Abstract
Discovering the means to control the increasing dissemination of pathogenic vibrios driven by recent climate change is challenged by the limited knowledge of the mechanisms in charge of Vibrio spp. persistence and spread in the time of global warming. To learn about physiological and gene expression patterns associated with the long-term persistence of V. harveyi at elevated temperatures, we studied adaptation of this marine bacterium in seawater microcosms at 30 °C which closely mimicked the upper limit of sea surface temperatures around the globe. We found that nearly 90% of cells lost their culturability and became partly damaged after two weeks, thus suggesting a negative impact of the combined action of elevated temperature and shortage of carbon on V. harveyi survival. Moreover, further gene expression analysis revealed that major adaptive mechanisms were poorly coordinated and apparently could not sustain cell fitness. On the other hand, elevated temperature and starvation promoted expression of many virulence genes, thus potentially reinforcing the pathogenicity of this organism. These findings suggest that the increase in disease outbreaks caused by V. harveyi under rising sea surface temperatures may not reflect higher cell fitness, but rather an increase in virulence enabling V. harveyi to escape from adverse environments to nutrient rich, host-pathogen associations.
Introduction
Vibrio harveyi is a heterotrophic Gram-negative luminous bacterium inhabiting marine environments and showing a preference for temperate and tropical waters. In its natural habitats, Vibrio spp. can be found attached to either biotic or abiotic surfaces1, in a free living state as well as in symbiotic2 or host-pathogen3 interactions with other organisms, occasionally causing diseases or even death of the infected organisms4.
The most recent studies have revealed a wider spread of pathogenic vibrios and this phenomenon was documented along with a gradual increase in sea surface temperature taking place in the time of global warming (GW)5. Consistent with the above trends, there have been a concomitant increase in the cholera or/and gastroenteritis outbreaks6–8 apparently caused by the consumption of water contaminated with pathogenic vibrios9–12. As the average temperature of water increases towards the equator, more cases of Vibrio-related diseases were reported in countries such as India or those in the Caribbean13,14. Moreover, a number of recent reports clearly point to the emergence of Vibrio-associated diseases in Europe including some countries where the presence of virulent variants of V. vulnificus and other pathogenic strains could barely been detected previously15. This alarming statistics includes discovery of pathogenic Vibrio species in several unexpected locations such as finding of V. cholerae in estuaries of Portugal16, V. parahaemolyticus on the French Atlantic coast17 and the highly pathogenic strain V. parahaemolyticus serovar O3:K6 in Spain18–21.
In accordance with the overall increase in the occurrence of Vibrio-associated diseases22, several highly virulent V. harveyi strains able to provoke mass mortality in both marine invertebrates23,24 and various fishes25,26 have been reported as well. V. harveyi is notoriously known for being an important pathogen of cultured penaeid shrimp27 and its wider occurrence in aquaculture farms causes serious problems for the seafood industry, especially in tropical countries where temperatures are generally higher. As the forecast is not promising, it is conceivable that the ocean is going to experience even more devastating consequences of GW in the coming years, which could likely lead to further spread of Vibrio-associated infections.
The ubiquitous presence and further spread of Vibrio spp. indicate that these marine bacteria are able to easily adapt to changing environmental conditions including those caused by GW. Several studies have previously examined their adaptation by analyzing phenotypical and gene expression changes taking place in Vibrio species facing different stress conditions28,29. Analysis of V. harveyi adaptation revealed that deprivation of nutrients (starvation) can trigger profound morphological changes leading to reduction of cell size and conversion of rod-shaped bacteria into their coccoid-like variants30–32. In addition, when environmental conditions become unfavorable (e.g. limitation of nutrients, low temperature, low salinity or ultra violet (UV) radiation), V. harveyi can occasionally acquire the viable but nonculturable (VBNC) phenotype33,34 and preserve it until the adverse conditions are eliminated, subsequently being able to recover from this state of dormancy and resume growth.
An important characteristics that enables vibrios to successfully strive in marine ecosystems is their metabolic versatility, in particular, their capacity to use a wide variety of organic substances (e.g. D-glucose, N-acetyl-D-glucosamine, L-asparagine aconitate etc.) as a primary carbon source. Moreover, the use of different carbon sources is, in turn, facilitated by the ability of Vibrio spp. to secrete various hydrolytic enzymes (amylases, gelatinases, lipases and chitinases) converting the naturally occuring biopolymers (e.g. proteins, polysaccharides, chitin etc.) into smaller, easily metabolizable compounds35.
Although changing environments are known to have a great impact on marine bacteria, little is known about how the physiology, metabolic preferences and adaptation mechanisms of Vibrio spp. are affected by the main environmental factors (elevated temperature, ocean acidification, suboptimal salinity concentration, etc.) caused by the ongoing GW. In order to learn more about Vibrio spp. responses to GW and their possible contribution to survival and pathogenicity of Vibrio spp., we studied phenotypical and gene expression changes that occur during the time-dependent adaptation of V. harveyi in seawater microcosm at 30 °C which is close to the upper limit of sea surface temperature (SST) recently observed in some regions of the globe36,37.
Experimental procedures
Survival assays, bacterial cell count and size measurements
To assess the combined effect of carbon limitation and elevated temperature on physiology and appearance of Vibrio harveyi ATCC® 14126™, cells were aerobically grown overnight at 26 °C (optimal temperature for V. harveyi growth) in Marine broth (MB) (Panreac) and three 20 mL aliquots of the stationary phase cultures were independently diluted (1:20) with sterile natural seawater to obtain 400 mL suspensions with a final density of 108 cells mL−1. The resulting inoculates of V. harveyi were incubated in dark with shaking (90 rpm) at 30 °C within 21 days. The experiments were carried out in 1 L glass flasks beforehand cleaned with H2SO4 (97%, v/v), rinsed with deionized water and heated at 250 °C for 24 h to avoid any presence of residual organic matter.
The seawater was sampled in the Port of Armintza in the Bay of Biscay, 43°26′24″N and 2°54′24″W and then was filter sterilized and autoclaved. The concentration of dissolved organic carbon (DOC) previously measured in the same area is in the range of 0.2–0.4 mM38, which is nearly 200 times lower than the concentration of organic carbon in MB (https://www.nature.com/articles/nrmicro1752/tables/1).
Periodically, samples were collected for further analysis. The total number of cells was determined by filtering aliquots of V. harveyi suspensions (withdrawn after 3, 6, 9, 13 and 21 days of incubation) through 0.22 μm pore-size polycarbonate membrane filters (Millipore), followed by staining of the attached cells with acridine orange and direct counting individual cells using epifluorescence microscopy39. The microscope (Eclipse E400, Nikon) was equipped with a video camera Hamamatsu 2400 (Hamamatsu Photonics, Japan) enabling to obtain high resolution images for their subsequent analysis by Scion Image 1.62a software to determine the size of individual cells as described by40. Approximately 150–200 cells were measured in each sample, and, according to their length, the cells fell into three groups:≤1.2 µm, >1.2–2 µm and >2 µm32.
Cultivability expressed as colony-forming units (CFU) was evaluated by spreading aliquots (withdrawn after 2, 3, 5, 7, 9, 12, 15, 17, 19 and 21 days of incubation) obtained by consecutive dilutions of V. harveyi suspensions on Marine Agar (MA, Oxoid) followed by incubation for 24 h at 26 °C.
Scanning electron microscopy
V. harveyi cells present in the control (overnight culture) and test samples (i.e after 3, 6 and 21 days of incubation in seawater at 30 °C) were fixed by adding 50 µl of 3% formalin to each sample (1 mL) and were further stored at 4 °C. The suspensions of the fixed cells were filtered through 0.22 μm pore-size membrane filters (GTTP filters, Millipore) and the cells attached to the filters were further examined by scanning electron microscopy (SEM) at the Advance Research Core Facility Unit (SGIker) of the University of the Basque Country. Briefly, the filters with the attached V. harveyi cells were dehydrated by sequentially immersing them (for 5 min each time) in water/ethanol solutions containing increasing concentrations of ethanol (30, 50, 70, 90, and 100%, respectively) followed by overlaying with hexamethyldisilazane, incubation for 5 min and drying on air. Finally, the samples were coated with a layer of 15 nm gold by using an Emitech K550X Sputter Coater, and imaging was further carried out by examining samples in a Hitachi S4800 scanning electron microscope.
