Abstract
Properties of Lactobacillus plantarum group strains isolated from two kinds of Japanese post-fermented teas, Ishizuchi-kurocha and Awa-bancha, were compared. Although lactic acid bacteria isolated from the fermented teas were identified as L. plantarum via homology comparison of 16S ribosomal RNA gene sequences, classification of L. plantarum based on ribosomal proteins showed that the strains isolated from Ishizuchi-kurocha and Awa-bancha were different. According to classification by the ribosomal protein typing, Ishizuchi-kurocha-derived strains belong to the same group as L. plantarum subsp. plantarum JCM 1149T. Awa-bancha-derived strains were assigned to a different group. This pattern was also applicable to strains isolated more than 10 years ago. A further analysis based on recA and a dnaK gene showed that Awa-bancha-derived strains were closely related to L. pentosus. The interactions with cultured cells were different between strain JCM 1149T and the Ishizuchi-kurocha-derived strains. The Ishizuchi-kurocha-derived strains showed strong adhesion to Caco-2 cells. In contrast, strain JCM 1149T and the Awa-bancha-derived strains hardly adhered to Caco-2 cells. According to the ribosomal protein typing, sugar utilization, and interaction with Caco-2 cells, although these properties were dependent on the strain strictly speaking, the L. plantarum group strains in this study can be subdivided into two groups: (1) type strain JCM 1149T and Ishizuchi-kurocha-derived strains and (2) Awa-bancha-derived strains. A regionally unique microorganism may persist in each traditional fermented drink.
Keywords: Lactobacillus plantarum, postfermented tea, ribosomal protein, Lactobacillus pentosus
INTRODUCTION
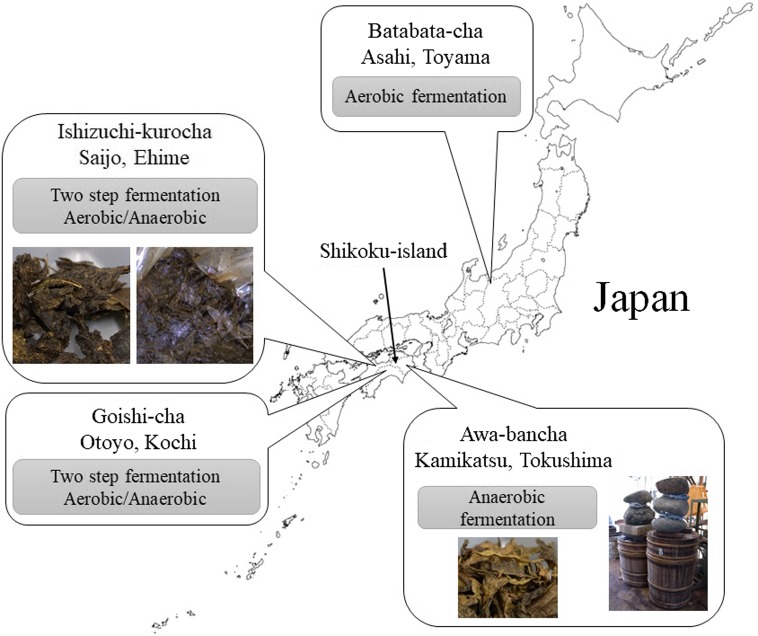
There are several kinds of post-fermented tea in East Asia such as Pu-erh tea in Yunnan, China, Miang in Thailand, and Lahpet in Myanmar. There are also four kinds of traditional post-fermented tea in Japan: Ishizuchi-kurocha (Ehime), Goishi-cha (Kochi), Awa-bancha (Tokushima), and Batabata-cha (Toyama) (Fig. 1). Three of these post-fermented teas are produced from tea plants on Shikoku Island, and the fermentation procedures are different for all post-fermented teas in Japan. Ishizuchi-kurocha and Goishi-cha are prepared via a two-step fermentation process. This process is unlike any other procedure for fermented-tea production in the world. Primary fermentation is fungi-driven aerobic fermentation, and secondary fermentation is lactic acid bacteria-driven anaerobic fermentation. Awa-bancha is produced via anaerobic fermentation only by lactic acid bacteria such as Miang. Batabata-cha is produced via aerobic fermentation only by fungi as in the case of Pu-erh tea. Ishizuchi-kurocha, Goishi-cha, and Awa-bancha have distinctive acidic tastes associated with lactate-generating fermentation. Compared with green tea, the concentrations of caffeine and catechins in these fermented teas are low [1]. The change in catechins in post-fermented tea may be caused by degradation and polymerization during fermentation. Nonetheless, details of the fermentation processes of Japanese post-fermented teas are still unclear.
Fig. 1.
The regions of production and origin of Japanese post-fermented teas.
It has been reported that Aspergillus sp. and Lactobacillus plantarum are mainly involved in the aerobic and anaerobic fermentation of tea leaves in the processes of fermentation of Goishi-cha and Ishizuchi-kurocha, respectively [2,3,4]. L. plantarum is one of the major lactic acid bacteria species obtained from plant material and is frequently found in fermented foods of plant origin. L. plantarum is found in plant materials and the gastrointestinal tracts of animals and is known as one of the probiotic species of lactic acid bacteria [5,6,7]. For example, some strains of L. plantarum alleviate irritable bowel syndrome [8, 9]. L. plantarum strain JSA22 inhibits infection of intestinal epithelial cells by Salmonella Typhimurium [6]. L. plantarum CICC 23174 has an immunostimulatory activities such as enhancement of the phagocytic activity of macrophages [7]. L. plantarum can reduce soy allergy [10]. Furthermore, some L. plantarum strains produce bacteriocin [11, 12]. For instance, plantaricin KL-1Y from L. plantarum KL-1 shows bactericidal activity against Bacillus cereus and growth-inhibitory activities toward Listeria innocua, Staphylococcus aureus, and Escherichia coli [13]. Extracellular proteins from the L. plantarum BMCM 12 strain inhibit adhesion of E. coli and S. enterica subsp. enterica to mucin [14]. Thus, L. plantarum is one of the beneficial species of lactic acid bacteria for probiotic use.
Japanese fermented teas prepared by lactate-generating fermentation are produced only on Shikoku Island. Although L. plantarum is the predominant species in these fermented teas, the regional characteristics of L. plantarum among the abovementioned fermented teas are unknown. Therefore, in this study, we compared properties of the lactobacilli isolated from two kinds of Japanese fermented tea, Ishizuchi-kurocha and Awa-bancha. Particularly, we focused on L. plantarum strains. We isolated and classified Lactobacillus strains from the post-fermented teas using sequences of the 16S rRNA gene and ribosomal proteins. Next, we evaluated their sugar utilization. In addition, we assessed the interaction between the L. plantarum and animal cells.
