Abstract
Sensitization to prodrugs via transgenic expression of suicide genes is a leading strategy for the selective elimination of potentially tumorigenic human pluripotent stem cells (hPSCs) in regenerative medicine, but transgenic modification poses safety risks such as deleterious mutagenesis. We describe here an alternative method of delivering suicide-inducing molecules explicitly to hPSCs using virus-like particles (VLPs), and demonstrate its use in eliminating undifferentiated hPSCs in vitro. VLPs were engineered from Qβ bacteriophage capsids to contain enhanced green fluorescent protein (EGFP) or cytosine deaminase (CD), and to simultaneously display multiple IgG-binding ZZ domains. After labeling with antibodies against the hPSC-specific surface glycan SSEA-5, EGFP-containing particles were shown to specifically bind undifferentiated cells in culture, and CD-containing particles were able to eliminate undifferentiated hPSCs with virtually no cytotoxicity to differentiated cells upon treatment with the prodrug 5-fluorocytosine.
Keywords: human pluripotent stem cells, protein delivery, virus-like particles
INTRODUCTION
Due to their virtually unlimited self-renewal capacity and differentiation potential in vitro, human pluripotent stem cells (hPSCs)—comprising human induced pluripotent stem cells (hiPSCs) and human embryonic stem cells (hESCs)—remain leading candidates for cell-based regenerative therapies.1–5 Despite substantial advances in protocol efficiencies, directed differentiation of hPSCs to a desired cell type usually yields a population containing residual undifferentiated and potentially tumorigenic cells. Before widespread clinical adoption of hPSC-based therapies can be achieved, effective strategies for the removal of hPSCs are needed to eliminate the risk of post-transplantation teratoma formation.6–8 To this end, making potentially tumorigenic hPSCs sensitive to prodrugs via expression of suicide-inducing genes is considered a promising approach.9–14 Transgenic expression of the bacterial gene encoding cytosine deaminase (CD), which enables the conversion of the negligibly toxic prodrug 5-fluorocytosine (5-FC) to the highly cytotoxic 5-fluorouracil (5-FU), has been used to selectively eradicate pluripotent stem cells derived from mice12 and primates.14 However, since transgenic modification of cells poses the risk of deleterious mutagenesis, a safer alternative system for specific intracellular delivery of suicide-inducing macromolecules such as CD to viable hPSCs is desirable for clinical applications.
In this regard, virus-like particles (VLPs) offer a promising way to provide both targeting and cytotoxic functions. VLPs are non-infectious, self-assembling nanostructures derived from the recombinant expression of viral capsid proteins.15 VLPs derived from the Qβ bacteriophage capsid can be engineered to encapsulate specific molecular cargoes16 and to incorporate multiple ZZ domains on the particle exterior as extensions of the coat protein.17 Derived from Staphylococcus aureus protein A, the Z domain binds the Fc region of IgG antibodies; we used a dimeric version (approximately 200 amino acids) first reported by Nilsson, et al.18 hence the “ZZ” designation. VLPs engineered to display ZZ domains can be labeled with multiple copies of any antibody bearing a protein A-binding Fc domain by simple mixing, and are thereby rendered capable of selectively targeting cells with specific cell-surface markers.19, 20 Here we describe the use of this targeting method to bring a prodrug-converting enzyme specifically to the undifferentiated stem cells that must be removed prior to administration.
RESULTS AND DISCUSSION
SSEA-5-labeled Qβ(ZZ)60@EGFP4 VLPs Selectively Deliver EGFP Intracellularly to hPSCs.
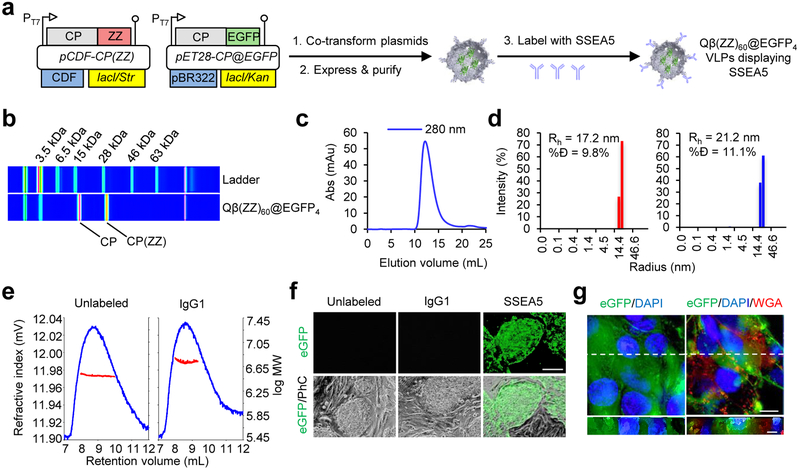
VLPs are named here as previously described: for example, Qβ(ZZ)n@EGFPm denotes a Qβ VLP in which an average of n of the 180 capsid protein subunits have the ZZ domain appended to the N-terminus for display on the external surface, and an average of m copies of the EGFP protein are encapsidated inside the particle.21 In addition, if the surface ZZ domain on the VLPs is bound to an antibody, a + sign appears followed by the antibody’s abbreviation. EGFP packaging was engineered by co-transformation of E. coli cells with two plasmids, one [designated pET28-CP@EGFP] coding for both the standard capsid protein (CP) and EGFP fused to the Rev peptide to assist in packaging,21 and the other [pCDF-CP(ZZ)] coding for the CP-ZZ fusion (Figure 1a). IPTG-triggered expression from both plasmids routinely yielded particles in a yield of 10–15 mg per L of culture with n = 60 and m = 4 (Figure 1b, 1c). Characterization by dynamic light scattering revealed a homogeneous hydrodynamic radius (Rh) of 17.2 nm, somewhat larger than the wild-type VLP (14 nm), as expected. Antibody SSEA-5, identified by Weissman, Drukker, and colleagues as a binder of an hPSC-specific fucosylated H-1-type carbohydrate antigen,22 was mixed with these particles to give Qβ(ZZ)60@EGFP4+SSEA-530, which showed an increase in apparent Rh to 21.2 nm (Figure 1d), consistent with the presence of no more than a single layer of IgG antibodies on the particle surface. Multiple angle light scattering (MALS) analysis indicated an increase in particle molecular weight from 3.2 MDa to 5.5 MDa, corresponding to occupancy of approximately half of the ZZ domains by IgG molecules (Figure 1e).
Figure 1. SSEA-5-labeled Qβ(ZZ)60@EGFP4 VLPs selectively deliver EGFP intracellularly to hPSCs.
