Abstract
Caspase-2 is a highly conserved cysteine protease with roles in apoptosis and tumor suppression. Our recent findings have also demonstrated that the tumor suppression function of caspase-2 is context specific. In particular, while caspase-2 deficiency augments lymphoma development in the EμMyc mouse model, it leads to delayed neuroblastoma development in Th-MYCN mice. However, it is unclear how caspase-2 mediates these differential outcomes. Here we utilized RNA sequencing to define the transcriptomic changes caused by caspase-2 (Casp2−/−) deficiency in tumors from EμMyc and Th-MYCN mice. We describe key changes in both lymphoma and neuroblastoma-associated genes and identified differential expression of the EGF-like domain-containing gene, Megf6, in the two tumor types that may contribute to tumor outcome following loss of Casp2. We identified a panel of genes with altered expression in Th-MYCN/Casp2−/− tumors that are strongly associated with neuroblastoma outcome, with roles in melanogenesis, Wnt and Hippo pathway signaling, that also contribute to neuronal differentiation. In contrast, we found that key changes in gene expression in the EμMyc/Casp2−/− tumors, are associated with increased immune signaling and T-cell infiltration previously associated with more aggressive lymphoma progression. In addition, Rap1 signaling pathway was uniquely enriched in Casp2 deficient EμMyc tumors. Our findings suggest that Casp2 deficiency augments immune signaling pathways that may be in turn, enhance lymphomagenesis. Overall, our study has identified new genes and pathways that contribute to the caspase-2 tumor suppressor function and highlight distinct roles for caspase-2 in different tissues.
Introduction
The role of caspases in tumor suppression has been largely ascribed to their established function in cell death1. Recent experimental evidence also suggests that non-apoptotic roles for caspases, including cell proliferation, inflammation, migration, or invasion, can contribute to tumorigenesis2,3. These features are well-established hallmarks of cancer, both independently and in co-operation with cell death evasion4, and may determine caspase contribution to both tumor suppression and tumor promotion in certain contexts3. While the mechanisms that regulate caspase functions in tumorigenesis are still unclear, there is evidence to suggest that caspases mediate their differential functions in a developmental and tissue-specific manner1,2.
Caspase-2 is the most evolutionarily conserved member of the mammalian caspase family5, with roles in apoptosis6, tumor suppression7–11, genomic stability12, DNA damage response12,13, oxidative stress response, metabolism, and ageing6,14–18. While mice deficient for the caspase-2 gene (Casp2−/−) do not develop spontaneous age-related tumors19, they show enhanced genomic instability and increased tumorigenesis in different mouse models, including EμMyc lymphomas7,20, Atm−/−thymomas8; MMTV/c-neu mammary tumors11, K-Ras-induced lung tumors10, and DEN-induced hepatocellular carcinoma (HCC)21. Recent studies suggest that caspase-2 mediates its tumor suppressor function by inducing senescence and/or apoptosis of aneuploid cells22,23. As a consequence, loss of caspase-2 leads to the survival of abnormal, multinucleated, and aneuploid cells, which are features of mitotic catastrophe and susceptibility to tumorigenesis15,21,22.
Correlative evidence supports a tumor suppressor function for caspase-2. The human CASP2 gene on Ch7q34, is part of a region frequently deleted in hematological malignancies24. In addition, reduced CASP2 expression has been reported in lymphoma and leukemia and correlates with poor prognosis in AML and ALL25,26. Somatic mutations in CASP2, although rare, are found in cases of high-grade colon and gastric cancer, lung, skin, and breast cancer19,27,28. Furthermore, in colorectal cancer reduced CASP2 expression is caused by loss of its transcriptional regulator BCL9L, and is a key cause of aneuploidy tolerance, tumor progression, and resistance29. In contrast, Casp2 deficiency in mice does not affect tumor onset or progression following 3-methylcholanthrene (3-MC)-induced fibrosarcoma or irradiation-driven lymphoma20. Our previous studies have also demonstrated that Casp2 loss delays neuroblastoma development in the Th-MYCN mouse model, and that low CASP2 expression correlates with increased survival in human neuroblastoma30. These findings suggest that caspase-2 has a context/tissue-specific function in tumor suppression and that both tissue and genomic variability can cooperate with caspase-2 to determine tumor outcome. To understand how caspase-2 mediates its differential functions in tumor suppression, it is important to determine if disruption of additional genes co-operate with caspase-2 in tissue-specific contexts.
In this study, we comparatively analyzed the transcriptomes of Casp2−/− tumors from EμMyc and Th-MYCN mice to identify changes in transcriptional tumor networks that are influenced by caspase-2 deficiency. While we identified several unique genes that were aberrantly expressed in the Casp2−/− tumors, we found that Megf6/EGFL3 was the only gene that was differentially expressed in the two tumor types. Our study also identified several enriched pathways and gene signatures specific to EμMyc/Casp2−/− or Th-MYCN/Casp2−/− tumors that may be associated with enhanced EμMyc-induced lymphomas and/or delayed Th-MYCN-mediated neuroblastoma. The cross-tumor-specific transcriptional aberrations are highly indicative of distinct roles for caspase-2 during neuronal and B-cell development that perhaps influence its tumor suppressor function.
Results
RNA-seq analysis of Th-MYCN and EμMyc tumors from Casp2−/− mice
To identify the transcriptomic differences in tumors from Casp2−/− mice that are associated with delayed Th-MYCN induced neuroblastoma30 and/or enhanced EμMyc lymphoma development7, we carried out RNA sequencing on tumor tissue isolated from Th-MYCN and Th-MYCN/Casp2−/− or from EμMyc and EμMyc/Casp2−/− mice (Fig. 1a, Supplementary Tables S1a-b). Multidimensional scaling plots showed that the EμMyc and Th-MYCN tumor transcriptomes form distinct clusters (Supplementary Figure S1a), highlighting differences in gene expression changes between the two tumor types.
Fig. 1. Experimental design and transcriptome profile of tumors from Th-MYCN/Casp2−/− and EµMyc/Casp2−/− mice.
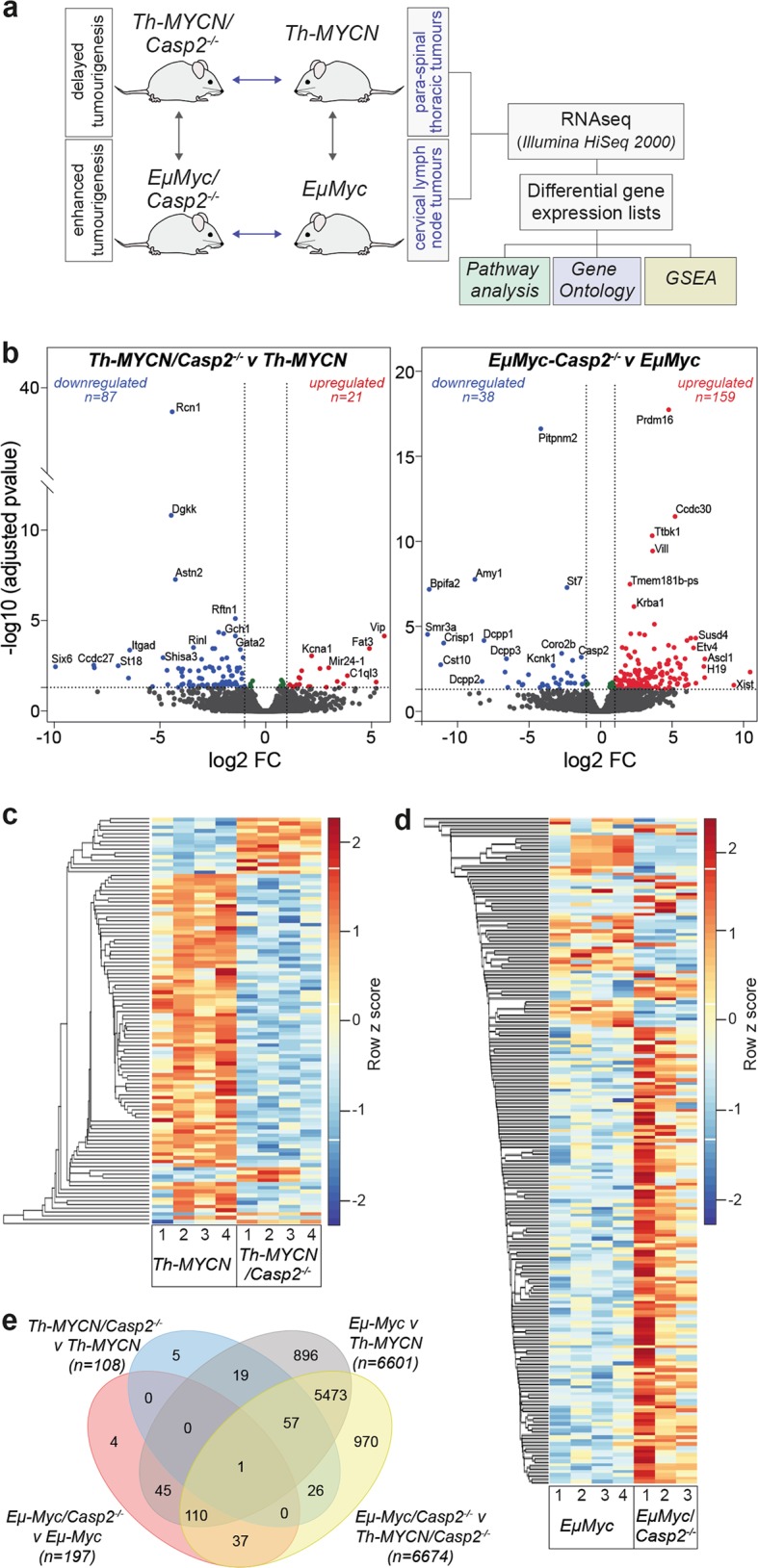
a Experimental design for RNA-seq and analysis of differentially expressed genes (DEGs) from tumor tissue. b Volcano plot illustrating DEGs in a comparison of Th-MYCN/Casp2−/− and Th-MYCN tumor samples (top) or EµMyc/Casp2−/− and EµMyc tumor samples. Colored points represent all DEGs (cut-off FDR < 0.05) with fold change > 2, that are either overexpressed (red) or underexpressed (blue) in Casp2−/− compared with WT tumor counterparts. For a complete list of DEGs, see Supplementary Tables S2a and S2b. c, d Heat-maps of DEGs when comparing c Th-MYCN/Casp2−/− and Th-MYCN and d EµMyc/Casp2−/− and EµMyc tumor transcriptomes. Heat-maps display the number of increased (red) or decreased (blue) genes. For gene lists associated with heat-maps see Supplementary Tables S3a and S3b. e Four-way Venn diagram summary of unique and overlapping DEGs in the indicated comparison groups. The list of unique and overlapping genes in each group is provided in Supplementary Table S4a
Comparisons of the differentially expressed genes (DEGs) were performed between (i) WT and Casp2−/− tumors to identify genes associated with tumorigenesis following Casp2 loss and (ii) between the EμMyc/Casp2−/− and Th-MYCN/Casp2−/− tumor samples, to narrow down candidate genes affected by Casp2 deficiency and/or that enhance lymphomagenesis in EμMyc/Casp2−/− mice. These DEGs are summarized as volcano plots (Fig. 1b, Supplementary Tables S2a-f). From the heat-maps, it is clear that Casp2 deficiency changes the expression of many genes, with more downregulated genes in Th-MYCN/Casp2−/− compared to Th-MYCN tumors (Fig. 1c) and more upregulated genes in EμMyc/Casp2−/− versus EμMyc tumors (Fig. 1d, Supplementary Tables S3a-b).