MATERIALS AND METHODS
Isolation and identification of Lactobacillus spp. from fermented tea
Lactobacillus spp. were isolated from Ishizuchi-kurocha and Awa-bancha. Pictures of these fermented teas are shown in Fig. 1. Approximately 0.5 g of wet tea leaves was added to 10 ml of 10% skim milk (Megmilk Snow Brand Co., Ltd., Tokyo, Japan) containing 2% of glucose (Wako Pure Chemical Industries, Ltd., Osaka, Japan), and the medium was incubated at 30°C for 1–2 days. Before the procedure, glucose-supplemented skim milk was sterilized at 60°C for 30 min. Skim milk will curdle due to a decrease in pH if lactic acid bacteria are growing in it. Curdled skim milk was streaked on MRS (Sigma-Aldrich) agar plates containing 2% of agar (Wako Pure Chemical Industries), and the plates were incubated at 30°C for 1–2 days under anaerobic conditions (atmosphere consisting of 20% of carbon dioxide and 80% of nitrogen). To test a different procedure, wet tea leaves were mixed with sterile distilled water at a concentration of 0.1 mg/ml. The suspension was serially diluted with distilled water and spread on MRS agar plates or LBS agar plates (Becton, Dickinson and Company, Le Pont-de-Claix, France). After that, the plates were incubated at 30°C for 1–2 days under anaerobic conditions. Anaerobic condition were provided by AnaeroPack-Anaero (Mitsubishi Gas Chemical Co., Inc., Tokyo, Japan). Stand-alone colonies were picked and inoculated into 10 ml of MRS broth in a test tube with a screw cap. Each isolate was cultured at 30°C for 1–2 days. Stock cultures were stored at –80°C in MRS broth containing 30% of glycerol. Frozen stocks of fresh isolates were prepared from the cultures within three passages after isolation. The isolates were identified by homology analysis of the 16S rRNA gene sequence, cultured in MRS broth overnight, and collected by centrifugation at 11,000 × g for 10 min. Pellets were resuspended in elution buffer, and the bacterial cells were lysed by sonication. Then, DNA was extracted from the isolates with a DNeasy Tissue & Blood Kit (Qiagen, Hilden, Germany). Polymerase chain reaction (PCR) amplification was performed on a TaKaRa PCR Thermal Cycler (Takara Bio Inc., Kusatsu, Japan) with the following set of bacterial universal primers and TaKaRa Ex Taq DNA polymerase (Takara Bio): 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) [15] or 10F (5′-GTTTGATCCTGGCTCA-3′) and 800R (5′-TACCAGGGTATCTAATCC-3′). The PCRs were conducted in a total volume of 50 µl containing 1 µl of template DNA, 0.5 μmol/l each primer, Ex Taq polymerase, 5 µl of PCR buffer, and 0.2 mmol/l each deoxynucleotide triphosphate. All the primers were synthesized by Eurofins Genomics K.K. (Tokyo, Japan). The following thermal cycling conditions were used for the Taq polymerase: 2 min at 93°C for initial denaturation; then 30 sec at 93°C, 30 sec at the annealing temperature described in Table 2, and 30 sec at 72°C for 30 cycles; and finally 1 min at 72°C. The PCR products were analyzed via electrophoresis in a 1.5% agarose gel followed by ethidium bromide staining. The sequence of the primer used for DNA sequencing was as follows: LAB-seqF (5′-TCCTGGCTCAGGACGAACGCT-3′), 23F, or 10F. Determination of the DNA sequence was performed by Macrogen Japan Corp. (Kyoto, Japan). Sequence identification was carried out by means of the Standard Nucleotide BLAST of the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi). L. plantarum JCM 1149T, which is a type strain of L. plantarum isolated from pickled cabbage, was purchased from Riken BioResource Center (Tsukuba, Japan). Multiplex PCR amplification of the recA gene was carried out as described in a previous paper [16]. The sequences of the primers were as follows: paraF (5′-GTCACAGGCATTACGAAAAC-3′), pentF (5′-CAGTGGCGCGGTTGATATC-3′), planF (5′-CCGTTTATGCGGAACACCTA-3′), and pREV (5′-TCGGGATTACCAAACATCAC-3′). The PCR amplifications were conducted in a total volume of 50 µl containing 1 µl template DNA, 1.5 mM MgCl2, 0.25 μM the primers paraF, pentF, and pREV, 0.12 μM planF, deoxynucleotide triphosphates mixture (0.2 mM each), and 1.25 U of Ex Taq. The following thermal cycling conditions were used: 3 min at 94°C for initial denaturation; then 30 sec at 94°C, 10 sec at 56°C, and 30 sec at 72°C for 30 cycles; and finally 5 min at 72°C. The PCR products were visualized in 2% agarose gel (Nippon Gene Co., Ltd., Tokyo, Japan). PCR amplification of the dnaK gene was carried out with TaKaRa Ex Taq DNA polymerase as described in a previous paper [17]. The sequences of the primers were as follows: Lpdnak-500F3 (5′-CCGTTCTTRTCRATRTCRAA-3′) and Lpdnak-1710R5 (5′-GAAAYYCAAGTYGGHGAAGT-3′). The Lpdnak-500F3 was also used as the sequence primer for DNA sequencing. The following thermal cycling conditions were used: 5 min at 94°C for initial denaturation; then 1 min at 94°C, 1 min at 58°C, and 1 min at 72°C for 35 cycles; and finally 7 min at 72°C. A phylogenetic analysis was carried out with ClustalW through the website of the DNA Data Bank of Japan (DDBJ) (http://clustalw.ddbj.nig.ac.jp/). Reference strains were chosen from GenBank for inclusion in the phylogenetic analysis.
Table 2. Availabilities of sugars by Lactobacillus plantarum group strains isolated from fermented tea.
| JCM1149T | ISC13 | IYO1401 | IYO1511 | IYO1615 | IYO1651 | AWA0701 | AWA1401 | AWA1501 | AWA1601 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Control | – | – | – | – | – | – | – | – | – | – |
| Glycerol | – | – | – | – | – | – | + | – | – | ± |
| Erythritol | – | – | – | – | – | – | – | – | – | – |
| D-Arabinose | – | – | – | – | – | – | – | – | – | – |
| L-Arabinose | + | + | + | – | + | + | – | + | – | + |
| D-Ribose | + | + | + | + | + | + | + | + | + | + |
| D-Xylose | – | – | + | – | – | – | + | ± | + | – |
| L-Xylose | – | – | – | – | – | – | – | – | – | – |
| D-Adonitol | – | – | – | – | – | – | – | – | – | – |
| Methyl-βD-Xylopyranoside | – | – | – | – | – | – | – | – | – | – |
| D-Galactose | + | + | + | + | + | + | + | + | + | + |
| D-Glucose | + | + | + | + | + | + | + | + | + | + |
| D-Fructose | + | + | + | + | + | + | + | + | + | + |
| D-Mannose | + | + | + | + | + | + | + | + | + | + |
| L-Sorbose | – | – | – | – | + | – | – | – | – | – |
| L-Rhamnose | – | ± | ± | ± | ± | ± | – | ± | – | ± |
| Dulcitol | – | – | – | – | – | – | + | ± | – | – |
| Inositol | – | – | – | – | – | – | – | – | – | – |
| D-Mannitol | + | + | + | + | + | + | + | + | + | + |
| D-Sorbitol | + | + | + | + | – | + | + | + | + | + |
| Methyl-αD-Mannopyranoside | + | + | + | + | + | + | – | – | – | – |
| Methyl-αD-Glucopyranoside | – | – | + | – | – | – | – | + | – | + |
| N-Acetyl Glucosamine | + | + | + | + | + | + | – | + | + | + |
| Amygdalin | + | + | + | + | + | + | + | + | + | + |
| Areutin | + | + | + | + | + | + | + | + | + | + |
| Esculin ferric citrate | + | + | + | + | – | + | + | + | + | + |
| Salicin | + | + | + | + | + | + | + | + | + | + |
| D-Cellobiose | + | + | + | + | + | + | + | + | + | + |
| D-Maltose | + | + | + | + | + | + | + | + | + | + |
| D-Lactose | – | + | + | + | + | + | – | + | + | + |
| D-Melibiose | + | + | + | + | + | – | + | + | – | + |
| D-Sucrose | + | + | + | + | + | + | + | + | + | + |
| D-Trehalose | + | + | + | + | + | + | + | + | + | + |
| Inulin | – | – | – | – | – | – | – | – | – | – |
| D-Melezitose | + | + | + | + | + | – | – | – | – | – |
| D-Raffinose | + | + | + | + | + | – | ± | – | – | + |
| Starch | – | – | – | – | – | – | – | – | – | – |
| Glycogen | – | – | – | – | – | – | – | – | – | – |
| Xylitol | – | – | – | – | – | – | – | – | – | – |
| Gentibiose | ± | ± | ± | ± | ± | ± | ± | ± | + | + |
| D-Turanose | + | + | + | + | + | + | – | – | – | – |
| D-Lyxose | – | – | – | – | – | – | – | – | – | – |
| D-Tagatose | – | – | – | – | – | – | – | – | – | – |
| D-Fucose | – | – | – | – | – | – | – | – | – | – |
| L-Fucose | – | – | – | – | – | – | – | – | – | – |
| D-Arabitol | – | – | – | – | – | – | – | – | – | – |
| L-Arabitol | – | – | – | – | – | – | – | – | – | – |
| Gluconate | ± | ± | ± | ± | ± | ± | ± | ± | ± | ± |
| 2-Keto Gluconate | – | – | – | – | – | – | – | – | – | – |
| 5-Keto Gluconate | – | – | – | – | – | – | – | – | – | – |
Classification of L. plantarum group strains by ribosomal proteins
Sample preparation and matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) were carried out as described in our previous papers [18,19,20] with slight modifications. In brief, each bacterial culture resuspended in TMA-1 buffer (10 mM Tris-HCl; 30 mM NH4Cl; 10 mM MgCl2; and 6 mM 3-mercapto-1,2-propanediol, an alternative to toxic β-mercaptoethanol) was ground up by bead beating with zirconia silica beads in a Beads Crusher μT-12 (Taitec, Koshigaya, Japan) to obtain cell lysates. After removing debris by centrifugation, the resulting supernatant mixed with a matrix (sinapinic acid) was applied to MALDI-MS measurements. All the reagents were purchased from Wako Pure Chemical Industries (Osaka, Japan) and were used as received. MALDI mass spectra in the range of m/z 4,000 to 20,000 were examined in positive linear mode by averaging 500 individual laser shots on an AXIMA-CFR plus time-of-flight mass spectrometer (Shimadzu/Kratos, Kyoto, Japan). Peak assignment was performed as per the method described in another paper [18].
