f, distorts the intermolecular potential of a ligand-receptor complex, leading to a lowering of the activation energy and an increase in the dissociation rate k(f) as follows:
| 1 |
where k0 is the dissociation rate constant in the absence of a pulling force, γ is the position of the transition state, T is the absolute temperature, and kB is the Boltzmann constant. For a constant loading rate, rf, the model can be described as:
| 2 |
hence, as predicted by the model, the most probable unbinding force f* is a linear function of the logarithm of the loading rate. Experimentally, f* was determined from the mode of the unbinding force histograms (Supplementary Figs S1 and S2). Fitting the DFS of TIM-1–PS interaction to the Bell-Evans model (Eq. 2) yielded a dissociation rate in the absence of force (k0) of 0.76 s−1, and an activation barrier width (γ) (i.e, the distance to the transition state) of 0.28 nm. The fitted curves are overlaid on the DFS, and the best-fit parameters, ko, and γ are tabulated in Table 1. Within fitting uncertainties, the fitted Bell-Evans model parameters for the TIM-1–VLP, and the TIM-1–EBOV GP/VSV interactions are similar to those of the TIM-1–PS interaction.
Table 1.
Bell-Evans model parameters of the TIM-ligand interaction. Uncertainties are the standard error of the fits. Asterisks indicate significant differences (p < 0.05) between TIM-1 and TIM-4 group values.
| TIM-ligand pairs | k0 (s−1) | τ0 (s) | γ (Å) |
|---|---|---|---|
| TIM-1 vs. PS | 0.76 ± 0.14 | 1.32 ± 0.25 | 2.8 ± 0.1* |
| TIM-1 vs. VLP | 1.01 ± 0.29 | 0.99 ± 0.28 | 2.7 ± 0.3* |
| TIM-1 vs. EBOV GP/VSV | 1.16 ± 0.18 | 0.86 ± 0.14 | 2.5 ± 0.2* |
| TIM-4 vs. PS | 1.21 ± 0.36 | 0.77 ± 0.10 | 1.8 ± 0.2* |
| TIM-4 vs. VLP | 1.26 ± 0.50 | 0.79 ± 0.31 | 1.6 ± 0.3* |
| TIM-4 vs. EBOV GP/VSV | 0.90 ± 0.45 | 1.11 ± 0.55 | 1.9 ± 0.3* |
Using a similar approach, we have studied the interactions between TIM-4 and ligands (PS and VLP) (Fig. 2C). Notably, unbinding forces for TIM-4–ligand interactions were higher than those of the TIM-1–ligand interactions (Supplementary Fig. S2), indicating that TIM-4–ligand bonds have greater mechanical strength than TIM-1–ligand bonds. Fitting the DFS to equation (2) indicates that TIM-4–ligand interactions have insignificantly different dissociation rates (all approximately 1 s−1), but significantly shorter barriers (around 0.2 nm) compared to those of TIM-1–ligand interactions (Table 1). The shorter barrier width suggests that the TIM-4–ligand bond is more resistant to mechanical pulling. Similarly, Dobrowsky et al., reported that a shorter barrier width yielded stronger binding for human HIV gp120–receptor interactions46.
In addition to using the Bell-Evans model, we also fitted the unbinding data to the Dudko-Hummer-Szabo model47. The distributions of unbinding forces at different loading rates (Supplementary Figs S1 and S2) were first fitted to a statistical model developed by Dudko et al.47 to obtain the force dependent lifetimes (Supplementary Fig. S3). The average lifetimes for six different TIM-ligand interactions were then fitted to the Dudko-Hummer-Szabo model (Supplementary Fig. S3). The fitted results were summarized in Table 2. Consistently, the Dudko-Hummer-Szabo model fit yielded comparable zero-force lifetimes (τ0) among all the tested TIM-ligand interactions, and that TIM-4–ligand bonds had shorter distance to transition state compared to TIM-1–ligand bonds. Notably, compared to the Bell-Evans model, the Dudko-Hummer-Szabo model showed a 2- to 5- fold greater zero-force lifetimes (τ0) (Tables 2 and 3).
Table 2.
Dudko-Hummer-Szabo model parameters of the TIM-ligand interaction. Uncertainties are the standard error of the fits.
| τ0 (s) | Δx (Å) | |
|---|---|---|
| TIM-1 vs. PS | 2.8 ± 2.5 | 4.0 ± 0.9 |
| TIM-1 vs. VLP | 3.7 ± 2.1 | 4.2 ± 0.7 |
| TIM-1 vs. EBOV GP/VSV | 3.3 ± 2.4 | 4.4 ± 1.0 |
| TIM-4 vs. PS | 3.8 ± 2.4 | 3.3 ± 0.6 |
| TIM-4 vs. VLP | 3.2 ± 2.8 | 2.6 ± 0.9 |
| TIM-4 vs. EBOV GP/VSV | 3.5 ± 2.5 | 2.7 ± 1.0 |
Table 3.