In total, 13,714 genes were included for analysis in each comparison group (Supplementary Tables S2a-f). Of these, 108 DEGs were identified in the Th-MYCN/Casp2−/− versus Th-MYCN tumor comparison and 197 DEGs in the EμMyc/Casp2−/− versus EμMyc tumors (fold change [FC] > 2 and < −2; FDR < 0.05). As expected, there were extensive differences in gene expression when comparing the two different tumor types, including 6601 DEGs when comparing EμMyc to Th-MYCN tumors and 6674 DEGs in the EμMyc/Casp2−/− versus Th-MYCN/Casp2−/− tumor comparison. A 4-way Venn diagram identified several exclusive and common DEGs between each comparison group (Fig. 1e, Supplementary Table S4a). In particular, the Th-MYCN/Casp2−/− comparison identified four unique downregulated genes (Ncor2, Diras2, Ednrb, Shb) and one upregulated (Eya2) gene, with roles in transcription (Ncor2), signal transduction (Ednrb, Shb), melanogenesis (Ednrb), and DNA damage repair (Eya2). Two of the unique EμMyc/Casp2−/− genes have associated roles in transcription regulation (Six5) and calmodulin binding (Nrgn) and have been previously associated with lung squamous cell carcinoma, breast cancer (Six531,32), and T-cell lymphoma (Nrgn33). Tdrd5 was excluded as it was only increased in one out of three biological replicates. Significant downregulation of Casp2 was also verified in the EμMyc/Casp2−/− comparison and Casp2 gene deletion was confirmed by genotyping and immunoblotting in all tumor samples.
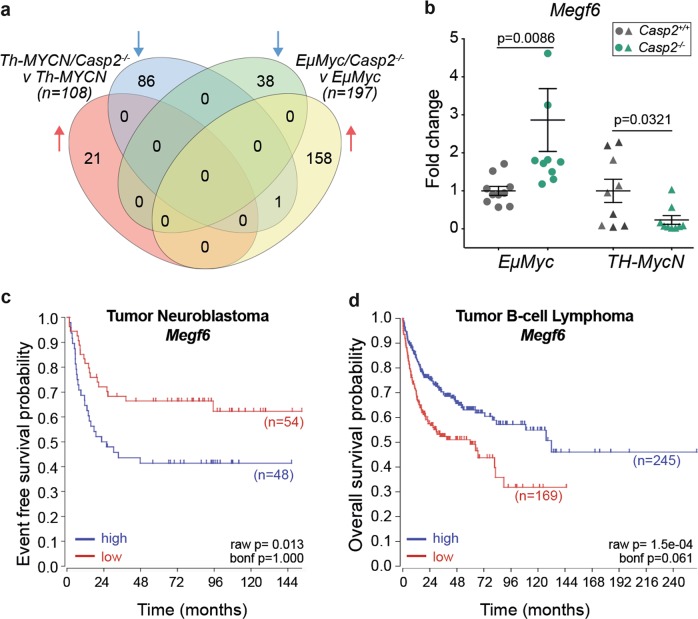
To identify common genes that are significantly altered in the Casp2−/− samples, the DEGs were subdivided into upregulated and downregulated lists, illustrated in a Venn diagram (Fig. 2a). This identified 86 uniquely downregulated and 21 upregulated genes in Th-MYCN/Casp2−/− versus Th-MYCN tumors and 38 downregulated and 158 upregulated genes in EμMyc/Casp2−/− versus EμMyc tumors (Fig. 2a, Supplementary Table S4b). There was also a single overlapping gene; Megf6 (Multiple EGF-Like Domains 6) differentially altered in EμMyc/Casp2−/− and Th-MYCN/Casp2−/− tumors, and this was validated by quantitative PCR in additional tumor samples (Fig. 2b).
Fig. 2. Comparison of upregulated and downregulated genes in Th-MYCN/Casp2−/−and EµMyc/Casp2−/−tumors.
a Four-way Venn diagram summary illustrating common and exclusive upregulated and downregulated genes when comparing Th-MYCN/Casp2−/− versus Th-MYCN and EµMyc/Casp2−/− versus EµMyc tumor transcriptomes (FC > 2 and < −2; FDR < 0.05). Complete gene lists are provided in Supplementary Table S4b. b Quantitative PCR analysis of differentially expressed Megf6 gene from the indicated tumor groups. Values represent means ± S.E.M, with P-values indicated (n = 9/group). c, d Kaplan–Meier plots showing the association of Megf6 expression with patient survival in Neuroblastoma (c) and B-cell lymphoma (d)
We used R2: Genomics Analysis and Visualization Platform, to examine the correlation between Megf6 transcript expression and clinical outcome in various human neuroblastoma and B-cell lymphoma publicly available expression array data sets. Our previous data showed that Casp2 levels correlated with clinical outcome in a subset of MYCN non-amplified human neuroblastomas30. Here, we also found that higher Megf6 transcript levels are associated with poor outcome in this neuroblastoma subset (Fig. 2c), and this is consistent with our RNA-seq data and the delayed tumorigenesis observed in the Th-MYCN/Casp2−/− mouse model30. We did not find any significant association in MYCN-amplified neuroblastoma. In contrast, we found there was a trend for lower Megf6 expression being predictive of poorer B-cell lymphoma outcome (e.g., Xiao – 420, fRMA-u133p2 dataset; P = 0.061) (Fig. 2d). Notably, high Megf6 expression in B-cell lymphomas also showed a somewhat poor survival outcome, indicating that Megf6 levels are probably not predictive of lymphoma progression.
Functional enrichment analysis of genes in Th-MYCN/Casp2−/− tumors
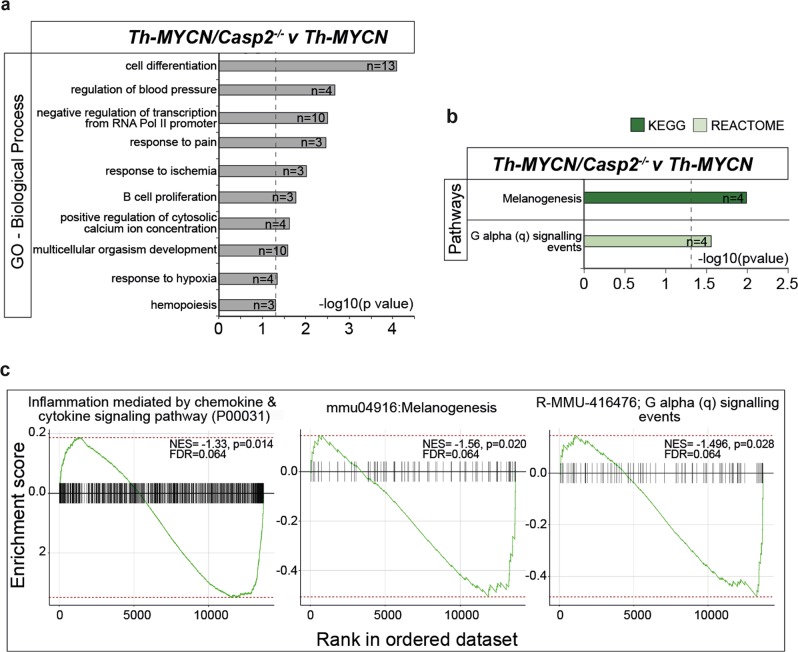
We next identified significantly altered gene ontology processes that were enriched in Casp2−/− tumors, compared to their WT counterparts. We firstly analyzed the DEGs in the Th-MYCN/Casp2−/− versus Th-MYCN tumor comparison. While there were no detected ontology terms associated with the small upregulated gene list (n = 21), the 87 downregulated genes were associated with 10 categories of ‘Biological Process’ (BP), 4 categories of ‘Molecular Function’ (MF), and 9 categories of ‘Cellular Component’ (CC) (P < 0.05, Fisher’s exact test) (Fig. 3a, Supplementary Table S5a). The top BP was cell differentiation (GO:0030154) (Fig. 3a, Supplementary Table S5a), with many of these genes also being associated with the BPs of transcriptional regulation from RNA polymerase II promoter (GO:0000122) and multicellular organismal development (GO:0007275). This indicates that Casp2 loss can affect neuronal differentiation and gene transcription in neuroblastoma. Of note, response to hypoxia (GO:0001666) and ischemia (GO:0002931), processes previously associated with caspase-2 function34–37 were also enriched.
