Abstract
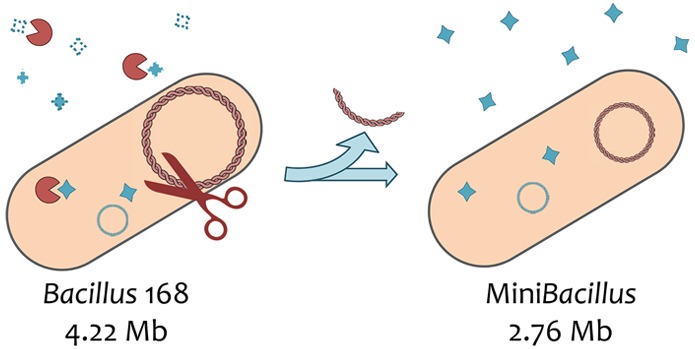
The availability of complete genome sequences and the definition of essential gene sets were fundamental in the start of the genome engineering era. In a recent study, redundant and unnecessary genes were systematically deleted from the Gram-positive bacterium Bacillus subtilis, an industrial production host of high-value secreted proteins. This culminated in strain PG10, which lacks about 36% of the genome, thus representing the most minimal Bacillus chassis currently available. Here, we show that this “miniBacillus” strain has synthetic traits that are favorable for producing “difficult-to-produce proteins”. As exemplified with different staphylococcal antigens, PG10 overcomes several bottlenecks in protein production related to the secretion process and instability of the secreted product. These findings show for the first time that massive genome reduction can substantially improve secretory protein production by a bacterial expression host, and underpin the high potential of genome-engineered strains as future cell factories.
Keywords: Bacillus subtilis, PG10, genome engineering, cell factory, staphylococcal proteins, secretion
In 1997 during the ninth International Conference on Bacilli, the complete genome sequence of the widely appreciated bacterial cell factory Bacillus subtilis 168 was announced.1 This represented one of the early milestones in bacterial genomics, and it was the starting point for many subsequent systems and synthetic biological studies. These included a first systematic analysis of essential gene functions and the genome-wide elucidation of gene regulatory and metabolic networks.2,3 Importantly, the genome sequence of a bacterium that is naturally competent for DNA uptake opened up unprecedented possibilities for synthetic biology. Accordingly, several subsequent studies explored to what extent the genome of B. subtilis can be engineered, for instance, by the removal of prophages, AT-rich islands, and otherwise dispensable genes.4,5
From a biotechnological perspective, engineering the Bacillus genome is very attractive, as it allows the elimination of unwanted features like the production of surfactants and the redirection of cellular metabolism toward the production of proteins and vitamins, which are major Bacillus products.6 Nonetheless, most Bacillus genome engineering studies to date were more focused on understanding the complexity of a living cell from a fundamental point of view than on industrial applications of genome-reduced bacteria.4,5,7 In fact, the possibility of redesigning Bacillus as a cell factory was so far only explored for the production of industrial enzymes and nucleosides such as guanosine and thymidine from bacilli.8 As shown by Ogasawara and colleagues, reducing the B. subtilis genome by 20.7% (strain MGB874) allowed improved production of the secreted alkaline cellulase Egl237 by about 2-fold.9 Yet, another B. subtilis strain (MG1M) with a 23.3% reduced genome secreted an alkaline cellulase and a subtilisin-like alkaline protease to comparable levels as the parental 168 strain.10 While these findings were encouraging, they did not yet represent radical improvements in terms of the overall productivity of B. subtilis.
In a recent study, Reuβ et al. presented the smallest engineered B. subtilis genomes known to date.11 Starting from the previously constructed B. subtilis Δ6 strain, which lacks 332 prophage- and AT-rich island-encoded genes (i.e., 7.7% of the wild-type genome), the total number of genes was reduced to 2700 in strain PG10 and 2648 in strain PS38. This represented a genome reduction of ∼36% compared to the wild-type genome, which includes 4253 genes. Importantly, compared to other genome-engineered B. subtilis strains, the PG10 and PS38 strains lacked the genes for eight major secreted Bacillus proteases, which were previously identified as major bottlenecks for heterologous protein production.12−15 In addition, Reuβ et al. overcame the loss of genetic competence that was previously observed upon genome reduction by introducing a cassette for enhanced expression of the competence transcription factor ComK. While the “miniBacillus” strains PG10 and PS38 were characterized in much detail with respect to overall physiological features, their application potential had not been addressed. Thus, we asked the question whether these genome-reduced strains might be advantageous for the production of “difficult proteins” that are highly susceptible to proteases and poorly secreted by the B. subtilis strains generally used in the laboratory or industry, including the prototype strain 168. In this respect, it is noteworthy that secretory protein production systems are preferable over cellular production systems, as the downstream processing of secreted proteins is generally easier and more cost-effective. Here we show that the genome-reduced B. subtilis PG10 strain allows the production of secreted heterologous proteins that cannot be obtained with the 168 strain. Briefly, the beneficial changes in the PG10 strain relate both to reduced proteolysis and enhanced translation. This represents an important step forward in the secretory production of difficult proteins.
Results and Discussion
Susceptibility of Model Staphylococcal Proteins to Particular Secreted Proteases
To test the application potential of miniBacillus strains for production of difficult proteins, we selected four heterologous secreted reporter proteins that we (i) could not produce in B. subtilis 168 or the previously developed protease mutant B. subtilis WB800, (ii) could produce and purify from another expression host, in this case Lactococcus lactis, and (iii) could detect either with a specific antibody or a tag.16−18 These are four secreted proteins of S. aureus, namely, the chemotaxis inhibitory protein (CHIPS), the staphylococcal complement inhibitor (SCIN), the immunodominant staphylococcal antigen A (IsaA), and the S. aureus nuclease (Nuc). In the present study, these proteins served primarily as read out for improved secretion, but they also have potential applications in antistaphylococcal immunotherapies or as diagnostic markers.17−19 In addition, we focused our study on the PG10 strain because it still contains the amyE gene that can serve as a facile chromosomal expression platform.20
As a first approach to determine the overall feasibility of producing CHIPS, SCIN, IsaA and Nuc in B. subtilis, we produced these proteins in L. lactis as previously described, and then tested their stability in spent culture media of different B. subtilis mutant strains. In addition to B. subtilis 168, these strains included mutants lacking combinations of the nprB, aprE, epr, bpr, nprE, mpr, vpr, wprA, htrA, and htrB protease genes as well as the genome-engineered Δ6 and PG10 strains. As shown in Figure 1, within 2 h of incubation, the four S. aureus reporter proteins were degraded in spent media of B. subtilis, with the exception of media from strains that lack the wprA gene for the “wall protease A” (including the miniBacillus PG10). Of note, all four reporter proteins remained stable for up to 24 h in spent growth media, as long as the respective cultured cells were WprA-deficient (not shown). WprA was thus far considered to be active at the membrane-cell wall interface of B. subtilis, and the present findings show for the first time that the secreted fraction of this enzyme has strong proteolytic activity as well (Figure 1). Taken together, these observations show that degradation by extracellular proteases can be a major limiting factor in the production of CHIPS, SCIN, IsaA, and Nuc in B. subtilis.