(a) Schematic of Qβ(ZZ)60@EGFP4 viral coat protein expression and particle assembly, followed by VLP labeling with SSEA-5 antibodies. (b) Electrophoretic analysis of Qβ(ZZ)60@EGFP4 VLPs. (c) Fast protein liquid chromatography analysis of Qβ(ZZ)60@EGFP4 VLPs. (d) Dynamic light scattering histograms of Qβ(ZZ)60@EGFP4 VLPs (red) and Qβ(ZZ) 60@EGFP4+SSEA-530 VLPs (blue); %Đ = % dispersity; Rh = hydrodynamic radius. (e) Multiple angle light scattering curves show the change in refractive index (blue peak) of the VLPs as a function of the elution volume through a size-exclusion column. A comparison between the log MW (red trace) of unlabeled (left) and antibody-labeled (right) VLPs indicates approximately half of the ZZ domains are occupied by antibodies. (f) Representative microscopy images of hPSC colonies grown on mouse embryonic fibroblasts incubated with 8 nM unlabeled, IgG1-labeled, or SSEA-5-labeled Qβ(ZZ)60@EGFP4 VLPs for 30 min at 37°C. Only hPSCs treated with SSEA-5-labeled Qβ(ZZ)60@EGFP4 showed detectable EGFP fluorescence. Scale bars = 100 μm. (g) Live-cell deconvolution fluorescence microscopy of hPSCs incubated for 30 min at 37°C with 8 nM Qβ(ZZ)60@EGFP4+SSEA-530, blue DAPI nuclear probe, and membrane-specific red wheat germ agglutinin (WGA). Left = EGFP and DAPI only; right = EGFP, DAPI, and WGA. XY optical sections are shown above orthogonal YZ cross-sections corresponding to the white dashed line in the XY plane. Cross-sections show prominent cytoplasmic co-localization of EGFP and DAPI along with punctate expression of the membrane-specific WGA dye. XY plane scale bars = 15 μm; YZ scale bars = 5 μm.
Qβ(ZZ)60@EGFP4+SSEA-530 particles were incubated (5, 10, and 15 nM in particle, 37°C) with hESC colonies cultured on mouse embryonic fibroblasts (MEFs). After 30 min, EGFP was detected in all the undifferentiated stem cell colonies at all particle concentrations (data not shown). To confirm the antibody-directed nature of EGFP delivery, a blinded experiment was performed in which hESC colonies were again cultured on MEFs and incubated for 30 min at 37°C with 8 nM of unlabeled, IgG1-labeled (κ isotype control), or SSEA-5 labeled Qβ(ZZ)60@EGFP4. Since MEFs do not express the SSEA-5 binding motif, they were used as a negative cell control. After 30 min, the excess particle-containing supernatant was removed and the cells were rinsed before imaging. Fluorescence images showed green fluorescent stem cell colonies in the wells treated with Qβ(ZZ)60@EGFP4+SSEA-530 particles, but virtually no green fluorescence in either the MEFs or the hESCs treated with the unlabeled Qβ(ZZ)60@EGFP4 or Qβ(ZZ)60@EGFP4+IgG1 isotype control particles (Figure 1f). In addition, live-cell deconvolution fluorescence microscopy with three-dimensional optical sectioning showed substantial diffuse and intracellular EGFP distribution, when live hESC colonies were incubated with DAPI as a nuclear probe (blue), a membrane-specific wheat germ agglutinin (WGA) dye (red), and 8 nM of Qβ(ZZ)60@EGFP4+SSEA-530 particles (green) (Figure 1g). These results demonstrate that Qβ(ZZ)60@EGFP4+SSEA-530 particles selectively target, and appear to be taken up by, undifferentiated stem cells while exhibiting negligible non-specific binding.
SSEA-5-labeled Qβ(ZZ)42@CD13 VLPs Selectively Kill hPSCs in the Presence of 5-FC.
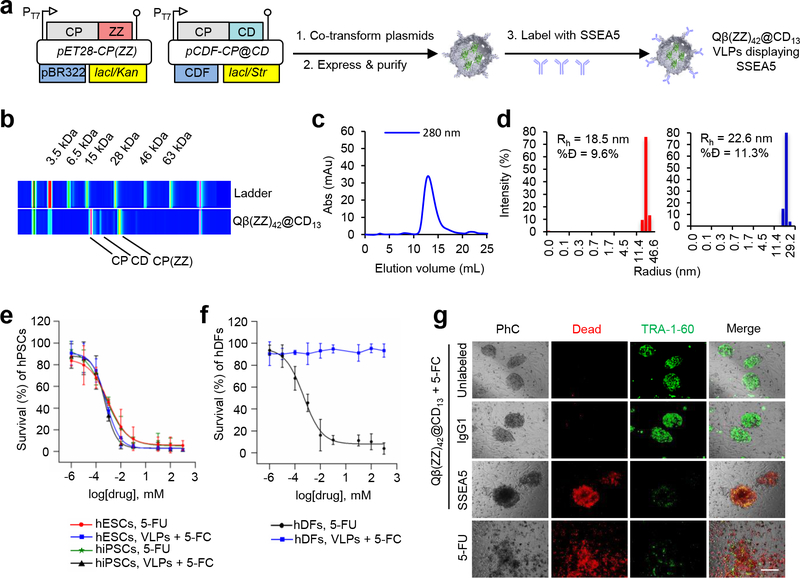
Targeted VLPs with cytotoxic capability were generated as above, replacing the packaged EGFP protein with a thermostable variant of yeast cytosine deaminase (CD).23 The resulting Qβ(ZZ)42@CD13 particles were addressed with SSEA-5 in the same manner, resulting in very similar yield and physical parameters as the EGFP-containing particles (Figure 2a-2d).
Figure 2. SSEA-5-labeled Qβ(ZZ)42@CD13 VLPs selectively kill hPSCs in the presence of 5-FC.
(a) Schematic of Qβ(ZZ)42@CD13 viral coat protein expression and particle assembly, followed by VLP labeling with SSEA-5 antibodies. (b) Electrophoretic, (c) FPLC, and (d) dynamic light scattering analyses of Qβ(ZZ)42@CD13 VLPs. (%Đ = % dispersity; Rh = hydrodynamic radius). (e) EC50 curves showing dose-dependent decreases in human induced pluripotent stem cell (hiPSC) and human embryonic stem cell (hESC) survival after 24 h treatment with both 8 nM Qβ(ZZ)42@CD13+SSEA-521 VLPs in the presence of the prodrug 5-fluorocytosine (5-FC) or the cytotoxin 5-fluorouracil (5-FU). (f) EC50 curves show dose-dependent decreases in human dermal fibroblast (hDF) survival percentages after 24 h treatment with 5-FU, but negligible cell death after treatment with 8 nM Qβ(ZZ)42@CD13+SSEA-521 VLPs in the presence of 5-FC. (g) Representative microscopy images show co-cultures of hPSC colonies and MEFs that were treated for 15 h with 8 nM of unlabeled, IgG1-labeled (isotype control), and SSEA-5-labeled Qβ(ZZ)42@CD13 VLPs in the presence of 100 μM 5-FC as test conditions, and 100 μM 5-FU as a positive control. Red fluorescence indicates dead cells and green fluorescence indicates expression of the pluripotent stem cell-specific TRA-1–60 marker.