Fig. 3. Functional category enrichment analysis of differentially expressed genes in Th-MYCN/Casp2−/−tumors.
a Gene Ontology annotation analysis summary of enriched biological processes associated with unique downregulated genes in Th-MYCN/Casp2−/− tumors (n = 87). Complete lists are provided in Supplementary Table S5a. b Pathway enrichment analysis (KEGG and REACTOME), of the differentially expressed downregulated genes in Th-MYCN/Casp2−/− tumors (n = 87). Complete lists are provided in Supplementary Table S6a. The number of genes associated with each biological process and pathway is indicated. Dotted line indicates significance cut off (-log10 (P-value), Fisher's exact test). c Illustration of GSEA score curves. Enrichment plots are shown for the identified upregulated and downregulated pathways from (b) in the Th-MYCN/Casp2−/− tumor comparison group. Black bars represent the position of members of the category in the ranked list together with the running enrichment score (plotted in green). NES = normalized enrichment score, FDR = false discovery rate
KEGG pathway analysis identified melanogenesis as the only enriched pathway, associated with four downregulated genes (Ednrb, Mitf, Lef1, Camk2a) (Fig. 3b, Supplementary Table S6a). Further analysis using REACTOME and PANTHER classification systems also identified G-alpha (q) signaling events (P = 0.03), inflammation mediated by chemokine and cytokine signaling pathway (P = 0.018) (Supplementary Table S6a). Gene set enrichment analysis (GSEA)38, using the gene list ranked by the FDR score, confirmed significant enrichment of each of these pathways (Fig. 3c).
Functional enrichment analysis of genes in EμMyc/Casp2−/− tumors
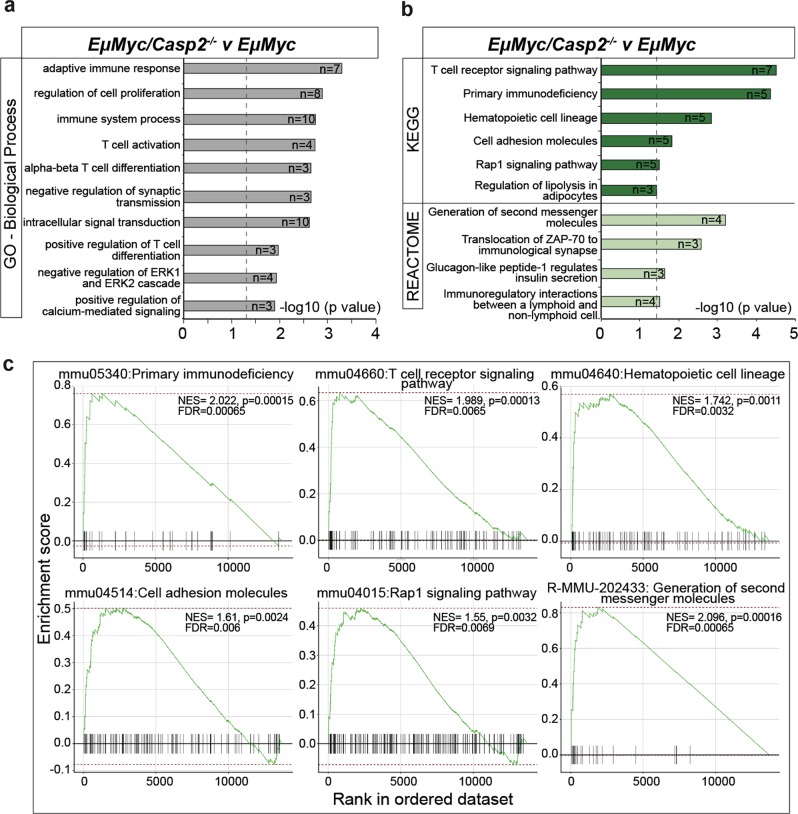
Analysis of GO terms in the EμMyc/Casp2−/− versus EμMyc tumor comparison identified several BPs associated the 159 upregulated genes, including immune response (GO:0002250, GO:0006955), immune system process (GO:0002376), T-cell activation (GO:0042110), T-cell differentiation (GO:0046632, GO:0045582, GO:0033077, GO:0030217), and inflammatory response (GO:0006954) (Fig. 4a, Supplementary Table S5b), indicative of a significant immune responses in EμMyc/Casp2−/− tumors. Increased processes commonly associated with tumorigenesis including signal transduction (GO:0035556, GO:0007165, GO:0009966, GO:0007169, GO:0070373), cell proliferation (GO:0042127, GO:0008285), and negative regulation of cell adhesion (GO:0007162) were also enriched (Fig. 4a and Supplementary Table S5b).
Fig. 4. Functional category enrichment analysis of differentially expressed genes in EµMyc/Casp2−/−tumors.
a Gene Ontology annotation analysis summary of the top 10 enriched biological processes associated with unique upregulated genes in EµMyc/Casp2−/− tumors (n = 159). Complete lists are provided in Supplementary Table S5b. b Pathway enrichment analysis (KEGG and REACTOME), of the differentially expressed upregulated genes in EµMyc/Casp2−/− tumors (n = 159). Complete lists are provided in Supplementary Table S6b. The number of genes associated with each biological process and pathway is indicated. Dotted line indicates significance cut off (-log10 (P-value), Fisher's exact test). c Illustration of gene set enrichment analysis (GSEA) score curves. Enrichment plots are shown for the identified upregulated and downregulated pathways from the EµMyc/Casp2−/− tumor comparison group. Black bars represent the position of members of the category in the ranked list together with the running enrichment score (plotted in green). NES = normalized enrichment score, FDR = false discovery rate
Consistent with the enriched BPs, KEGG pathway analysis identified T-cell receptor signaling and primary immunodeficiency as the most significantly altered pathways followed by cell adhesion molecules and Rap1 signaling (Fig. 4b, Supplementary Table S6b). All pathways were significantly enriched when analyzed using GSEA (Fig. 4a, c), suggesting that caspase-2 may have a role in regulating T-cell signaling and/or Rap1 signaling to influence lymphoma development in EμMyc mice.
Differential expression of genes in caspase-2 deficient tumors associated with increased survival in human neuroblastoma
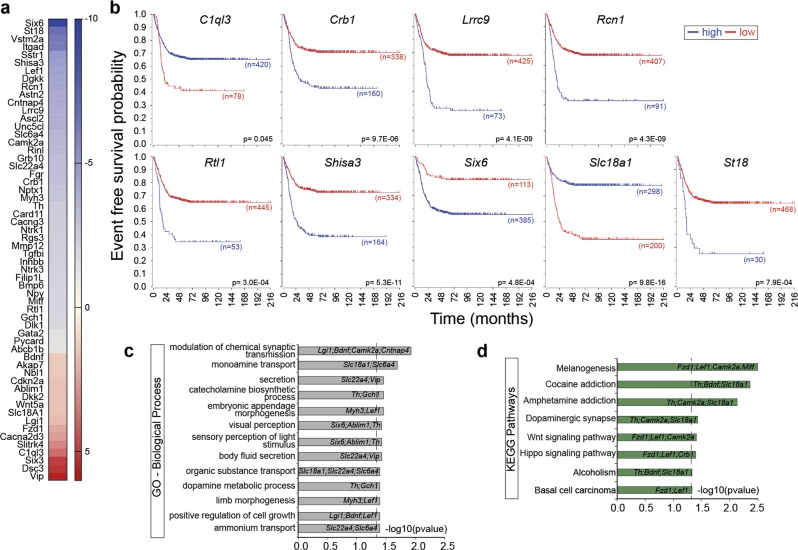
To identify genes associated with delayed neuroblastoma development in Th-MYCN/Casp2−/− mice, we examined various molecular biomarkers of neuroblastoma development and prognosis39,40. The expression levels of several favorable neuroblastoma genes were found to be significantly altered in Th-MYCN/Casp2−/− tumors, including increased expression of Slc18A1, Vip, and reduced expression of Gata2, Gch1, Fgr, and Six6. While the decrease in tyrosine hydroxylase (Th) expression was not significant in the biological replicates (FDR =0.584) (Fig. 5a, Supplementary Table S7a), we did note that several genes adjacent to Th on chromosome 7 (Ascl2, Igf2, and H19) were also somewhat decreased (Supplementary Table S7b). This suggests this gene region may be affected in some cells.
