Abstract
We aimed to determine whether the composition of the fecal microbiota changes under hyperbaric conditions. In this study, we collected fecal samples from 6 healthy divers at three points during deep diving training (before, 2.1 MPa, end). The frequency of Clostridium cluster XVIII tended to be increased after compression. The frequencies of Clostridium cluster IV and subcluster XIVa were inversely correlated with that of Bacteroides. The compositional changes in the fecal microbiota exhibited interindividual variability. These findings suggest that hyperbaric conditions affect the fecal microbiota.
Keywords: alteration, fecal microbiota, Clostridium cluster, Bacteroides, T-RFLP analysis
Fecal microbiota play important roles in nutrient absorption, immune system function, and disease development [1,2,3,4]. The microbiota colonize every surface exposed to the external environment, such as the skin and intestinal lumina, and the immune system has developed various ways of cohabiting with the microbiota. There are many factors that can affect fecal microbiota, but they have not been fully elucidated. Recently, NASA reported that fecal microbiota markedly changed in a zero-gravity environment. A few studies have examined the changes in the frequencies of bacterial species in hyperbaric environments [5,6,7] using a rodent model. However, there have been no studies of the changes in the frequencies of intestinal bacterial species that occur in the human body under hyperbaric conditions. Some changes in human immune function due to exposure to hyperbaric environments have been reported [8], and fecal microbiota have been shown to influence the human immune system. Thus, we hypothesized that intestinal microbial flora may change under hyperbaric conditions. At the Maritime Self-Defense Force Undersea Medical Center, we regularly perform diving training under hyperbaric conditions such as deep-diving training using a simulator. During such training, the divers stay in a completely enclosed diving training module at 2.1 MPa, which is equal to the pressure of seawater at a depth of 200 m. The deep-diving module can pressurize the inner chamber up to approximately 6 MPa. The simulator is very small, and the trainees have no private space. If, while at 2.1 MPa, trainees need care from the medics, they must stay in the simulator for 10 days before they can leave. We aimed to determine whether the composition of fecal microbiota changes under hyperbaric conditions.
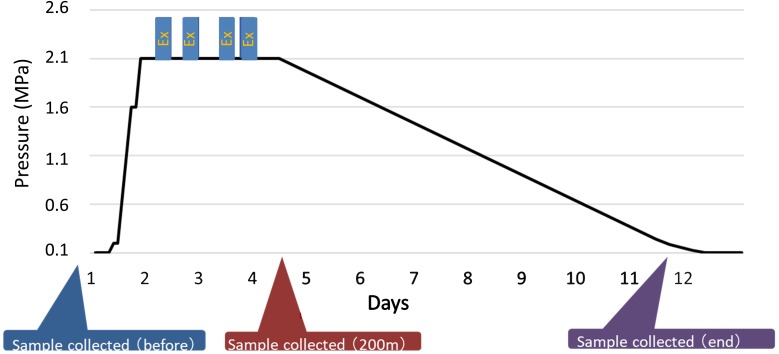
The study population consisted of six healthy Japanese males (subjects A–F; mean age, 35.2 ± 8.4 years old: range, 25 to 48 years old). None of the subjects regularly used any drugs or had been diagnosed with a digestive disease. An outline of the diving training procedure is shown in Fig. 1. All of the subjects stayed in the deep-diving module during the training period. They were gradually pressurized to hyperbaric conditions at 2.1 MPa and then were subjected to decompression. The first fecal samples were collected just before the training period (before). The second samples were collected on day 4 (2.1 MPa), and the third samples were collected on the last day of the training (end). The divers underwent medical checkups every day, during which their health, including their abdominal symptoms and the conditions of their stools, was assessed. During this study, all of the subjects received the same diet. None had any antibiotics, probiotics, or prebiotics before or during this study. None received any medication. This study protocol was approved by the ethics committee of JSDF Hospital Yokosuka. Written informed consent was obtained from all subjects.
Fig. 1.
Profile of the 200-m saturation diving exercise and the timing of sample collection.
Ex: Excursion.