The cytotoxic activity of the CD-containing particles was verified first on monocultures of hiPSCs and hESCs, which were incubated for 24 h with titrating concentrations of either 8 nM Qβ(ZZ)42@CD13+SSEA-521 VLPs in the presence of the prodrug 5-fluorocytosine (5-FC), or its product, 5-fluorouracil (5-FU), alone as a positive cytotoxicity control. Survival percentages for each treatment were determined using a tetrazolium salt-based assay, and decreased in a dose-dependent manner in both hiPSCs and hESCs (Figure 2e). The mean EC50 values of 5-FC in the presence of CD-containing particles and 5-FU alone for hESCs and hiPSCs were similar (approximately 1.5 μM), with negligible cell survival observed at 100 μM of either small molecule. To test targeting specificity, monocultures of primary human dermal fibroblasts (hDFs) were treated for 24 h with 8 nM Qβ(ZZ)42@CD13+SSEA-521 VLPs and either 5-FC or 5-FU at 0.1 μM to 2 mM. 5-FU was found to kill these differentiated cells with efficiency comparable to the killing of stem cells, but no dose-dependent cell death was observed in the presence of 5-FC and the SSEA-5-coated particles containing packaged CD (Figure 2f).
The use of this system requires the elimination of residual undifferentiated hPSCs in a mixed population. Thus, co-cultures of hPSC colonies and MEFs were treated for 15 h with 8 nM of unlabeled, IgG1-labeled (isotype control), and SSEA-5-labeled Qβ(ZZ)42@CD13 VLPs in the presence of 100 μM 5-FC, and 100 μM of 5-FU as a positive control. Subsequent staining with ethidium homodimer-1 (a specific red marker of dead cells) and a green fluorescent antibody against the hPSC-specific TRA-1–60 surface marker gave the results shown in Figure 2g. Negligible cell death was induced by unlabeled and isotype control-labeled Qβ(ZZ)42@CD13 VLPs in the presence of 5-FC; in these cases, hPSC colonies remained intact with bright, flat morphology and abundant TRA-1–60 expression. In contrast, hPSC colonies in co-cultures treated with Qβ(ZZ)42@CD13+SSEA-521 and 5-FC showed extensive ethidium staining; a dark, barely adherent, mounted morphology; and minimal detection of TRA-1–60, likely due to antibody binding to loose fragments of dead hPSC membranes. The surrounding MEFs were unaffected, demonstrating the specificity of VLP-mediated hPSC killing. Co-cultures treated with 5-FU exhibited indiscriminate cell death.
SSEA-5-labeled Qβ(ZZ)42@CD13 VLPs Specifically Eliminate Residual hPSCs without Impeding Cardiomyogenesis.
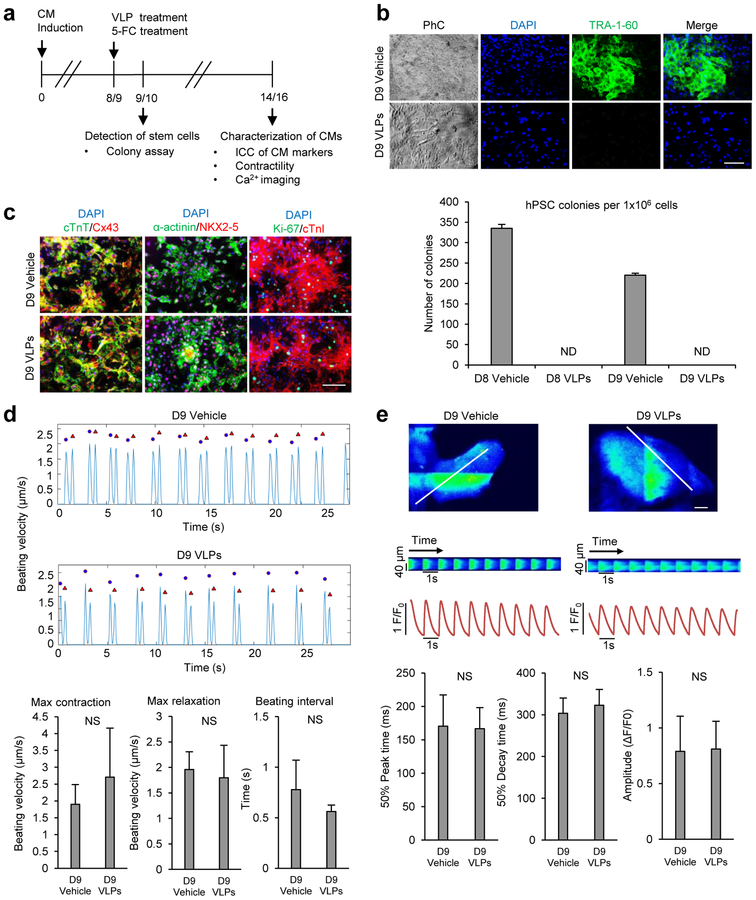
Currently it is still unclear what stages of hPSC-derived cardiomyocytes are most efficacious for therapeutic application.24 Clinical application of this methodology may require the killing of undifferentiated cells at an early stage to selectively eliminate residual hPSCs without affecting cardiomyocyte differentiation. To test this, such treatment (8 nM VLPs for 1 h + 100 μM 5-FC for 24 h) was performed at differentiation day 8 or 9 (Figure 3a and Supporting Information Figure 1a). From initial 1×106 cells seeded, at least 200–300 colonies with stem cell morphology and positive for the pluripotent stem cell marker TRA-1–60 were detected in cells expanded from progenitors treated with vehicle as a control. In contrast, no hPSC colonies were detected in otherwise identical cultures treated with the antibody-directed VLP+prodrug (Figure 3b and Supporting Information Figure 1b).
Figure 3. SSEA-5-labeled Qβ(ZZ)42@CD13 VLPs selectively kill residual hPSCs during CM differentiation in the presence of 5-FC.