Fig. 5. Differentially expressed genes in Th-MYCN/Casp2−/−tumors associated with survival in neuroblastoma patients.
a Heat-map illustrating the increase (red) and decrease (blue) in expression (log2-fold change) of various neuroblastoma-associated genes in Th-MYCN/Casp2−/− tumors compared to Th-MYCN tumors. Gene lists and pathway associations are available in Supplementary Table S7a. b Event-free survival curves for neuroblastoma patients from publicly available expression array data showing the expression correlation of the indicated genes with survival outcome in neuroblastoma. P-values using the method of Kaplan–Meier (Bonferroni correction) are shown. c Gene Ontology annotation analysis of the significantly enriched biological processes (Benjamini adjusted P-value < 0.05), associated with the genes listed in (a), that show altered gene expression (FDR < 0.1) and are strongly associated with neuroblastoma outcome in (b) (n = 41). Complete gene ontology lists are provided in Supplementary Table S7c. d Pathway enrichment analysis (KEGG) associated with neuroblastoma-associated genes above (n = 41). Complete pathway enrichment lists are provided in Supplementary Table S7d. The genes associated with each biological process and pathways are indicated. Dotted line indicates significance cut off [-log10(P-value), Benjamini corrected]
To determine the correlation between the expression of these genes and neuroblastoma survival, we analyzed data from published studies along with publicly available expression array data sets from primary human neuroblastoma tumor samples (R2 database). Overall, we found that downregulation of 7 genes (Crb1, Lrrc9, Rcn1, Rtl1, Shisa3, Six6, St18) and upregulation of 2 genes (C1ql3, Slc18A1), are strongly predictive of favorable neuroblastoma outcome (Fig. 5b, Supplementary Table S7a), consistent with delayed tumor development in Th-MYCN/Casp2−/− mice30.
Our analysis also identified altered expression of several known neuroblastoma-associated genes that would be predictive of unfavorable neuroblastoma outcome (Supplementary Figure S3b, Supplementary Table S7a). These include decreased levels of Ntrk1, Ntrk3 that are associated with poorly differentiated tumors and poor prognosis in neuroblastoma40,41 (Fig. 5a, Supplementary Table S7a). We also did not find significant changes in other neuroblastoma marker genes (e.g., Bcl2, Cd44, Dcc, Ddx1, Lmo1, Max, Mdr1/Abcb1, Mrp/ Abcc1, Nf1, Ngf, Ntf3, Ntrk2, Nme1, Odc1, Rb1, Trp53)39 (Supplementary Table S2a). These findings importantly show that these genes do not necessarily have a role in delayed tumor onset in Th-MYCN/Casp2−/− and/or their role in neuroblastoma onset may be counteracted by loss of Casp2.
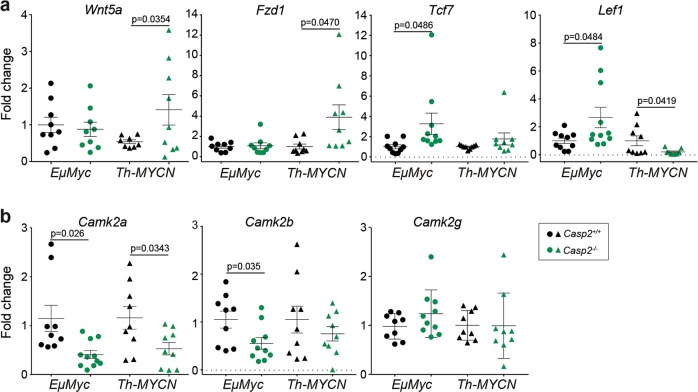
Gene ontology analysis of the significantly altered neuroblastoma genes indicated their involvement in synaptic transmission, monoamine transport and cell growth regulation (Fig. 5c, Supplementary Table S7b). Interestingly, many of these genes are associated with melanogenesis, dopaminergic synapse, Wnt, and Hippo signaling (Fig. 5d, Supplementary Tables S7a and S7c). Downregulation of melanogenesis is associated with reduced expression of the transcription factor Mitf, along with Wnt signaling pathway components (Fzd1, Lef1, Camk2a). We noted a trend of altered expression of other Wnt signaling components (Wnt5a, Dkk2, Bmp6; FDR > 0.1) and their regulators, including Gata2 and its target homeobox transcription factors Six3 and Hesx1 (Supplementary Tables S2a, S7a), which also have roles in neural determination42,43. Increased expression of Wnt5a and Fzd1 and decreased expression of Lef1 and Camk2a were validated in different tumor samples by quantitative PCR (Fig. 6a, b). Together, these data identify altered expression levels of several Wnt-associated genes and suggest a fine-tuning of Wnt signaling components in Th-MYCN/Casp2−/− tumors that may determine neural crest cell (NCC) specification to influence neuroblastoma onset.
Fig. 6. Validation of the differential expression of Wnt pathway associated genes in Casp2−/− tumors.
Quantitative PCR analysis of the Wnt pathway associated genes Wnt5a, Fzd1, Tcf7, and Lef (a) and CamkII family members Camk2a, Camk2b, Camk2g (b) in EµMyc/Casp2−/− and Th-MYCN/Casp2−/− tumors as indicated. Values represent mean ± S.E.M, with P-values indicated (n = 8–11/group)
Differential expression of genes in caspase-2 deficient tumors associated with enhanced EuMyc lymphoma
To identify candidate oncogenes and tumor suppressor genes differentially expressed in the EμMyc/Casp2−/− tumors that may contribute to enhanced lymphomagenesis, we interrogated several published cancer gene lists44–46 and databases (Sanger Cancer Gene Consensus; Bushman Lab ‘allOnco’ list; Oncogene Tumor Suppressor Gene Resource). This analysis identified a 147-gene signature in the EμMyc/Casp2−/− tumors that included increased expression of a set of 72 oncogenes, decreased expression of nine genes with known or predicted tumor suppressor functions and 12 genes reported to have both oncogenic and tumor suppressor functions (Figure S7a, Supplementary Table S8a). Many oncogenes have previously been associated with EμMyc-mediated tumor progression and/or B-cell lymphoma47–50. Several of these genes are clustered on nearby chromosome regions, including Ch.4 (Megf6-Prdm1, Arhgef16), Ch.6 (Cd4-Lag3, Cd8b-Cd8a), Ch.9 (Cd3d, Cd3g, Cd3e), Ch.10 (Dgka-Pme1), and Ch.11 (Sgsh, Slc26a11, Card14) (Supplementary Table S8b) indicative of possible common regulatory elements or amplification of these regions in the EμMyc/Casp2−/− tumors. These findings highlight changes in the expression of multiple cancer-associated genes and/or gene regions that likely contribute to enhanced EμMyc lymphomagenesis onset.
The majority of the genes that make up the Myc core signature (MCS), associated with lymphoma malignancy and progression51 were not further altered in EμMyc/Casp2−/− compared to EμMyc tumors (Fig. 7, Supplementary Table S8a). Interestingly, we found nine common upregulated genes, which form part of the Bcor expression signature in EμMyc mice52, (Bcl6, Camkk1, Cr2, Hivep3, Kcnb1, Lef1, Slamf1, St3gal5, Ube2l6) (Supplementary Table S8a). However, there was no evidence of an associated increase in TGFβ signaling components in EμMyc/Casp2−/− tumors, suggesting that Bcor-associated TGFβ signaling may not contribute to enhanced lymphomagenesis in EμMyc/Casp2−/− mice.
Fig. 7. Differentially expressed genes in EµMyc/Casp2−/− tumors associated with cancer progression.