Figure 1.
Stability of staphylococcal proteins to particular secreted proteases. Culture supernatants from L. lactis overexpressing the staphylococcal proteins IsaA, CHIPS, Nuc, or SCIN were mixed with spent culture media from B. subtilis 168, the protease mutants BRB02 to BRB14 or the genome-engineered strains Δ6 or PG10. Names of strains that lack the wprA gene are indicated in bold. Proteins were TCA-precipitated after 2 h of incubation at 37 °C, and their degradation was assessed by Western blotting. Immunodetection was performed with anti-his6 antibodies to detect CHIPS and Nuc, the human monoclonal antibody 6D4 against SCIN, or the human monoclonal antibody 1D9 against IsaA.
Secretion of Staphylococcal Proteins by B. subtilis PG10
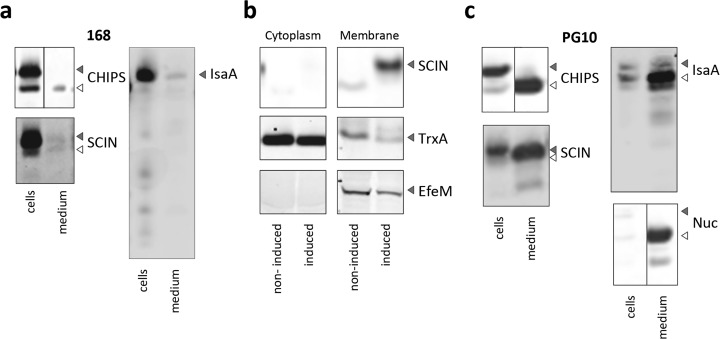
To express the staphylococcal reporter proteins CHIPS, SCIN, IsaA, and Nuc in B. subtilis we used the “subtilin-regulated expression system” (SURE).20 This inducible high-level expression system is based on sensing of the bacteriocin subtilin by the SpaRK two-component regulatory system which subsequently drives the transcription of the PspaS promoter. Therefore, the spaRK genes were inserted into the amyE gene of both the 168 and PG10 strains. To direct secretion of the staphylococcal reporters into the growth medium, we initially selected the N-terminal signal peptide of the α-amylase AmyQ (SPAmyQ) of Bacillus licheniformis.21 While this signal peptide directed some secretion of SCIN in the 168 strain, it did not facilitate any secretion of CHIPS, IsaA, or Nuc. Yet, the CHIPS and IsaA proteins fused to this signal peptide were detectable in the cell fraction and the same applied to SCIN (Figure 2A and data not shown). In fact, 168 cells producing the SPAmyQ-SCIN fusion accumulated this precursor in substantial amounts (Figure 2A). Since ineffective protein secretion may relate to the choice of signal peptide, the CHIPS, IsaA, and Nuc proteins were also fused to the signal peptide of the xylanase XynA (SPXynA) of B. subtilis,22 which did allow some secretion of CHIPS and IsaA in the 168 strain (Figure 2A). However, in this case also the CHIPS and IsaA proteins accumulated in the cells, mostly in a precursor form (Figure 2A). Of note, no secretion of Nuc was detectable when fused to SPXynA (not shown). To pinpoint potential bottlenecks in the secretion of SCIN and IsaA by the 168 strain, the respective cells were fractionated, and the localization of these two staphylococcal proteins was assessed by Western blotting, using the native cytoplasmic protein TrxA and the membrane-associated lipoprotein EfeM as controls (Figure 2B). This showed that SCIN and IsaA accumulated in the membrane fraction, while these proteins were not detectable in the cytoplasmic fraction (Figure 2B) or in the cell wall (not shown). The accumulation of SCIN and IsaA in the membrane fraction is indicative of aberrant translocation by the Sec secretion machinery and ineffective processing of the signal peptide by signal peptidase. Further, the absence of these proteins from the cell wall and growth medium suggests that SCIN and IsaA either do not reach these destinations due to defective membrane translocation or that they are subject to degradation by proteases like WprA as soon as they appear at the membrane-cell wall interface.
Figure 2.
Induced overproduction of staphylococcal proteins in B. subtilis 168 and PG10. The spaRK genes were introduced in the amyE locus of B. subtilis 168 and PG10 to allow subtilin-inducible expression of reporter proteins with the aid of the spaS promoter on plasmids pRAG3::chp (SPxynA), pRAG1::scn (SPamyQ), pRAG3::isaA (SPxynA) and pRAG3::nuc (SPxynA). Protein expression was induced with subtilin and culture samples were collected 2 h post induction. (a) The cellular (cells) and extracellular (medium) levels of the expressed staphylococcal proteins were assessed by Western blotting with an anti-his6 antibody to detect CHIPS, the monoclonal antibody 6D4 against SCIN, or the monoclonal antibody 1D9 against IsaA. (b) The subcellular localization of SCIN produced in B. subtilis 168 was assessed by fractionation. As a negative control, noninduced cells were used. TrxA and EfeM were used as markers for cytoplasmic and membrane-bound proteins, respectively. (c) CHIPS, SCIN, IsaA, and Nuc were produced and secreted by the miniBacillus strain PG10. Precursor forms of CHIPS, SCIN, IsaA and Nuc, and full-size TrxA and EfeM are marked with filled arrow heads; mature forms of CHIPS, SCIN, IsaA, and Nuc are marked with open arrow heads.
Next, we investigated the secretion of CHIPS, SCIN, IsaA and Nuc in the genome-reduced strain PG10. To this end, secretion of SCIN was directed by SPAmyQ, whereas secretion of CHIPS, IsaA and Nuc was driven by SPXynA. As shown in Figure 2C, effective secretion of all four staphylococcal proteins by the PG10 strain could be demonstrated, albeit that the cells did accumulate some precursor forms of CHIPS, SCIN and IsaA. Of note, all the Nuc produced by strain PG10 was secreted. On this basis, we conclude that the PG10 strain displays improved secretion and substantially reduced extracytoplasmic degradation of CHIPS, SCIN, IsaA and Nuc. The lowered degradation of the four staphylococcal proteins is in line with the absence of eight secreted Bacillus proteases from the PG10 strain, and the absence of CHIPS, SCIN, IsaA and Nuc degradation in spent medium of this strain as shown in Figure 1. Having demonstrated production and secretion of staphylococcal proteins by PG10, we next examined the amount of IsaA protein secreted into the growth medium because, of all four staphylococcal proteins, IsaA was produced at the highest level. As shown by LDS-PAGE and a standard curve with bovine serum albumin, IsaA was secreted to a concentration of about 7 mg/L after 2 h of induction (data not shown). While this is a relatively low yield compared to the gram/liter yields of industrial enzymes obtained in industrial fermentations, it is still a considerable yield compared to the close-to-zero yield observed for the 168 strain. Nonetheless, this raised the question whether genome engineering had somehow affected the full potential of the PG10 strain for protein secretion compared to the parental 168 strain. This possibility was tested by expressing the α-amylase AmyQ in the PG10 strain. The level of AmyQ secretion by PG10 was about 2-fold lower compared to the 168 strain (Figure 3). This may relate in part to the degradation by an as yet unidentified protease as evidenced by the presence of AmyQ degradation products in the growth medium of strain PG10.