RNA isolation, sequencing and processing of RNAseq data
V. harveyi ATCC® 14126™ cultures were grown in triplicate overnight (12–16 h) at 26 °C in MB, and 20 mL of each culture were individually diluted (1:20) with sterile seawater (originated from the same batch of sterile seawater used in the survival assays) and further incubated in 1 L glass flasks with shaking (90 rpm) at 30 °C. Aliquots of V. harveyi suspensions were withdrawn after 12 h, 3 and 6 days of incubation, mixed (8:1) with stop solution (5% phenol in ethanol) and then incubated on ice for 15–20 min. The cells were collected by centrifugation (15 min, 4 °C, 4400 g) and the resulting cell pellets were further used to isolate total RNA by employing TRIzol reagent and PureRNA mini Kit (Invitrogen) following the vendor’s instructions.
After verifying the quality of the purified RNA by using Bioanalyzer (Agilent), it was further processed with the assistance of the General Genomic Service (SGIker) of the University of the Basque Country. The libraries were prepared from total RNA by using ScriptSeq Complete kit (Illumina) optimized for bacterial RNA. The complete kit included the Ribo-Zero rRNA Removal Kit and ScriptSeq v2 Kit with ScriptSeq Index PCR Primers Set. The Ribo-Zero rRNA Removal kit was used to deplete 275–400 ng of total RNA with ribosomal RNA following the low input protocol. The samples depleted with rRNA were then concentrated with RNA Clean & Concentrator-5 kit (Zymo Research) for recovery and preservation of large RNA fragments (>200 nt) and afterwards were used to prepare stranded RNA-Seq libraries according to the manufacturer’s instructions. Further RNA sequencing and bioinformatics analysis were carried out at the National Center for Genomic Analysis (CNAG) in Barcelona integrated with the Center for Genomic Regulation (CRG). The libraries were sequenced on HiSeq. 4000 (Illumina) in a paired-end mode with a read length of 2 × 76 bp using HiSeq. 4000 SBS kit in a fraction of a HiSeq. 4000 PE Cluster kit sequencing flow cell lane following the standard protocol.
Image analysis, base calling and quality scoring of the run were processed using the manufacturer’s software Real Time Analysis (RTA 2.7.6) and followed by generation of FASTQ sequence files by CASAVA. The processed RNA-seq reads were mapped against V. harveyi ATCC® 43516™ reference genome (available at ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/; accession number GCA_001558435.1) with STAR41 and were further quantified with RSEM42. Information about some sRNAs initially absent in the annotation of V. harveyi ATCC® 43516™ genome was added to the annotation file by performing a highly restrictive BLAST search by using the sequences of V. campbellii ATCC® BAA-1116™ sRNAs retrieved from RFAM and BSRD databases as queries. Functional annotation of other genes was performed by blasting each assembled gene sequence to find homology to other V. harveyi genes already annotated in the NCBI database and the functional annotations of the best hits was used for the final annotations of transcripts revealed by RNA-seq. Differential expression analysis was performed with DESeq. 2 R43 and genes with false discovery rate (FDR)<5% and fold change >2 were considered significant and were further used to construct temporal clusters. Time series cluster analysis according to the soft clustering algorithm was performed with Mfuzz bioconductor package44 with k = 6 found to be optimal to obtain the minimal average cluster overlap. Gene ontology enrichment of the differentially expressed genes was performed with GOstats45. All raw and processed RNA-seq data were deposited into the GEO archive (https://www.ncbi.nlm.nih.gov/geo/) and are publicly available under accession number GSE113564.
Results
Effects of prolonged incubation of V. harveyi cells in seawater at 30 °C on their culturability and morphology
Experiments were done with three independent microcosms (biological replicates) prepared by diluting aliquots of the stationary phase cultures (control) with sterile seawater and analyzing the appearance and culturability of V. harveyi populations after 12 hours, 3, 6, 14 and 21 days of incubation. While determining culturable populations in aliquots withdrawn at different time points and analyzed by spreading on MA plates, a 10-fold decrease in the number of culturable cells was observed after three weeks of incubation (Fig. 1).
Figure 1.
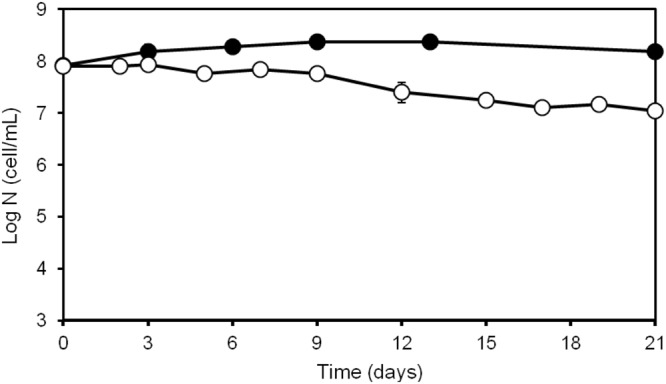
Vibrio harveyi counts obtained during its persistence in seawater at 30 °C. The number of total (•) and culturable (○) bacteria were estimated as described in Materials and Methods. The data are mean values from three independent experiments with errors bars representing the standard deviations calculated.
Moreover, the loss of culturability was accompanied by a marked reduction of cell size that became readily distinguishable already after 3 days of incubation. Furthermore, a gradual increase of the fractions of smaller cells was observed until the last day of the measurements (Fig. 2). In other words, while 75.5% of the cells in the control sample represented by the overnight culture (day 0) had size between 1.2 to 2 µm, this number has decreased to 27.4% after 3 days of incubation and the percentage of longer cells continued decreasing progressively until day 6, when nearly all cells (i.e. 95%) were shorter than 1.2 µm.
Figure 2.
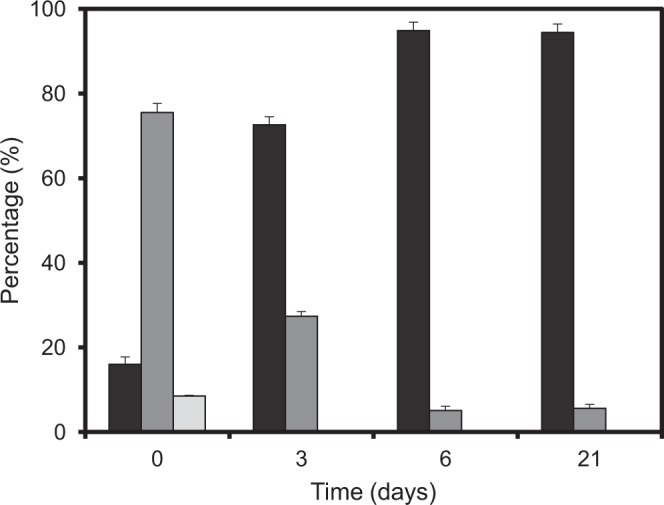
Size distribution of Vibrio harveyi cells during their persistence in seawater at 30 °C. The values were obtained by analyzing cell images captured by epifluorescence microscopy and processed as described in Materials and Methods. The bars show the percentage of cell within each size range (■ ≤1.2 µm;  >1.2 − ≤ 2 µm and
>1.2 − ≤ 2 µm and  >2 µm, respectively).
>2 µm, respectively).
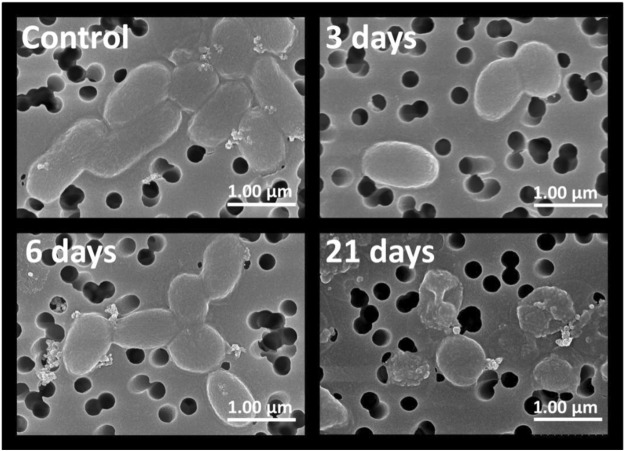
The observed size reduction was further corroborated by the results of scanning electron microscopy (SEM). SEM confirmed (Fig. 3) the appearance of cells with a coccoid-like morphology (i.e. with a nearly spherical shape) after 3 to 6 days of incubation. Moreover, the electron microscopy revealed a considerable number of damaged cells, thus indicating that prolong incubation (>6 days) under carbon limitation at 30 °C can lead to the loss of cell integrity.