(a) Schematic of experimental design for treatments of cardiac progenitors with VLPs. (b) Colony assay for the detection of undifferentiated hPSCs. Day 9 differentiated cells were treated with 8 nM Qβ(ZZ)42@CD13+SSEA-521 or PBS (vehicle as a control) for 1 h and washed to remove excess VLPs. Both groups of cells were then treated with 5-FC (100 μM) for 24 h, washed to remove excess prodrug, and cultured on MEF feeders for the detection of residual hPSCs. After 10 more days, cell were examined by immunocytochemistry (top panels: blue, DAPI nuclear stain; green, TRA-1–60; scale bars = 100 μm). ND = not detected. (c) Expression of cardiomyocyte proteins. Beating cells derived from day 9 vehicle- and VLP-treated cultures were examined by immunocytochemistry for their expression of cardiac transcription factor NKX2–5, gap junction protein connexin 43 (Cx43), cardiac structural proteins including cardiac troponin T (cTnT), α-actinin, cardiac troponin I (cTnI), and proliferation marker Ki-67. Scale bars = 100 μm. (d) Video-based analysis of contractility. Representative traces show beating velocity recording of vehicle- and VLP-treated cells. Contraction parameters are presented as mean ± standard deviation of n=3 biological samples. Blue dots denote contraction, and red triangles denote relaxation. (e) Ca2+ transient analyses. Representative line-scan images show calcium transients in cardiomyocytes derived from day 9 vehicle- and VLP-treated cells from the area indicated by the white lines. Scale bars = 5 μm. Ca2+ transient parameters are presented as mean ± standard deviation of n=20 cells. NS = no significant difference between groups (p value >0.05). D8 = Day 8; D9 = Day 9.
To examine if the treatment affected cardiomyocyte differentiation, we maintained the treated cells in RPMI/B27 medium, monitored cell morphology, and examined the expression of cardiomyocyte genes. At differentiation day 14, beating cardiomyocytes were observed in cultures derived from cells that were treated with either the VLPs or vehicle (Supporting Information Movies 1, 2). Similar to cells derived from control cultures, the majority of the cells derived from the VLP-treated cultures were positive for cardiac transcription factor NKX2–5, gap junction protein connexin 43, and cardiac structural proteins including cardiac troponin I, cardiac troponin T and α-actinin as detected by immunocytochemistry (Figure 3c and Supporting Information Figure 1c). In addition, these cells also expressed similar levels of Ki-67, a marker associated with cells at active phases of cell cycle.(Figure 3c and Supporting Information Figure 1c).
The contractility of cardiomyocytes derived from VLP-treated cultures was found to be indistinguishable from that of vehicle-treated control cultures by video-based analysis25 (Figure 3d), including maximum contraction, maximum relaxation, and beating intervals. Furthermore, intracellular Ca2+ transients (observed with the aid of calcium indicator dye Fluo-4) of cardiomyocytes derived from VLP-treated cultures were also very similar in Ca2+ transient amplitude, 50% peak time, and 50% decay time to those from control cultures (Figure 3e and Supporting Information Figure 1d). These results suggest that the VLP-based elimination of residual hPSCs from early-stage differentiation cultures did not significantly affect cardiomyocyte differentiation.
SSEA-5-labeled Qβ(ZZ)42@CD13 VLPs Specifically Eliminate Residual hPSCs in Co-cultures with Late-stage Cardiomyocytes without Affecting Cardiomyocyte Function.
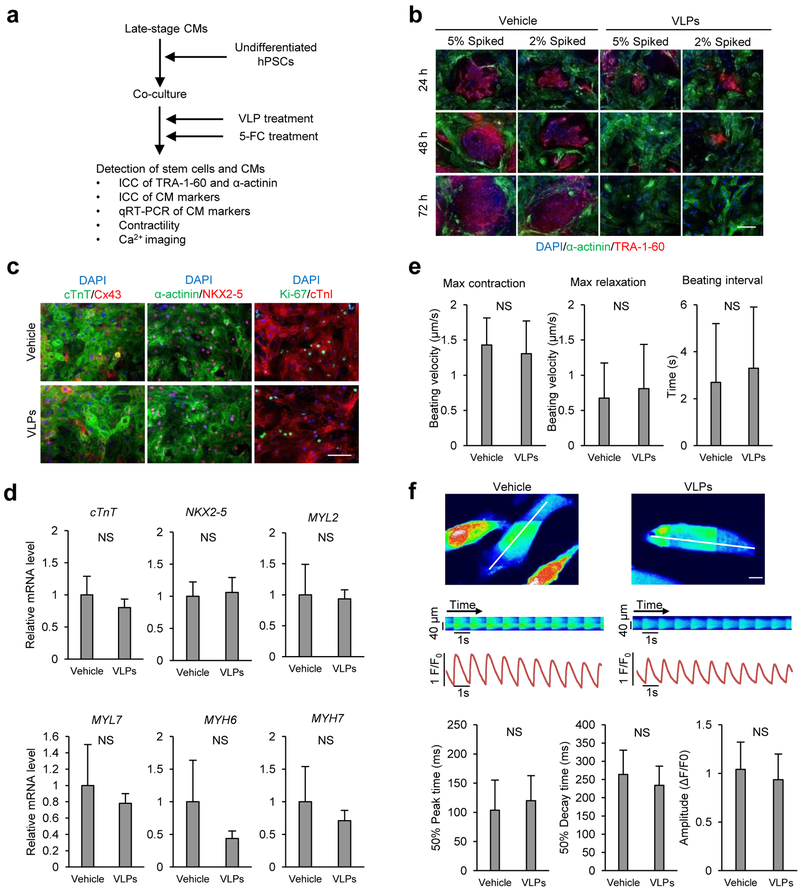
To mimic a high level of residual hPSCs in a differentiation culture at the late stage, we co-cultured 1–5% undifferentiated hPSCs with late-stage cardiomyocyte differentiation culture from hPSCs, conditions previously used to evaluate specific elimination of stem cells in a mixed population.26 The co-culture was treated with the prodrug-converting VLPs (8 nM) for 1 h and then with 5-FC (100 μM) for 24 h (Figure 4a). Immediately after these treatments or following further culture, the presence of hPSCs and cardiomyocytes was examined by staining with antibodies against markers for stem cells (TRA-1–60) and cardiomyocytes (α-actinin). As expected, TRA-1–60-positive hPSC colonies/cells were detectable in co-cultures treated with vehicle at all time points examined. The amount of TRA-1–60 positive cells decreased over time in the VLP-treated cultures, and the signal for TRA-1–60 was undetectable 3 days after the VLP treatment by fluorescence microscopy (Figure 4b) or by flow cytometry (Supporting Information Figure 2). In contrast, the signal for α-actinin was detectable in the majority of TRA-1–60 negative cells in all cultures and was comparable in the vehicle- and VLP-treated cultures (Figure 4b). Similarly, no significant loss of hPSC-derived cardiomyocytes was observed by flow cytometry following VLP treatments (Supporting Information Figure 2).
Figure 4. SSEA-5-labeled Qβ(ZZ)42@CD13 VLPs selectively kill hPSCs in co-cultures of hPSC and cardiomyocytes in the presence of 5-FC.