Evaluation of sugar utilization by L. plantarum group strains
Sugar utilization by L. plantarum strains was examined by means of the API 50 CH system (bioMérieux, Inc., Marcy-I’Etoile, France). L. plantarum strains were preincubated in MRS broth overnight and collected by centrifugation at 5,400 × g for 10 min. After the bacteria were washed with PBS once, each pellet was resuspended in the API 50 CHL medium (bioMérieux) at McFarland Standard No. 2. Each suspension was inoculated into a test tube containing API 50 CH and covered by mineral oil. Next, the bacteria were cultured for 48 h at 37°C. Sugar utilization was detected by examining the color of the medium.
Evaluation of survival of L. plantarum group strains under acidic conditions
The survival rate of L. plantarum under acidic conditions was determined by the method described by Takiguchi and Suzuki [21]. L. plantarum strains were preincubated in MRS broth (pH 6.2) overnight. Preculture suspension was added at 10% (v/v) into MRS broth with a pH of 2 or 3. The pH level of the MRS broth was adjusted to 2.0 or 3.0 with hydrochloric acid, and then a 4% pepsin (Wako Pure Chemical Industries) solution was added at 1:100 (v/v). Preculture suspension was also added into normal MRS broth (pH 6.2) at 10%. Each culture was incubated for 2 hr at 37°C; then, the cultures were serially diluted 10-fold with PBS from 101 to 106, and the diluted cultures were spread on MRS plates. Surviving bacteria were counted by plating serial 10-fold dilutions of the suspension on fresh MRS agar plates. After anaerobic incubation overnight at 37°C, colony-forming units (CFUs) were estimated by counting.
Assay of adhesion to epithelial cells
Human colon carcinoma Caco-2 cells were acquired from the Riken BioResource Center (Tsukuba, Ibaraki, Japan). The cells were cultured in the minimum essential medium (MEM; Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 20% of heat-inactivated fetal bovine serum (FBS; HyClone Laboratories, Inc., South Logan, UT, USA), 100 U/ml penicillin, 100 µg/ml streptomycin, 250 ng/ml amphotericin B (Nacalai Tesque Inc., Kyoto, Japan), and a 0.1 mM solution of nonessential amino acids in MEM (Gibco). The cells were seeded in 75-cm2 flasks (Violamo, As One Corporation, Osaka, Japan) and cultured at 37°C in an atmosphere containing 5% of CO2. The adhesion assay was performed by the method of Todoriki et al. [22]. For adhesion assays, Caco-2 cell monolayers were prepared in 6-well tissue culture plates (As One Corporation). The cells were seeded at a density of 4 × 105/well to ensure confluence and cultured for 24 hr prior to the adhesion assay. L. plantarum strains were precultured in 10 ml of MRS broth at 30°C for 24 hr. After the bacteria were washed with PBS once, bacterial suspensions were prepared at a concentration of 107 cells/ml in MEM without antibiotics. The culture media for Caco-2 monolayers were changed to the MEM-bacterial suspensions at 2 ml per well, and the cells were incubated for 2 hr at 37°C and 5% CO2. Next, the wells were washed three times with PBS for 5 min by means of a mild mixer (Taitec Corporation, Koshigaya, Japan). After the last wash, Caco-2 cell monolayers were covered with 2 ml of distilled water and were mechanically agitated by vigorous pipetting to resuspend the Caco-2 cells and bacteria. Adherent bacterial cells were counted by plating the serial 10-fold dilutions of the suspensions on fresh MRS agar plates and were incubated anaerobically.
Assay of adhesion to extracellular matrix (ECM) proteins
These assays were performed based on another study [23]. Fibronectin-, laminin-, or type IV collagen-coated multiwell glass slides were purchased from Nippi, Inc. (Tokyo, Japan). L. plantarum strains were precultured in 10 ml of MRS broth at 30°C for 24 hr. After the bacteria were washed with PBS once, the bacteria in suspension were counted using a bacterial counting chamber (Sunlead Glass Corp., Koshigaya, Japan). Bacterial suspensions were prepared at a concentration of 108 cells/ml in PBS. Next, 20 µl of the bacterial cell suspensions were applied to the wells of ECM-coated multiwell glass slides. The slide was incubated for 2 hr at room temperature. After three washes with PBS, the slide was stained with Gram’s crystal violet solution (Kanto Chemical Co., Inc., Tokyo, Japan). Randomly chosen visual fields were photographed under a light microscope. The bacteria in each of four microscopic fields of view were counted, and the results were expressed as the mean ± standard deviation.
Evaluation of the cytotoxicity of isolates
Caco-2 cells were seeded in 24-well tissue culture plates (As One Corporation) at a concentration of 2 × 105 cells/ml and cultured for 24 hr. L. plantarum strains were precultured in 10 ml of MRS broth at 37°C for 24 hr. After the bacteria were washed with PBS once, bacterial suspensions were prepared at a concentration of 107 cells/ml in MEM. The culture media for Caco-2 monolayers were changed to the MEM-bacterial suspensions at 1 ml per well, and the cells were then incubated for 6 hr, washed with PBS three times, and covered with fresh MEM without bacteria. The cells were incubated further for 18 hr in 5% CO2. In total, the cells were incubated with bacteria for 24 hr. After that, the cell culture supernatants were collected, and the bacterial cells were pelleted by centrifugation at 10,000 rpm for 10 min. The supernatants were subjected to measurement of cell membrane damage and IL-8 secretion. Cell membrane damage was determined according to the release of lactate dehydrogenase (LDH) from the cells into the culture medium at 6 hr of incubation. The LDH release was measured with a tetrazolium salt using the Cytotoxicity Detection KitPLUS (LDH) (Roche Diagnostics GmbH, Mannheim, Germany). The amount of formazan salt formed was measured at 492 nm on an Infinite 200 PRO microplate reader (Tecan Group Ltd., Männedorf, Switzerland). The maximum amount of LDH released was determined by incubating the cells with a lysis solution provided in the kit. The cytotoxicity was calculated as follows: cytotoxicity (%) = [(experimental value – low control)/(high control – low control)] × 100. The low control value, which corresponds to a spontaneous LDH release, was determined as the LDH released from untreated normal cells. The high control value, which corresponds to the maximum LDH release, was determined as the LDH released from the cells lysed by surfactant treatment. IL-8 secretion into the culture supernatant was determined by enzyme-linked immunosorbent assay (ELISA) with a Human IL-8 ELISA Ready-SET-Go! Kit (Thermo Fisher Scientific).
Evaluation of the antibiotic resistance of L. plantarum group strains
L. plantarum group strains were preincubated in MRS broth overnight. The preculture suspensions were added at 0.1% (v/v) into MRS broth including penicillin G (MP Biomedicals, LLC, Solon, OH, USA) at concentrations of 100, 50, 25, 12.5, 6.25, 3.13, 1.56, and 0.78 units/ml. Each culture was incubated for 24 hr at 37°C. Then, the OD600 of each culture was measured by BioPhotometer plus (Eppendorf AG, Hamburg, Germany). If bacteria did not grow after 24 hr of incubation, the culture was added at 1% (v/v) into fresh MRS broth without penicillin G. The culture was then incubated for 24 hr at 37°C, and the OD600 of the culture was subsequently measured.