Figure 3.
Morphology and integrity of Vibrio harveyi populations incubated in seawater at 30 °C. Scanning electron microscopy images were obtained for the cells present in the initial inoculate (control) as well as those incubated at 30 °C for 3, 6 and 21 days, respectively.
Gene expression analysis
Besides monitoring cell culturability and morphological changes, V. harveyi adaptation was also analyzed at the whole transcriptome level. To carry out this analysis, total RNA was isolated from the initial inoculate (control) and from V. harveyi cells after their incubation in sterile seawater for 12 h, 3, 6, 14 and 21 days. Total RNA samples were extracted from replicate cultures, i.e. they were originated from three independent cell cultures (biological replicates) grown in parallel. Electrophoretic profiles of RNA samples isolated from cells incubated for 14 and 21 days revealed that they contained a considerable fraction of degraded RNA (apparently accumulating due to cell damage beginning after prolong incubation of V. harveyi cells in seawater (Fig. 3). RNA purified from the control (t = 0) and microcosms at early time points (12 h, 3 and 6 days) were used for RNA sequencing.
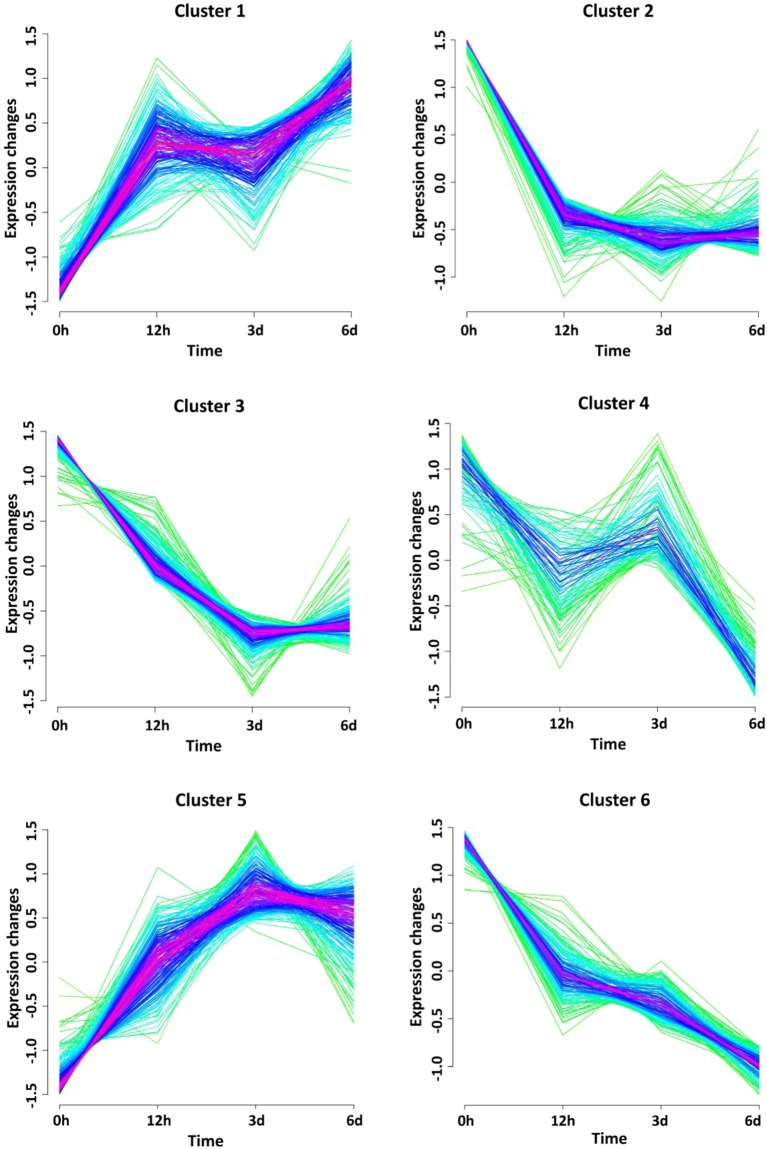
Based on the results of RNA sequencing and their processing (see Materials and Methods), a list of the differentially expressed genes with false discovery rate less than 5% and fold change >2 was compiled (see Table S1). Further bioinformatics analysis of this list made it possible to reveal six clusters of genes with similar expression profiles (Fig. 4). Two of them (clusters 1 and 5) were gradually upregulated during the course of the experiment and included a number of genes (see Table 1) primarily related to transport, virulence (clusters 1 and 5) and stress responses (cluster 5). The other four clusters (i.e. clusters 2, 3, 4 and 6) contained numerous genes controlling various cellular functions. The expression of these genes either was initially decreased and then remained low (clusters 2, 3 and 6) or was transitionally upregulated (cluster 4) at later time points. While clusters 2 and 3 were primarily represented by metabolic genes (e.g. genes controlling carboxylic acid and nucleotide metabolism, respectively), the other two (clusters 4 and 6) comprised the genes playing important roles in the processing of genetic information (namely, transcription and translation, respectively).
Figure 4.
Gene co-expression clusters. Temporal expression patterns were obtained for the time-points corresponding to Vibrio harveyi incubation in seawater at 30 oC for 12 hours, 3 days and 6 days. Gene membership values are color-encoded with red and green shades denoting high and low membership values of genes, respectively. Scaled expression values (z-scores) are used for cluster visualization.
Table 1.
Some examples of V. harveyi genes and their products highly up- or down-regulated during persistence in seawater microcosm at 30 °C.