(a) Schematic of experimental design for the VLP treatment for co-cultures of hPSCs and cardiomyocytes. (b) Detection of hPSCs and cardiomyocytes in co-cultures. Co-cultures of hPSCs and cardiomyocytes were treated with 8 nM Qβ(ZZ)42@CD13+SSEA-521 (test) or PBS (control) for 1 h and washed to remove excess VLPs. After the treatment of both groups with 5-FC (100 μM), cells were examined by immunocytochemistry of hPSC-specific marker TRA-1–60 (red) and cardiomyocyte-associated protein α-actinin (green). Scale bars = 100 μm. (c) Detection of cardiomyocyte proteins. Cells from vehicle- and VLP-treated co-cultures were examined for the expression of cardiac transcription factor NKX2–5, gap junction protein connexin 43 (Cx43), cardiac structural proteins including cardiac troponin T (cTnT), α-actinin and cardiac troponin I (cTnI), and proliferation marker Ki-67. Scale bars = 100 μm. (d) Detection of the expression of cardiomyocyte genes by qRT-PCR. Data are presented as mean ± standard deviation of n=3 biological samples. (e) Video-based analysis of contractility. Contraction parameters are presented as mean ± standard deviation of n=6 biological samples. (f) Ca2+ transient analyses. Representative line-scan images show calcium transients in vehicle- and VLP-treated cells in co-cultures from the area indicated by the white lines. Scale bars = 5 μm. Ca2+ transient parameters are presented as mean ± standard deviation of n=22 cells. NS = no significant difference between groups (p value>0.05).
A kill curve for 5-FC dose-response was performed in a co-culture of hPSCs and hPSC-derived cardiomyocytes to determine the cytotoxic range of 5-FC. The co-culture was treated for 1 h with the VLPs followed by 24 h incubation with titrated concentrations of 5-FC (from 0.1 μM to 1 mM). Immunochemical analysis showed 100 μM 5-FC to be the minimal lethal concentration for elimination of hPSCs, while hPSC-CMs were not affected even when treated with 1 mM 5-FC (Supporting Information Figure 3). These results show that the VLP+prodrug combination can specifically eliminate undifferentiated hPSCs in the mixture.
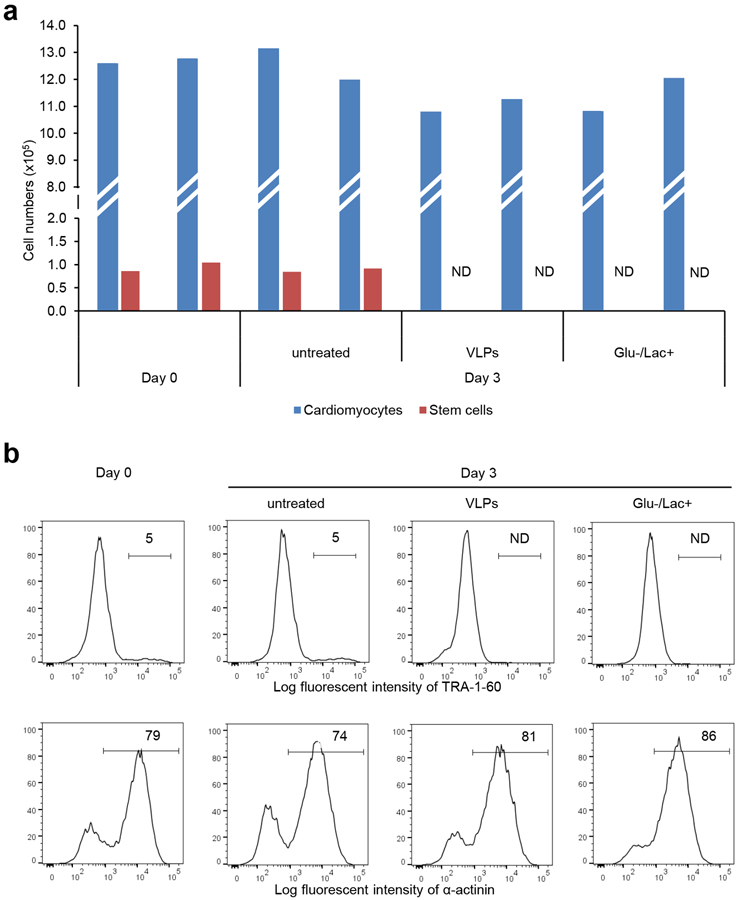
In our hands, metabolic selection – the use of glucose-depleted and lactate-supplemented media to purify late-stage hPSC-derived cardiomyocytes from differentiation culture27, 28 – was also able to eliminate all detectable TRA-1–60-positive stem cells from co-cultures with hPSC-derived cardiomyocytes (Figure 5). Both VLP+prodrug treated and metabolically selected co-cultures had similar amounts of hPSC-derived cardiomyocytes (Figure 5). Thus, while both are effective in the presence of late-stage cardiomyocytes, only the VLP-based method is also applicable to early-stage cardiomyocyte differentiation cultures.
Figure 5. Comparison of SSEA-5-labeled Qβ(ZZ)42@CD13 VLPs with metabolic selection method.
Flow cytometry was performed to compare metabolic selection with VLP treatment. hPSC-CMs at day 18 were dissociated using Trypsin/EDTA and plated onto a Matrigel-coated 24 well plate at a density of 600,000 cells per well, cultured until they recovered beating activity. Undifferentiated hPSCs were then plated at a density of 2% compared to cardiomyocytes (12,000 cells per well) and further cultured in MEF-conditioned medium. For the VLP method, cells were treated for 1 h with medium containing 8 nM SSEA-5-labeled Qβ(ZZ)@CD VLPs followed by 24 h incubation with 100 μM 5-FC in RPMI/B27 medium. After 5-FC incubation, cells were maintained in RPMI/B27 medium for 2 more days before harvesting on Day 3. Medium was changed on alternate days. For metabolic selection, the co-culture of hPSCs and hPSC-CMs was cultured in RPMI/B27 medium without glucose and with 5 mM sodium DL-lactate 60% syrup (Glu-/Lac+). Medium was changed on alternate days, and cells were harvested after 3 days of metabolic selection. In controls wells, cells were maintained in RPMI/B27 medium, which was changed on alternated days, and collected at Day 0 (before the start of both VLP-treatment and metabolic selection) and Day 3. Following harvesting, cell numbers were estimated by the hemocytometer, and ~1.5×106 cells aliquot for each treated group was prepared for flow cytometry analysis of cardiac marker α-actinin and stem cell marker TRA-1–60. Summary (a) and representative histograms (b) of all groups for each marker are shown. ND = Not detectable.