The fecal microbiota were analyzed by terminal resection fragment length polymorphism (T-RFLP) at Techno Suruga Lab. The 16S rDNA gene was amplified from the fecal bacterial DNA of the patients using the fluorescently labeled 516f (5′-TGC CAG CAG CCG GGGTA-3′, Escherichia coli positions 516 to 532) and 1510r (5′-GGT TAC CTT GTT ACG ACT T-3′, E. coli positions 1510 to 1492) primers. The amplified DNA was purified and analyzed by electrophoresis. The restriction enzymes were selected according to Nagashima et al. [9]. The resultant DNA fragments, namely, fluorescently labeled terminal restriction fragments (T-RFs), were analyzed using a next-generation sequencing genetic analyzer, and their lengths and peak areas were determined using the GeneMapper genotyping software. The T-RFs were divided into 29 operational taxonomic units (OTUs). The bacteria were predicted for each classification unit, and the corresponding OTU was identified according to reference human fecal microbiota profiles.
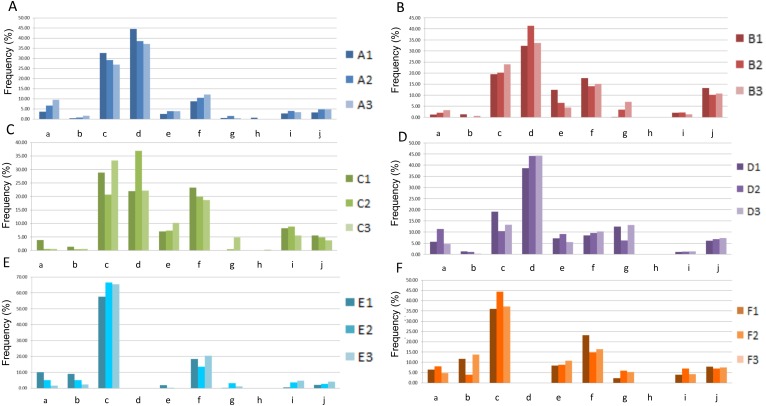
The obtained results were as follows: (1) None of the 6 divers exhibited abdominal symptoms or other health abnormalities. After being kept at 2.1 MPa, subject C exhibited watery diarrhea on day 2, but he recovered without any medication and produced normal stools on day 3. The other subjects did not display any abnormalities and did not receive any medication during the training period. At the end of the training period, all of the subjects were in a healthy condition. (2) The frequencies of each type of bacterial species in subjects A–F are shown in Fig. 2. Whereas Prevotella species were predominantly found in subjects A, B, and D, Bacteroides species were predominantly detected in subjects E and F, in which Prevotella species were not present. In subject C, the frequencies of Prevotella and Bacteroides species exhibited an inverse relationship under hyperbaric conditions. The frequency of Clostridium cluster XVIII was higher in subject C than in the other subjects. At 2.1 MPa, the frequency of Clostridium cluster XVIII increased in all subjects. In addition, the frequency of Clostridium cluster IX increased, and the frequency of Lactobacillales decreased in 5 of the 6 (83.3%) subjects. Furthermore, the frequencies of Bifidobacterium species and Clostridium cluster IV increased, and the frequency of Clostridium subcluster XIVa decreased in 4 of the 6 (66.7%) subjects. The patterns of change in the frequencies of Clostridium cluster IV and subcluster XIVa were inversely correlated with the pattern of change in the frequency of Bacteroides species. At the end of the training, the frequency of Clostridium cluster XVIII showed an identical pattern of change in 5 of the 6 (83.3%) subjects. The frequencies of Lactobacillus species, Clostridium cluster IV, Clostridium cluster IX, and Clostridium subcluster XIVa showed identical patterns of change in 3 of the 6 (50%) subjects. Clostridium cluster XI was detected in subjects A and C. In subject C, the frequency of Clostridium cluster XI was increased.
Fig. 2.
The frequency of each type of bacterial species in subjects A–F.
The left, middle, and right bars indicate the frequencies of microbiota before training, at 200 m depth, and at the end of training, respectively.
a: Bifidobacterium; b: Lactobacillales; c: Bacteroides; d: Prevotella; e: Clostridium cluster IV; f: Clostridium subcluster XIV; g: Clostridium cluster IX; h: Clostridium cluster XI; i: Clostridium cluster XVIII; j: others.