RESULTS
Identification and classification of Lactobacillus spp. isolated from the fermented teas
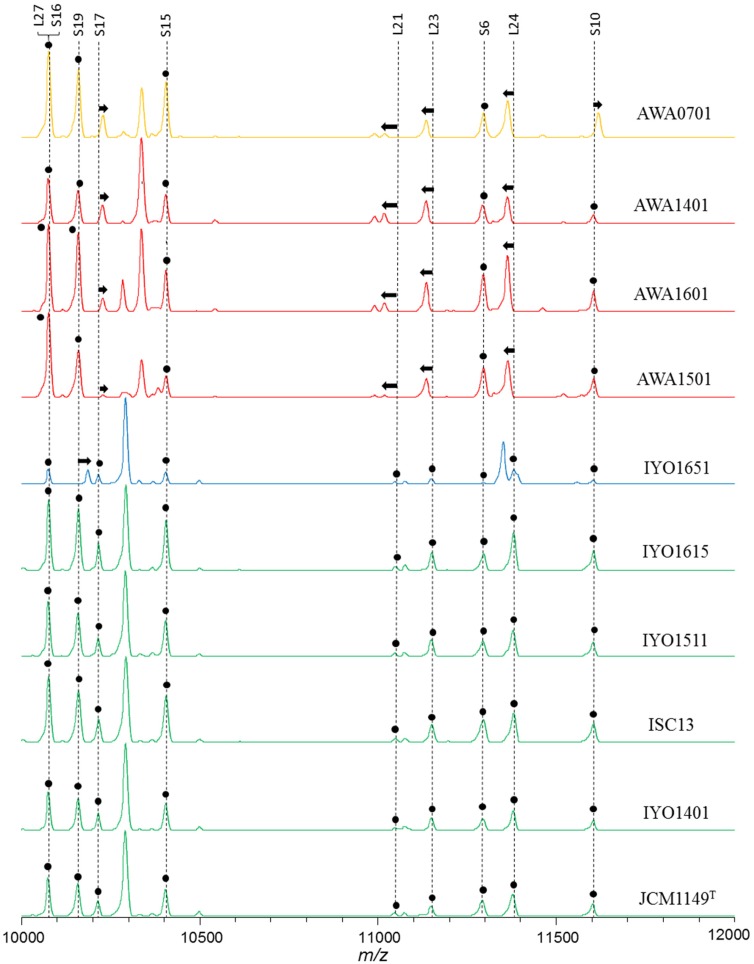
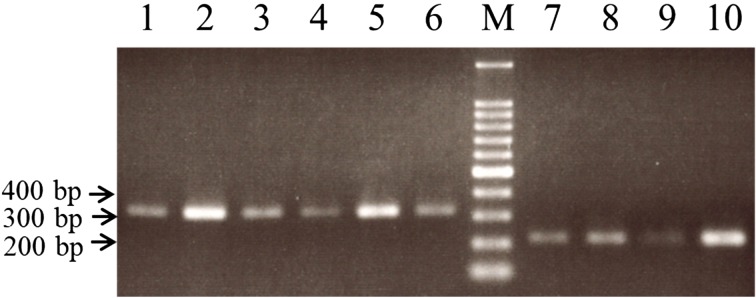
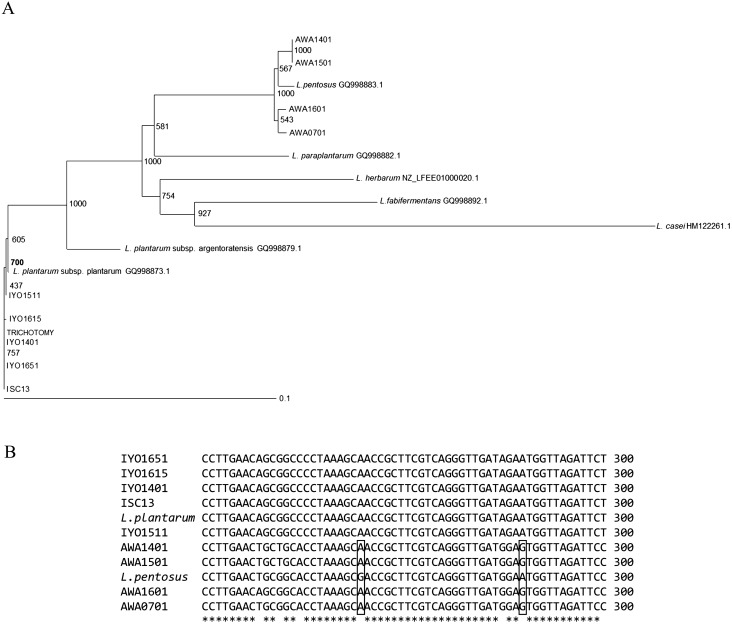
Lactobacilli were isolated from fresh leaves of Ishizuchi-kurocha and Awa-bancha in 2014, 2015, and 2016. Six strains of Lactobacillus spp. were isolated from Ishizuchi-kurocha, and four strains of Lactobacillus spp. were isolated from Awa-bancha. Based on 16S ribosomal RNA sequence homology, seven strains were identified as L. plantarum. The results concerning identification of Lactobacillus spp. are presented in Table 1. To isolate various Lactobacillus species, we employed three kinds of medium for isolation: skim milk, MRS agar, and LBS agar. Isolated Lactobacillus species were similar regardless of isolation media. Mainly, L. plantarum was isolated. L. plantarum was present among lactic acid bacteria isolates from both Ishizuchi-kurocha and Awa-bancha. Additionally, L. plantarum was present among isolates every year. Two Lactobacillus strains that were isolated from Ishizuchi-kurocha had high homology to L. pantheris and L. brevis. One Lactobacillus strain isolated from Awa-bancha shared high homology with L. fermentum. On the other hand, the species in the L. plantarum group are difficult to identify based on only 16S rRNA gene sequencing. L. plantarum, L. pentosus, L. paraplantarum, and L. herbarum are closely related species with 16S rRNA gene sequence similarities higher than 99%. Among these lactobacilli, a newly isolated L. plantarum strain, L. plantarum JCM 1149T, and previously isolated L. plantarum strain, ISC13 [3], and AWA0701 were compared with respect to the types of ribosomal subunit proteins (Fig. 2). According to the ribosomal protein typing, L. plantarum JCM 1149T and Ishizuchi-kurocha-derived strains belong to the same group. On the other hand, Awa-bancha-derived strains were assigned to another group. We examined further identification of newly isolated “L. plantarum” strains. PCR amplification products of the recA gene of L. plantarum, L. pentosus, and L. paraplantarum were of different sizes. The sizes of the amplicons for L. plantarum, L. pentosus, and L. paraplantarum were 318 bp, 218 bp, and 107 bp, respectively [16]. PCR amplification products of the recA gene of Ishizuchi-kurocha-derived strains were observed in the same position as L. plantarum JCM1149T (Fig. 3). On the other hand, the sizes of the amplicons of Awa-bancha-derived strains were different from that of L. plantarum JCM 1149T. The sizes of the amplicons of Awa-bancha-derived strains were similar to that of L. pentosus. Additionally, we performed phylogenetic analysis based on the dnaK gene (Fig. 4A). The Ishizuchi-kurocha-derived strains were closely related to L. plantarum subsp. plantarum, and the Awa-bancha-derived strains were closely related to L. pentosus. However, some single nucleotide polymorphisms between the L. pentosus and the Awa-bancha-derived strains were observed in the dnaK gene (Fig. 4B).