To determine if the VLP+prodrug treatment affected molecular and functional properties of cardiomyocytes, the expression of cardiomyocyte genes, cardiomyocyte contractility and intracellular Ca2+ transients were examined after the co-cultures were treated with vehicle or VLP + 5-FC. In both vehicle- and VLP-treated co-cultures, beating cells were observed (Supporting Information Movies 3, 4), which expressed cardiomyocyte transcription factor NKX2–5, connexin 43, cardiac troponin I, cardiac troponin T, α-actinin, and proliferation marker Ki-67 to similar extents (Figure 4c). In addition, the expression levels of genes associated with cardiomyocytes (cTnT, NKX2–5, MYH6, MYH7, MYL2 and MYL7) were comparable between the vehicle- and VLP-treated samples (Figure 4d). Measurements of cardiac contractility were also similar between vehicle- and VLP-treated cells with no significant differences observed in maximum contraction, maximum relaxation, and beating interval as analyzed by a video-based method25 (Figure 4e). Key parameters of intracellular Ca2+ transient recordings were similar between vehicle- and VLP-treated cells (Figure 4f). These results suggest that the VLP treatment does not alter molecular and functional characteristics of hPSC-derived cardiomyocytes.
Overall, prodrug-converting VLPs were able to selectively kill tumorigenic stem cells in mixed differentiation cultures without harming surrounding non-stem cells. The resulting stem cell-derived cardiomyocytes were apparently unaffected by this treatment, rendering it an interesting possibility for improving the safety of stem cell-based therapies. This combination of cytotoxic efficiency and selectivity derives from the production of only small amounts of toxic molecule overall but high enough concentrations to the target cells by virtue of localized delivery and enzymatic turnover. Operationally, this method for the targeted elimination of stem cells is simple to perform, requiring no gene transfection or editing and only a simple procedure involving medium change. In addition, the modular nature of the system should allow one to address different cell targets by simply loading different antibodies on the particle surface. The VLP protects the cargo enzymes from degradation by proteases and other factors, and they can be produced easily and at large scale.
METHODS
Cell Culture.
Human induced pluripotent stem cells [iPS(IMR90)-1, WiCell]29 and human embryonic stem cells (H7, WiCell)30 were cultured either on a feeder layer of irradiated mouse embryonic fibroblasts (MEFs), where they were maintained in hPSC medium (KnockOut DMEM, 20% KnockOut Serum Replacement, 2 mM L-glutamine, 0.1 mM non-essential amino acids, 0.1 mM β-mercaptoethanol, 100 U/mL Penicillin-Streptomycin) supplemented with 8 ng/mL human basic fibroblast growth factor (bFGF), or in feeder-free hPSC conditions as previously described.31 Primary human dermal fibroblasts (hDFs) were obtained with informed consent from a patient skin biopsy in a safe and ethical manner as approved by the Emory University Institutional Review Board (IRB). Isolated hDFs were cultured in fibroblast growth medium (DMEM, 10% FBS, 4 mM L-glutamine, 100 U/mL Penicillin-Streptomycin). All cell culture reagents were from Fisher Scientific unless specified.
Protein Expression and Purification of Qβ(ZZ) VLPs.
The hybrid VLP constructs were produced and characterized as previously described.17 Briefly, ClearColi® BL21(DE3) electrocompetent E. coli cells (Lucigen) were co-transformed with one of the following plasmid combinations: pET28-CP@EGFP/pCDF-CP(ZZ) or pET28-CP(ZZ)/pCDF-CP@CD according to the manufacturer’s recommended protocol. The ZZ domain was connected to the C-terminal end of the Qβ capsid sequence by an 8-amino acid linker; the full sequence of the extended coat protein [(Qβ)(linker)(Z)(Z)] was as follows, noting that the N-terminal methionine required for bacterial expression is processed off the capsid structure by cellular proteases:
(AKLETVTLGNIGKDGKQTLVLNPRGVNPTNGVASLSQAGAVPALEKRVTVSVSQPSRNRKNYKVQVKIQNPTACTANGSCDPSVTRQAYADVTFSFTQYSTDEERAFVRTELAALLASPLLIDAIDQLNPAY)(GGASESGA)(AMVDNKFNKEQQNAFYEILHLPNLNEEQRNAFIQSLKDDPSQSANLLAEAKKLNDAQAPKNLNEEQRNAFIQSLKDDPSQSANLLAEAKKLNDAQAPK)(AMVDNKFNKEQQNAFYEILHLPNLNEEQRNAFIQSLKDDPSQSANLLAEAKKLNDAQAPKNLNEEQRNAFIQSLKDDPSQSANLLAEAKKLNDAQAPK).
Cells were plated on selective super optimal broth (SOB) agar; after 24 h, single colonies were isolated and transferred to selective SOB media (1% NaCl; w/v) for overnight growth at 37°C. The following morning, cultures were diluted under the same conditions and growth was monitored via optical density (600 nm) until the cultures reached mid-log phase (i.e., O.D.600 ~ 0.9). Protein expression was induced by the addition of IPTG at a final concentration of 1 mM. Cultures were maintained at 30°C for 24 h, after which time cells were harvested by centrifugation (6,000 rpm, 10 min). Cell pellets were suspended in 1X Tris-buffered saline (TBS; pH 7.06), and cells were lysed with a probe sonicator in an ice bath (10 min total, 5 s pulse intervals). Cell debris was removed by centrifugation (14,000 rpm, 10 min) and the supernatant was collected. VLPs were precipitated by the addition of 30% (NH4)2SO4 (w/v) for 2 h at 4°C, and the precipitate was pelleted by centrifugation (14,000 rpm, 10 min). The pellet was dissolved in 1x TBS and extracted with a solution of n-BuOH:CHCl3 (1:1, v/v) to remove lipids and aggregates. The aqueous phase was collected following centrifugation (14,000 rpm, 10 min) and VLPs were further purified on 10–40% sucrose density gradients (28,000 rpm, 4 h). VLP bands were isolated via syringe and pelleted by ultracentrifugation (68,000 rpm, 2 h). Subsequent pellets were dissolved in 0.1M potassium phosphate buffer and characterized.
Characterization of Qβ(ZZ) VLPs.
VLP concentration was quantified using Bradford assay with BSA standards according to the manufacturer’s protocol (ThermoFisher) with average yields of 10–20 mg/L. Particle purity was assessed by fast protein liquid chromatography (FPLC) with a Superose 6 GL column and monitored by absorbance (280 nm). Size of the VLPs was determined by dynamic light scattering (DLS) using a Dynapro II plate reader (Wyatt Technology), and particle composition was determined by microfluidic gel electrophoresis using an Agilent 2100 Bioanalyzer with Series II Protein 80 chips. The relative particle composition was determined by comparing peak integrations from the electropherograms using the Bioanalyzer software. Due to overlap of the CP(ZZ) and EGFP bands, EGFP concentration was calculated by measuring the sample absorbance at 488 nm and using the Beer-Lambert law (Abs = εcl; εEGFP = 56,000 M−1cm−1). On average, 4–8 molecules of EGFP were packaged, 13–18 molecules of CD were packaged, and 40–60 ZZ-domain subunits were incorporated per particle, respectively.