| Systematic name | Gene product (specific biological pathway) | Log2 Fold change (time after exposure to seawater) | Gene expression cluster | ||
|---|---|---|---|---|---|
| 12 h | 3 d | 6 d | |||
| Amino acid metabolism | |||||
| AL538_RS23670 | Methylmalonate-semialdehyde dehydrogenase (CoA acylating) | −2.325 | −3.377 | −2.733 | 2 |
| AL538_RS15645 | Alanine dehydrogenase | −2.877 | −2.910 | −3.289 | 2 |
| AL538_RS01790 | Ornithine cyclodeaminase | −2.714 | −4.302 | −3.209 | 2 |
| AL538_RS07820 | Argininosuccinate synthase | −2.225 | −2.890 | −2.258 | 2 |
| AL538_RS07290 | Aspartate carbamoyltransferase | −1.401 | −1.345 | −1.897 | 6 |
| AL538_RS07295 | Aspartate carbamoyltransferase regulatory subunit | −1.251 | −1.976 | −1.289 | 2 |
| AL538_RS25000 | Glutamate synthase | −0.858 | −1.233 | −1.597 | 6 |
| AL538_RS11745 | Glutamate synthase subunit beta | −1.595 | −2.359 | −1.828 | 2 |
| AL538_RS01370 | Alkaline serine protease | −1.378 | −3.486 | −2.372 | 4 |
| AL538_RS14635 | Serine protein kinase PrkA | −1.385 | −1.333 | −2.215 | 6 |
| AL538_RS01665 | 4-hydroxyphenylpyruvate dioxygenase | −2.624 | −2.794 | −3.415 | 2 |
| AL538_RS03245 | Ribonucleotide-diphosphate reductase subunit beta | −1.553 | −2.794 | −2.503 | 3 |
| AL538_RS14510 | Uridine phosphorylase | −1.682 | −2.946 | −3.119 | 3 |
| Carbon metabolism | |||||
| Citrate, glyoxylate & 2-methylcitrate cycles | |||||
| AL538_RS17025 | Citrate synthase/methylcitrate synthase | −2.020 | −1.656 | −2.315 | 6 |
| AL538_RS06380 | Aconitate hydratase B | −1.588 | −2.634 | −2.372 | 3 |
| AL538_RS10235 | Phosphoenolpyruvate carboxykinase (ATP) | −1.458 | −3.015 | −2.504 | 3 |
| Glycolysis and gluconeogenesis | |||||
| AL538_RS06930 | Phosphoglycerate kinase | −1.341 | −2.545 | −2.503 | 3 |
| AL538_RS10235 | Phosphoenolpyruvate carboxykinase (ATP) | −1.458 | −3.015 | −2.504 | 3 |
| AL538_RS04660 | Type I glyceraldehyde-3-phosphate dehydrogenase | −1.325 | −2.871 | −2.660 | 3 |
| Lipid biogenesis | |||||
| AL538_RS16785 | Lipid A biosynthesis lauroyl acyltransferase | 0.664 | 0.993 | 1.203 | 1 |
| AL538_RS25885 | Choline dehydrogenase | 1.176 | 1.370 | 0.993 | 5 |
| AL538_RS03425 | Acyl-CoA thioesterase | 0.878 | 1.704 | 1.073 | 5 |
| AL538_RS06930 | Phosphoglycerate kinase | −1.341 | −2.545 | −2.503 | 3 |
| AL538_RS26865 | Glycerophosphoryl diester phosphodiesterase | −2.629 | −3.534 | −2.947 | 2 |
| AL538_RS23675 | Acyl-CoA dehydrogenase | −2.053 | −2.768 | −2.308 | 2 |
| AL538_RS23680 | Enoyl-CoA hydratase | −2.034 | −2.947 | −2.470 | 2 |
| AL538_RS27060 | Beta-ketoacyl-ACP reductase | −1.347 | −2.564 | −2.983 | 3 |
| AL538_RS03900 | Malonyl CoA-acyl carrier protein transacylase | −1.815 | −2.683 | −2.368 | 3 |
| Acetyl-CoA-dependent metabolism | |||||
| AL538_RS14965 | Peptidoglycan-associated lipoprotein | −1.663 | −2.943 | −2.801 | 3 |
| AL538_RS04550 | Bifunctional acetaldehyde-CoA/alcohol Dehydrogenase | −1.470 | −3.161 | −2.871 | 3 |
| AL538_RS08465 | Acetyl-CoA synthetase | −1.601 | −2.312 | −2.177 | 3 |
| Processing of genetic information | |||||
| AL538_RS06035 | Elongation factor G | −1.316 | −2.547 | −2.559 | 3 |
| AL538_RS12075 | Energy-dependent translational throttle protein EttA | −1.550 | −2.247 | −2.244 | 3 |
| AL538_RS02125 | Translation initiation factor IF-3 | −1.436 | −2.093 | −2.476 | 6 |
| Ribosome biogenesis | |||||
| AL538_RS07900 | 30S ribosomal protein S12 | −1.454 | −2.108 | −2.716 | 6 |
| AL538_RS10850 | 50S ribosomal protein L22 | −0.954 | −2.066 | −2.065 | 6 |
| AL538_RS03245 | Ribonucleotide-diphosphate reductase subunit beta | −1.553 | −2.794 | −2.503 | 3 |
| Transporters and ancillary factors | |||||
| Energy-dependent transport TonB-ExbB-ExbD complex | |||||
| AL538_RS26550 | Biopolymer transporter ExbB | 1.623 | 2.108 | 1.740 | 5 |
| AL538_RS19445 | TonB-system energizer ExbB | 0.201 | 1.186 | 1.025 | 5 |
| ABC transporters | |||||
| AL538_RS26870 | ABC transporter substrate-binding protein | −2.683 | −4.749 | −3.279 | 2 |
| AL538_RS19380 | Amino acid ABC transporter | −1.236 | −1.698 | −1.907 | 3 |
| AL538_RS00950 | Amino acid ABC transporter substrate-binding protein | −1.188 | −2.888 | −2.524 | 3 |
| AL538_RS01680 | Peptide ABC transporter substrate-binding protein | −2.146 | −4.524 | −3.021 | 3 |
| AL538_RS01780 | Polyamine ABC transporter ATP-binding protein | −2.676 | −3.963 | −3.018 | 2 |
| AL538_RS01795 | Spermidine/putrescine ABC transporter substrate-binding protein | −2.751 | −4.780 | −3.382 | 2 |
| Other transporters | |||||
| AL538_RS01055 | EamA family transporter | 0.950 | 1.603 | 1.870 | 5 |
| AL538_RS25015 | Long-chain fatty acid transporter | 0.640 | 0.909 | 1.212 | 1 |
| AL538_RS21850 | MFS transporter | 1.249 | 1.445 | 1.537 | 1 |
| AL538_RS17670 | PTS mannitol transporter subunit IIA | 1.036 | 1.597 | 1.373 | 5 |
| Protein turnover and folding | |||||
| Protein degradation | |||||
| AL538_RS01370 | Alkaline serine protease | −1.378 | −3.486 | −2.372 | 4 |
| AL538_RS12810 | Aminoacyl-histidine dipeptidase | −1.960 | −2.991 | −2.215 | 2 |
| AL538_RS10020 | Oligopeptidase A | −1.105 | −1.895 | −2.198 | 6 |
| AL538_RS01150 | Peptidase A24 | 0.249 | 1.061 | 1.581 | 1 |
| AL538_RS01675 | Peptidase M20 | 0.859 | 0.886 | 1.368 | 1 |
| AL538_RS24140 | Peptidase M50 | 1.118 | 0.888 | 1.354 | 1 |
| AL538_RS25255 | Peptidase S8 | 0.698 | 0.813 | 1.005 | 1 |
| AL538_RS18135 | D-alanyl-D-alanine carboxypeptidase | −1.318 | −2.530 | −2.336 | 3 |
| AL538_RS01705 | Xaa-Pro aminopeptidase | −2.220 | −4.042 | −3.019 | 3 |
| Protein folding | |||||
| AL538_RS25315 | Molecular chaperone | 0.594 | 1.004 | 1.011 | 5 |
| AL538_RS05430 | Molecular chaperone | −1.777 | −1.784 | −2.056 | 2 |
| AL538_RS00890 | RNA chaperone ProQ | −1.198 | −1.487 | −2.016 | 6 |
| Energy production | |||||
| AL538_RS03195 | Cytochrome c nitrite reductase subunit NrfD | 0.565 | 1.438 | 0.764 | 5 |
| AL538_RS00765 | Cytochrome-c oxidase, cbb3-type subunit III | −1.764 | −2.531 | −2.308 | 2 |
| ATP synthesis coupled proton transport | |||||
| AL538_RS09605 | F0F1 ATP synthase subunit delta | −1.768 | −3.170 | −2.672 | 3 |
| AL538_RS09595 | F0F1 ATP synthase subunit gamma | −1.825 | −3.548 | −2.778 | 3 |
| Fumarate reductase (complex II) | |||||
| AL538_RS08265 | Fumarate reductase flavoprotein subunit | −1.862 | −2.041 | −1.584 | 2 |
| AL538_RS08280 | Fumarate reductase subunit D | −1.508 | −2.045 | −1.847 | 2 |
| Stress responses | |||||
| Cell envelope stress: Phage-shock-protein response | |||||
| AL538_RS15985 | Phage shock protein PspA | −0.908 | 0.819 | −1.526 | 4 |
| AL538_RS15995 | Envelope stress response membrane protein PspC | −1.042 | −0.263 | −1.819 | 4 |
| Stringent response | |||||
| AL538_RS11565 | Stringent starvation protein B | −1.121 | −0.789 | −1.168 | 2 |
| Antioxidative defence | |||||
| AL538_RS17640 | Catalase | 1.260 | 2.075 | 1.668 | 5 |
| AL538_RS21845 | Glutaredoxin, GrxB family | 0.713 | 1.418 | 0.700 | 5 |
| Nitric oxide detoxification | |||||
| AL538_RS24635 | Nitrite reductase large subunit | 1.472 | 1.227 | 1.433 | 1 |
| DNA damage, repair and synthesis | |||||
| AL538_RS18185 | Deoxyribodipyrimidine photolyase | 1.446 | 1.734 | 1.280 | 5 |
| AL538_RS23750 | DNA mismatch repair protein MutT | 1.004 | 0,776 | 1.593 | 1 |
| Iron uptake, storage & utilization | |||||
| AL538_RS26680 | Ferric reductase | 0.938 | 1.185 | 1.140 | 5 |
| Iron uptake/transport | |||||
| AL538_RS21155 | Iron permease | 0.959 | 1.295 | 1.249 | 1 |
| AL538_RS23515 | Iron ABC transporter | 1.203 | 1.009 | 0.937 | 1 |
| Iron storage | |||||
| AL538_RS07875 | Bacterioferritin | −2.859 | −2.868 | −3.496 | 2 |
| AL538_RS12375 | Ferredoxin, 2Fe-2S type, ISC system | −0.365 | −0.256 | −1.152 | 4 |
| Iron cluster biogenesis | |||||
| AL538_RS16405 | Cysteine desulfurase | −2.740 | −3.362 | −3.312 | 2 |
| Virulence factors involved in V. harveyi pathogenicity | |||||
| AL538_RS26540 | Putative heme utilization radical SAM enzyme HutW | 1.028 | 1.301 | 0.458 | 5 |
| AL538_RS20930 | virK protein | 0.708 | 1.912 | 1.755 | 5 |
| Quorum Sensing | |||||
| AL538_RS06465 | LuxR family transcriptional regulator | −1.543 | −2.549 | −2.843 | 3 |
| AL538_RS19310 | Sensor histidine kinase | 1.253 | 1.080 | 1.148 | 1 |
| AL538_RS25660 | TonB-dependent siderophore receptor | 1.338 | 1.520 | 1.380 | 5 |
| Chemotaxis, Motility & Biofilm | |||||
| AL538_RS25425 | Chemotaxis protein | 0.892 | 1.340 | 1.235 | 5 |
| AL538_RS21340 | Flagellar biosynthesis protein FlhB | 0.891 | 1.557 | 1.256 | 5 |