Table 1. Lactobacillus species isolated and used in this study.
| Strain No. | Species | Primers for PCR amplification of the 16S rRNA gene | DDBJ Accession No. |
Isolation medium | Culture temperature | Gas-forming | Source | Year isolated | |
|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA gene | dnaK gene | ||||||||
| JCM 1149T | Lactobacillus plantarum subsp. plantarum | Not applicable | 30 | – | Pickled cabbage | ||||
| ISC13 | Lactobacillus plantarum subsp. plantarum | 27F/1491R | LC333405 | LC416083 | 30 | – | Ishizuchi-Kurocha | 1993 | |
| IYO1401 | Lactobacillus plantarum subsp. plantarum | 27F/1492R | LC199957 | LC416084 | Skim milk | 30 | – | 2014 | |
| IYO1404 | Lactobacillus pantheris | 27F/1492R | LC199956 | Skim milk | 30 | – | 2014 | ||
| IYO1511 | Lactobacillus plantarum subsp. plantarum | 27F/1492R | LC199963 | LC416085 | MRS agar | 30 | – | 2015 | |
| IYO1516 | Lactobacillus brevis | 27F/1492R | LC199964 | MRS agar | 30 | + | 2015 | ||
| IYO1615 | Lactobacillus plantarum subsp. plantarum | 27F/1492R | LC333406 | LC416087 | LBS agar | 30 | – | 2016 | |
| IYO1651 | Lactobacillus plantarum subsp. plantarum | 27F/1492R | LC333407 | LC416086 | MRS agar | 30 | – | 2016 | |
| AWA0701 | Lactobacillus pentosus | 27F/1491R | LC333409 | LC416088 | 30 | – | Awa-Bancha | 2007 | |
| AWA1401 | Lactobacillus pentosus | 27F/1492R | LC199958 | LC416080 | Skim milk | 30 | – | 2014 | |
| AWA1501 | Lactobacillus pentosus | 27F/1492R | LC199959 | LC416081 | MRS agar | 30 | – | 2015 | |
| AWA1504 | Lactobacillus fermentum | 10F/800R | LC199960 | MRS agar | 30 | + | 2015 | ||
| AWA1601 | Lactobacillus pentosus | 27F/1492R | LC333408 | LC416082 | MRS agar | 30 | – | 2016 | |
L. plantarum JCM 1149T parchased from the RIKEN BioResource Center (JCM).
Fig. 2.
Classification of Lactobacillus plantarum group strains based on the MALDI mass spectra of ribosomal subunit proteins.
The peak labels indicate the names of assigned ribosomal subunit proteins. The peaks marked with a filled circle are common among the strains. Arrows indicate that peak shifts occurred in the strains. The strains show four patterns as indicated by different colors of the mass spectra.
Fig. 3.
Classification of Lactobacillus plantarum group strains based on the recA gene.
Amplification products obtained from the recA gene multiplex PCR were revealed. Lane 1, L. plantarum JCM 1149T; 2, ISC13; 3, IYO1401; 4, IYO1511; 5, IYO1615; 6, IYO1651; 7, AWA0701; 8, AWA1401; 9, AWA1501; 10, AWA1601. Lane M contained a 100 bp DNA ladder (TaKaRa Bio).
Fig. 4.
DnaK phylogenic tree for Lactobacillus plantarum group bacteria.
A) DnaK phylogenic tree. Reference sequences of L. plantarum subsp. plantarum, L. plantarum subsp. argentoratensis, L. paraplantarum, L. pentosus, L. herbarum, and L. fabifermentans were obtained from GenBank. L. casei was employed as outgroup species of L. plantarum. The accession numbers of the reference species are indicated in the figure. Bootstrap values are recorded on the nodes. B) Alignment of partial dnaK gene sequences of L. plantarum subsp. plantarum JCM 1149T, L. pentosus, and the isolates. The boxed bases indicate a single nucleotide polymorphism. *indicates that all bases are homology.
Evaluation of sugar utilization by L. plantarum strains
This property of L. plantarum strains isolated from the fermented teas was examined with the API 50 CH system (Table 2). The sugar utilization was loosely categorized into two groups: L. plantarum JCM 1149T and Ishizuchi-kurocha-derived strains utilized methyl-αD-mannopyranoside, D-melezitose, D-raffinose, and D-turanose. In contrast, Awa-bancha-derived strains could not use these sugars.
Adhesion of L. plantarum to Caco-2 cells
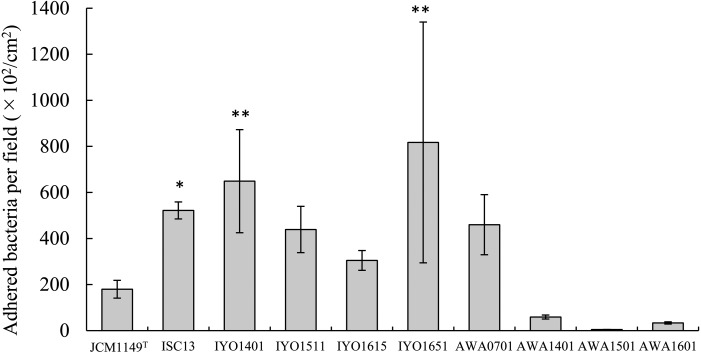
Among the new Lactobacillus strains isolated from fermented teas, adhesion properties of L. plantarum toward human colon epithelial cells were evaluated. L. plantarum IYO1401, which was obtained from Ishizuchi-kurocha, showed strong adhesion to Caco-2 cells (Fig. 5). Additionally, L. plantarum JCM 1149T showed weak adhesion to Caco-2 cells. The number of adherent bacterial cells in strains isolated from Ishizuchi-kurocha was 1.7-4.5-fold greater than that of strain JCM 1149T. By contrast, L. plantarum isolated from Awa-bancha showed weak adhesion to Caco-2 cells except strain AWA0701. The numbers of adherent bacterial cells in strains AWA1401, AWA1501, and AWA1601 were approximately 0.3, 0.02, and 0.2-fold greater than that of strain JCM 1149T, respectively. Thus, strains isolated from Ishizuchi-kurocha and Awa-bancha showed the opposite properties in terms of adhesion to Caco-2 cells.
Fig. 5.
Adhesion of Lactobacillus plantarum group strains to Caco-2 cells.
The Lactobacillus strains were resuspended at a concentration of 107 cells/ml in MEM containing 20% of FBS. The bacterial suspensions were added to Caco-2 cell monolayers with incubation for 2 hr. Then, the cells were washed three times with PBS for 5 min. After the last wash, the Caco-2 cell monolayers were disrupted with distilled water. Adherent bacteria were counted by plating the serial 10-fold dilutions of the suspension on MRS agar plates incubated anaerobically. **p<0.01 (compared with JCM1149T; Dunnett’s test, ANOVA).
Adhesion of L. plantarum to ECM proteins
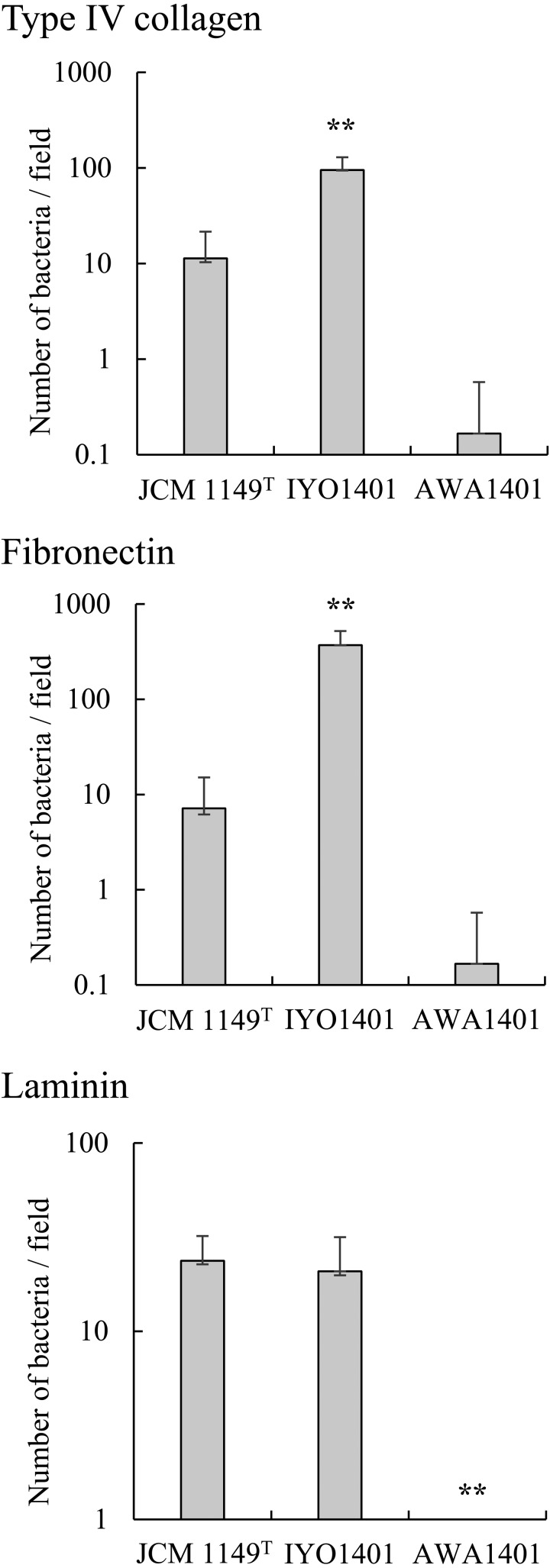
Some Lactobacillus spp. adhere not only to epithelial cells but also to ECM proteins [23]. Therefore, we analyzed the adhesion of L. plantarum strains to type IV collagen, laminin, and fibronectin, which are components of the basement membrane (Fig. 6). Among L. plantarum strains, JCM 1149T, IYO1401, and AWA1401 were subjected to the adhesion test. Strain IYO1401 revealed strong adhesion to fibronectin and type IV collagen, and strain IYO1401 also adhered to laminin. Strain JCM 1149T adhered to fibronectin, laminin, and type IV collagen. The adhesion ability of strain JCM 1149T was weaker than that of strain IYO1401. In contrast, L. plantarum strain AWA1401 did not adhere to the ECM proteins under study.