Continuum model variables and parameters.
| Variables and Parameters | Definition | Value | Normalized Variable |
|---|---|---|---|
| β | Free energy of each TIM/PS bond | ||
| R | EBOV radius | ||
| Density of TIM bonds | |||
| Cell membrane bending rigidity | |||
| Half of the EBOV/host cell contact width | |||
| Half of the characteristic distance over which adhesion occurs | |||
| δ | EBOV indentation depth | ||
| External force | |||
| Total energy (per unit length out of plane) | |||
| Normalized bending constant |
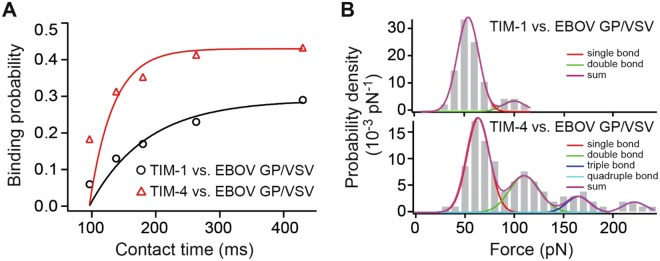
The kinetics for a receptor-ligand interaction (i.e., bonding and de-bonding) is characterized by the interaction’s on- and off-rates. We have estimated on-rates (kon) of TIM-1−EBOV GP/VSV and TIM-4−EBOV GP/VSV interactions, using a method established by the Hinterdorfer group23,48. By assuming the interactions follow a pseudo first-order kinetics, the on-rates can be estimated using kon = (τ Ceff)−1, where τ refers to the interaction time, and Ceff denotes the effective concentration of the binding partner (in this case TIM-1 or TIM-4 on the surface) within an effective volume, Veff. The Veff can be approximated by the volume of a sphere, whose radius reff is the sum of the size of the virus and cross-linker48. Therefore, the equation can be rewritten as: kon = NA·(4/3)·π·reff3·nb−1·τ−1, where NA is the Avogadro constant, and nb is the number of the binding partner48. Fig. 3A shows binding probabilities (P) of TIM-1–EBOV GP/VSV and TIM-4–EBOV GP/VSV interactions as a function of contact time (t). The P vs. t curves were fitted to a monoexponential decay function, yielding the time constant τ of the interaction at 89 ± 18 ms for TIM-1 and 37 ± 17 ms for TIM-4, respectively. nb can be estimated from probability density curves for unbinding forces at the longest contact time23. Shown in Fig. 3B, at a contact time of 430 ms, both TIM-1 and TIM-4 unbinding forces showed multimodal distributions. Multiple Gaussian peak analyses showed two peaks for TIM-1–EBOV GP/VSV interactions: at 58 and 101 pN, and four peaks for TIM-4–EBOV GP/VSV interactions: at 69, 116, 170, and 221 pN. Therefore, the available binding partners for the EBOV GP/VSV to engage, estimated by the maximum bond numbers, were two and four for TIM-1 and TIM-4, respectively. Finally, using the 107.8 nm reported size of VSV49, and the 3 nm cross-linker length23, the kon is estimated as 1.9 × 107 M−1s−1 for TIM-1, and as 2.2 × 107 M−1s−1 for TIM-4. Using the k0off values estimated from the Bell-Evans model (Table 1), the equilibrium dissociation constant Kd, was then estimated to be approximately 50 nM for both TIM-1–EBOV GP/VSV and TIM-4–EBOV GP/VSV interactions. If using the kinetics values estimated from the Dudko-Hummer-Szabo model (Table 2), the Kd would be approximately 15 nM. These two Kd values are comparable with the 24 nM Kd reported by Rankl et al.23 for human rhinovirus − host cell interactions.
Figure 3.
On-rate measurements. (A) The binding probabilities (P) of TIM-1–EBOV GP/VSV and TIM-4–EBOV GP/VSV interactions are plotted as a function of the contact time (t). The solid line is the result of least-squares fits of equation P = A(1 − (1−exp(−(t − t0)/τ)), where A and t0 are the maximal observed binding probability and the shortest contact time tested, respectively. The binding time constant is τ. Equation kon = 1/(τCeff) was used to estimation the on-rates. (B) Unbinding force distributions of TIM-1–EBOV GP/VSV (upper panel) and TIM-4–EBOV GP/VSV (lower panel) interactions at 430 ms contact time. Multiple-peak Gaussians were fitted to the curves. Each Gaussian peak and the sum of fitted Gaussians are overlaid on the distribution curve.
How do the biophysical parameters identified from single-molecule studies relate to the mechanics of virus adhesion? To address this question, we have developed a simple model for EBOV-host cell interaction driven by adhesive interactions between the virus and cell-membrane receptors. The model parameters are summarized in Table 3. The inset in Fig. 4 shows the model schematically. The virus is assumed to be cylindrical and relatively stiff so that it maintains its circular cross-section. Because its length can be large compared to the radius, we model the interaction in the two-dimensions of the virus cross-section. The viral particle is about 80 nm in diameter, much smaller than the size of the cell it infects. Therefore, we assume that the cell membrane (Fig. 4, blue line) is originally flat but deforms under contact with the virus. The contact width is 2a and the cell membrane is supported some distance l = a + b away from the center of the virus attachment point; this represents a characteristic distance over which macropinocytosis occurs.
Figure 4.
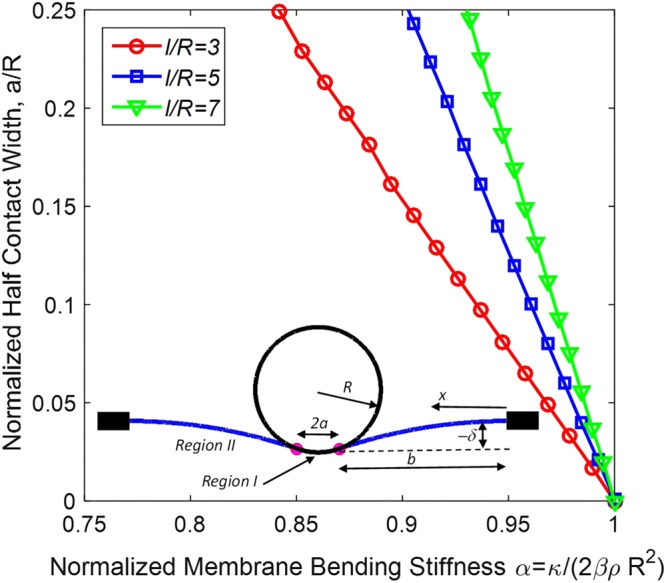
Mechanical model of EBOV-host cell attachment driven by adhesion and resisted by membrane bending. Our principal result is that if a dimensionless parameter representing the ratio of bending and adhesion energies has value greater than one, then there is no adhesion. If it assumes values less than one, contact width grows rapidly, leading to strong adhesion.