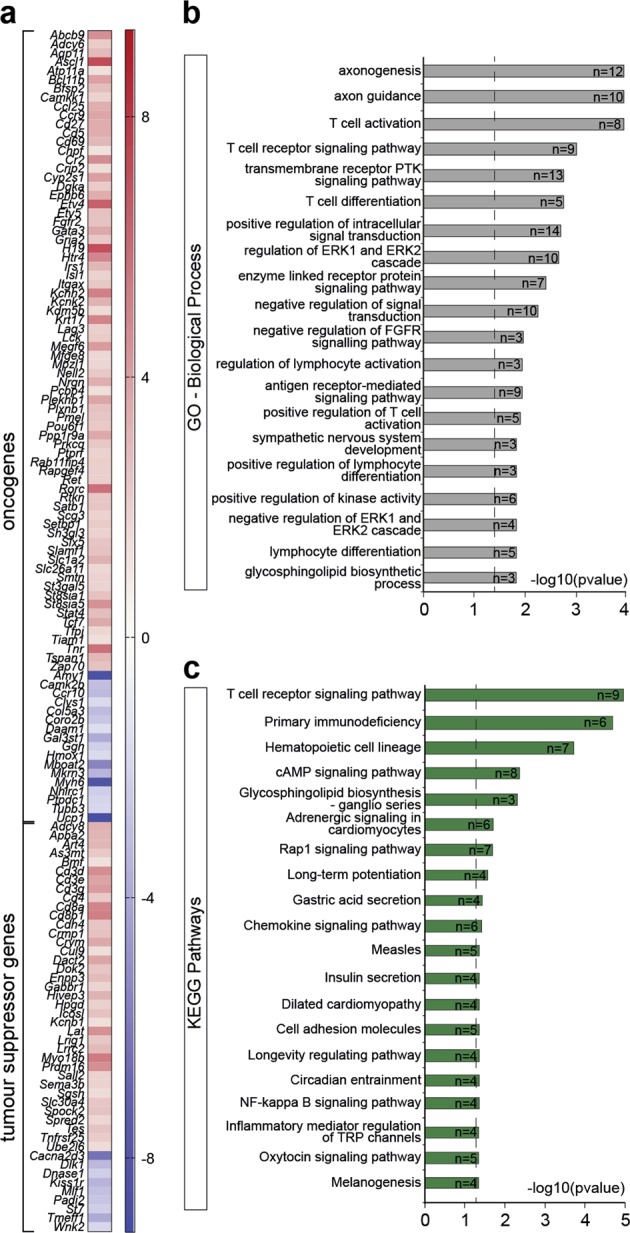
a Heat-map illustrating the increase (red) and decrease (blue) in expression (log2-fold change) of various cancer-associated genes in EµMyc/Casp2−/− tumors. b Gene Ontology annotation analysis of the top 20 significantly enriched biological processes (Benjamini adjusted P < 0.05), associated with the genes listed in (a), that show changes in gene expression (FDR < 0.1) (n = 147). The number of genes associated with each biological process is indicated. Complete gene ontology lists are provided in Supplementary Table S8c. c Pathway enrichment analysis (KEGG and REACTOME), of the cancer-associated genes differentially expressed in EµMyc/Casp2−/− tumors (n = 147). The top 20 enriched pathways are shown. Complete enriched pathway lists are provided in Supplementary Table S8d. The genes associated with each pathway are indicated. Dotted line indicates significance cut off (-log10(P-value), Benjamini corrected)
Analysis of the 147-gene signature in EμMyc/Casp2−/− tumors, identified axonogenesis, T-cell signaling, protein kinase B signaling, and signal transduction as the top enriched BPs and MFs (Fig. 7b, Supplementary Table S8c). Consistent with this, T-cell receptor signaling, primary immunodeficiency and hematopoietic cell lineage were the top enriched KEGG pathways (Fig. 7c, Supplementary Table S8d), with Rap1 signaling and cell adhesion molecules also identified from our total DEG list (Fig. 4e). This indicates the DEGs in EμMyc/Casp2−/− tumors are largely established cancer-associated genes.
Interestingly, the pathways melanogenesis and Wnt signaling were identified in the DEGs from both [EμMyc/Casp2−/− v EμMyc] (Fig. 7c, Supplementary Table S8d) and [Th-MYCN/Casp2−/− v Th-MYCN] (Fig. 5d) tumor comparisons. Increased expression of Tcf7, and its co-operating transcription factor Lef1, were further confirmed by quantitative PCR analysis in different EμMyc/Casp2−/− tumor samples (Fig. 6a).
Comparison of DEGs in EμMyc/Casp2−/− compared to Th-MYCN/Casp2−/− tumors
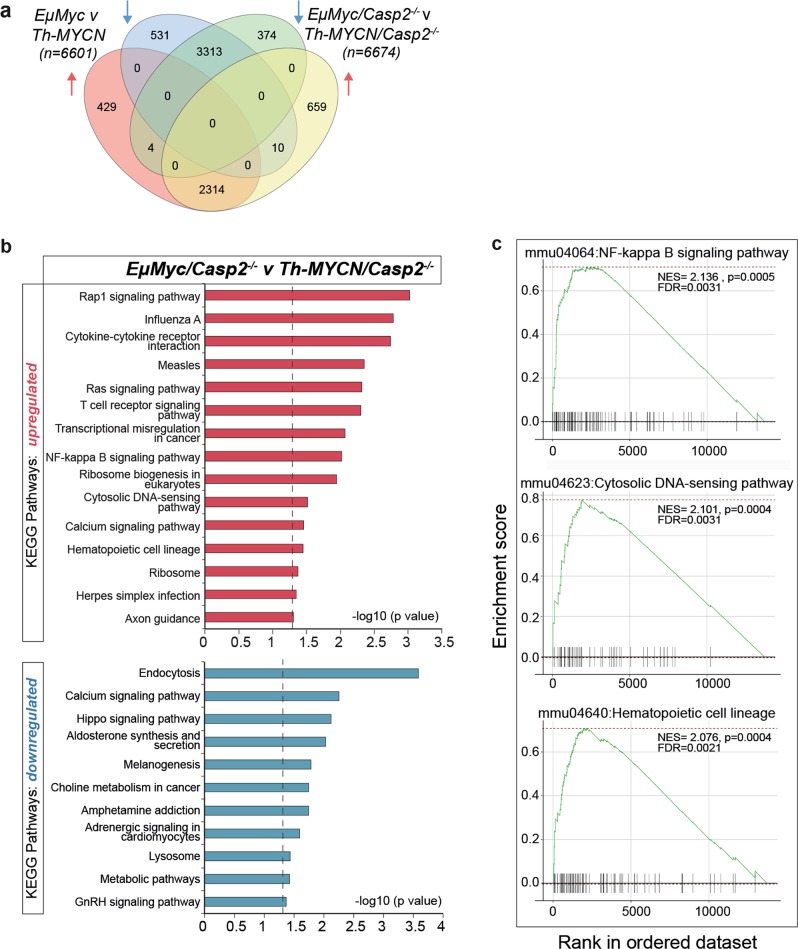
We compared the DEGs from EμMyc/Casp2−/− to Th-MYCN/Casp2−/− tumors in aim to define unique components and pathways associated with enhanced EμMyc-mediated lymphomagenesis. A 4-way Venn diagram identified 659 uniquely upregulated and 374 downregulated genes in the EμMyc/Casp2−/− compared to Th-MYCN/Casp2−/− tumors (Fig. 8a). An additional 14 genes (Abcc2, Ccdc27, H19, Itgax, Krt17, Megf6, Mitf, Mmp12, Rcn1, Rorc and Amy1, Col5a3, Kcnk1, Pitpnm2) were differentially regulated compared to the EμMyc v Th-MYCN DEGs (Supplementary Table S4c), with some genes significantly associated with extracellular matrix organization (Mmp12, Col5a3, Itgax) (GO:0030198, Benjamini corrected P = 0.047), highlighting an important difference in the EμMyc/Casp2−/− tumors.
Fig. 8. Comparison of differentially expressed genes and enriched pathways in EµMyc/Casp2−/− and Th-MYCN/Casp2−/− tumors.
a Four-way Venn diagram summary of unique and overlapping upregulated and downregulated genes in the indicated comparison groups. The list of unique and overlapping genes in each group is provided in Supplementary Tables S4c and S4d. b KEGG Pathway enrichment analysis of upregulated (red) and downregulated (blue) pathways in EµMyc/Casp2−/− tumors compared to Th-MYCN/Casp2−/− tumors. Number of genes associated with each pathway is indicated. Complete enriched pathway lists are provided in Supplementary Table S6d. Dotted line indicates significance cut off (-log10(P-value) Fisher's exact test). c GSEA of the upregulated and downregulated pathways from (b). Black bars represent the position of members of the category in the ranked list together with the running enrichment score (plotted in green). NES = normalized enrichment score, FDR = false discovery rate
Gene ontology analysis of the exclusively upregulated genes (n = 659) identified the BPs immune response, inflammatory response, angiogenesis, and cell migration (Benjamini corrected P < 0.05) (Supplementary Figures S4a-b, Supplementary Tables S5c-d). Interestingly, regulation of apoptosis, response to hypoxia, reactive oxygen species, metabolic process and hematopoietic progenitor cell differentiation are processes consistent with loss of caspase-2 function6 (Supplementary Figure S4b, Supplementary Table S5d). Analysis of the downregulated genes (n = 374) identified 13 unique BPs, with neuron maturation being the most significantly decreased, perhaps consistent with its primary role in Th-MYCN/Casp2−/− tumors (Benjamini corrected P < 0.05). Other notable downregulated pathways (e.g., response to fatty acid, ubiquitination, negative regulation of epithelial cell proliferation, and cell cycle arrest) highlight multiple uniquely deregulated processes, previously been linked to Casp2 loss, and suggest their role in contributing to enhanced lymphomagenesis in EμMyc/Casp2−/−mice.
KEGG pathway analysis identified increased Rap1 signaling and cytokine-cytokine receptor interaction as the most significantly enriched and unique pathways (P<0.001, Fisher's exact test) (Fig. 8b, Supplementary Tables S6c-d). In addition to T-cell receptor signaling identified from the above analysis (Fig. 4b), enrichment of NFκB signaling and cytosolic-DNA sensing were also confirmed by GSEA (Fig. 8c) (Supplementary Table S6d). Interestingly, endocytosis was the top pathway associated with downregulated genes (Benjamini corrected P = 0.049), a process that has emerged as a key hallmark of cancer, by affecting cell surface expression of critical molecules and signaling processes53. Calcium signaling, and Hippo signaling pathways were also uniquely enriched (Fig. 8b, Supplementary Table S6d). Consistent with this, reduced expression of Camk2a and Camk2b was validated in different EμMyc/Casp2−/− tumor samples (Fig. 6b). These findings highlight several new pathways that are deregulated in EμMyc/Casp2−/− tumors that can contribute to tumorigenesis.