Figure 3.
Overproduction of AmyQ in the miniBacillus strain PG10. To express the B. amyloliquefaciens α-amylase AmyQ, B. subtilis PG10 was transformed with plasmid pKTH10. (a) Production of AmyQ by B. subtilis strains 168 and PG10. Mature AmyQ retained in the cells or secreted into the growth medium was detected by Western blotting using specific antibodies. The filled arrowhead indicates the position of mature AmyQ. Detectable dominant degradation products of AmyQ are marked with open arrow heads. (b) Amylase activity of AmyQ secreted by B. subtilis strains 168 and PG10 was visualized by spotting 20 μL aliquots of the growth medium fraction of the respective cultures on LB agar plates supplemented with 1% starch. Zones of starch degradation were detected after overnight incubation at 37 °C.
Enhanced Secretion of IsaA in B. subtilis PG10
To assess whether the addition of subtilin has a detrimental effect on the PG10 strain, we compared the growth curves of the 168 and PG10 strains carrying pRAG3::isaA when subtilin was added during the exponential growth phase (Figure 4). The Figure 4 shows that addition of subtilin, as was done in the experiments presented in Figure 2, slowed down the growth of both strains, but it certainly did not impair growth or lead to severe cell lysis.
Figure 4.

Growth of B. subtilis 168 and PG10 carrying pRAG3::isaA in the presence or absence of subtilin induction. Overnight cultures of B. subtilis 168 and PG10 carrying pRAG3::isaA were diluted 1:50 in 100 μL of fresh LB medium in a 96-well microtiter plate, and incubated with shaking at 37 °C in a Biotek synergy 2 plate reader. OD600 readings were recorded every 10 min. When the strains reached the midexponential phase, 1% subtilin was added to the cultures (indicated with arrows) to induce IsaA production.
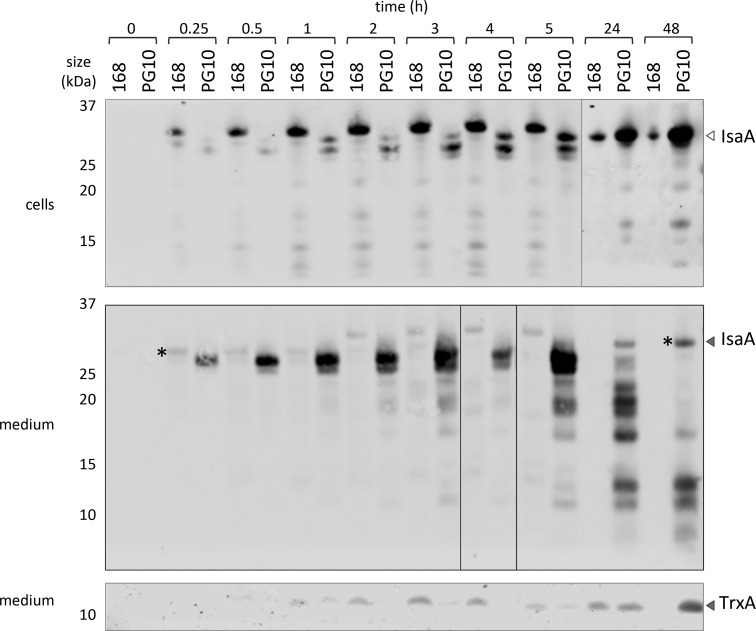
To investigate whether the improved performance of strain PG10 relates to changes in the balance between induced protein secretion and degradation, we performed a time course experiment where the production and secretion of IsaA in the PG10 and 168 strains was assayed as a function of the time of induction with subtilin. Indeed, the amount of IsaA detectable in the medium at 15 min post induction was higher in PG10 than in 168, which implies that IsaA translation was more effective in the PG10 strain. In addition, the time course experiment highlighted distinctive features of the 168 and PG10 strains in terms of IsaA secretion. In particular, induction of IsaA production in the 168 strain led to rapid accumulation of full-size IsaA in the cells until 5 h post induction (Figure 5, upper panel), while at later time points (from 24 h onward) IsaA production was strongly reduced. The secreted full-size IsaA (marked *) was mostly detectable but at very low amounts in the growth medium of the 168 strain at 15 min post induction after which it started to disappear; concomitantly, IsaA forms with aberrant mobility started to appear in the medium. Between 30 min and 4 h post induction of IsaA production, strain 168 mounted a high secretion stress response as was evidenced by relatively high levels of the secretion stress-responsive HtrA and HtrB proteins in the medium (Supplementary Figure S1). Since it was previously shown that the secretion stress response in B. subtilis is elicited by malfolded secretory proteins at the membrane-cell wall interface,23,24 our present findings for strain 168 are indicative of saturation of the secretion pathway with IsaA molecules at this subcellular location, ultimately leading to a block in secretion and production of IsaA. Compared to 168, the levels of IsaA detectable in the PG10 cells remained low, and the full-size IsaA form (*) as observed in strain 168 was not detectable at all in cells of the PG10 strain. Yet, this form was gradually secreted into the growth medium and clearly accumulated at late time points post induction (from 24 h). Of note, PG10 did secrete smaller-sized forms of IsaA at earlier time points, especially until 5 h post induction (Figure 5, middle panel). The latter forms may represent incompletely synthesized IsaA as the full-size mature IsaA was 100% stable when incubated in spent medium of the PG10 strain (Figure 1). Of note, the detected smaller-sized forms of IsaA are possibly not derived from IsaA degradation by HtrA and HtrB in the cells and growth medium, because the levels of these proteases were comparable in samples with high or low IsaA degradation levels (Supplementary Figure S1). Moreover, at those time points where the 168 strain decreased the production of IsaA (i.e., 24 and 48 h post induction), the HtrA and HtrB proteins were barely detectable.
Figure 5.