Physically, attachment of the virus to the cell membrane is driven by adhesion between the two and we assume it is resisted by energy required to bend the cell membrane. (We assume that the membrane tension is sufficiently small such that bending dominates over tension. In Supporting Information, we provide a quantitative criterion for this condition, as well as results for the limit where tension dominates over bending.) To express this mathematically, we write the total energy of the system as a sum of contributions from the elastic bending of the membrane and adhesion between the membrane and viral particle (by symmetry, we model only the right half of the region shown in Fig. 4):
| 3 |
where Uelastic is the contribution due to membrane bending, and is the contribution due to adhesion between TIM and the viral surface33,50. The elastic bending energy is a sum of contributions from two regions, region I where the membrane is in contact with the virus and region II where it is free of lateral loads. The deflection w(x) is prescribed in region I by the circular shape of the virus cross-section and, according to small-deflection plate theory51 w(x) is governed in region II by the differential equation
| 4 |
The elastic energy (per unit length out of plane) in terms of the displacements is given by
| 5 |
where is the bending rigidity of the cell membrane (in Joules; typically 10–100 kBT52–54). The adhesion energy is given by
| 6 |
where is the number of bonds per unit area, and is the free energy of binding of each bond. We proceed as follows. (See SI for details.) For specified values of parameters b,l,δ,R,β, and , we solve equation [4] subject to the conditions that deflection matches the circular profile of the virus in region I; w and w' are both zero at x = 0; and both are continuous at x = b. We then impose a condition of configurational energy balance, that is, energy is minimized with respect to location of the contact edge:
| 7 |
The minimum energy condition provides the equilibrium value of b (and hence of a) eliminating b as a variable. Finally, we vary δ until the value of the net normal force, which is proportional to w'' evaluated at x=b, equals zero. This situation corresponds to adhesion of the virus to the cell membrane in the absence of a net external force, and thus eliminates δ as a variable.
The following normalization simplifies the analysis and reveals the important dimensionless parameters that govern the adhesion behavior. We normalize all distances and sizes by the radius of the virus:
| 8 |
and force and energy as
| 9 |
With normalization we find that the solution depends solely on two dimensionless parameters:
| 10 |
(This dimensionless parameter has previously been identified by Deserno33,50). In particular, the equilibrium contact width,. The first parameter, , represents the ratio of bending energy () for one radian and adhesion energy .
Figure 4 shows results for normalized half contact width versus normalized membrane bending stiffness for three different values of . The most important and definitive conclusion of our analysis is that a condition for any adhesion between the virus particle and the cell is:
| 11 |
This condition is necessary regardless of the value of . If this condition is not met, the equilibrium contact width is zero; there is no adhesion. The second important conclusion is that for relevant values of , the normalized contact width rises rapidly with decrease of membrane stiffness below the value needed to have any adhesion. We show results only over a range of contact width for which the linear Euler-Bernoulli beam model used here should be accurate. However, it does show the importance of the parameter : if it exceeds unity there is no adhesion; if it is only somewhat smaller than unity, the contact width increases rapidly.
In order to connect this result with the force-spectroscopy measurements, we estimate β in terms of the dissociation constant (for the usual reference concentration of 1 Mol/L)
| 12 |
which yields a value of about 17 when using the 50 nM Kd estimated from single-molecule experiments for both TIM-1/PS and TIM-4/PS interactions. The EBOV virus has a radius of about 40 nm. Therefore the range of l/R used in Fig. 4 matches the range of expected dimensions. Picking reasonable values for 55, = 40 kBT56; R = 40 nm, we find α = 0.69, i.e., having a value sufficiently small to result in strong adhesion.
In SI we present an analysis of the limiting case in which tension dominates over bending. If T is the tension in the membrane, we show that if the parameter, , we predict strong adhesion of the viral particle.
Discussion
Here, using single-molecule force spectroscopy, we have quantified the mechanical strength between the following TIM-ligand bonds: TIM-1–PS, TIM-4–PS, TIM-1–EBOV GP/VSV, TIM-1–VLP, TIM-4–VLP, and TIM-4–EBOV GP/VSV. Our DFS data revealed that under the loading rates ranging from 200 to 4,000 pN/s, unbinding forces ranged, respectively, from 40 to 80 pN for TIM-1–ligand bonds, and from 50 to 100 pN for TIM-4–ligand bonds. The loading rate range was chosen based on the estimated physiological loading rates (i.e., 125 to 2500 pN/s.) of cellular tethered bonds in the vasculature57. Similar level of unbinding forces within the loading rates have been reported for several adhesion receptor-ligand systems such as integrin-ligand, selectin-ligand, and antibody-antigen interactions, suggesting that the mechanical strength of TIM-ligand bond is comparable to these adhesion molecule-ligand interactions that are known to withstand mechanical loads. In line with this observation, TIM-1 has been shown to interact with P-selectin and mediate T-lymphocyte tethering and rolling on vascular endothelium58.