Discussion
We used transcriptome profiling to characterize gene expression changes and pathways affected by Casp2 deficiency, in tumors from Th-MYCN neuroblastoma30 and EμMyc lymphoma transgenic mouse models7. Our study has identified specific caspase-2 expression signatures in the different tumor types that likely contribute to the distinct tumor outcomes7,30. The diversity in gene expression highlights the (1) heterogeneity in tumor cell populations and/or clonal co-operation between sub-clones within the same tumor (2) tumor type-specificity (3) aberrations in components of several different signaling pathways, and/or (4) subtle differences that make it difficult to detect single causative pathways.
While the changes in gene expression between the Th-MYCN/Casp2−/− and EμMyc/Casp2−/− comparison groups were distinct, it was interesting that Megf6 was the only gene differentially expressed in the Casp2−/− tumors. Megf6 is implicated in neural system disorders, such as ataxia54 and recently, the epithelial-to-mesenchyme transition (EMT) to promote metastasis in colorectal cancer via TGFβ signaling55. Consistent with a tumor progression function, we found higher expression of Megf6 in EμMyc/Casp2−/− and lower expression in Th-MYCN/Casp2−/− tumors. Lower levels of Megf6 expression also correlated with better survival outcome in human neuroblastoma, suggesting it may contribute to delayed neuroblastoma in the Th-MYCN/Casp2−/− mouse30. The lack of clinical correlation with Megf6 expression in B-cell lymphoma may be indicative of a tissue-specific function for Megf6 and/or that Casp2 loss can cooperate with high Megf6 expression to augment lymphomagenesis in the EμMyc/Casp2−/− model. Nevertheless, these findings provide premise to further investigate Megf6 role and potential co-operation with caspase-2 function in tumorigenesis.
The delayed neuroblastoma onset in Th-MYCN/Casp2−/− mice, previously highlighted the tissue/context specific role for caspase-2 in tumor suppression30. Our study has now identified cell differentiation as the most significantly altered biological process in Th-MYCN/Casp2−/− tumors, associated with downregulation of genes that regulate neuronal differentiation and cell fate determination (Ascl2, Gldn, Mitf, Notch3, Shb, Syt17) and many being transcription factors (Ascl2, Batf3, Cbfa2t3, Elf4, Gata2, Mitf). A role for caspase-2 in neuronal differentiation, remodeling and protection against ischemic injury, has been previously implicated34,56,57. Silencing Casp2 can increase expression of neuronal differentiation markers57, and increased differentiation in Casp2−/− neurons, would be consistent with favorable neuroblastoma outcome40 and delayed tumor onset in Th-MYCN/Casp2−/− mice30. Increased neuronal differentiation is also associated with high ROS levels58, a feature of Casp2−/− cells14,15, that can further contribute to neuronal differentiation58. Identification of caspase-2 substrate(s) and/or interacting partners in differentiating NCCs will be key to defining its role in neurons and elucidate its function in neuroblastoma.
Our analysis identified a 41-gene signature in Th-MYCN/Casp2−/− tumors, associated with neuroblastoma outcome, and involved in altered melanogenesis, Wnt and Hippo pathway signaling. Melanogenesis is known to be inhibited by excessive ROS59 and may therefore be affected by the increased oxidative stress and ROS in Casp2−/− cells14,15. Melanogenesis is also associated with aggressive neuroblastoma, characterized by higher tyrosinase activity and associated increase in DOPA synthesis60–62. The key regulator of tyrosinase is Mitf, was significantly reduced in Th-MYCN/Casp2−/− compared to Th-MYCN tumors, and may suggest a defect in DOPA synthesis in Th-MYCN/Casp2−/− tumors, consistent with the reduction in Dopaminergic synapse signaling. In addition, Wnt signaling regulates melanogenesis by stabilization and signaling through Mitf59. While a role for Wnt signaling in neuroblast differentiation has been described43,63, it has diverse functions in cell proliferation, polarity and migration, so its role in neuroblastoma is likely complex63. Nevertheless, the finding here, show that loss of Casp2 affects several components regulating the Wnt signaling pathway in neuroblastoma, that could impact neuronal differentiation and possibly neuroblastoma onset in Th-MYCN/Casp2−/− mice.
Our previous studies demonstrated that loss of Casp2 accelerates EμMyc-driven lymphomagenesis7. This study has now identified aberrant expression of 147 cancer-associated genes including increased expression of 72 oncogenes and reduced expression of 9 TSG in the EμMyc/Casp2−/− tumor samples. While many of these genes may play co-operating roles with Casp2 loss to enhance EμMyc-mediated lymphomagenesis, aberrant expression of some genes are likely a consequence of enhanced tumor growth caused by Casp2 loss. Interestingly, the key pathways associated with EμMyc/Casp2−/− tumors, was increased immune response, in particular T-cell activation and signaling pathways. Higher expression of CD4, CD25/IL2ra and Lag3 in EμMyc/Casp2−/− tumors, are indicative of activated regulatory T-cells (Tregs) at the tumor site, which are associated with both favorable and unfavorable prognosis in B-cell lymphomas64. They can facilitate tumor immune evasion by directly suppressing B-lymphoma cells, and also function to inactivate tumor specific CD4+/CD8+ T-cells, leading to an immunosuppressive network associated with increased T-cell tolerance and reduced T-cell mediated killing64. This is a key factor in uncontrolled tumor growth and poor clinical outcome in many solid tumors64. Several clinical studies have also described the presence of T-cell markers (including CD3, CD4, CD8) in diffuse large B-cell lymphoma (DLBCL), but the prognostic significance of this is not clear65. Our data are highly suggestive that the active immune response plays a key role in augmenting tumorigenesis in EμMyc/Casp2−/− mice. A role for caspase-2 in regulating inflammasome signaling and the innate immune response has been previously reported6. However, it is also likely that the potential role of caspase-2 in hematopoietic cell differentiation, (e.g., myeloid cell differentiation)66 may contribute to immunosurveillance mechanisms that determine tumor outcomes.
The comparison of gene expression profiles in EμMyc/Casp2−/− tumors to Th-MYCN/Casp2−/− tumors identified a significant enrichment of genes associated with Rap1 signaling. Rap1 regulates cell adhesion; a process also enriched in the EμMyc/Casp2−/− tumors, and perhaps indicates a link between these two processes. While Rap1 can promote tumor cell migration, invasion, and metastasis67, these features have not been observed in EμMyc/Casp2−/− mice, indicating possible alternative roles for Rap1 signaling in EμMyc/Casp2−/− tumors. This may include Rap1 roles in regulating cytoskeletal dynamics and/or cell proliferation, previously associated with loss of caspase-2 function6,12. Alternatively, Rap1 signaling plays diverse roles in hematopoietic cells, including lymphocyte activation, migration/trafficking, immunological synapse67, B-cell development, and self-tolerance68. While the specific and diverse functions of Rap1 are context dependent, a role for caspase-2 in regulating Rap1 signaling is highly relevant to lymphoma onset and progression and will be important to further define the contributing roles of Rap1 and caspase-2 in B-cell lymphomagenesis.
In summary, this is the first description of tissue specific and differential gene expression signatures associated with caspase-2 deficiency that can influence tumorigenesis. In particular, our findings indicating that loss of Casp2 impacts neuronal differentiation, is likely a key contributor to delaying neuroblastoma onset and reduced adrenal tumor development in Th-MYCN/Casp2−/− mice. In contrast, increased immune processes, including hematopoietic cell lineage determination, are potential effectors in EμMyc/Casp2−/− lymphomagenesis. In addition, we have identified aberrant expression of several oncogenes and tumor suppressor genes that potentially co-operate with Casp2 loss, to determine tumor outcome. These findings reinforce the distinct contribution of signaling pathways associated with tumor onset and progression in both Th-MYCN/Casp2−/− and in EμMyc/Casp2−/− mice.
Methods
Tumor samples
Lymphomas from EuMyc and EuMyc/Casp2−/− mice were taken from cervical lymph nodes7. Tumor samples from Th-MYCN and Th-MYCN/Casp2−/− mice were selected from paraspinal thoracic tumors based on absence or reduced blood vascularization30. Four individual tumors per genotype were used for mRNA-seq analysis.
RNA extraction
Total RNA was extracted from frozen tumor tissue using Trizol® reagent (Life Technologies) according to the manufacturer’s protocol. RNA was resuspended in diethylpyrocarbonate-(DEPC) treated water and quantified using a NanoDrop1000™ spectrophotometer (Thermo Scientific). RNA samples were quality tested using a 2100 Bioanalyser (Agilent Technologies) to confirm RNA integrity number (RIN) > 7.
mRNA sequencing and bioinformatics analysis
RNA-seq and bioinformatics analysis was carried out at the ACRF Cancer Genomics Facility (Centre for Cancer Biology, UniSA). Briefly, polyA+ mRNA was enriched using oligo-dT beads and samples were barcoded for pooled sequencing on an Illumina HiSeq 2000 (Agilent Technologies). Short, single-end reads were carried out (1 × 50 bp flow cells) with four samples per lane. This yielded ~20–30 million raw reads per sample.