Fig. 6.
Adhesion of Lactobacillus plantarum group strains to ECM proteins.
L. plantarum AWA1401, IYO1401, and JCM1149T were resuspended in PBS at a concentration of 108 cells/ml. Then, 20 µl of the bacterial suspensions were added into wells coated with type IV collagen, laminin, or fibronectin in a multiwell glass slide. The slides were incubated for 2 hr at room temperature. After three washes with PBS, the adherent bacteria were counted under a microscope. **p<0.01 (compared with JCM1149T; Dunnett’s test, ANOVA).
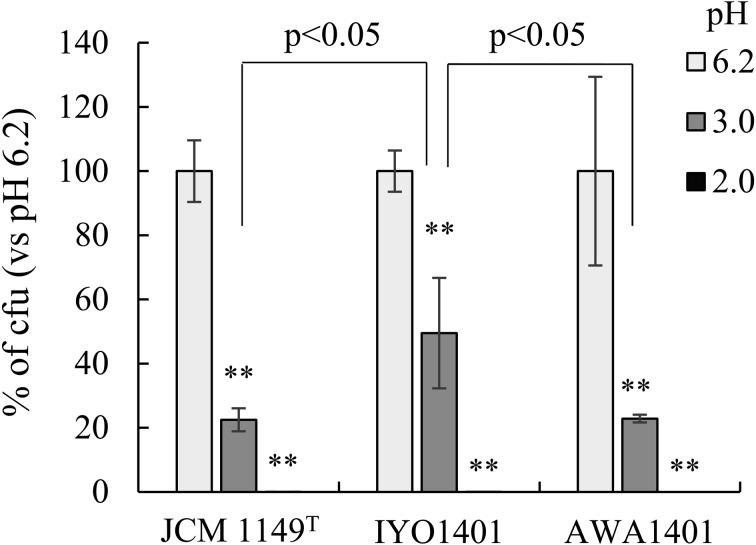
Survival of L. plantarum strains under acidic conditions
To evaluate survival of L. plantarum in gastric juice, survival of L. plantarum strains under acidic conditions was examined (Fig. 7). Each L. plantarum strain was cultured for 2 hr in MRS broth with a pH of 6.2, 3.0, or 2.0, and then colony formation on MRS agar plates was assessed. At pH 2.0, colony formation was not observed for any strain. Compared with the normal pH of MRS broth (6.2), colony formation of strains JCM 1149T and AWA1401 was decreased to approximately 20% at pH 3.0. Colony formation by strain IYO1401 was decreased to 45% at pH 3.0. Therefore, the acid resistance of strain IYO1401 was slightly stronger those that of the other strains.
Fig. 7.
Survival of Lactobacillus plantarum group strains under acidic conditions.
L. plantarum strains AWA1401, IYO1401, and JCM1149T precultured in MRS broth (pH 6.2) were added at 10% (v/v) into MRS broth with a pH of 2 or 3. The preculture suspensions were also added to normal MRS broth (pH 6.2) at 10%. Each mixture was incubated for 2 hr at 37°C, and then serially diluted cultures were spread on MRS plates. After anaerobic incubation overnight at 37°C, colony-forming units (CFUs) were estimated by counting. The survival rate (calculated from CFUs) in a pH 6.2 medium was set as 100%. **p<0.01 (compared with pH 6.2; Dunnett’s test, ANOVA).
Antibiotic resistance of L. plantarum
The antibiotic resistance of L. plantarum was examined (Table 3). Specifically, the antibiotic resistance of L. plantarum strains JCM 1149T, ISC13, IYO1401, and AWA1401 to penicillin G was examined. L. plantarum strains ISC13 and IYO1401 showed stronger resistance to penicillin G than strains JCM 1149T and AWA1401. The minimum inhibitory concentrations (MICs) of strains JCM1149T, ISC13, IYO1401, and AWA1401 were estimated to be 6.25, 100, 50, and 3.125 units/ml, respectively.
Table 3. Antibiotics resistance of Lactobacillus plantarum group strains isolated from fermented tea.
| Penicillin G concentration (units/ml) | Strain |
|||
|---|---|---|---|---|
| JCM 1149T | ISC13 | IYO1401 | AWA1401 | |
| 0 | 4.24 | 7.65 | 7.65 | 5.67 |
| 0.78 | 0.09 | 2.68 | 2.60 | 0.52 |
| 1.56 | 0.08 | 1.90 | 1.80 | 0.04 |
| 3.13 | 0.00 | 0.72 | 0.87 | 0.00 |
| 6.25 | 0.00 | 0.17 | 0.17 | 0.00 |
| 12.5 | 0.00 | 0.08 | 0.12 | 0.00 |
| 25.0 | 0.00 | 0.04 | 0.04 | 0.00 |
| 50.0 | 0.00 | 0.01 | 0.00 | 0.00 |
| 100 | 0.00 | 0.00 | 0.00 | 0.00 |
Values represent the OD600 of culture fluid of L. plantarum which was cultured for 24 hr at 37°C in MRS broth including penicillin G.
Effects of L. plantarum strains on animal cells
Possible toxic effects of L. plantarum strains on colon epithelial Caco-2 cells were examined next. We conducted two kinds of experiments. In Experiment 1, Caco-2 monolayers were incubated with each L. plantarum strain at a concentration of 107 cells/ml for 6 hr. After that, the bacterial suspension was removed, the cells were washed with PBS three times, and then the cells were cultured further for 18 hr in fresh MEM. This experimental model simulated adhesion and subsequent colonization by the bacteria. Cell membrane damage was examined by analyzing LDH release into the culture supernatant at 6 hr of incubation. No LDH release was detected from any groups of cells incubated with L. plantarum. Secretion of IL-8 into the culture supernatant from Caco-2 cells was examined at 6 and 24 hr of incubation. IL-8 is a cytokine and a marker of inflammation. The IL-8 concentration in the culture supernatant was under the measurement limit (2 pg/ml). An increase in IL-8 secretion was not detected in L. plantarum strains.
DISCUSSION
Most of the lactobacilli isolated from Ishizuchi-kurocha and Awa-bancha were identified as L. plantarum by 16S rRNA gene sequence. Other studies also revealed that among lactic acid bacteria, mostly L. plantarum group bacteria are isolated from Japanese post-fermented tea based on sugar utilization [3]. L. plantarum has been a dominant lactic acid bacterium in the fermented teas and may be involved in their fermentation. Therefore, L. plantarum is an important species for flavor formation of these post-fermented teas. Although the lactobacilli isolated from the two kinds of Japanese post-fermented teas here were identified as L. plantarum by 16S rRNA gene sequence, the strains showed different patterns of ribosomal subunit proteins (Fig. 2). According to classification by the ribosomal protein typing, Ishizuchi-kurocha-derived strains belong to the same group as JCM 1149T does, whereas Awa-bancha-derived L. plantarum strains were assigned to a different group. This pattern is also applicable to the strains isolated more than 10 years ago. L. plantarum, L. pentosus, L. paraplantarum, and L. herbarum are closely related species. The Awa-bancha-derived strains were closely related to L. pentosus by a further analysis based on recA and a dnaK gene (Figs. 3, 4).