Both TIM-1 and TIM-4 are known to mediate EBOV entry via binding of PS15,16. However, a discrepancy exists as to whether TIM-1 or TIM-4 also binds with EBOV surface GP59. For both TIM-1 and TIM-4, the DFS among TIM-PS, TIM-VLP, and TIM-EBOV GP/VSV are almost indistinguishable (Fig. 2). This observation is consistent with previous force spectroscopy studies of single virus−host cell interactions. For instance, Rankl et al.23 reported similar reaction kinetics (i.e., kon, koff, and γ) for the interactions between human rhinovirus and two forms of low-density lipoprotein receptor: cell membrane-bound and soluble recombinant. Consistently, Chang, et al.60 and Dobrowsky et al.46 conducted single-molecule analyses of the interactions between type-1 human immunodeficiency virus gp120 and T-cell CD4 receptor. The authors observed similar reaction kinetics in both transmembrane and soluble forms of CD4. In addition, since the both the VLPs and EBOV GP/VSVs used here express GP, and the DFS provides a biomechanical signature of different TIM-ligand interaction, our data suggests that TIM-1 and TIM-4 interact with the PS on the VLP or EBOV GP/VSV, not the GP.
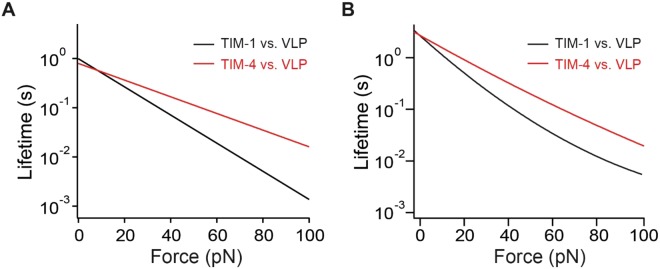
Studies suggest that both murine and human TIM-1 serve as the most important plasma membrane receptors for EBOV in epithelial cells12. EBOV can also infect dendritic cells and macrophages, causing disseminated intravascular coagulation61,62. Due to its expression on some macrophages, TIM-4 is likely a membrane receptor for EBOV16,20 on these cells. Our study provides an estimation of the effects of pulling force on bond lifetimes (Fig. 5). A comparison of the lifetimes of the TIM-1–VLP and TIM-4–VLP bonds, given by both Bell-Evans and Dudko-Hummer-Szabo models, consistently revealed that the lifetimes for the two bonds are comparable under no force (Fig. 5). However, when force is applied, TIM-4–VLP bond exhibits greater mechanical resistance and longer lifetime than the TIM-1–VLP bond. At higher force (60–100 pN), the differences of lifetime between the bonds are close to, or exceed, one order of magnitude (Fig. 5). Therefore, the TIM-4 is more suitable than TIM-1 to mediate attachment of EBOV under mechanical disturbance. Since TIM-4 expresses on leukocytes (such as macrophages) that may be exposed to larger forces in the blood, the higher force resistance of TIM-4–ligand interaction could help EBOV to remain adherent to host cells in the vasculature.
Figure 5.
(A) The force-dependent lifetimes of TIM-1–VLP and TIM-4–VLP bonds given by Bell-Evans model fit (Equation 1). (B) The force-dependent lifetimes of TIM-1–VLP and TIM-4–VLP bonds given by Dudko-Hummer-Szabo model fit (taken from Supplementary Fig. S3).
Both TIM-1 and TIM-4 interact with PS through their amino terminal (N-terminal) immunoglobulin variable (IgV)-like domain16,63. The PS-binding pocket is located between two loops of the IgV domain64,65, the upper FG and the lower CC’ loop, which form a conserved pocket known as a metal ion-dependent ligand-binding site (MILIBS)66. The stronger mechanical strength in TIM-4–ligand interaction may be explained by a cooperative binding among TIM-4’s PS-binding residues when they interact with PS. A recent study suggests that TIM-4 has more IgV residues interacting with EBOV than those of TIM-116. Tietjen et al. has reported that these additional PS-binding residues located outside the MILIBS promote cooperative binding of TIM-4 to PS67, which could explain the enhancement of mechanical strength in TIM-4–PS complex. To understand the contributions of different structural components of IgV on the mechanical strength of the binding, further force spectroscopy studies using various IgV mutants11,16 of TIM-1 and TIM-4 are needed.
We have developed a simple model for the mechanics of attachment of viral particles to a cell membrane. The purpose of the model is to show how single molecule measurements can be combined with other physical properties of the system, such as density of ligand-receptor pairs and membrane stiffness, to predict whether and to what extent a viral particle will adhere to the cell membrane. We model attachment as being driven by TIM–PS adhesion and resisted by membrane bending. (In some cells, membrane tension will be the dominant player resisting deformation.) The parameters that govern the process of adhesion in this model: adhesion free energy β, viral radius R, bending stiffness k, length l, tension, T, TIM surface density β, combine into three dimensionless groups. These are α, a ratio of bending and adhesion energies; γ, a ratio of tension and adhesion energy, and a normalized distance. When resistance to deformation is dominated by bending, if α exceeds unity in value, our continuum model predicts that no adhesion is possible. That is, membrane bending stiffness (and/or tension) can act as an agent blocking adhesion. For values of α less than one, the size of the contact increases rapidly, leading to strong adhesion of the apoptotic or viral particle. If tension dominates over bending as the agent resisting deformation, the process of adhesion is dominated by the value of a single parameter representing the ratio of works of tension and adhesion. In order to connect the mechanical model to the AFM experiments, we show that viral particle adhesion to the membrane occurs for TIM-ligand adhesion free energy calculated using values of and determined experimentally. Because experiments have established that is very small (<<mM), the probability of association of even a single bond is nearly unity. Thus, our model shows that the interplay between adhesion and bending energies sets up equilibrium states that either have association probability of essentially ‘1’ (α < 1) or ‘0’ (α > 1).