| AL538_RS25995 | Pilus assembly protein CpaF | 1.152 | 1.522 | 1.409 | 5 |
| Secretion systems | |||||
| AL538_RS15160 | EscJ/YscJ/HrcJ family type III secretion inner membrane ring protein (T3SS) | 1.077 | 1.212 | 1.477 | 1 |
| AL538_RS23415 | Type VI secretion protein | 1.769 | 1.838 | 1.741 | 5 |
| Drug efflux & Antibiotic resistance | |||||
| AL538_RS20560 | Bcr/CflA family drug resistance efflux transporter | 0.695 | 1.286 | 1.148 | 5 |
| AL538_RS25145 | Polyketide cyclase | 0.622 | 1.104 | 1.046 | 5 |
Discussion
Ongoing GW profoundly affects the physicochemical parameters and biological variety of natural aquatic systems. The relatively low level of dissolved organic matter in marine environments was shown to constrain bacterial growth in seawater microcosms (e.g31,46–48.) and is believed to be one of the major factors accountable for the presence of coccoid-like bacterioplankton in marine ecosystems47,49. To investigate the combined impact of elevated temperatures and limitation of carbon on Vibrio species, we used V. harveyi as a model organism and studied its persistence in seawater microcosms at 30 °C known to be within the upper limit of sea surface temperature observed in some areas of the global oceans50.
Analysis of cell morphology by fluorescence and electron microscopy revealed a discernible reduction of cell size by 3 day after exposure to microcosm conditions. In silico processing of cell images corroborated the overall reduction of cell size until the sixth day of incubation with no significant changes afterwards (Figs 2 and 3). The transition to the coccoid-like phenotype is consistent with the previously documented response of V. harveyi to carbon limitation32. Similar changes in morphology (i.e. size reduction and shape alterations) have been reported in old cell cultures of V. parahaemolyticus and V. angustum subjected to carbon starvation in marine mineral medium under laboratory conditions51–53. In natural aquatic systems, carbon concentration varies and brief periods of carbon abundance are normally interspersed with long periods of its scarcity. Therefore, it is quite common that free-living bacteria have reduced size in their natural habitats due to the shortage of carbon sources54. Comparison of three independent experiments performed with the same V. harveyi strain and under the same laboratory settings at 4 °C32, 20 °C31 and 30 °C (present work), demonstrates that reduction of cell size and occasional acquisition of a coccoid-like phenotype occur in a wide range of temperatures (i.e. from 4 °C to 30 °C). Moreover, elevated temperature (i.e. 30 °C) and shortage of carbon appear to limit the cell’s capacity to cope with stress and ultimately lead to the appearance of damaged cells seen in 21-day-old populations (Fig. 3). The appearance of damaged cells is congruent with a higher percentage of degraded RNA detected in V. harveyi cells after 14 and 21 days of incubation (data not shown). Moreover, an increase in the number of damaged cells is also consistent with a 90% loss of culturability within the entire population (Fig. 1) observed during the same period of time (i.e. after 14 days). Thus, in contrast to V. harveyi persistence and preservation of culturability at 20 °C31, its exposure to limitation of carbon at 30 °C leads to the higher vulnerability of the stressed cells to damage, which apparently causes their faster injury and potential death.
To learn about the major adaptation mechanisms that V. harveyi and likely other Vibrio species trigger to cope with elevated temperatures and limitation of nutrients in their natural habitats, we used RNA sequencing to obtain transcriptome profiles of V. harveyi following its incubation in seawater microcosm at 30 °C for 12 hours, 3 and 6 days. Comparison of these transcriptome data with those obtained for the control samples (initial inoculate) revealed a large number of genes that were differentially expressed during the incubation. Time series clustering analysis of the expression data (see Experimental procedures) revealed six distinct co-expressed clusters (Fig. 4), each comprising a set of genes with similar time-dependent expression profiles. The differentially expressed genes were further grouped according to their biological functions and presented in Table S1.
The fast morphological changes and accumulation of cells with damaged membranes (see above) indicate that V. harveyi was unable to properly control expression of genes involved in cell envelope biogenesis and associated metabolic pathways. The expression of phage shock proteins PspA, PspB and PspC (cluster 4) (Table 1), known for their role in relieving membrane stress55, is consistent with this explanation. In contrast to the previously described gradual upregulation of these genes in V. harveyi grown at 20 °C31, in this work, expression of these genes under limitation of carbon at 30 °C decreases after 6 day of incubation. This indicates the failure of V. harveyi to engage the products of the phage shock operon in the adaptation process.
Another example of “unexpected” response concerns lipid biogenesis and transport. Although the shrinkage of V. harveyi cell envelope associated with the reduction of cell size is anticipated to lead to a discharge of extra lipids and other membrane components for further recycling, we surprisingly found that expression of genes controlling lipid turnover decreased. A number of genes that are involved in lipid degradation via β-oxidation pathway (i.e. gene encoding phosphoglycerate kinase, glycerophosphoryl diester phosphodiesterase, acyl-CoA dehydrogenase, enoyl-CoA hydratase, beta-ketoacyl-ACP reductase, malonyl CoA-acyl carrier protein transacylase) (clusters 2 and 3) were strongly downregulated. The downregulation of genes controlling β-oxidation pathway as well as those involved in further conversion of acetyl-CoA, the key cellular metabolite generated via β-oxidation pathway, by the enzymes of the glyoxylate cycle (clusters 2 and 3) was unexpected and, in fact, was opposite to their upregulation at 20 °C observed in our previous work31. Thus, it seems likely, that, despite the considerably fast shrinkage of cell envelope and concomitant release of free lipids under carbon limiting conditions at 30 °C, V. harveyi fails to upregulate the key genes, the products of which are involved in lipid recycling. Nevertheless, the upregulation of some genes coding for long-chain fatty acid transporters (AL538_RS25015) could indicate that V. harveyi might try to deal with an excess of lipids accumulated due to reduction of cell size by changing their subcellular localization or exporting them outside of cells. In addition to some anticipated problems with lipid recycling, the shrinkage of cell envelope could additionally be hindered by discoordinate expression of major enzymes (for example, murein L,D-transpeptidase (cluster 4) and D-alanyl-D-alanine carboxypeptidase (cluster 3)) involved in peptidoglycan remodeling. Besides alterations in expression of genes controlling cell envelope biogenesis, another large group of the affected genes included those related to the V. harveyi central carbon metabolism. Analysis of their regulation revealed a fast downregulation of many genes involved in carboxylic acid metabolism (e.g. aconitate hydratase B (acnB), phosphoenolpyruvate carboxykinase; see Table S1 in Supplementary Data) as well as those controlling carbon storage such as the one encoding CsrB known for its roles in post-transcriptional control of gene expression. The downregulation of these genes is likely caused by limitation of carbon in seawater and it was observed already after 12 hours of incubation. Moreover, the initial rate of downregulation was considerably faster in the present study (i.e. at 30 °C) than that previously observed at 20 °C31, thus pointing to the overall increase in the rate of the cellular response to carbon scarcity at elevated temperatures. In addition, we found that, while decreasing the intensity of the central carbon metabolism, V. harveyi apparently attempts to switch to utilization of alternative carbon sources by upregulating a number of genes (e.g. alpha-amylase (cluster 1) and several alpha- and beta-mannosidase genes (belonging to clusters 1 and 5) encoding various hydrolases able to convert some polysaccharides naturally present in seawater into smaller metabolizable sugars.