Secretion of IsaA by B. subtilis 168 or PG10. The production of IsaA by exponentially growing cells of B. subtilis 168 and PG10 containing the spaRK genes in amyE and carrying plasmid pRAG3::isaA was induced with 1% subtilin (t = 0). Subsequently, samples were withdrawn at the indicated time points, and cells were separated from the growth medium by centrifugation. After correction for the respective culture OD600, cellular proteins and proteins secreted into the growth medium were separated by LDS-PAGE and analyzed by Western blotting with the IsaA-specific monoclonal antibody 1D9. The levels of the cytoplasmic protein TrxA in growth medium fractions were assessed by Western blotting with a specific polyclonal antibody and used as readout for cell lysis. The two IsaA-specific bands marked* have the same electrophoretic mobility.
The appearance of the marker for cell lysis, TrxA, in the growth medium of the 168 strain will depend on the combined effects of cell lysis and extracellular proteolysis by secreted proteases.27 As evidenced by the TrxA levels, within the first 5 h post induction, extracellular proteolysis was probably still low in the 168 strain, whereas some cells started to lyse. At later stages, extracellular proteolysis led to complete disappearance of secreted IsaA and, ultimately, also extracellular TrxA became degraded (Figure 5, lower panel). In contrast, extracellular proteolysis was largely suppressed in the PG10 strain as shown in Figure 1 and, hence, in this strain the extracellular appearance of TrxA was probably largely due to cell lysis.
To assess a possible effect on protein production owing to differences in the copy number of plasmid pRAG3::isaA in the 168 and PG10 strains, a whole-genome sequencing analysis was performed. On the basis of the ratios of the average coverage of plasmid and chromosomal reads, it was inferred that pRAG3::isaA has relative copy numbers of ∼107 in the PG10 strain and ∼80 in the 168 strain (Supplementary Table S3). This difference is too small to account for the strong difference in IsaA productivity in the two strains.
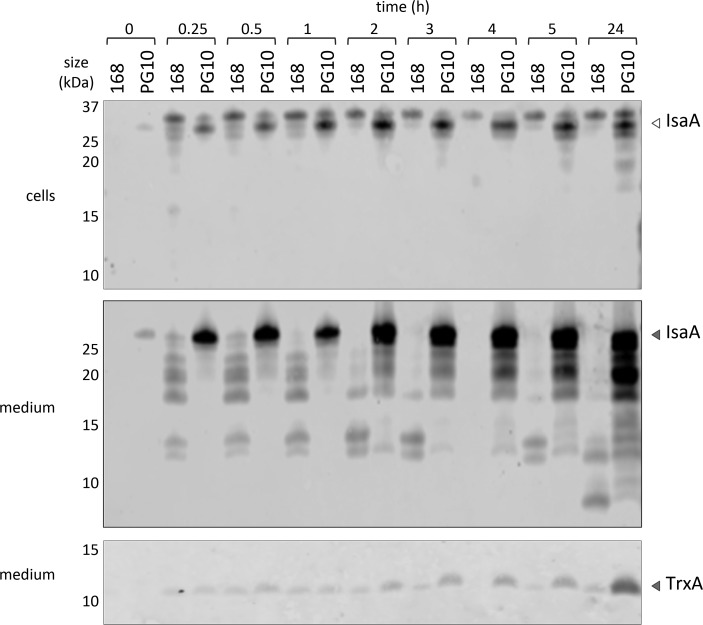
Since even in the PG10 strain some IsaA degradation was detectable, as was the case for AmyQ (Figure 3), we investigated whether this residual protease activity could be inhibited by adding protease inhibitors to the media of growing bacteria. Indeed, this was the case for IsaA produced by the PG10 strain, where IsaA degradation was substantially reduced for the first 5 h postinduction (Figure 6). On the other hand, the protease inhibitors were incapable of rescuing IsaA produced by the 168 strain. Still, especially at 24 h postinduction IsaA degradation did increase, while the levels of HtrA and HtrB remained the same (Supplementary Figure S2). In fact, the latter is indicative of effective protease inhibition as it was previously shown that the secreted forms of HtrA and HtrB are sensitive for degradation by other secreted proteases of B. subtilis.14,25,26 This raises the question, which residual proteases are still released into the growth medium of the PG10 strain. An intriguing option is that these are cytoplasmic proteases, because the degradation of IsaA secreted by strain PG10 after 24 h post induction coincided with increased levels of cell lysis as visualized by increased extracellular levels of the cytoplasmic protein TrxA (Figure 5, bottom panel). Of note, in a previous study, we have shown that autolysis can be increased in protease mutant strains as the extracellular proteases downregulate the levels of autolysins like LytD.27
Figure 6.
Secretion of IsaA by B. subtilis 168 or PG10 in the presence of protease inhibitors. Production of IsaA in B. subtilis 168 and PG10 was assessed as described for Figure 5, but in this case, the growth medium was supplemented with the Complete protease inhibitor without EDTA cocktail from Roche. Production of IsaA was induced with 1% subtilin (t = 0), and samples were collected as a function of the time indicated. The cytoplasmic protein TrxA was used as a marker for cell lysis. The positions of TrxA and the precursor and mature forms of IsaA are indicated with arrow heads.
While reduced extracellular proteolysis explains part of the improved extracellular production of staphylococcal antigens by strain PG10, it explains neither the drastic differences in the observed precursor levels of CHIPS and IsaA in the cells, nor the accumulation of SCIN in the membrane (Figure 2b). Instead, this would suggest that there may also be differences between the 168 and PG10 strains in the secretion efficiency of these proteins. Such differences could relate to the total amount of secreted proteins competing for the same pathway or to the overall rates of translation. In fact, the idea that the translation rates in strains 168 and PG10 differ would be consistent with the previous observation that cells of the PG10 strain produce about 25% less ribosomal proteins than the 168 strain.11 To test whether the rates of translation in strains 168 and PG10 differ, we employed a synthetic module where GFP transcription is coupled to the expression of the bmrCD genes for a drug efflux pump of B. subtilis. In a previous study, we have shown that the expression of bmrCD is regulated via transcriptional attenuation that is modulated by translation of the BmrB leader peptide.28 When BmrB translation is slowed down by ribosome-targeted antibiotics, such as lincomycin, the expression of bmrCD is triggered. Therefore, we introduced the transcriptional bmrC-GFP fusion into the bmrBCD locus of strains 168 and PG10, and assessed the levels of GFP expression during growth in the presence or absence of lincomycin. Under both conditions, the expression of GFP in the PG10 strain remained much lower than in the 168 strain where the presence of lincomycin elicited strong induction of GFP transcriptional activity (Figure 7).
Figure 7.