Although we are not aware of direct experimental evidence for the predicted blocking effect of membrane stiffness (or tension), there is significant indirect evidence of the kind of adhesive process we have modeled34. See Fery et al.68 for direct visual evidence of adhesion between a bead and a vesicle; Malsam et al.69 and LeBihan et al.70 for evidence of adhesion between vesicles. Several micrographs in Hernandez et al.71 indicate equilibrium shapes of the sort we have calculated, evidently based on balance between an adhesive driving force and resistance either by bending or tension.
Our theoretical model uses a continuum description of the main physical agents: membrane bending stiffness, distributed adhesion energy, and tension. By its simplicity, it has allowed us to highlight the importance of two dimensionless groups of parameters and their potential ability to block adhesion. By the same token, our model suffers from limitations, addressing which will require more complex models. For example, in the continuum model, we identify vanishing contact width with lack of adhesion whereas it is conceivable that a single receptor-ligand pair of sufficient strength57 might tether the virus to the cell membrane for long enough to initiate processes leading to engulfment. (A line of contacting pairs is deemed unlikely given the distribution of TIM location on the cell membrane). That would represent a breakdown of the continuum assumptions we have made. The model also does not explicitly account for thermal fluctuations or the discrete nature of receptor-ligand interactions. The former means, for example, that the coupling between tension and bending fluctuations is not captured; the latter can have a significant effect on effective work of adhesion72,73 if individual ligand-receptor strength is strong but their surface density is small.
In conclusion, the study shows the biomechanical parameters important for Ebola attachment to host cells. The study has demonstrated experimentally that TIM-EBOV interactions are mechanically comparable to adhesion molecule (e.g., selectin)−ligand interactions. Through a simple mechanical model, we further demonstrate how molecular binding parameters determine whether they are sufficient for viral adhesion. The study may provide new information to aid in the development of new antiviral therapeutics for the prevention and treatment of EBOV disease.
Materials and Methods
Cell culture
Immortalized HEK 293 T cells purchased from American Type Culture Collection (ATCC) were cultured in DMEM medium (ATCC), and supplemented with 4 mM L-glutamine, 4500 mg/L glucose, 1 mM sodium pyruvate, 1500 mg/L sodium bicarbonate, 1% penicillin streptomycin, and 10% fetal bovine serum. The cells were grown in T-25 flasks (Corning) at 37 °C in a 5% CO2 atmosphere and plated for experiments in 35 mm Cyto-One culture dishes (USA Scientific).
VLP generation
EBOV GP-pseudotyped VLPs were generated by co-transfecting HEK 293 T cells with a plasmid expressing EBOV VP40 fused to green fluorescent protein (GFP)74, and a plasmid expressing EBOV GP, at a 1:1 ratio15. Supernatants were collected 48 and 72 h after transfection. VLPs were concentrated by centrifuging supernatants overnight at 5,380 × g at 4 °C. Pellets were resuspended in 1 × PBS and purified through a sucrose cushion. Pellets were resuspended in 1 × PBS, filtered through a 0.45-μm syringe filter, aliquoted, and stored at −80 °C.
EBOV GP/VSV pseudovirion generation
As previously described15, vesicular stomatitis virus (strain Indiana) virions with genomes in which the G glycoprotein gene is replaced with enhanced green fluorescent protein (EGFP), HEK 293 T cells were transfected with plasmids expressing EBOV GP and transduced 24 h later with EBOV GP/VSV ΔG-EGFP pseudovirions. After 4 h of virus uptake, the plates were washed, and medium was repleted. Pseudotyped virions were collected in supernatant 48 and 72 h following transduction, pooled, and filtered through a 0.45-μm filter. The pseudovirions were concentrated by centrifuging supernatants at 5,380 × g overnight at 4 °C and resuspending pellet in fresh medium to achieve higher-titer stocks.
TIM-1 and TIM-4 Transfection
The HEK 293 T cells were split the day before transfection in 35 mm Cyto-One culture dishes (US Scientific) at 50% confluence. 200 µl of Opti-MEM (1×) reduced serum medium (Life Technologies) was added to a 0.5 ml tube. 1 µg of TIM-1 or TIM-4 expression plasmid15,16 was transfected into the cells using MegaTran 1.0 transfection reagent (Origene Technologies) with a ratio of 1:3 (DNA:MegaTran) according to manufacturer’s instructions.
Cantilever preparation/ coverslip preparation
To functionalize AFM cantilevers (MLTC, Bruker Nano) with PS, the cantilever was first silanized with 3-(trimethoxysilyl)propyl methacrylate to obtain surface thiol groups. Phosphatidylserine (PS)-PEG-Maleimide (NANOCS) was coupled to the cantilever23. VLPs15,16 of EBOV, or EBOV GP/VSV were immobilized onto a (3-aminopropyl)-triethoxysilane sinalized AFM cantilever (MLTC, Bruker Nano) using a heterobifunctional polyethylene glycol (PEG) crosslinker, Acetal-PEG-NHS (Institute of Biophysics, Johannes Kepler University), according to the protocol developed by Dr. Hermann J. Gruber, Johannes Kepler University (http://www.jku.at/biophysics/content)23. Soluble recombinant TIM-1 or TIM-4 (R&D Systems) was attached to the silanized glass coverslips using the same crosslinking approach. Functionalized cantilevers and glass surfaces were stored in PBS (3 × 5 min) and used for AFM experiment within 8 hours.
Single-molecule force measurements
All single-molecule force measurements were conducted using a custom-designed AFM apparatus. AFM measurements were collected at cantilever retraction speeds ranging from 0.19 to 3.7 µm/s to achieve the desired loading rate (200–4,000 pN/s) or 1.53 to 3.7 µm/s when the proteins were being separated from HEK 293 T cells. This was necessary to compensate for softness of the cell surface, which would act to lower the loading rate. All measurements were conducted at 25 °C in Tris-buffered saline (TBS), supplemented with 5 mM CaCl2. The contact time and indentation force between the cantilever and the sample were minimized to obtain measurements of the unitary unbinding force.