An additional option to scavenge carbon sources from a carbon-limiting environments consists in secretion of proteinases able to degrade polypeptides potentially present in the environment (e.g. within the infected host cells56) into oligopeptides or amino acids for their further uptake by the starved cells. This strategy is less costly then de novo protein synthesis and is likely preferred by V. harveyi unable to maintain the required level of amino acids during its persistence in seawater microcosm under carbon-limiting conditions. Consistent with this scenario, the key enzymes involved in amino acid biosynthesis (i.e. arginine-succinate synthase, hydroxyproline-2-epimerase or ornithine cyclodeaminase from cluster 2; carbamoyl-phosphate synthase small subunit, cluster 3; glutamate synthase, cluster 6) were downregulated. Concomitantly, V. harveyi upregulated a number of genes (cluster 1) coding for various proteinases such as the subtilisin-serine peptidase S8 and protease M20. Most members of the peptidase S8 family are secreted and they are known to be implicated in the pathogenesis along with other metalloproteases by damaging host tissues57,58. The periplasmic peptidases belonging to M20 family are carboxy-metallo-proteases59 appear to assist the uptake of peptides by converting them into free amino acids in the periplasm. In contrast to the above examples, several protease genes were found to be downregulated. Their repression could be caused by different factors including the high cost of their operation (such as ATP-dependence of oligopeptidase A), reduced cellular needs in some specialized enzymes (e.g. proteases (HtpX and metalloprotease RseP) involved in protein quality control) or simply due to their redundancy (see Table S1).
Under carbon limitation and elevated temperature, V. harveyi also up- and downregulates a large number of transporter-encoding genes primarily found in clusters 1 and 5 (Fig. 4). Some upregulated genes control (i) the energy-dependent transport mediated by the TonB-ExbB-ExbD complex, (ii) multidrug efflux transporters that belong to the multidrug and toxic compound extrusion (MATE) family and (iii) membrane transport proteins of the major facilitator superfamily (MFS) that assist movement of small solutes across cell membranes. Many genes coding for ABC- and PTS-type transporters (clusters 2, 3 and 5) were downregulated, thus indicating that the transport of the corresponding molecules (certain sugars and amino acids) is likely taken over by alternative transport systems or is deactivated due to its redundancy.
Marine bacteria in the open ocean experience limitation of iron60, an essential metal vital for all living organisms. Consistent with the necessity to deal with low concentrations of iron in seawater microcosms, we observed that a number of genes involved in iron uptake/transport and storage were up- and downregulated, respectively. Moreover, incubation in seawater seems to activate some adaptive mechanisms aimed at protecting V. harveyi cells from the possible damage potentially caused by reactive oxygen species (ROS) and/or nitric oxide (NO). Consistently, we found that V. harveyi upregulated several stress-responsive genes (cluster 5) encoding catalase, alkyl hydroperoxide reductase subunit F, glutaredoxin as well as some nitrate reductases and cytochrome c nitrite reductase subunit NrfD able to minimize (or reduce) the damaging effects of ROS and / or NO.
Incubation at elevated temperature also affected regulation of V. harveyi genes controlling its virulence and ancillary mechanisms (i.e. production and secretion of virulence factors, biofilm formation and motility). We found that, in addition to increased expression of lytic enzymes including various glycosidases and proteinases, V. harveyi upregulated a number of genes encoding the Type III61, Type I62, Type IV63 and Type VI64 secretion systems involved in transport of virulence factors.
Similarly, upregulation was observed for structural proteins of flagella and pilus (flagellar biosynthesis proteins FlhB and FliR from cluster 5; pilus assembly proteins CpaB, CpaF, PapD, PilN, PilZ, TadB and TadE FliR from cluster 1 and 5) as well as ancillary enzymes such as peptidase A24 (gene AL538_RS01150) required for the type IV pilus formation, secretion of toxin and enzymes, gene transfer and biofilm formation65. The upregulation of the above genes can play an important role in cell motility and attachment during the initials steps of infection. Moreover, the overall increase in motility and capacity to attach to biotic surfaces could also increase V. harveyi chances to find new sources of food (e.g. via attachment to sea shells whose components (e.g. amino polysaccharides) can further be processed by secreted hydrolyzes to yield amino sugars in carbon-limiting environments.
Finally, we found that the gene coding for LuxR, a transcription factor mediating the quorum sensing (QS) response in a large variety of Vibrio spp66. was markedly downregulated, thereby suggesting that QS might not play a significant role in V. harveyi response to elevated SST and carbon limitation. Meanwhile, many genes encoding for proteins of two-component (sensor histidine kinase/response regulator) systems (CreC, UhpB and others, cluster 1 and 5) playing essential roles in sensing environmental signals and accordingly adjusting gene expression67 were upregulated. Several two-component systems previously described in vibrios and shown to be upregulated in the present study are involved in pathogenicity68–70. Therefore, their upregulation might enhance the pathogenic potential of V. harveyi exposed to elevated temperature and carbon limitation in seawater microcosm.
In summary, we observed a significant size reduction and acquisition of coccoid-like morphology by V. harveyi cells in seawater microcosms during their long-term incubation under carbon limitation at 30 °C. Since morphological changes, loss of culturability and cell damage are more pronounced at 30 °C than at 20 °C31, our data suggest that shortage of carbon at elevated temperature has a negative effect on V. harveyi. Despite its adverse effect on V. harveyi survival due to counter adaptive downregulation of some metabolic pathways (e.g. those controlling β-oxidation pathway), incubation in sea water microcosm at 30 °C readily enhances expression of many known virulence factors (i.e. lytic enzymes, components of the T3SS secretion system and iron-chelating compounds; reviewed in71) apparently accountable for the increased presence of pathogenic Vibrio species in the microbiota of marine invertebrate during the warm seasons72 and therefore potentially important for the spread of Vibrio-associated diseases in response to GW9–12. As the morphological, physiological and gene expression changes were observed in seawater microcosms, the rate and extent of Vibrio spp. responses to elevated temperature and limitation of carbon in their natural habitats are not necessarily the same and can likely be modulated by protozoan grazing, intensity of solar radiation and other environmental factors influencing the dynamics of marine ecosystems. Therefore, it is conceivable that some of the conclusions are limited by the experimental design.
Electronic supplementary material
Acknowledgements
This work was supported by the Spanish Ministry of Economy and Competitiveness grant CGL2015-70929-R, Ikerbasque (Basque Foundation for Science) and pre-doctoral grants PRE-2013-1-901 (I.M.) from the Basque Government and PIF15/101 (E.O.) from the University of the Basque Country. A.E.-C. is funded by ISCIII of the MINECO (Ref: PT17/0009/0019) and cofinanced by FEDER.
Author Contributions
I.M. and V.R.K. conceived the experiment(s), I.M., E.O., M.O. and I.A. conducted the experiment(s), I.M., E.O., A.H.P., A.E.-G. and J.G.-G. analyzed the results, I.M. and V.R.K. wrote the manuscript. I.M., I.A. and A.H.P. prepared the figures. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-36483-0.