The interactions with cultured animal cells were different between strain JCM 1149T and the Ishizuchi-kurocha-derived strains. The Ishizuchi-kurocha-derived L. plantarum strains manifested strong adhesion to Caco-2 cells (Fig. 5). By contrast, strain JCM 1149T and the Awa-bancha-derived strains weakly adhered to Caco-2 cells.
According to the ribosomal protein typing, analysis of recA and the dnaK gene, sugar utilization (Table 2), and interaction with Caco-2 cells, although these properties were dependent on the strain strictly speaking, L. plantarum group strains in this study can be subdivided into two groups: type strain JCM 1149T and Ishizuchi-kurocha-derived strains and Awa-bancha-derived strains which were identified as L. pentosus by recA and the dnaK gene.
Neither Ishizuchi-kurocha nor Awa-bancha involve a starter strain. The lactobacilli involved in their fermentation are present naturally. They may live on the tea leaves. Unique and beneficial regional Lactobacillus strains may persist in a traditional fermented drink. The areas where Ishizuchi-kurocha and Awa-bancha are produced by fermentation are located on the same island. The aerial distance between the two areas is approximately 120 km. Nevertheless, these areas are located deep in the mountains of Shikoku and interactions between them are scarce. Therefore, the unique strains of L. plantarum have persisted in each area, and these strains may have contributed to the unique flavor of the post-fermented teas. The results of this study suggest that there are regional L. plantarum strains in fermented teas. There is a possibility that the properties of the Lactobacillus strains depend on the geographic areas where the strains are isolated. Regionality may be one of the important factors for application of Lactobacillus strains to the food industry.
On the other hand, there is also the possibility that the difference in two kinds of properties of L. plantarum group species is dependent on the fermention processes of Ishizuchi-kurocha and Awa-bancha. Ishizuchi-kurocha is produced by 2-step fermentation, whereas Awa-bancha is produced by only anaerobic fermentation. Aerobic fermentation by fungi may affect the selection of L. plantarum strains in Ishizuchi-kurocha. In fact, L. plantarum strains isolated from Ishizuchi-kurocha had stronger antibiotic resistance than L. plantarum JCM 1149T and L. pentosus strains isolated from Awa-bancha (Table 3). One of the causes of the difference in the antibiotics resistance may be selection of lactic acid bacteria by a fungus. The Ishizuchi-kurocha-derived Lactobacillus spp. might be screened by antibiotics produced by fungi employed in the first-step fermentation.
There are many reports about probiotic properties of L. plantarum. For example, some L. plantarum strains improve immune responses. L. plantarum CGMCC 1.557 improves functioning of the spleen and enhances the phagocytic activity of macrophages [7]. Some L. plantarum strains can prevent food poisoning by inhibiting the growth of pathogenic bacteria such as E. coli, Shigella flexneri, S. Typhimurium, Vibrio parahaemolyticus, Campylobacter jejuni, and S. aureus [6, 24,25,26,27]. Oral administration of L. plantarum to rats improves lipid metabolism and alleviates oxidative stress in the liver [28]. It is reported that some L. plantarum strains can survive under gastric and intestinal conditions [29].
Besides, the adhesion ability of Lactobacillus to intestinal epithelial cells is one of the important characteristics for probiotics. Colonization of intestines by L. plantarum leads to continuous probiotic effects. As a step preceding the colonization, the ability of probiotic bacteria to adhere to the intestinal wall is also important [30]. There are many studies on the adhesion of Lactobacillus strains, including L. plantarum, to intestinal epithelial cells [22, 26, 31,32,33]. Moreover, some Lactobacillus strains adhere to ECM proteins such as fibronectin, laminin, and collagens [23, 34, 35]. One of the L. plantarum strains can adhere to type I collagen, and the adhesin in this case was identified as the EnoA1 protein [36]. These adhesion abilities depend on the strain [37].
On the other hand, in some cases, L. plantarum has shown a pathogenic potential [38]. It was reported that L. plantarum is involved in infective endocarditis [39]. Safety is important for probiotic use of L. plantarum. No L. plantarum group strains isolated from fermented teas in this study exerted cytotoxicity or induced cytokines. The L. plantarum group strains evaluated here did not manifest notable pathogenicity.
Differences in properties between the L. plantarum group strains isolated from Ishizuchi-kurocha and Awa-bancha were not be affected by the isolation year. They were dependent only on the geographic area of origin. Probiotic properties have been studied in most bacterial strains. It may also be necessary to estimate some biological characteristics of lactobacilli (including their probiotic ability) by regional affiliation. In the future, it will be worthwhile to elucidate the relation between the environment and the properties of Lactobacillus.
Acknowledgments
We thank Ms. Kyoko Shinozuka, Mr. Yusuke Ide (Social Welfare Corporation Peace, Saijo, Ehime, Japan), Mr. Yoshifumi Yamanouchi (Gutokusan natural farm, Saijo, Ehime, Japan), and Ms. Hisami Toda (Satsuki-Kai, Saijo, Ehime, Japan) for kindly giving us Ishizuchi-kurocha samples. We also appreciate the assistance provided by Mr. Daichi Hyakuno (Irodori-Bancha, Kamikatsu, Tokushima, Japan).
REFERENCES
- 1.Horie M, Nara K, Sugino S, Umeno A, Yoshida Y. 2016. Comparison of antioxidant activities among four kinds of Japanese traditional fermented tea. Food Sci Nutr 5: 639–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okada S, Takahashi N, Ohara N, Uchimura T, Kozaki M. 1996. Microorganisms in fermentation of Goishi-cha, Japanese fermented tea leaves. Nippon Shokuhin Kagaku Kogaku Kaishi 43: 1019–1027. [Google Scholar]
- 3.Tamura A, Kato M, Omori M, Nanba A, Miyagawa K. 1994. Characterization of microorganisms in post-heating fermented teas in Japan. Journal of Home Economics of Japan 45: 1095–1101. [Google Scholar]
- 4.Shintani T, Matsumoto Y, Bessyo Y. 2000. Analysis of microflora in Ishizuchi-kurocha by using ribosome RNA gene and BiOLOG system. Bulletin of Industrial Research Institutes of Ehime Prefecture 38: 73–79. [Google Scholar]
- 5.Sadeghi-Aliabadi H, Mohammadi F, Fazeli H, Mirlohi M. 2014. Effects of Lactobacillus plantarum A7 with probiotic potential on colon cancer and normal cells proliferation in comparison with a commercial strain. Iran J Basic Med Sci 17: 815–819. [PMC free article] [PubMed] [Google Scholar]
- 6.Eom JS, Song J, Choi HS. 2015. Protective effects of a novel probiotic strain of Lactobacillus plantarum JSA22 from traditional fermented soybean food against infection by Salmonella enterica Serovar Typhimurium. J Microbiol Biotechnol 25: 479–491. [DOI] [PubMed] [Google Scholar]
- 7.Ren D, Li C, Qin Y, Yin R, Du S, Liu H, Zhang Y, Wang C, Rong F, Jin N. 2015. Evaluation of immunomodulatory activity of two potential probiotic Lactobacillus strains by in vivo tests. Anaerobe 35 Pt B: 22–27. [DOI] [PubMed] [Google Scholar]
- 8.Molin G. 2001. Probiotics in foods not containing milk or milk constituents, with special reference to Lactobacillus plantarum 299v. Am J Clin Nutr 73 Suppl: 380S–385S. [DOI] [PubMed] [Google Scholar]