Statistical Analysis
For each pulling speed, over 500 force curves were recorded, which yielded 40 to 200 unbinding forces. Curve fitting was performed using IGOR Pro or Origin software by minimizing the chi-square statistic for the optimal fit. Unless otherwise stated, the data is reported as the mean and the standard error of the estimate. Statistical analyses between groups were performed using an unpaired t-test or ANOVA, with a p-value less than 0.05 considered to be statistically significant.
Electronic supplementary material
Supplementary figures and modeling information
Acknowledgements
This work was supported in part by an NIH grant R15AI133634 and an NSF grant 1804117 to A.J. and X.F.Z., start-up funding from Lehigh University to X.F.Z, and an NIH grant R01AI077519 to W. M. The authors thank M. Yehia for proofreading the manuscript. K.S., M.S., and D.M.M. received research support from the Lehigh Department of Bioengineering.
Author Contributions
M.A.D. designed and performed single-molecule force measurements, analyzed data, prepared figures and wrote the manuscript; N.F. and A.J. developed the model and performed simulation, analyzed data, prepared figures and wrote the manuscript; W.Z., Y.X., K.S., M.S., and D.M.M. performed single-molecule force measurements and analyzed data; S.M.-T. purified the VLP, EBOV GP/VSV and prepared TIM plasmids; W.M. helped design the study and edited the manuscript; X.F.Z. designed study, analyzed data, prepared figures and wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36449-2.
References
- 1.CDC. 2014 Ebola Outbreak in West Africa - Cumulative Reported Cases Graphs, Released on January 8, 2015 (2015).
- 2.WHO. Ebola data and statistics, published on 05 January 2015 (2015).
- 3.Meyers L, Frawley T, Goss S, Kang C. Ebola Virus Outbreak 2014: Clinical Review for Emergency Physicians. Annals of emergency medicine. 2015;65:101–108. doi: 10.1016/j.annemergmed.2014.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Hunt CL, Lennemann NJ, Maury W. Filovirus entry: a novelty in the viral fusion world. Viruses. 2012;4:258–275. doi: 10.3390/v4020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takada A. Filovirus tropism: cellular molecules for viral entry. Frontiers in microbiology. 2012;3:34. doi: 10.3389/fmicb.2012.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geisbert TW, Hensley LE. Ebola virus: new insights into disease aetiopathology and possible therapeutic interventions. Expert reviews in molecular medicine. 2004;6:1–24. doi: 10.1017/S1462399404008300. [DOI] [PubMed] [Google Scholar]
- 7.Towner JS, et al. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS pathogens. 2008;4:e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nanbo, A. et al. Ebolavirus Is Internalized into Host Cells via Macropinocytosis in a Viral Glycoprotein-Dependent Manner. PLoS pathogens6, 10.1371/journal.ppat.1001121 (2010). [DOI] [PMC free article] [PubMed]
- 9.Noda T, et al. Assembly and budding of Ebolavirus. PLoS pathogens. 2006;2:864–872. doi: 10.1371/journal.ppat.0020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bharat, T. A. M. et al. Cryo-Electron Tomography of Marburg Virus Particles and Their Morphogenesis within Infected Cells. Plos Biol9, 10.1371/journal.pbio.1001196 (2011). [DOI] [PMC free article] [PubMed]
- 11.Moller-Tank S, Albritton LM, Rennert PD, Maury W. Characterizing functional domains for TIM-mediated enveloped virus entry. Journal of virology. 2014;88:6702–6713. doi: 10.1128/JVI.00300-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondratowicz AS, et al. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8426–8431. doi: 10.1073/pnas.1019030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feigelstock D, Thompson P, Mattoo P, Zhang YA, Kaplan GG. The human homolog of HAVcr-1 codes for a hepatitis A virus cellular receptor. Journal of virology. 1998;72:6621–6628. doi: 10.1128/jvi.72.8.6621-6628.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meertens L, et al. The TIM and TAM Families of Phosphatidylserine Receptors Mediate Dengue Virus Entry. Cell Host Microbe. 2012;12:544–557. doi: 10.1016/j.chom.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moller-Tank S, Kondratowicz AS, Davey RA, Rennert PD, Maury W. Role of the Phosphatidylserine Receptor TIM-1 in Enveloped-Virus Entry. Journal of virology. 2013;87:8327–8341. doi: 10.1128/Jvi.01025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhein BA, Brouillette RB, Schaack GA, Chiorini JA, Maury W. Characterization of Human and Murine T-Cell Immunoglobulin Mucin Domain 4 (TIM-4) IgV Domain Residues Critical for Ebola Virus Entry. Journal of virology. 2016;90:6097–6111. doi: 10.1128/JVI.00100-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jemielity, S. et al. TIM-family Proteins Promote Infection of Multiple Enveloped Viruses through Virion-associated Phosphatidylserine. PLoS pathogens9, 10.1371/journal.ppat.1003232 (2013). [DOI] [PMC free article] [PubMed]
- 18.Mercer J, Helenius A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science. 2008;320:531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- 19.Morizono K, Chen IS. Role of phosphatidylserine receptors in enveloped virus infection. Journal of virology. 2014;88:4275–4290. doi: 10.1128/JVI.03287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moller-Tank S, Maury W. Phosphatidylserine receptors: enhancers of enveloped virus entry and infection. Virology. 2014;468-470:565–580. doi: 10.1016/j.virol.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinterdorfer P, Dufrene YF. Detection and localization of single molecular recognition events using atomic force microscopy. Nat. Methods. 2006;3:347–355. doi: 10.1038/nmeth871. [DOI] [PubMed] [Google Scholar]