References
- 1.Yildiz FH, Visick KL. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 2009;17:109–118. doi: 10.1016/j.tim.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell S, Harada RM, DeFelice SV, Bienfang PK, Li QX. Bacterial production of tetrodotoxin in the pufferfish Arothron hispidus. Nat Prod Res. 2009;23:1630–1640. doi: 10.1080/14786410903003780. [DOI] [PubMed] [Google Scholar]
- 3.Owens, L., Busico-Salcedo, N. Vibrio harveyi: Pretty problems in paradise. In: Thompson, FL, Austin, B, Swings, J (eds) The Biology of Vibrios. ASM Press., Washington, D. C., 266–280 (2006)
- 4.Ruwandeepika HA, et al. Presence of typical and atypical virulence genes in vibrio isolates belonging to the Harveyi clade. J Appl Microbiol. 2010;109:888–899. doi: 10.1111/j.1365-2672.2010.04715.x. [DOI] [PubMed] [Google Scholar]
- 5.Le Roux F, et al. The emergence of Vibrio pathogens in Europe: ecology, evolution, and pathogenesis (Paris, 11-12th March 2015) Front Microbiol. 2015;6:830. doi: 10.3389/fmicb.2015.00830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones MK, Oliver JD. Vibrio vulnificus: disease and pathogenesis. Infect Immun. 2009;77:1723–1733. doi: 10.1128/IAI.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris JG., Jr. Cholera and other types of vibriosis: a story of human pandemics and oysters on the half shell. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2003;37:272–280. doi: 10.1086/375600. [DOI] [PubMed] [Google Scholar]
- 8.Baker-Austin C, et al. Vibrio spp. infections. Nat Rev Dis Primers. 2018;4:8. doi: 10.1038/s41572-018-0005-8. [DOI] [PubMed] [Google Scholar]
- 9.Martínez-Urtaza J, Bowers JC, Trinanes J, DePaola A. Climate anomalies and the increasing risk of Vibrio parahaemolyticus and Vibrio vulnificus illnesses. Food Res Int. 2010;43:1780–1790. doi: 10.1016/j.foodres.2010.04.001. [DOI] [Google Scholar]
- 10.McMichael AJ, Woodruff RE, Hales S. Climate change and human health: present and future risks. Lancet. 2006;367:859–869. doi: 10.1016/S0140-6736(06)68079-3. [DOI] [PubMed] [Google Scholar]
- 11.Paz S, Bisharat N, Paz E, Kidar O, Cohen D, Climate change and the emergence of Vibrio vulnificus disease in Israel. Environ Res. 2007;103:390–396. doi: 10.1016/j.envres.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Vezzulli L, Colwell RR, Pruzzo C. Ocean warming and spread of pathogenic vibrios in the aquatic environment. Microb Ecol. 2013;65:817–825. doi: 10.1007/s00248-012-0163-2. [DOI] [PubMed] [Google Scholar]
- 13.The update, Global Health Workforce Statistics, World Health Organization, Geneva, (http://www.who.int/hrh/statistics/hwfstats/)2017).
- 14.Letchumanan V, Chan KG, Lee LH. Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front Microbiol. 2014;5:705. doi: 10.3389/fmicb.2014.00705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vezzulli L, et al. Climate influence on Vibrio and associated human diseases during the past half-century in the coastal North Atlantic. Proc Natl Acad Sci USA. 2016;113:E5062–5071. doi: 10.1073/pnas.1609157113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albuquerque A, Cardoso H, Pinheiro D, Macedo G. Vibrio cholerae non-O1 and non-O139 bacteremia in a non-traveler Portuguese cirrhotic patient: first case report. Gastroenterol Hepatol. 2013;36:309–310. doi: 10.1016/j.gastrohep.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Julie D, et al. Ecology of pathogenic and non-pathogenic Vibrio parahaemolyticus on the French Atlantic coast. Effects of temperature, salinity, turbidity and chlorophyll a. Environ Microbiol. 2010;12:929–937. doi: 10.1111/j.1462-2920.2009.02136.x. [DOI] [PubMed] [Google Scholar]
- 18.Baker-Austin C, et al. Emerging Vibrio risk at high latitudes in response to ocean warming. Nature Climate Change. 2013;3:73–77. doi: 10.1038/nclimate1628. [DOI] [Google Scholar]
- 19.Martinez-Urtaza J, et al. Spread of Pacific Northwest Vibrio parahaemolyticus strain. N Engl J Med. 2013;369:1573–1574. doi: 10.1056/NEJMc1305535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Urtaza J, et al. Environmental determinants of the occurrence and distribution of Vibrio parahaemolyticus in the rias of Galicia, Spain. Appl Environ Microbiol. 2008;74:265–274. doi: 10.1128/AEM.01307-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez-Urtaza J, et al. Epidemiological investigation of a foodborne outbreak in Spain associated with U.S. West Coast genotypes of Vibrio parahaemolyticus. Springerplus. 2016;5:87. doi: 10.1186/s40064-016-1728-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvell CD, et al. Climate warming and disease risks for terrestrial and marine biota. Science. 2002;296:2158–2162. doi: 10.1126/science.1063699. [DOI] [PubMed] [Google Scholar]
- 23.Martin GG, Rubin N, Swanson E. Vibrio parahaemolyticus and V. harveyi cause detachment of the epithelium from the midgut trunk of the penaeid shrimp Sicyonia ingentis. Dis Aquat Organ. 2004;60:21–29. doi: 10.3354/dao060021. [DOI] [PubMed] [Google Scholar]
- 24.Soto-Rodriguez SA, Roque A, Lizarraga-Partida ML, Guerra-Flores AL, Gomez-Gill B. Virulence of luminous vibrios to Artemia franciscana nauplii. Dis Aquat Organ. 2003;53:231–240. doi: 10.3354/dao053231. [DOI] [PubMed] [Google Scholar]
- 25.Lee KK, Liu PC, Chuang WH. Pathogenesis of gastroenteritis caused by Vibrio carchariae in cultured marine fish. Mar Biotechnol (NY) 2002;4:267–277. doi: 10.1007/s10126-002-0018-9. [DOI] [PubMed] [Google Scholar]
- 26.Liu P-C, Lin J-Y, Chuang W-H, Lee K-K. Isolation and Characterization of Pathogenic Vibrio harveyi (V. carchariae) From the farmed marine cobia fish Rachycentron canadum L. with Gastroenteritis Syndrome. World J Microbiol Biotechnol. 2004;20:495–499. doi: 10.1023/B:WIBI.0000040402.44340.0e. [DOI] [Google Scholar]
- 27.Austin B, Zhang XH. Vibrio harveyi: a significant pathogen of marine vertebrates and invertebrates. Lett Appl Microbiol. 2006;43:119–124. doi: 10.1111/j.1472-765X.2006.01989.x. [DOI] [PubMed] [Google Scholar]
- 28.Lutz C, Erken M, Noorian P, Sun S, McDougald D. Environmental reservoirs and mechanisms of persistence of Vibrio cholerae. Front Microbiol. 2013;4:375. doi: 10.3389/fmicb.2013.00375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Townsley L, Sison-Mangus MP, Mehic S, Yildiz FH. Response of Vibrio cholerae to low-temperature shifts: CspV regulation of Type VI secretion, biofilm formation, and association with zooplankton. Appl Environ Microbiol. 2016;82:4441–4452. doi: 10.1128/AEM.00807-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farmer, J. J. The Family Vibrionaceae. In: Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E. (eds) The Prokaryotes 3rd edn. Springer Science and Business Media, New York, 495–507 (2006).
- 31.Kaberdin VR, et al. Unveiling the metabolic pathways associated with the adaptive reduction of cell size during Vibrio harveyi persistence in seawater microcosms. Microb Ecol. 2015;70:689–700. doi: 10.1007/s00248-015-0614-7. [DOI] [PubMed] [Google Scholar]
- 32.Parada C, et al. Changes in the Vibrio harveyi cell envelope subproteome during permanence in cold seawater. Microb Ecol. 2016;72:549–558. doi: 10.1007/s00248-016-0802-0. [DOI] [PubMed] [Google Scholar]
- 33.Orruno M, Kaberdin VR, Arana I. Survival strategies of Escherichia coli and Vibrio spp.: contribution of the viable but nonculturable phenotype to their stress-resistance and persistence in adverse environments. World J Microbiol Biotechnol. 2017;33:45. doi: 10.1007/s11274-017-2218-5. [DOI] [PubMed] [Google Scholar]
- 34.Ramaiah N, Ravel J, Straube WL, Hill RT, Colwell RR. Entry of Vibrio harveyi and Vibrio fischeri into the viable but nonculturable state. J Appl Microbiol. 2002;93:108–116. doi: 10.1046/j.1365-2672.2002.01666.x. [DOI] [PubMed] [Google Scholar]
- 35.Chróst, R. J. Microbial Ectoenzymes in Aquatic Environments. In: Overbeck J., Chróst R. J. (eds) Aquatic Microbial Ecology. Springer, New York, N. Y (1990).