Sequence quality was analyzed using FastQC and the resulting data sets were aligned using the STAR alignment algorithm (https://www.ncbi.nlm.nih.gov/pubmed/23104886) to the UCSC mm10/GRCm38 version of the Mus musculus reference genome. Gene counts were obtained from the second strand of the STAR output and the resulting files were transferred to the R statistical environment. The edgeR Bioconductor package69 was used to fit a linear model and samples were log transformed for normalization and ranking and then expressed as log2-fold change of expression (Log2FC). Data are available in Gene Expression Omnibus (GEO) under accession GSE124051.
Multidimensional scaling plots were generated to explore the relationships in the data before and after processing. (Supplementary Figure S1a and Supplementary Table S1c). All biological replicates were similar to each other except for EμMyc/Casp2−/− sample #212, which fell outside the EμMyc cluster. Normalization of data was able to correct for this, however a scatterplot matrix analysis of the EμMyc samples indicated that sample #212 was a strong outlier (Supplementary Figure S1a) and did not correlate well with other biological replicates (Supplementary Figure S1b). For this reason, EμMyc/Casp2−/− sample #212 was removed from further differential expression analysis.
Differential expression analysis of individual genes
Differential expression analysis of individual genes was carried out using edgeR. We divided samples into the two tumor types EµMyc and MYCN groups, and also into two genotype groups Casp2+/+ and Casp2−/−. Four-way comparisons in gene expression levels were made, including: (1) Th-MYCN/Casp2−/− versus Th-MYCN; (2) EuMyc/Casp2−/− versus EuMyc; (3) EuMyc versus Th-MYCN, and (4) EuMyc/Casp2−/− versus Th-MYCN/Casp2−/−. Lastly we carried out an additional analysis of the differentially expressed factors caused by Casp2 loss in EµMyc compared to Th-MYCN tumors; (EµMyc v EµMyc/Casp2−/−) versus (Th-MYCN v Th-MYCN/Casp2−/−). Raw counts were extracted for each of these samples and edgeR was used to calculate the DEGs between the two phenotypic groups and expressed as log2-fold change (Log2FC) of expression between conditions being tested. Significant changes in gene expression levels were determined using adjusted P-values according to the Benjamini–Hochberg method to control for the false discovery rate (FDR) (Supplementary Tables S1c and S2a-f). For heat-maps, samples were clustered based on gene expression levels and false discovery rate (FDR < 0.05), and the most variable genes plotted (Fig. 1c, d, Supplementary Figures S2d-f, Supplementary Tables S3a-e).
Enrichment analysis of biological processes and pathways
Analysis of Gene Ontology and enriched biological pathways was carried out on DEGs with log2FC > 1 and ≤ −1 (i.e., 2^(1); equivalent to an actual fold change of 2).(FDR < 0.05), using the Functional Annotation Tool in the Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/home.jsp)70,71. Enriched pathways were analyzed from the KEGG (Kyoto Encyclopedia of Genes and Genomes) and REACTOME database in DAVID. The threshold value of gene counts was set at 3, and the EASE score was set at 0.1 and pathways with Fisher's exact P < 0.05, were considered to be enriched. Where indicated, pathways were also determined using PANTHER72 (http://www.pantherdb.org/about.jsp) and EnRichR (http://amp.pharm.mssm.edu/Enrichr/)73,74 databases. Gene Set Enrichment Analysis (GSEA) of pathways and genes was performed using Bioconductor fast pre-ranked GSEA (fgsea) package in R.
Real-time quantitative PCR (qPCR)
Total RNA extracted from frozen tumor tissue, was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) with MultiScribe™ Reverse Transcriptase and oligo-dT primers. Gene expression was normalized to the housekeeping gene β-actin and then expressed as fold change compared to EµMyc or Th-MYCN/Casp2+/+ tumor samples, using the 2−∆∆Ct method. See Supplementary Table S9 for primer sequences.
Statistical analyses
Statistical analysis was carried out using GraphPad Prism software (v 6.0, San Diego, CA, USA). Data are expressed as mean ± SEM and two-tailed unpaired t-test with Welch’s correction was used for pair-wise comparisons, unless otherwise indicated. Heat-maps were generated using R software, from natural log2 transformed data. Venn diagrams were generated using online software jvenn (http://jvenn.toulouse.inra.fr/app/example.html)75.
Supplementary information
Acknowledgements
We thank staff at the ACRF Cancer Genomics Facility, in particular Joel Geoghegan and Andreas Schreiber for their genomics and bioinformatics expertise and thank Claire Wilson for advice and discussions. This work was supported by a Worldwide Cancer Research Project Grant (13-1015), the National Health and Medical Research Council (NHMRC) of Australia project grants (1021456 and 1043057), and a NHMRC Senior Principal Research Fellowship (1103006).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by M. Piacentini
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Emily Hackett-Jones, Andrej Nikolic
Contributor Information
Loretta Dorstyn, Phone: +61 8 83027914, Email: loretta.dorstyn@unisa.edu.au.
Sharad Kumar, Phone: +61 8 8302 7918, Email: sharad.kumar@unisa.edu.au.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41419-018-1296-0).
References
- 1.McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb. Perspect. Biol. 2013;5:a008656. doi: 10.1101/cshperspect.a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shalini S, Dorstyn L, Dawar S, Kumar S. Old, new and emerging functions of caspases. Cell Death Differ. 2015;22:526–539. doi: 10.1038/cdd.2014.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jager R, Zwacka RM. The enigmatic roles of caspases in tumor development. Cancers (Basel) 2010;2:1952–1979. doi: 10.3390/cancers2041952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Kumar S, Kinoshita M, Noda M, Copeland NG, Jenkins NA. Induction of apoptosis by the mouse Nedd2 gene, which encodes a protein similar to the product of the Caenorhabditis elegans cell death gene ced-3 and the mammalian IL-1 beta-converting enzyme. Genes Dev. 1994;8:1613–1626. doi: 10.1101/gad.8.14.1613. [DOI] [PubMed] [Google Scholar]
- 6.Miles M, Kitevska-Ilioski T, Hawkins CJ. Old and novel functions of caspase-2. Int. Rev. Cell. Mol. Biol. 2017;332:155–212. doi: 10.1016/bs.ircmb.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Ho LH, et al. A tumor suppressor function for caspase-2. Proc. Natl Acad. Sci. USA. 2009;106:5336–5341. doi: 10.1073/pnas.0811928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puccini J, et al. Loss of caspase-2 augments lymphomagenesis and enhances genomic instability in Atm-deficient mice. Proc. Natl Acad. Sci. USA. 2013;110:19920–19925. doi: 10.1073/pnas.1311947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren K, Lu J, Porollo A, Du C. Tumor-suppressing function of caspase-2 requires catalytic site Cys-320 and site Ser-139 in mice. J. Biol. Chem. 2012;287:14792–14802. doi: 10.1074/jbc.M112.347625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Terry MR, et al. Caspase-2 impacts lung tumorigenesis and chemotherapy response in vivo. Cell Death Differ. 2015;22:719–730. doi: 10.1038/cdd.2014.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons MJ, et al. Genetic deletion of caspase-2 accelerates MMTV/c-neu-driven mammary carcinogenesis in mice. Cell Death Differ. 2013;20:1174–1182. doi: 10.1038/cdd.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorstyn L, et al. Caspase-2 deficiency promotes aberrant DNA-damage response and genetic instability. Cell Death Differ. 2012;19:1288–1298. doi: 10.1038/cdd.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliver TG, et al. Caspase-2-mediated cleavage of Mdm2 creates a p53-induced positive feedback loop. Mol. Cell. 2011;43:57–71. doi: 10.1016/j.molcel.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shalini S, et al. Impaired antioxidant defence and accumulation of oxidative stress in caspase-2-deficient mice. Cell Death Differ. 2012;19:1370–1380. doi: 10.1038/cdd.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shalini S, et al. Caspase-2 protects against oxidative stressin vivo. Oncogene. 2015;34:4995–5002. doi: 10.1038/onc.2014.413. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, et al. Caspase-2 deficiency enhances aging-related traits in mice. Mech. Ageing Dev. 2007;128:213–221. doi: 10.1016/j.mad.2006.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson CH, et al. Sex-specific alterations in glucose homeostasis and metabolic parameters during ageing of caspase-2-deficient mice. Cell Death Discov. 2016;2:16009. doi: 10.1038/cddiscovery.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson CH, et al. Age-related proteostasis and metabolic alterations in Caspase-2-deficient mice. Cell Death Dis. 2015;6:e1615. doi: 10.1038/cddis.2014.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S. Caspase 2 in apoptosis, the DNA damage response and tumour suppression: enigma no more? Nat. Rev. Cancer. 2009;9:897–903. doi: 10.1038/nrc2745. [DOI] [PubMed] [Google Scholar]