- 22.Dobrowsky TM, et al. Adhesion and fusion efficiencies of human immunodeficiency virus type 1 (HIV-1) surface proteins. Scientific reports. 2013;3:3014. doi: 10.1038/srep03014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rankl C, et al. Multiple receptors involved in human rhinovirus attachment to live cells. Proc Natl Acad Sci USA. 2008;105:17778–17783. doi: 10.1073/pnas.0806451105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sieben C, et al. Influenza virus binds its host cell using multiple dynamic interactions. Proc Natl Acad Sci USA. 2012;109:13626–13631. doi: 10.1073/pnas.1120265109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyubartsev AP. Multiscale modeling of lipids and lipid bilayers. European biophysics journal: EBJ. 2005;35:53–61. doi: 10.1007/s00249-005-0005-y. [DOI] [PubMed] [Google Scholar]
- 26.Cooke IR, Deserno M. Solvent-free model for self-assembling fluid bilayer membranes: stabilization of the fluid phase based on broad attractive tail potentials. The Journal of chemical physics. 2005;123:224710. doi: 10.1063/1.2135785. [DOI] [PubMed] [Google Scholar]
- 27.Cooke IR, Kremer K, Deserno M. Tunable generic model for fluid bilayer membranes. Physical review. E, Statistical, nonlinear, and soft matter physics. 2005;72:011506. doi: 10.1103/PhysRevE.72.011506. [DOI] [PubMed] [Google Scholar]
- 28.Shi X, Von dem Bussche A, Hurt RH, Kane AB, Gao H. Cell entry of one-dimensional nanomaterials occurs by tip recognition and rotation. Nat Nanotechnol. 2011;6:714–719. doi: 10.1038/nnano.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atilgan E, Sun SX. Shape transitions in lipid membranes and protein mediated vesicle fusion and fission. The Journal of chemical physics. 2007;126:095102. doi: 10.1063/1.2483862. [DOI] [PubMed] [Google Scholar]
- 30.Noguchi H, Gompper G. Shape transitions of fluid vesicles and red blood cells in capillary flows. Proc Natl Acad Sci USA. 2005;102:14159–14164. doi: 10.1073/pnas.0504243102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou T. Stochastic entry of enveloped viruses: fusion versus endocytosis. Biophys J. 2007;93:1116–1123. doi: 10.1529/biophysj.107.106708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipowsky R, Döbereiner HG. Vesicles in contact with nanoparticles and colloids. Europhysics Letters (EPL) 1998;43:219–225. doi: 10.1209/epl/i1998-00343-4. [DOI] [Google Scholar]
- 33.Deserno, M. Elastic deformation of a fluid membrane upon colloid binding. Phys Rev E69, 10.1103/PhysRevE.69.031903 (2004). [DOI] [PubMed]
- 34.Zhang S, Gao H, Bao G. Physical Principles of Nanoparticle Cellular Endocytosis. ACS Nano. 2015;9:8655–8671. doi: 10.1021/acsnano.5b03184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fortoul N, Singh P, Hui CY, Bykhovskaia M, Jagota A. Coarse-Grained Model of SNARE-Mediated Docking. Biophys J. 2015;108:2258–2269. doi: 10.1016/j.bpj.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stahelin RV. Membrane binding and bending in Ebola VP40 assembly and egress. Frontiers in microbiology. 2014;5:300. doi: 10.3389/fmicb.2014.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Licata JM, Johnson RF, Han Z, Harty RN. Contribution of ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. Journal of virology. 2004;78:7344–7351. doi: 10.1128/JVI.78.14.7344-7351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kondratowicz AS, Hunt CL, Davey RA, Cherry S, Maury WJ. AMP-activated protein kinase is required for the macropinocytic internalization of ebolavirus. Journal of virology. 2013;87:746–755. doi: 10.1128/JVI.01634-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agnandji ST, et al. Phase 1 Trials of rVSV Ebola Vaccine in Africa and Europe. The New England journal of medicine. 2016;374:1647–1660. doi: 10.1056/NEJMoa1502924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Craig SE, Kirby H, Humphries MJ, Moy VT. Molecular basis for the dynamic strength of the integrin alpha4beta1/VCAM-1 interaction. Biophys J. 2004;87:3470–3478. doi: 10.1529/biophysj.104.045690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Wojcikiewicz E, Moy VT. Force spectroscopy of the leukocyte function-associated antigen-1/intercellular adhesion molecule-1 interaction. Biophys J. 2002;83:2270–2279. doi: 10.1016/S0006-3495(02)73987-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dragovich MA, et al. Mechanotransduction of the endothelial glycocalyx mediates nitric oxide production through activation of TRP channels. American journal of physiology. Cell physiology. 2016;311:C846–C853. doi: 10.1152/ajpcell.00288.2015. [DOI] [PubMed] [Google Scholar]
- 43.Chesla SE, Selvaraj P, Zhu C. Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys J. 1998;75:1553–1572. doi: 10.1016/S0006-3495(98)74074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evans E. Probing the relation between force–lifetime–and chemistry in single molecular bonds. Annu Rev Biophys Biomol Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 45.Evans E, Ritchie K. Dynamic strength of molecular adhesion bonds. Biophys J. 1997;72:1541–1555. doi: 10.1016/S0006-3495(97)78802-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobrowsky TM, Zhou Y, Sun SX, Siliciano RF, Wirtz D. Monitoring early fusion dynamics of human immunodeficiency virus type 1 at single-molecule resolution. Journal of virology. 2008;82:7022–7033. doi: 10.1128/JVI.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dudko OK, Hummer G, Szabo A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc Natl Acad Sci USA. 2008;105:15755–15760. doi: 10.1073/pnas.0806085105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rankl C, et al. Determination of the kinetic on- and off-rate of single virus-cell interactions. Methods Mol Biol. 2011;736:197–210. doi: 10.1007/978-1-61779-105-5_13. [DOI] [PubMed] [Google Scholar]
- 49.Akpinar F, Yin J. Characterization of vesicular stomatitis virus populations by tunable resistive pulse sensing. Journal of Virological Methods. 2015;218:71–76. doi: 10.1016/j.jviromet.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bahrami AH, et al. Wrapping of nanoparticles by membranes. Advances in Colloid and Interface Science. 2014;208:214–224. doi: 10.1016/j.cis.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 51.Timoshenko, S. & Woinowsky-Krieger, S. Theory of plates and shells. (McGraw-Hill, 1959).