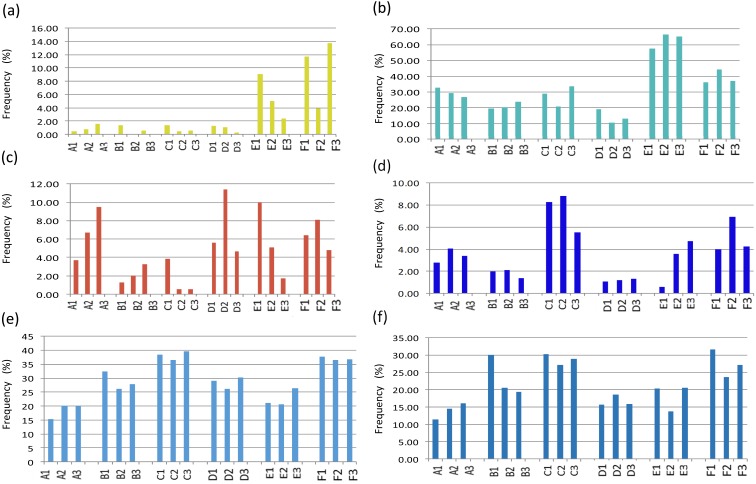
In this study, Lactobacillus and Bacteroides species exhibited marked variability among the subjects (Fig. 3). This variability was not similar to previous studies in terms of changes in the frequencies of bacterial species in hyperbaric environments [5,6,7]. Concerning the changes in mouse fecal microbiota, Bornside et al. reported that the frequency of Lactobacillus species was greater at 0.3 MPa than at 0.1 MPa [6]. Daily et al. reported that Bacteroides was the most frequently identified bacterial genus in mice exposed to 3.5 MPa and 0.1 MPa [7].
Fig. 3.
Changes in the frequency distribution patterns of each type of bacterial species.
(a) Lactobacillales, (b) Bacteroides, (c) Bifidobacterium, (d) Clostridium cluster XVIII, (e) all Clostridium clusters, and (f) Clostridium cluster IV plus Clostridium subcluster XIVa.
The pattern of change in the frequency of Bifidobacterium species showed marked variability among the subjects, whereas the frequency of Clostridium cluster XVIII showed the smallest amount of variability among the subjects. There have been some reports that the frequency of Bifidobacterium and Lactobacillus species decreases due to emotional stress [10,11,12]. The results of this study were not in agreement with previous studies despite the subjects being under extreme conditions. The reason for the difference was not clear, but hyperbaric oxygenation of tissue might reduce the decrease in Bifidobacterium and Lactobacillus species. Further evaluations of this issue are necessary because many factors are implicated in the compositional changes in the fecal microbiota, for example, emotional stress associated with deep diving or other types of stress, such as from hyperbaric conditions.
In this study, depth-dependent changes in the frequencies of the Clostridium clusters in the fecal microbiota were seen. The total frequency of all Clostridium clusters displayed similar patterns of change in subjects B–F, whereas the frequencies of cluster IV and subcluster XIVa showed various patterns of change among the subjects. However, the tendencies exhibited by cluster IV and subcluster XIVa were inversely correlated with that exhibited by Bacteroides species in 5 of the 6 subjects. Clostridium cluster IV and Clostridium subcluster XIVa activate intestinal epithelial cells and dendritic cells by producing short-chain fatty acids and inducing regulatory T cells to activate intestinal immunity. These clusters have important anti-inflammatory properties [13] and may improve glucose metabolism [14] in the human body. Bacteroides species were reported to be involved in the induction of regulatory T cells [15]. This suggests that interactions between some Clostridium clusters and Bacteroides species may preserve intestinal homeostasis and immunity. Under hyperbaric conditions, granulocytes and lymphocytes were reported to increase in number, and they also exhibited greater activity and adhesive capacity [16]. This may be one of the reasons why immunological changes involving Clostridium cluster IV, Clostridium subcluster XIVa, and Bacteroides species were seen in the fecal microbiota in our study. All of the subjects exhibited consistent increases in the frequency of cluster XVIII at 2.1 MPa. These bacteria were reported to be associated with colonic cancer [17], obesity [18], and fatty liver [19]. Therefore, hyperbaric conditions may increase the risk of intestinal conditions related to Clostridium cluster XVIII.
In conclusion, our study was the first bacterial metagenomic analysis by T-RFLP to evaluate the composition of fecal microbiota under hyperbaric conditions, and it showed that the changes in the frequencies in Clostridium cluster IV and subcluster XIVa species seen under hyperbaric conditions were inversely correlated with the changes in the frequency of Bacteroides species. Interactions between certain Clostridium clusters and Bacteroides species may play an important role in preserving intestinal homeostasis and immunity, although further studies are required to clarify the effects of the changes in fecal microbiota observed under stress conditions on the human body. We investigated the fecal microbiota of saturation divers who were pressurized to 2.1 MPa to confirm the effects of such diving on fecal microbiota. The changes in the fecal microbiota induced by hyperbaric conditions exhibited significant interindividual variability, although marked compositional differences in the fecal microbiota were observed between 0.1 and 2.1 MPa. It was suggested that the changes in fecal microbiota induced by hyperbaric conditions may consequently affect physiological conditions in vivo.