Translational efficiency in B. subtilis 168 and PG10 based on lincomycin-inducible bmrCD expression. The expression of bmrCD gene transcription in B. subtilis 168 and PG10 was measured in real time using a bmrC::GFP fusion. (a) GFP transcriptional activity (TAU) in B. subtilis 168 bmrC-GFP and PG10 bmrC-GFP was assessed in the presence or absence of lincomycin. (b) GFP transcriptional activity in B. subtilis PG10 bmrC-GFP in the presence or absence of lincomycin.
We also assessed the effects of subinhibitory concentrations of lincomycin for the 168 and PG10 strains carrying the bmrC-GFP fusion. As previously shown for the 168 strain, subinhibitory concentrations of lincomycin between 0.02 and 0.2 μg/mL still induced bmrC-GFP, but the level of induction decreased with the reduction of the lincomycin concentration.28 In line with this previous observation, in the PG10 strain a very mild bmrC-GFP induction was observed in the presence of the subinhibitory lincomycin concentration of 0.2 μg/mL, but not for a lincomycin concentration of 0.02 μg/mL (data not shown).
These observations imply that, despite the lower levels of ribosomal proteins, the translational efficiency of BmrB is higher in strain PG10 than in the 168 strain, irrespective of the presence of lincomycin. Accordingly, it appears that translation is more effective in strain PG10 than in the 168 strain. It is tempting to speculate that the apparently enhanced translational activity in the miniBacillus PG10 relates to the substantially decreased number of translatable mRNAs as a consequence of the significant genome reduction in this strain.
Conclusion
In conclusion, the present study provides proof-of-principle that genome engineering can open a window of unprecedented possibilities for the production of “difficult proteins” in the bacterial cell factory B. subtilis. This was exemplified with the miniBacillus strain PG10 producing four different staphylococcal antigens that cannot be produced with currently applied B. subtilis strains. Of note, the 168 and PG10 strains are far from isogenic due to the deletion of 36% of the genome in strain PG10. This makes it challenging to pinpoint the particular mutations and their relative contributions that resulted in improved protein production by the PG10 strain. Nonetheless, the present study underlines the benefits of genomic streamlining as an approach to potentiate and increase the valuable properties of Bacillus strains for protein production. Clearly, to reach the full potential of such miniBacillus strains they need to be further optimized, especially with respect to potential product degradation, reduced cell lysis and ease of use in large-scale fermentation. We are confident that this will deliver new-generation production strains for a wide spectrum of proteins including not only enzymes but also many dearly needed biopharmaceuticals.
Materials and Methods
Strains and Plasmids
Bacterial strains and plasmids used in this study are listed in Supplementary Table S1. L. lactis was grown in M17 medium (Oxoid Limited) containing 0.5% glucose (GM17) at 30 °C without shaking. Medium was supplemented with 5 μg mL–1 chloramphenicol, or 5 μg mL–1 erythromycin as appropriate. B. subtilis was grown in LB medium (Becton Dickinson) at 37 °C with shaking. Where needed, medium was supplemented with 2 μg mL–1 erythromycin, or 20 μg mL–1 kanamycin. S. aureus N315 was grown at 37 °C as standing culture in tryptone soy broth (Oxoid Limited).
Protein Stability Tests
To assess the stability of CHIPS, SCIN, IsaA and Nuc, aliquots of L. lactis cultures producing these staphylococcal proteins were added to spent growth media of different B. subtilis strains. To produce the staphylococcal proteins, overnight cultures of L. lactis carrying pNG4210::scn, pNG4210::chp, pNG4210::isaA or pNG400::nuc were diluted 1:20 in GM17 medium with chloramphenicol. Upon reaching an optical density of 600 nm (OD600) of ∼0.5, the production of CHIPS, SCIN, IsaA or Nuc was induced with nisin by addition of the culture supernatant from an overnight culture of the nisin-producing L. lactis strain NZ9700 at a 1:1000 dilution. After 16 h induction, the cultures were centrifuged and growth medium fractions were harvested. 500 μL growth medium aliquots containing either CHIPS, SCIN, IsaA, or Nuc were mixed with 500 μL spent growth medium of different protease-deficient B. subtilis strains. The latter spent media were obtained by growing B. subtilis strains overnight in LB medium, dilution of the overnight cultures into fresh LB medium, and continued growth until 2 h after entry into the stationary phase. At this point in growth, cells were separated from the growth medium by centrifugation and the supernatant fractions were collected and applied to assess the stability of CHIPS, SCIN, IsaA, or Nuc by incubation at 37 °C for 2 or 24 h. The presence or absence of CHIPS, SCIN, IsaA, or Nuc was visualized by LDS-PAGE and Western blotting with specific antibodies against SCIN, IsaA or a hexa-histidine tag.
Construction of Expression Plasmids
All plasmids in this study were constructed using conventional cloning techniques. The oligonucleotides listed in Supplementary Table S2 were obtained from Eurogentec. Phusion-HF DNA polymerase, restriction endonucleases, and T4 DNA ligase were obtained from New England Biolabs. L. lactis MG1363 was transformed by electrotransformation and used as an intermediate cloning host for plasmid amplification and verification. Constructions were verified by Sanger sequencing (Eurofins Genomics) prior to introduction into B. subtilis through transformation of competent cells. The spaRK genes, integrated in the amyE gene of B. subtilis 168 (also referred to as pNZ8900), were transferred to strain PG10 by transformation of competent cells. To this end, the competence of strain PG10 was induced with mannitol at a final concentration of 0.25%. Subsequently, the PG10 strain carrying spaRK was transformed with plasmids for the expression of CHIPS, SCIN, IsaA or Nuc.
To create the plasmid pRAG1 for subtilin-inducible expression of C-terminally his6-tagged staphylococcal proteins, the sequence for SPAmyQ was amplified from pKTH10 with primers SPamyQ_F and SPamyQ_R, thereby introducing a BspHI cleavage site 5′ to the SPAmyQ-coding sequence. A multiple cloning site was copied from plasmid pNG4210 using primers MCS4210_F and MCS4210_R, thereby adding the sequence encoding a C-terminal his6-tag as well as a 3′ HindIII cleavage site. Both PCR fragments were merged by overlap extension PCR with primers SPamyQ_F and MCS4210_R. The resulting PCR fragment was cut with BspHI and HindIII and ligated into the NocI and HindIII sites of the receiving plasmid pNZ8910.
The plasmid pRAG1 was used as a backbone for the insertion of staphylococcal genes. The chp, scn, and isaA, genes were amplified using primers pNG42ins_F and pNG42ins_R. The chp gene was amplified from pNG4210::chp, scn was amplified from pNG4210::scn and the isaA gene was amplified from pNG4210::isaA. The nuc gene was amplified from S. aureus N315 with primers NucN315_F and NucN315_R. The amplified PCR products and receiving plasmid pRAG1 were digested with BamHI and NotI, and T4-ligated. The resulting plasmids were verified by sequencing and named pRAG1::chp, pRAG1::scn, pRAG1::isaA and pRAG1::nuc.