- 9.Niedzielin K, Kordecki H, Birkenfeld B. 2001. A controlled, double-blind, randomized study on the efficacy of Lactobacillus plantarum 299V in patients with irritable bowel syndrome. Eur J Gastroenterol Hepatol 13: 1143–1147. [DOI] [PubMed] [Google Scholar]
- 10.Frias J, Song YS, Martínez-Villaluenga C, González de Mejia E, Vidal-Valverde C. 2008. Immunoreactivity and amino acid content of fermented soybean products. J Agric Food Chem 56: 99–105. [DOI] [PubMed] [Google Scholar]
- 11.Song DF, Zhu MY, Gu Q. 2014. Purification and characterization of Plantaricin ZJ5, a new bacteriocin produced by Lactobacillus plantarum ZJ5. PLoS One 9: e105549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu X, Zhao Y, Sun Y, Gu Q. 2014. Purification and characterisation of plantaricin ZJ008, a novel bacteriocin against Staphylococcus spp. from Lactobacillus plantarum ZJ008. Food Chem 165: 216–223. [DOI] [PubMed] [Google Scholar]
- 13.Rumjuankiat K, Perez RH, Pilasombut K, Keawsompong S, Zendo T, Sonomoto K, Nitisinprasert S. 2015. Purification and characterization of a novel plantaricin, KL-1Y, from Lactobacillus plantarum KL-1. World J Microbiol Biotechnol 31: 983–994. [DOI] [PubMed] [Google Scholar]
- 14.Sánchez B, Urdaci MC. 2012. Extracellular proteins from Lactobacillus plantarum BMCM12 prevent adhesion of enteropathogens to mucin. Curr Microbiol 64: 592–596. [DOI] [PubMed] [Google Scholar]
- 15.Lane D. 1991. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics, Stackebrandt E, Goodfellow M (eds). J Wiley and Sons, Chichester, pp. 115–175. [Google Scholar]
- 16.Torriani S, Felis GE, Dellaglio F. 2001. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl Environ Microbiol 67: 3450–3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang CH, Chang MT, Huang MC, Lee FL. 2011. Rapid identification of Lactobacillus plantarum group using the SNaPshot minisequencing assay. Syst Appl Microbiol 34: 586–589. [DOI] [PubMed] [Google Scholar]
- 18.Sun L, Teramoto K, Sato H, Torimura M, Tao H, Shintani T. 2006. Characterization of ribosomal proteins as biomarkers for matrix-assisted laser desorption/ionization mass spectral identification of Lactobacillus plantarum. Rapid Commun Mass Spectrom 20: 3789–3798. [DOI] [PubMed] [Google Scholar]
- 19.Teramoto K, Sato H, Sun L, Torimura M, Tao H. 2007. A simple intact protein analysis by MALDI-MS for characterization of ribosomal proteins of two genome-sequenced lactic acid bacteria and verification of their amino acid sequences. J Proteome Res 6: 3899–3907. [DOI] [PubMed] [Google Scholar]
- 20.Teramoto K, Sato H, Sun L, Torimura M, Tao H, Waguri S, Hayashi T, Haga S. 2007. Rapid identification and classification of psychrotrophic lactic acid bacteria by matrix-assisted laser desorption/ionization mass spectrometry. Bunseki Kagaku 56: 1063–1070. [Google Scholar]
- 21.Takiguchi R, Suzuki Y. 2000. Survival of lactic acid bacteria in simulated digestive juice. Chonai Saikingaku Zasshi 14: 11–18. [Google Scholar]
- 22.Todoriki K, Mukai T, Sato S, Toba T. 2001. Inhibition of adhesion of food-borne pathogens to Caco-2 cells by Lactobacillus strains. J Appl Microbiol 91: 154–159. [DOI] [PubMed] [Google Scholar]
- 23.Horie M, Murakami T, Sato T, Tarusawa Y, Nakamura S, Toba T. 2005. Anaerobic induction of adherence to laminin in Lactobacillus gasseri strains by contact with solid surface. Curr Microbiol 51: 275–282. [DOI] [PubMed] [Google Scholar]
- 24.Al-Madboly LA, Abdullah AK. 2015. Potent antagonistic activity of Egyptian Lactobacillus plantarum against multiresistant and virulent food-associated pathogens. Front Microbiol 6: 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davoodabadi A, Soltan Dallal MM, Rahimi Foroushani A, Douraghi M, Sharifi Yazdi MK, Amin Harati F. 2015. Antibacterial activity of Lactobacillus spp. isolated from the feces of healthy infants against enteropathogenic bacteria. Anaerobe 34: 53–58. [DOI] [PubMed] [Google Scholar]
- 26.Satish Kumar R, Kanmani P, Yuvaraj N, Paari KA, Pattukumar V, Arul V. 2011. Lactobacillus plantarum AS1 binds to cultured human intestinal cell line HT-29 and inhibits cell attachment by enterovirulent bacterium Vibrio parahaemolyticus. Lett Appl Microbiol 53: 481–487. [DOI] [PubMed] [Google Scholar]
- 27.Nishiyama K, Seto Y, Yoshioka K, Kakuda T, Takai S, Yamamoto Y, Mukai T. 2014. Lactobacillus gasseri SBT2055 reduces infection by and colonization of Campylobacter jejuni. PLoS One 9: e108827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li C, Nie SP, Zhu KX, Ding Q, Li C, Xiong T, Xie MY. 2014. Lactobacillus plantarum NCU116 improves liver function, oxidative stress and lipid metabolism in rats with high fat diet induced non-alcoholic fatty liver disease. Food Funct 5: 3216–3223. [DOI] [PubMed] [Google Scholar]
- 29.Tokatlı M, Gülgör G, Bağder Elmacı S, Arslankoz İşleyen N, Özçelik F. 2015. In vitro properties of potential probiotic indigenous lactic acid bacteria originating from traditional pickles. BioMed Res Int 2015: 315819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spivey MA, Dunn-Horrocks SL, Duong T. 2014. Epithelial cell adhesion and gastrointestinal colonization of Lactobacillus in poultry. Poult Sci 93: 2910–2919. [DOI] [PubMed] [Google Scholar]
- 31.Kainulainen V, Tang Y, Spillmann T, Kilpinen S, Reunanen J, Saris PE, Satokari R. 2015. The canine isolate Lactobacillus acidophilus LAB20 adheres to intestinal epithelium and attenuates LPS-induced IL-8 secretion of enterocytes in vitro. BMC Microbiol 15: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garriga M, Rubio R, Aymerich T, Ruas-Madiedo P. 2015. Potentially probiotic and bioprotective lactic acid bacteria starter cultures antagonise the Listeria monocytogenes adhesion to HT29 colonocyte-like cells. Benef Microbes 6: 337–343. [DOI] [PubMed] [Google Scholar]
- 33.Sun Z, Huang L, Kong J, Hu S, Zhang X, Kong W. 2012. In vitro evaluation of Lactobacillus crispatus K313 and K243: high-adhesion activity and anti-inflammatory effect on Salmonella braenderup infected intestinal epithelial cell. Vet Microbiol 159: 212–220. [DOI] [PubMed] [Google Scholar]
- 34.Hsueh HY, Yueh PY, Yu B, Zhao X, Liu JR. 2010. Expression of Lactobacillus reuteri Pg4 collagen-binding protein gene in Lactobacillus casei ATCC 393 increases its adhesion ability to Caco-2 cells. J Agric Food Chem 58: 12182–12191. [DOI] [PubMed] [Google Scholar]
- 35.Yadav AK, Tyagi A, Kumar A, Saklani AC, Grover S, Batish VK. 2015. Adhesion of indigenous Lactobacillus plantarum to gut extracellular matrix and its physicochemical characterization. Arch Microbiol 197: 155–164. [DOI] [PubMed] [Google Scholar]
- 36.Salzillo M, Vastano V, Capri U, Muscariello L, Sacco M, Marasco R. 2015. Identification and characterization of enolase as a collagen-binding protein in Lactobacillus plantarum. J Basic Microbiol 55: 890–897. [DOI] [PubMed] [Google Scholar]
- 37.Ramos CL, Thorsen L, Schwan RF, Jespersen L. 2013. Strain-specific probiotics properties of Lactobacillus fermentum, Lactobacillus plantarum and Lactobacillus brevis isolates from Brazilian food products. Food Microbiol 36: 22–29. [DOI] [PubMed] [Google Scholar]
- 38.Harty DW, Oakey HJ, Patrikakis M, Hume EB, Knox KW. 1994. Pathogenic potential of lactobacilli. Int J Food Microbiol 24: 179–189. [DOI] [PubMed] [Google Scholar]
- 39.Struve J, Weiland O, Nord CE. 1988. Lactobacillus plantarum endocarditis in a patient with benign monoclonal gammopathy. J Infect 17: 127–130. [DOI] [PubMed] [Google Scholar]