- 36.Kizhakudan SJ, et al. Correlation between changes in sea surface temperature and fish catch along Tamil Nadu coast of India - an indication of impact of climate change on fisheries? Indian J Fish. 2014;61:111–115. [Google Scholar]
- 37.Salas-Perez J, González-Gándara C. Temporal and spatial fluctuations of sea surface temperature and chlorophyll a levels due to atmospheric forcing in a tropical coastal lagoon. Ciencias Marinas. 2016;42:49–65. doi: 10.7773/cm.v42i1.2551. [DOI] [Google Scholar]
- 38.Alonso-Sáez A, et al. Changes in marine prokaryotic community induced by varying types of dissolved organic matter and subsequent grazing pressure. J Plankton Research. 2009;31:1373–1383. doi: 10.1093/plankt/fbp081. [DOI] [Google Scholar]
- 39.Hobbie JE, Daley RJ, Jasper S. Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977;33:1225–1228. doi: 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Massana R, et al. Measurement of bacterial size via image analysis of epifluorescence preparations: description of an inexpensive system and solutions to some of the most common problems. Sci Mar. 1997;61:397–407. [Google Scholar]
- 41.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Futschik ME, Carlisle B. Noise-robust soft clustering of gene expression time-course data. J Bioinform Comput Biol. 2005;3:965–988. doi: 10.1142/S0219720005001375. [DOI] [PubMed] [Google Scholar]
- 45.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 46.Baker RM, Singleton FL, Hood MA. Effects of nutrient deprivation on Vibrio cholerae. Appl Environ Microbiol. 1983;46:930–940. doi: 10.1128/aem.46.4.930-940.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Felter RA, Colwell RR, Chapman GB. Morphology and round body fermation in Vibrio marinus. J Bacteriol. 1969;99:326–335. doi: 10.1128/jb.99.1.326-335.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Novitsky JA, Morita RY. Morphological characterization of small cells resulting from nutrient starvation of a psychrophilic marine vibrio. Appl Environ Microbiol. 1976;32:617–622. doi: 10.1128/aem.32.4.617-622.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ducklow, H. W. Bacterioplankton. In: Steele, J, Thorpe, S, Turekian, K (eds) Encyclopedia of Ocean Sciences. Elsevier Ltd, Amsterdam, 269–275 (2001).
- 50.Pierrehumbert RT. Climate change and the tropical Pacific: the sleeping dragon wakes. Proc Natl Acad Sci USA. 2000;97:1355–1358. doi: 10.1073/pnas.97.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen SY, Jane WN, Chen YS, Wong HC. Morphological changes of Vibrio parahaemolyticus under cold and starvation stresses. Int J Food Microbiol. 2009;129:157–165. doi: 10.1016/j.ijfoodmicro.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 52.Holmquist L, Kjelleberg S. Changes in viability, respiratory activity and morphology of the marine Vibrio sp. strain S14 during starvation of individual nutrients and subsequent recovery. FEMS Microbiol. Ecology. 1993;12:215–223. doi: 10.1111/j.1574-6941.1993.tb00034.x. [DOI] [Google Scholar]
- 53.Stretton S, Danon SJ, Kjelleberg S, Goodman AE. Changes in cell morphology and motility in the marine Vibrio sp. strain S14 during conditions of starvation and recovery. FEMS Microbiol Lett. 1997;146:23–29. doi: 10.1111/j.1574-6968.1997.tb10166.x. [DOI] [Google Scholar]
- 54.McDougald, D., Kjelleberg, S. Adaptative Responses of Vibrios. In: Thompson, FL, Austin, B, Swings, J (eds) The Biology of Vibrios. ASM Press., Washington, D. C., 133–155 (2006).
- 55.Joly N, et al. Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol Rev. 2010;34:797–827. doi: 10.1111/j.1574-6976.2010.00240.x. [DOI] [PubMed] [Google Scholar]
- 56.Liu PC, Lee KK. Cysteine protease is a major exotoxin of pathogenic luminous Vibrio harveyi in the tiger prawn, Penaeus monodon. Lett Appl Microbiol. 1999;28:428–430. doi: 10.1046/j.1365-2672.1999.00555.x. [DOI] [PubMed] [Google Scholar]
- 57.Saulnier D, et al. A large-scale epidemiological study to identify bacteria pathogenic to Pacific oyster Crassostrea gigas and correlation between virulence and metalloprotease-like activity. Microb Ecol. 2010;59:787–798. doi: 10.1007/s00248-009-9620-y. [DOI] [PubMed] [Google Scholar]
- 58.Zhang YZ, Ran LY, Li CY, Chen XL. Diversity, structures, and collagen-degrading mechanisms of bacterial collagenolytic proteases. Appl Environ Microbiol. 2015;81:6098–6107. doi: 10.1128/AEM.00883-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rawlings ND, Barrett AJ. Evolutionary families of metallopeptidases. Methods Enzymol. 1995;248:183–228. doi: 10.1016/0076-6879(95)48015-3. [DOI] [PubMed] [Google Scholar]
- 60.Johnson KS, Gordon RM, Coale KH. What controls dissolved iron concentrations in the world ocean? Marine Chemistry. 1997;57:137–161. doi: 10.1016/S0304-4203(97)00043-1. [DOI] [Google Scholar]
- 61.Henke JM, Bassler BL. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J Bacteriol. 2004;186:3794–3805. doi: 10.1128/JB.186.12.3794-3805.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Delepelaire P. Type I secretion in gram-negative bacteria. Biochim Biophys Acta. 2004;1694:149–161. doi: 10.1016/j.bbamcr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 63.Juhas M, Crook DW, Hood DW. Type IV secretion systems: tools of bacterial horizontal gene transfer and virulence. Cell Microbiol. 2008;10:2377–2386. doi: 10.1111/j.1462-5822.2008.01187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ho BT, Dong TG, Mekalanos JJ. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe. 2014;15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.LaPointe CF, Taylor RK. The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J Biol Chem. 2000;275:1502–1510. doi: 10.1074/jbc.275.2.1502. [DOI] [PubMed] [Google Scholar]
- 66.Bassler BL. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol. 1999;2:582–587. doi: 10.1016/S1369-5274(99)00025-9. [DOI] [PubMed] [Google Scholar]
- 67.Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu Rev Microbiol. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Geszvain K, Visick KL. The hybrid sensor kinase RscS integrates positive and negative signals to modulate biofilm formation in Vibrio fischeri. J Bacteriol. 2008;190:4437–4446. doi: 10.1128/JB.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martinez-Wilson HF, Tamayo R, Tischler AD, Lazinski DW, Camilli A. The Vibrio cholerae hybrid sensor kinase VieS contributes to motility and biofilm regulation by altering the cyclic diguanylate level. J Bacteriol. 2008;190:6439–6447. doi: 10.1128/JB.00541-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shikuma NJ, et al. Overexpression of VpsS, a hybrid sensor kinase, enhances biofilm formation in Vibrio cholerae. J Bacteriol. 2009;191:5147–5158. doi: 10.1128/JB.00401-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ruwandeepika HA, et al. Pathogenesis, virulence factors and virulence regulation of vibrios belonging to the Harveyi clade. Reviews in. Aquaculture. 2012;4:59–74. doi: 10.1111/j.1753-5131.2012.01061.x. [DOI] [Google Scholar]
- 72.King WL, et al. characterisation of the pacific oyster microbiome during a summer mortality event. Microb Ecol. 2018 doi: 10.1007/s00248-018-1226-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.