Further analysis with more subjects is required to understand the possible changes in fecal microbiota induced by hyperbaric conditions.
Conflict of interest
The authors have no conflicts of interest that are directly related to the content of this article.
References
- 1.Guarner F. 2006. Enteric flora in health and disease. Digestion 73 Suppl 1: 5–12. [DOI] [PubMed] [Google Scholar]
- 2.Servin AL. 2004. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol Rev 28: 405–440. [DOI] [PubMed] [Google Scholar]
- 3.Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, Zhang Y, Shen J, Pang X, Zhang M, Wei H, Chen Y, Lu H, Zuo J, Su M, Qiu Y, Jia W, Xiao C, Smith LM, Yang S, Holmes E, Tang H, Zhao G, Nicholson JK, Li L, Zhao L. 2008. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA 105: 2117–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hooper LV. 2004. Bacterial contributions to mammalian gut development. Trends Microbiol 12: 129–134. [DOI] [PubMed] [Google Scholar]
- 5.Hopkinson WI, Towers AG. 1963. Effects of hyperbaric oxygen on some common pathogenic bacteria. Lancet 2: 1361–1362. [DOI] [PubMed] [Google Scholar]
- 6.Bornside GH, Cherry GW, Myers MB. 1973. Intracolonic oxygen tension and in vivo bactericidal effect of hyperbaric oxygen on rat colonic flora. Aerosp Med 44: 1282–1286. [PubMed] [Google Scholar]
- 7.Daily OP, Gillmore JD. 1981. Alterations in mouse fecal flora associated with hyperbaric stress. Undersea Biomed Res 8: 23–31. [PubMed] [Google Scholar]
- 8.Brenner I, Shephard RJ, Shek PN. 1999. Immune function in hyperbaric environments, diving, and decompression. Undersea Hyperb Med 26: 27–39. [PubMed] [Google Scholar]
- 9.Nagashima K, Hisada T, Sato M, Mochizuki J. 2003. Application of new primer-enzyme combinations to terminal restriction fragment length polymorphism profiling of bacterial populations in human feces. Appl Environ Microbiol 69: 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liźko NN, Silov VM, Syrych GD. 1984. [Events in the development of dysbacteriosis of the intestines in man under extreme conditions]. Nahrung 28: 599–605 (in German). [DOI] [PubMed] [Google Scholar]
- 11.Logan AC, Venket Rao A, Irani D. 2003. Chronic fatigue syndrome: lactic acid bacteria may be of therapeutic value. Med Hypotheses 60: 915–923. [DOI] [PubMed] [Google Scholar]
- 12.Jin JS, Touyama M, Yamada S, Yamazaki T, Benno Y. 2014. Alteration of a human intestinal microbiota under extreme life environment in the Antarctica. Biol Pharm Bull 37: 1899–1906. [DOI] [PubMed] [Google Scholar]
- 13.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. 2008. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27: 104–119. [DOI] [PubMed] [Google Scholar]
- 14.De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, Bäckhed F, Mithieux G. 2014. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 156: 84–96. [DOI] [PubMed] [Google Scholar]
- 15.Atarashi K, Tanoue T, Honda K. 2017. [Dynamics of health and disease based on biological barrier mucosa and skin] (Chapter1) Molecular, cellular and tissue infrastructure supporting biological barrier. Immune barrier regulatory mechanism by intestinal flora. Experimental Medicine 35: 1129–1136 (In Japanese). [Google Scholar]
- 16.Strewe C, Crucian BE, Sams CF, Feuerecker B, Stowe RP, Choukèr A, Feuerecker M. 2015. Hyperbaric hyperoxia alters innate immune functional properties during NASA Extreme Environment Mission Operation (NEEMO). Brain Behav Immun 50: 52–57. [DOI] [PubMed] [Google Scholar]
- 17.Chen W, Liu F, Ling Z, Tong X, Xiang C. 2012. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One 7: e39743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031. [DOI] [PubMed] [Google Scholar]
- 19.Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA. 2011. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 140: 976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]