- 20.Manzl C, et al. PIDDosome-independent tumor suppression by Caspase-2. Cell Death Differ. 2012;19:1722–1732. doi: 10.1038/cdd.2012.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shalini S, et al. Caspase-2 deficiency accelerates chemically induced liver cancer in mice. Cell Death Differ. 2016;23:1727–1736. doi: 10.1038/cdd.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dawar S, et al. Caspase-2-mediated cell death is required for deleting aneuploid cells. Oncogene. 2017;36:2704–2714. doi: 10.1038/onc.2016.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fava LL, et al. The PIDDosome activates p53 in response to supernumerary centrosomes. Genes Dev. 2017;31:34–45. doi: 10.1101/gad.289728.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S, et al. Apoptosis regulatory gene NEDD2 maps to human chromosome segment 7q34-35, a region frequently affected in haematological neoplasms. Hum. Genet. 1995;95:641–644. doi: 10.1007/BF00209480. [DOI] [PubMed] [Google Scholar]
- 25.Hofmann WK, et al. Altered apoptosis pathways in mantle cell lymphoma detected by oligonucleotide microarray. Blood. 2001;98:787–794. doi: 10.1182/blood.V98.3.787. [DOI] [PubMed] [Google Scholar]
- 26.Holleman A, et al. Decreased PARP and procaspase-2 protein levels are associated with cellular drug resistance in childhood acute lymphoblastic leukemia. Blood. 2005;106:1817–1823. doi: 10.1182/blood-2004-11-4296. [DOI] [PubMed] [Google Scholar]
- 27.Kim MS, Chung NG, Yoo NJ, Lee SH. Somatic mutation of proapoptotic caspase-2 gene is rare in acute leukemias and common solid cancers. Eur. J. Haematol. 2011;86:449–450. doi: 10.1111/j.1600-0609.2011.01591.x. [DOI] [PubMed] [Google Scholar]
- 28.Kim MS, et al. Somatic mutations of caspase-2 gene in gastric and colorectal cancers. Pathol. Res. Pract. 2011;207:640–644. doi: 10.1016/j.prp.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Lopez-Garcia C, et al. BCL9L dysfunction impairs Caspase-2 expression permitting aneuploidy tolerance in colorectal cancer. Cancer Cell. 2017;31:79–93. doi: 10.1016/j.ccell.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorstyn L, et al. An unexpected role for caspase-2 in neuroblastoma. Cell Death Dis. 2014;5:e1383. doi: 10.1038/cddis.2014.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Q, et al. The expression profile and clinic significance of the SIX family in non-small cell lung cancer. J. Hematol. Oncol. 2016;9:119. doi: 10.1186/s13045-016-0339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu HX, et al. Expression profile of SIX family members correlates with clinic-pathological features and prognosis of breast cancer: a systematic review and meta-analysis. Med. (Baltim.) 2016;95:e4085. doi: 10.1097/MD.0000000000004085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen AA, et al. Activation of the brain-specific neurogranin gene in murine T-cell lymphomas by proviral insertional mutagenesis. Gene. 2009;442:55–62. doi: 10.1016/j.gene.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlsson Y, et al. Genetic inhibition of caspase-2 reduces hypoxic-ischemic and excitotoxic neonatal brain injury. Ann. Neurol. 2011;70:781–789. doi: 10.1002/ana.22431. [DOI] [PubMed] [Google Scholar]
- 35.Angelis D, Delivoria-Papadopoulos M. Effects of Src kinase inhibition on expression of pro-caspase-2 after brain hypoxia in a piglet animal model. Neuroreport. 2017;28:770–773. doi: 10.1097/WNR.0000000000000835. [DOI] [PubMed] [Google Scholar]
- 36.Park MS, Kim BS, Devarajan P. Hypoxia/re-oxygenation injury induces apoptosis of LLC-PK1 cells by activation of caspase-2. Pediatr. Nephrol. 2007;22:202–208. doi: 10.1007/s00467-006-0256-6. [DOI] [PubMed] [Google Scholar]
- 37.Araya R, Uehara T, Nomura Y. Hypoxia induces apoptosis in human neuroblastoma SK-N-MC cells by caspase activation accompanying cytochrome c release from mitochondria. FEBS Lett. 1998;439:168–172. doi: 10.1016/S0014-5793(98)01363-5. [DOI] [PubMed] [Google Scholar]
- 38.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maris JM, Matthay KK. Molecular biology of neuroblastoma. J. Clin. Oncol. 1999;17:2264–2279. doi: 10.1200/JCO.1999.17.7.2264. [DOI] [PubMed] [Google Scholar]
- 40.Matthay KK, et al. Neuroblastoma. Nat. Rev. Dis. Prim. 2016;2:16078. doi: 10.1038/nrdp.2016.78. [DOI] [PubMed] [Google Scholar]
- 41.Nakagawara A, et al. Association between high levels of expression of the TRK gene and favorable outcome in human neuroblastoma. N. Engl. J. Med. 1993;328:847–854. doi: 10.1056/NEJM199303253281205. [DOI] [PubMed] [Google Scholar]
- 42.Li Y, et al. Transcriptome analysis reveals determinant stages controlling human embryonic stem cell commitment to neuronal cells. J. Biol. Chem. 2017;292:19590–19604. doi: 10.1074/jbc.M117.796383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szemes M, et al. Wnt signalling drives context-dependent differentiation or proliferation in neuroblastoma. Neoplasia. 2018;20:335–350. doi: 10.1016/j.neo.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davoli T, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155:948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bailey MH, et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173:371–385 e318. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuldiner O, Benvenisty NA. DNA microarray screen for genes involved in c-MYC and N-MYC oncogenesis in human tumors. Oncogene. 2001;20:4984–4994. doi: 10.1038/sj.onc.1204459. [DOI] [PubMed] [Google Scholar]
- 48.Lossos IS, et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N. Engl. J. Med. 2004;350:1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 49.Cai YD, Huang T, Feng KY, Hu L, Xie L. A unified 35-gene signature for both subtype classification and survival prediction in diffuse large B-cell lymphomas. PLoS ONE. 2010;5:e12726. doi: 10.1371/journal.pone.0012726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reddy A, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171:481–494 e415. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ji H, et al. Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PLoS ONE. 2011;6:e26057. doi: 10.1371/journal.pone.0026057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lefebure M, et al. Genomic characterisation of Emu-Myc mouse lymphomas identifies Bcor as a Myc co-operative tumour-suppressor gene. Nat. Commun. 2017;8:14581. doi: 10.1038/ncomms14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: an emerging feature of cancer. Nat. Rev. Cancer. 2008;8:835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- 54.Lim J, et al. A protein–protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 55.Hu H, et al. MEGF6 promotes the epithelial-to-mesenchymal transition via the TGFbeta/SMAD signaling pathway in colorectal cancer metastasis. Cell. Physiol. Biochem. 2018;46:1895–1906. doi: 10.1159/000489374. [DOI] [PubMed] [Google Scholar]
- 56.Mukherjee A, Williams DW. More alive than dead: non-apoptotic roles for caspases in neuronal development, plasticity and disease. Cell Death Differ. 2017;24:1411–1421. doi: 10.1038/cdd.2017.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pistritto G, Papaleo V, Sanchez P, Ceci C, Barbaccia ML. Divergent modulation of neuronal differentiation by caspase-2 and -9. PLoS ONE. 2012;7:e36002. doi: 10.1371/journal.pone.0036002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oswald MCW, Garnham N, Sweeney ST, Landgraf M. Regulation of neuronal development and function by ROS. FEBS Lett. 2018;592:679–691. doi: 10.1002/1873-3468.12972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D’Mello SA, Finlay GJ, Baguley BC, Askarian-Amiri ME. Signaling pathways in melanogenesis. Int J Mol Sci. 2016;17:1144. doi: 10.3390/ijms17071144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aubert C, et al. Melanogenesis in cultured human neuroblastomas. Ann. Clin. Res. 1980;12:288–294. [PubMed] [Google Scholar]
- 61.Helson L, Johnson GA, Smith R. DOPA metabolism in neuroblastoma. Med. Pediatr. Oncol. 1980;8:317–322. doi: 10.1002/mpo.2950080402. [DOI] [PubMed] [Google Scholar]
- 62.LaBrosse EH, Comoy E, Bohuon C, Zucker JM, Schweisguth O. Catecholamine metabolism in neuroblastoma. J. Natl. Cancer Inst. 1976;57:633–638. doi: 10.1093/jnci/57.3.633. [DOI] [PubMed] [Google Scholar]
- 63.Becker J, Wilting J. WNT signaling, the development of the sympathoadrenal-paraganglionic system and neuroblastoma. Cell. Mol. Life Sci. 2018;75:1057–1070. doi: 10.1007/s00018-017-2685-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beyer M, Schultze JL. Regulatory T cells in cancer. Blood. 2006;108:804–811. doi: 10.1182/blood-2006-02-002774. [DOI] [PubMed] [Google Scholar]
- 65.Tsuyama N, et al. Clinical and prognostic significance of aberrant T-cell marker expression in 225 cases of de novo diffuse large B-cell lymphoma and 276 cases of other B-cell lymphomas. Oncotarget. 2017;8:33487–33500. doi: 10.18632/oncotarget.16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dawar S, et al. Impaired haematopoietic stem cell differentiation and enhanced skewing towards myeloid progenitors in aged caspase-2-deficient mice. Cell Death Dis. 2016;7:e2509. doi: 10.1038/cddis.2016.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang YL, Wang RC, Cheng K, Ring BZ, Su L. Roles of Rap1 signaling in tumor cell migration and invasion. Cancer Biol. Med. 2017;14:90–99. doi: 10.20892/j.issn.2095-3941.2017.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chu H, et al. Rap1b regulates B cell development, homing, and T cell-dependent humoral immunity. J. Immunol. 2008;181:3373–3383. doi: 10.4049/jimmunol.181.5.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 71.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mi H, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45:D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinforma. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bardou P, Mariette J, Escudie F, Djemiel C, Klopp C. jvenn: an interactive Venn diagram viewer. BMC Bioinforma. 2014;15:293. doi: 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.