- 52.Evans E, Rawicz W. Entropy-driven tension and bending elasticity in condensed-fluid membranes. Physical review letters. 1990;64:2094–2097. doi: 10.1103/PhysRevLett.64.2094. [DOI] [PubMed] [Google Scholar]
- 53.Brochard F, Lennon JF. Frequency spectrum of the flicker phenomenon in erythrocytes. Journal de Physique. 1975;36:1035–1047. doi: 10.1051/jphys:0197500360110103500. [DOI] [Google Scholar]
- 54.Boal, D. H. Mechanics of the Cell. (Cambridge University Press, 2012).
- 55.Romo GM, et al. The glycoprotein Ib-IX-V complex is a platelet counterreceptor for P-selectin. J Exp Med. 1999;190:803–814. doi: 10.1084/jem.190.6.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boal, D. Mechanics of the Cell, 2nd Edition. (Cambridge University Press, 2012).
- 57.Rinko LJ, Lawrence MB, Guilford WH. The molecular mechanics of P- and L-selectin lectin domains binding to PSGL-1. Biophys J. 2004;86:544–554. doi: 10.1016/S0006-3495(04)74133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Angiari S, et al. TIM-1 glycoprotein binds the adhesion receptor P-selectin and mediates T cell trafficking during inflammation and autoimmunity. Immunity. 2014;40:542–553. doi: 10.1016/j.immuni.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong S, et al. Insight into the Ebola virus nucleocapsid assembly mechanism: crystal structure of Ebola virus nucleoprotein core domain at 1.8 A resolution. Protein & cell. 2015;6:351–362. doi: 10.1007/s13238-015-0163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chang MI, Panorchan P, Dobrowsky TM, Tseng Y, Wirtz D. Single-molecule analysis of human immunodeficiency virus type 1 gp120-receptor interactions in living cells. Journal of virology. 2005;79:14748–14755. doi: 10.1128/JVI.79.23.14748-14755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Twenhafel NA, et al. Pathology of experimental aerosol Zaire ebolavirus infection in rhesus macaques. Vet Pathol. 2013;50:514–529. doi: 10.1177/0300985812469636. [DOI] [PubMed] [Google Scholar]
- 62.Geisbert TW, et al. Mechanisms underlying coagulation abnormalities in ebola hemorrhagic fever: overexpression of tissue factor in primate monocytes/macrophages is a key event. The Journal of infectious diseases. 2003;188:1618–1629. doi: 10.1086/379724. [DOI] [PubMed] [Google Scholar]
- 63.McIntire JJ, Umetsu DT, DeKruyff RH. TIM-1, a novel allergy and asthma susceptibility gene. Springer Semin Immun. 2004;25:335–348. doi: 10.1007/s00281-003-0141-3. [DOI] [PubMed] [Google Scholar]
- 64.Kobayashi N, et al. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity. 2007;27:927–940. doi: 10.1016/j.immuni.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miyanishi M, et al. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450:435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- 66.Freeman GJ, Casasnovas JM, Umetsu DT, DeKruyff RH. TIM genes: a family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity. Immunological reviews. 2010;235:172–189. doi: 10.1111/j.0105-2896.2010.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tietjen GT, et al. Molecular mechanism for differential recognition of membrane phosphatidylserine by the immune regulatory receptor Tim4. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1463–E1472. doi: 10.1073/pnas.1320174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fery A, Moya S, Puech P-H, Brochard-Wyart F, Mohwald H. Interaction of polyelectrolyte coated beads with phospholipid vesicles. Comptes Rendus Physique. 2003;4:259–264. doi: 10.1016/s1631-0705(03)00030-6. [DOI] [Google Scholar]
- 69.Malsam J, et al. Complexin arrests a pool of docked vesicles for fast Ca2+-dependent release. The EMBO journal. 2012;31:3270–3281. doi: 10.1038/emboj.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Le Bihan O, et al. Cryo-electron tomography of nanoparticle transmigration into liposome. Journal of Structural Biology. 2009;168:419–425. doi: 10.1016/j.jsb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 71.Hernandez JM, et al. Membrane Fusion Intermediates via Directional and Full Assembly of the SNARE Complex. Science. 2012;336:1581–1584. doi: 10.1126/science.1221976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Evans EA. Detailed mechanics of membrane-membrane adhesion and separation. I. Continuum of molecular cross-bridges. Biophys J. 1985;48:175–183. doi: 10.1016/S0006-3495(85)83770-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Evans EA. Detailed mechanics of membrane-membrane adhesion and separation. II. Discrete kinetically trapped molecular cross-bridges. Biophys J. 1985;48:185–192. doi: 10.1016/S0006-3495(85)83771-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin-Serrano J, Perez-Caballero D, Bieniasz PD. Context-dependent effects of L domains and ubiquitination on viral budding. Journal of virology. 2004;78:5554–5563. doi: 10.1128/JVI.78.11.5554-5563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and modeling information