To construct pRAG3, the sequence encoding SPxynA was amplified from B. subtilis 168 with the primers SPxynA_F and SPxynA_R, which introduced flanking 5′ EcoRV and 3′ BamHI restriction sites. The pRAG1 based plasmids carrying the chp, scn, isaA, or nuc genes were used as vector backbones, by amplification with primers woSP_F and woSP_R, thereby eliminating the sequences encoding SPAmyQ. The amplified PCR products were digested with BamHI and EcoRV and T4-ligated, resulting in the fusion of chp, scn, isaA, or nuc with sequences encoding SPxynA. The plasmids thus obtained were named, pRAG3::chp, pRAG3::scn, pRAG3::isaA and pRAG3::nuc. The Part ID’s of the plasmids constructed in this study are available from the ACS Synthetic Biology registry as specified in Supplementary Table S4.
Lithium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (LDS-PAGE) and Western Blotting
Protein samples were prepared and separated by LDS-PAGE as described previously.29 Before loading, samples were corrected for OD600. For Western blotting analysis, proteins separated by LDS-PAGE were blotted onto a nitrocellulose membrane (Protran). Subsequent immunodetection of bound proteins was performed with anti-his6 antibodies (Life Technologies), the human monoclonal antibody 6D4 against SCIN,18 or the human monoclonal antibody 1D9 against IsaA.17 For visualization of antibody binding, the 6D4 and 1D9 antibodies were directly labeled with IRDye 800CW (LiCor Biosciences), whereas bound anti-his6 antibodies were visualized with secondary antibodies labeled with IRDye 800CW. Fluorescence was recorded at 800 nm with an Odyssey Infrared Imaging System (LiCor Biosciences).
Protein Production, Localization, and Quantification
For production of staphylococcal proteins in B. subtilis strains, overnight cultures were diluted to a final OD600 of 0.15 and incubated until midexponential growth when protein expression was induced with 1:100 supernatant from B. subtilis ATCC6633, which contains subtilin. Cells were further incubated for 2 h, and samples were taken for LDS-PAGE and Western blotting. For the secretion of IsaA by B. subtilis 168 or PG10 in terms of time and protease activity, cells were grown in medium in the presence or absence of the Complete protease inhibitor cocktail without EDTA from Roche and samples were taken at different time points after induction with subtilin.
Subcellular localization of the staphylococcal proteins in B. subtilis 168 was performed by fractionation experiments as described previously.26 In short, cells from an overnight culture were harvested by centrifugation, resuspended in protoplast buffer (100 mM Tris-HCl, pH 8.2, 20 mM MgCl2, 20% sucrose, 1 mg/mL lysozyme, 0.01% DNase, and Complete protease inhibitors from Roche) and incubated for 30 min. The resulting protoplasts and liberated cell wall proteins were then separated by centrifugation. The protoplasts were resuspended in disruption buffer (50 mM Tris-HCl, pH 8.2, 2.5 mM EDTA) and disrupted with glass beads using a Precellys24 bead beater (Bertin Technologies, Montigny-le-Bretonneux, France). Cellular debris and unbroken protoplast were removed by low-speed centrifugation (10 min, 4000g, 4 °C), and the resulting supernatant fraction was subject to ultracentrifugation (30 min, 200 000g, 4 °C). The resulting supernatant fraction with cytosolic proteins was collected. The pelleted membranes were resuspended in solubilization butter (20 mM Tris, pH 8.0, 10% glycerol, 50 mM NaCl, 0.03% DDM) and incubated overnight at 4 °C. Subsequently, nonsolubilized and solubilized membrane proteins were separated by centrifugation (15 min, 100 000g, 4 °C). Lastly, the resulting supernatant fraction with solubilized membrane proteins was collected. The cytoplasmic protein TrxA and the lipoprotein EfeM were used as markers for the cytosolic and membrane fractions, respectively. Correct localization of EfeM, TrxA, and the staphylococal proteins was corroborated by Western blotting with rabbit polyclonal antibodies against TrxA or EfeM.
Quantification of staphylococcal proteins was done with a standard curve of bovine serum albumin. Protein samples were prepared for LDS-PAGE, and proteins were stained with SimplyBlue SafeStain (Novex). Proteins were visualized with an Odyssey Infrared Imaging System (LiCor Biosciences), and the signal in each lane was quantified using the ImageJ gel analyzer.
Growth Curves
Overnight cultures were diluted 1:50 in 100 μL of LB medium. Then cultures were incubated with shaking at 37 °C in a Biotek synergy 2 plate reader. When the cultures reached the midexponential phase, subtilin was added at a final concentration of 1%. OD600 readings were recorded every 10 min.
Live Cell Array Analyses
Live cell array analyses were performed as described previously.28,30 In short, overnight cultures were diluted 1:1000 in 100 μL culture supplemented with 2 μg/mL lincomycin or H2O in 96-well flat bottom microtiter plates (Greiner Bio-One). Then, cultures were grown at 37 °C in a Biotek synergy 2 plate reader, and OD600 and GFP fluorescence (excitation 485/20 nm, emission 528/20 nm) readings were taken every 10 min for 24 h. Background fluorescence was subtracted from control strains not expressing GFP. Finally, arbitrary transcriptional activity units (TAU) were calculated with the equation: (GFPt – GFPt–1)/OD600t. In this equation, t represents a specific time point and t – 1 the previous time point at which fluorescence was measured.
Plasmid Copy Number Determination
Next generation sequencing was used to determine the copy number of plasmid pRAG3::isaA in B. subtilis 168 and PG10 as described previously.31 Total DNA extraction for sequencing was performed from colonies using the Ultraclean Microbial DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, US) according to the manufacturer’s protocol. DNA concentrations were determined using a Qubit 2.0 fluorometer and the dsDNA HS and/or BR assay kit (Life technologies, Carlsbad, CA, US). DNA libraries were prepared using the Nextera XT DNA Library Preparation Kit (llumina, San Diego, CA, US). Sequence analysis was performed with an Illumina Miseq System generating paired-end reads of 300 bp. De novo assembly of paired-end reads was performed using CLC Genomics Workbench v11.0.1 (QIAGEN, Hilden, Germany). To approximate plasmid copy numbers, the ratio of the average coverage of plasmid and genomic reads was calculated for each strain.
Acknowledgments
We thank Rocky M. Cranenburgh for providing the BRB strains and Max den Uijl for experimental support. Part of this research was supported by the EU Horizon 2020 grant 720776 (JS) and a National Council of Science and Technology scholarship (CONACyT) (RAS).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssynbio.8b00342.