Antibody-labeling of Qβ(ZZ) VLPs.
Qβ(ZZ)@EGFP or Qβ(ZZ)@CD VLPs were labeled with anti-SSEA-5 antibodies. Briefly, VLPs were diluted to 0.1 mg/mL and 20 μg VLP was incubated with 50 μg purified anti-human SSEA-5 mouse IgG1, κ antibody (Clone 8e11, BioLegend) for 2 h at 37°C. A antibody:ZZ-domain mole ratio of 0.75 was used for all labeling reactions and was sufficient for uniform labeling of the particles. To serve as a control, the same procedure was repeated with purified mouse IgG1, κ isotype control antibody (Clone MG1–45, BioLegend).
Quantification of Antibody Loading by Multi-angle Light Scattering.
Empty Qβ(ZZ) VLPs were split into two samples at 0.1 mg/mL, and one sample was labeled with mouse IgG1, κ isotype control antibody as previously described. Samples were separated over a Superose 6 10/300 GL size exclusion column (GE Healthcare) using an Agilent HPLC to maintain a 0.2 mL/min flow rate of 0.1M KPO4 buffer (pH 7.4). Samples of 100 μL were injected onto the column. Samples were detected using a UV-vis detector (Agilent), a Viscotek SEC-MALS 20 multi-angle light scattering detector (Malvern), and a Viscotek VE3580 refractometer (Malvern). The number-average molecular weight, Mn, was calculated with OmniSEC 5.0 software and plotted across the elution peak. The increase in molecular weight for the antibody-labeled sample was related to the number of bound antibodies using the average molecular weight for IgG antibodies (~150 kDa).
Cardiomyocyte Differentiation.
hPSCs were induced for cardiac differentiation as described previously.32, 33 hPSCs were first dissociated using Versene and plated on Matrigel-coated wells plate at density of 1.6×105 cells/cm2. Cells were cultured in conditioned medium from MEF (MEF-CM) for 2 days and treated with 100 ng/mL activin A (R&D Systems) from differentiation day 0 till day 1 and 10 ng/mL bone morphogenic protein-4 (BMP4, R&D Systems) from day 1 till day 5 in RPMI/B27 insulin-free medium. Cells were cultured successively with RPMI/B27 medium with insulin and the medium was changed on alternate days. Cardiomyocytes showed beating activity at day 9/10 of differentiation.
Treatment of Cardiac Progenitors with VLPs.
One day before the treatment of cardiac progenitors derived from hPSCs (at day 7 or 8), the medium was replaced with a blocking solution (medium with 20% KnockOut Serum Replacement) to prevent non-specific binding of VLPs. After cultured for 24 h (at day 8 or 9), the cells were then treated for 1 h with medium containing 8 nM SSEA-5-labeled Qβ(ZZ)@CD VLPs or vehicle (PBS as a control) followed by 24 h incubation with 100 μM 5-FC.
Treatment of Co-culture of hPSCs and Cardiomyocytes with VLPs.
Cardiomyocytes at day 14 to 18 were plated on a Matrigel-coated plate at density of 1.6×105 cells/cm2 and cultured in RPMI/B27 medium until they regained beating activity. Undifferentiated hPSCs were then plated at density of 1–5% compared to cardiomyocytes (1.6×103 to 8×103 cells/cm2) and cultured in MEF-CM medium. Cells were treated for 1 h with medium containing 8 nM SSEA-5-labeled Qβ(ZZ)@CD VLPs or PBS (vehicle as a control) followed by 24 h incubation with 100 μM 5-FC.
Statistics.
For calcium imaging analysis and video-based analysis of contractility, significant differences between vehicle- and VLP-treated cells were assessed by two-sample t-test. All data were expressed as mean ± SD and p values <0.05 were considered as significant.
Supplementary Material
ACKNOWLEDGEMENTS
This project was supported in part by the NIH grant R21HL123928 to C.X, R01GM101421 to M.G.F., R01HL136345 to C.X., and a seed grant to C.X. and M.G.F. from the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR000454. S.N.C. was supported by a predoctoral research fellowship from the National Science Foundation. We thank Emory Pediatric Research Center Flow Cytometry Core, which is supported by Children’s Healthcare of Atlanta. We also thank Ms. Sandra Grijalva, Department of Biomedical Engineering at Emory/Georgia Tech, for her guidance on contractility analysis.
Abbreviations:
- VLP
virus-like particle
- EGFP
enhanced green fluorescent protein
- CD
cytosine deaminase
- 5-FC
5-fluorocytosine
- 5-FU
5-fluorouracil
- hPSC
human pluripotent stem cell
Footnotes
Supporting Information
The Supporting Information includes methods, 3 tables, 4 movies and 3 figures.