Supporting figures and tables (PDF)
Author Contributions
R.A.S. and J.M.v.D. performed, designed, and analyzed experiments. R.A.S. and J.M.v.D. conceived the study. R.A.S., J.S., and J.M.v.D. wrote the manuscript. R.A.S prepared the figures.
The authors declare no competing financial interest.
Supplementary Material
References
- Kunst F.; Ogasawara N.; Moszer I.; Albertini A. M.; Alloni G.; Azevedo V.; Bertero M. G.; Bessières P.; Bolotin A.; Borchert S.; Borriss R.; Boursier L.; Brans A.; Braun M.; Brignell S. C.; Bron S.; Brouillet S.; Bruschi C. V.; Caldwell B.; Capuano V.; Carter N. M.; Choi S. K.; Codani J. J.; Connerton I. F.; Danchin A. (1997) The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390, 249–256. 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Buescher J. M.; Liebermeister W.; Jules M.; Uhr M.; Muntel J.; Botella E.; Hessling B.; Kleijn R. J.; Le Chat L.; Lecointe F.; Mäder U.; Nicolas P.; Piersma S.; Rügheimer F.; Becher D.; Bessieres P.; Bidnenko E.; Denham E. L.; Dervyn E.; Devine K. M.; Doherty G.; Drulhe S.; Felicori L.; Fogg M. J.; Goelzer A.; Hansen A.; Harwood C. R.; Hecker M.; Hubner S.; Hultschig C.; Jarmer H.; Klipp E.; Leduc A.; Lewis P.; Molina F.; Noirot P.; Peres S.; Pigeonneau N.; Pohl S.; Rasmussen S.; Rinn B.; Schaffer M.; Schnidder J.; Schwikowski B.; Van Dijl J. M.; Veiga P.; Walsh S.; Wilkinson A. J.; Stelling J.; Aymerich S.; Sauer U. (2012) Global network reorganization during dynamic adaptations of Bacillus subtilis metabolism. Science 335, 1099–1103. 10.1126/science.1206871. [DOI] [PubMed] [Google Scholar]
- Nicolas P.; Mäder U.; Dervyn E.; Rochat T.; Leduc A.; Pigeonneau N.; Bidnenko E.; Marchadier E.; Hoebeke M.; Aymerich S.; Becher D.; Bisicchia P.; Botella E.; Delumeau O.; Doherty G.; Denham E. L.; Fogg M. J.; Fromion V.; Goelzer A.; Hansen A.; Härtig E.; Harwood C. R.; Homuth G.; Jarmer H. (2012) Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335, 1103–1106. 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- Westers H.; Dorenbos R.; Van Dijl J. M.; Kabel J.; Flanagan T.; Devine K. M.; Jude F.; Séror S. J.; Beekman A. C.; Darmon E.; Eschevins C.; De Jong A.; Bron S.; Kuipers O. P.; Albertini A. M.; Antelmann H.; Hecker M.; Zamboni N.; Sauer U.; Bruand C.; Ehrlich D. S.; Alonso J. C.; Salas M.; Quax W. J. (2003) Genome engineering reveals large dispensable regions in Bacillus subtilis. Mol. Biol. Evol. 20, 2076–2090. 10.1093/molbev/msg219. [DOI] [PubMed] [Google Scholar]
- Tanaka K.; Henry C. S.; Zinner J. F.; Jolivet E.; Cohoon M. P.; Xia F.; Bidnenko V.; Ehrlich S. D.; Stevens R. L.; Noirot P. (2013) Building the repertoire of dispensable chromosome regions in Bacillus subtilis entails major refinement of cognate large-scale metabolic model. Nucleic Acids Res. 41, 687–699. 10.1093/nar/gks963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann H., van Dijl J. M., Krishnappa L., and Pragái Z. (2016) Host Organisms: Bacillus subtilis. In Ind. Biotechnol. (Wittmann C. J. C. L., Ed.) Wiley-VCH Verlag GmbH, Weinheim, Germany. [Google Scholar]
- Reuß D. R.; Commichau F. M.; Gundlach J.; Zhu B.; Stülke J. (2016) The blueprint of a minimal cell: MiniBacillus. Microbiol. Mol. Biol. Rev. 80, 955–987. 10.1128/MMBR.00029-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Zhu X.; Zhang X.; Fu J.; Wang Z.; Chen T.; Zhao X. (2016) Characterization of genome - reduced Bacillus subtilis strains and their application for the production of guanosine and thymidine. Microb. Cell Fact. 15 (94), 1–15. 10.1186/s12934-016-0494-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe K.; Kageyama Y.; Morimoto T.; Ozawa T.; Sawada K.; Endo K.; Tohata M.; Ara K.; Ozaki K.; Ogasawara N. (2011) Combined effect of improved cell yield and increased specific productivity enhances recombinant enzyme production in genome-reduced Bacillus subtilis strain MGB874. Appl. Environ. Microbiol. 77, 8370–8381. 10.1128/AEM.06136-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ara K.; Ozaki K.; Nakamura K.; Yamane K.; Sekiguchi J.; Ogasawara N. (2007) Bacillus minimum genome factory: effective utilization of microbial genome information. Biotechnol. Appl. Biochem. 46, 169–178. 10.1042/BA20060111. [DOI] [PubMed] [Google Scholar]
- Reuß D. R.; Altenbuchner J.; Mäder U.; Rath H.; Ischebeck T.; Sappa P. K.; Thürmer A.; Guérin C.; Nicolas P.; Steil L.; Zhu B.; Feussner I.; Klumpp S.; Daniel R.; Commichau F. M.; Völker U.; Stülke J. (2017) Large-scale reduction of the Bacillus subtilis genome: consequences for the transcriptional network, resource allocation, and metabolism. Genome Res. 27, 289–299. 10.1101/gr.215293.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl S.; Bhavsar G.; Hulme J.; Bloor A. E.; Misirli G.; Leckenby M. W.; Radford D. S.; Smith W.; Wipat A.; Williamson E. D.; Harwood C. R.; Cranenburgh R. M. (2013) Proteomic analysis of Bacillus subtilis strains engineered for improved production of heterologous proteins. Proteomics 13, 3298–3308. 10.1002/pmic.201300183. [DOI] [PubMed] [Google Scholar]
- Stephenson K.; Harwood C. R. (1998) Influence of a cell-wall-associated protease on production of alfa-amylase by Bacillus subtilis. Appl. Environ. Microbiol. 64, 2875–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnappa L.; Monteferrante C. G.; Neef J.; Dreisbach A.; van Dijl J. M. (2014) Degradation of extracytoplasmic catalysts for protein folding in Bacillus subtilis. Appl. Environ. Microbiol. 80, 1463–1468. 10.1128/AEM.02799-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westers H.; Westers L.; Darmon E.; Van Dijl J. M.; Quax W. J.; Zanen G. (2006) The CssRS two-component regulatory system controls a general secretion stress response in Bacillus subtilis. FEBS J. 273, 3816–3827. 10.1111/j.1742-4658.2006.05389.x. [DOI] [PubMed] [Google Scholar]