REFERENCES
- 1.Gauthier M, Maury Y, Peschanski M, and Martinat C (2011) Human pluripotent stem cells for genetic disease modeling and drug screening, Regen Med 6, 607–622. [DOI] [PubMed] [Google Scholar]
- 2.Rajamohan D, Matsa E, Kalra S, Crutchley J, Patel A, George V, and Denning C (2013) Current status of drug screening and disease modelling in human pluripotent stem cells, Bioessays 35, 281–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bajada S, Mazakova I, Richardson JB, and Ashammakhi N (2008) Updates on stem cells and their applications in regenerative medicine, J Tissue Eng Regen Med 2, 169–183. [DOI] [PubMed] [Google Scholar]
- 4.Christou YA, Moore HD, Shaw PJ, and Monk PN (2007) Embryonic stem cells and prospects for their use in regenerative medicine approaches to motor neurone disease, Neuropathol Appl Neurobiol 33, 485–498. [DOI] [PubMed] [Google Scholar]
- 5.Tabar V, and Studer L (2014) Pluripotent stem cells in regenerative medicine: challenges and recent progress, Nat Rev Genet 15, 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malecki M (2014) ‘Above all, do no harm’: safeguarding pluripotent stem cell therapy against iatrogenic tumorigenesis, Stem Cell Res Ther 5, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malecki M, LaVanne C, Alhambra D, Dodivenaka C, Nagel S, and Malecki R (2013) Safeguarding stem cell-based regenerative therapy against iatrogenic cancerogenesis: transgenic expression of DNASE1, DNASE1L3, DNASE2, DFFB controlled by promoter in proliferating and directed differentiation resisting human autologous pluripotent induced stem cells leads to their death, J Stem Cell Res Ther Suppl 9 21559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee MO, Moon SH, Jeong HC, Yi JY, Lee TH, Shim SH, Rhee YH, Lee SH, Oh SJ, Lee MY, Han MJ, Cho YS, Chung HM, Kim KS, and Cha HJ (2013) Inhibition of pluripotent stem cell-derived teratoma formation by small molecules, Proc Natl Acad Sci U S A 110, E3281–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, and Xiang AP (2013) Safeguarding clinical translation of pluripotent stem cells with suicide genes, Organogenesis 9, 34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schuldiner M, Itskovitz-Eldor J, and Benvenisty N (2003) Selective ablation of human embryonic stem cells expressing a “suicide” gene, Stem Cells 21, 257–265. [DOI] [PubMed] [Google Scholar]
- 11.Hara A, Aoki H, Taguchi A, Niwa M, Yamada Y, Kunisada T, and Mori H (2008) Neuron-like differentiation and selective ablation of undifferentiated embryonic stem cells containing suicide gene with Oct-4 promoter, Stem Cells Dev 17, 619–627. [DOI] [PubMed] [Google Scholar]
- 12.Chen F, Cai B, Gao Y, Yuan X, Cheng F, Wang T, Jiang M, Zhou Y, Lahn BT, Li W, and Xiang AP (2013) Suicide gene-mediated ablation of tumor-initiating mouse pluripotent stem cells, Biomaterials 34, 1701–1711. [DOI] [PubMed] [Google Scholar]
- 13.Wu C, Hong SG, Winkler T, Spencer DM, Jares A, Ichwan B, Nicolae A, Guo V, Larochelle A, and Dunbar CE (2014) Development of an inducible caspase-9 safety switch for pluripotent stem cell-based therapies, Mol Ther Methods Clin Dev 1, 14053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong B, Watts KL, Gori JL, Wohlfahrt ME, Enssle J, Adair JE, and Kiem HP (2011) Safeguarding nonhuman primate iPS cells with suicide genes, Mol Ther 19, 1667–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeltins A (2013) Construction and characterization of virus-like particles: a review, Mol Biotechnol 53, 92–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhee JK, Hovlid M, Fiedler JD, Brown SD, Manzenrieder F, Kitagishi H, Nycholat C, Paulson JC, and Finn MG (2011) Colorful virus-like particles: fluorescent protein packaging by the Qbeta capsid, Biomacromolecules 12, 3977–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown SD, Fiedler JD, and Finn MG (2009) Assembly of hybrid bacteriophage Qbeta virus-like particles, Biochemistry (Mosc) 48, 11155–11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nilsson B, Moks T, Jansson B, Abrahmsen L, Elmblad A, Holmgren E, Henrichson C, Jones TA, and Uhlen M (1987) A synthetic IgG-binding domain based on staphylococcal protein A, Protein Eng 1, 107–113. [DOI] [PubMed] [Google Scholar]
- 19.Kickhoefer VA, Han M, Raval-Fernandes S, Poderycki MJ, Moniz RJ, Vaccari D, Silvestry M, Stewart PL, Kelly KA, and Rome LH (2009) Targeting vault nanoparticles to specific cell surface receptors, ACS Nano 3, 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braisted AC, and Wells JA (1996) Minimizing a binding domain from protein A, Proc Natl Acad Sci U S A 93, 5688–5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiedler JD, Brown SD, Lau JL, and Finn MG (2010) RNA-directed packaging of enzymes within virus-like particles, Angew Chem Int Ed Engl 49, 9648–9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang C, Lee AS, Volkmer JP, Sahoo D, Nag D, Mosley AR, Inlay MA, Ardehali R, Chavez SL, Pera RR, Behr B, Wu JC, Weissman IL, and Drukker M (2011) An antibody against SSEA-5 glycan on human pluripotent stem cells enables removal of teratoma-forming cells, Nat Biotechnol 29, 829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korkegian A, Black ME, Baker D, and Stoddard BL (2005) Computational thermostabilization of an enzyme, Science 308, 857–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skelton RJ, Brady B, Khoja S, Sahoo D, Engel J, Arasaratnam D, Saleh KK, Abilez OJ, Zhao P, Stanley EG, Elefanty AG, Kwon M, Elliott DA, and Ardehali R (2016) CD13 and ROR2 Permit Isolation of Highly Enriched Cardiac Mesoderm from Differentiating Human Embryonic Stem Cells, Stem Cell Reports 6, 95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huebsch N, Loskill P, Mandegar MA, Marks NC, Sheehan AS, Ma Z, Mathur A, Nguyen TN, Yoo JC, Judge LM, Spencer CI, Chukka AC, Russell CR, So PL, Conklin BR, and Healy KE (2015) Automated Video-Based Analysis of Contractility and Calcium Flux in Human-Induced Pluripotent Stem Cell-Derived Cardiomyocytes Cultured over Different Spatial Scales, Tissue Eng Part C Methods 21, 467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben-David U, Gan QF, Golan-Lev T, Arora P, Yanuka O, Oren YS, Leikin-Frenkel A, Graf M, Garippa R, Boehringer M, Gromo G, and Benvenisty N (2013) Selective elimination of human pluripotent stem cells by an oleate synthesis inhibitor discovered in a high-throughput screen, Cell Stem Cell 12, 167–179. [DOI] [PubMed] [Google Scholar]
- 27.Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, Egashira T, Seki T, Muraoka N, Yamakawa H, Ohgino Y, Tanaka T, Yoichi M, Yuasa S, Murata M, Suematsu M, and Fukuda K (2013) Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes, Cell Stem Cell 12, 127–137. [DOI] [PubMed] [Google Scholar]
- 28.Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, Plews JR, Abilez OJ, Cui B, Gold JD, and Wu JC (2014) Chemically defined generation of human cardiomyocytes, Nat Methods 11, 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, and Thomson JA (2007) Induced pluripotent stem cell lines derived from human somatic cells, Science 318, 1917–1920. [DOI] [PubMed] [Google Scholar]
- 30.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, and Jones JM (1998) Embryonic stem cell lines derived from human blastocysts, Science 282, 1145–1147. [DOI] [PubMed] [Google Scholar]
- 31.Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, and Carpenter MK (2001) Feeder-free growth of undifferentiated human embryonic stem cells, Nature Biotech. 19, 971–974. [DOI] [PubMed] [Google Scholar]
- 32.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, and Murry CE (2007) Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts, Nat Biotechnol 25, 1015–1024. [DOI] [PubMed] [Google Scholar]
- 33.Jha R, Xu RH, and Xu C (2015) Efficient differentiation of cardiomyocytes from human pluripotent stem cells with growth factors, Methods Mol Biol 1299, 115–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.