- Neef J.; Milder F. J.; Koedijk D. G. A. M.; Klaassens M.; Heezius E. C.; van Strijp J. A. G.; Otto A.; Becher D.; van Dijl J. M.; Buist G. (2015) Versatile vector suite for the extracytoplasmic production and purification of heterologous His-tagged proteins in Lactococcus lactis. Appl. Microbiol. Biotechnol. 99, 9037–9048. 10.1007/s00253-015-6778-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg S.; Bonarius H. P. J.; van Kessel K. P. M.; Elsinga G. S.; Kooi N.; Westra H.; Bosma T.; van der Kooi-Pol M. M.; Koedijk D. G. A. M.; Groen H.; van Dijl J. M.; Buist G.; Bakker-Woudenberg I. A. J. M. (2015) A human monoclonal antibody targeting the conserved staphylococcal antigen IsaA protects mice against Staphylococcus aureus bacteremia. Int. J. Med. Microbiol. 305, 55–64. 10.1016/j.ijmm.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Hoekstra H.; Romero Pastrana F.; Bonarius H. P. J.; van Kessel K. P. M.; Elsinga G. S.; Kooi N.; Groen H.; van Dijl J. M.; Buist G. (2018) A human monoclonal antibody that specifically binds and inhibits the staphylococcal complement inhibitor protein SCIN. Virulence 9, 262–272. 10.1080/21505594.2017.1294297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosman C. W. K.; Romero Pastrana F.; Buist G.; Heuker M.; Van Oosten M.; McNamara J. O.; Van Dam G. M.; Van Dijl J. M. (2018) Ex vivo tracer efficacy in optical imaging of Staphylococcus aureus nuclease activity. Sci. Rep. 8, 1–8. 10.1038/s41598-018-19289-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongers R. S.; Veening J.-W.; Van Wieringen M.; Kuipers O. P.; Kleerebezem M.; Van M.; Van Wieringen M. (2005) Development and characterization of a Subtilin-Regulated Expression System in Bacillus subtilis: strict control of gene expression by addition of subtilin. Appl. Environ. Microbiol. 71, 8818–24. 10.1128/AEM.71.12.8818-8824.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt B.; Antelmann H.; Albrecht D.; Ehrenreich A.; Maurer K.-H.; Evers S.; Gottschalk G.; van Dijl J. M.; Schweder T.; Hecker M. (2009) Cell physiology and protein secretion of Bacillus licheniformis compared to Bacillus subtilis. J. Mol. Microbiol. Biotechnol. 16, 53–68. 10.1159/000142894. [DOI] [PubMed] [Google Scholar]
- Gilbert C.; Howarth M.; Harwood C. R.; Ellis T. (2017) Extracellular self-assembly of functional and tunable protein conjugates from Bacillus subtilis. ACS Synth. Biol. 6, 957–967. 10.1021/acssynbio.6b00292. [DOI] [PubMed] [Google Scholar]
- Darmon E.; Noone D.; Masson A.; Bron S.; Kuipers O. P.; Devine K. M.; Van Dijl J. M. (2002) A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J. Bacteriol. 184, 5661–5671. 10.1128/JB.184.20.5661-5671.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyyrylainen H. L.; Bolhuis A.; Darmon E.; Muukkonen L.; Koski P.; Vitikainen M.; Sarvas M.; Pragai Z.; Bron S.; Van Dijl J. M.; Kontinen V. P. (2001) A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41, 1159–1172. 10.1046/j.1365-2958.2001.02576.x. [DOI] [PubMed] [Google Scholar]
- Antelmann H.; Darmon E.; Noone D.; Veening J. W.; Westers H.; Bron S.; Kuipers O. P.; Devine K. M.; Hecker M.; Van Dijl J. M. (2003) The extracellular proteome of Bacillus subtilis under secretion stress conditions. Mol. Microbiol. 49, 143–156. 10.1046/j.1365-2958.2003.03565.x. [DOI] [PubMed] [Google Scholar]
- Zweers J. C.; Wiegert T.; van Dijl J. M. (2009) Stress-responsive systems set specific limits to the overproduction of membrane proteins in Bacillus subtilis. Appl. Environ. Microbiol. 75, 7356–7364. 10.1128/AEM.01560-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnappa L.; Dreisbach A.; Otto A.; Goosens V. J.; Cranenburgh R. M.; Harwood C. R.; Becher D.; Van Dijl J. M. (2013) Extracytoplasmic proteases determining the cleavage and release of secreted proteins, lipoproteins, and membrane proteins in Bacillus subtilis. J. Proteome Res. 12, 4101–4110. 10.1021/pr400433h. [DOI] [PubMed] [Google Scholar]
- Reilman E.; Mars R. A. T. T.; Van Dijl J. M.; Denham E. L. (2014) The multidrug ABC transporter BmrC/BmrD of Bacillus subtilis is regulated via a ribosome-mediated transcriptional attenuation mechanism. Nucleic Acids Res. 42, 11393–11407. 10.1093/nar/gku832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef J.; Koedijk D. G. A. M.; Bosma T.; van Dijl J. M.; Buist G. (2014) Efficient production of secreted staphylococcal antigens in a non-lysing and proteolytically reduced Lactococcus lactis strain. Appl. Microbiol. Biotechnol. 98, 10131–10141. 10.1007/s00253-014-6030-y. [DOI] [PubMed] [Google Scholar]
- Botella E.; Fogg M.; Jules M.; Piersma S.; Doherty G.; Hansen A.; Denham E. L.; Le Chat L.; Veiga P.; Bailey K.; Lewis P. J.; Van Dijl J. M.; Aymerich S.; Wilkinson A. J.; Devine K. M. (2010) pBaSysBioII: An integrative plasmid generating gfp transcriptional fusions for high-throughput analysis of gene expression in Bacillus subtilis. Microbiology 156, 1600–1608. 10.1099/mic.0.035758-0. [DOI] [PubMed] [Google Scholar]
- Nepal S.; Bonn F.; Grasso S.; Stobernack T.; de Jong A.; Zhou K.; Wedema R.; Rosema S.; Becher D.; Otto A.; Rossen J. W.; van Dijl J. M.; Bathoorn E. (2018) An ancient family of mobile genomic islands introducing cephalosporinase and carbapenemase genes in Enterobacteriaceae. Virulence 9, 1377–1389. 10.1080/21505594.2018.1509666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.