Abstract
Purpose:
Nurses investigate reasons for variable patient symptoms and responses to treatments to inform how best to improve outcomes. Genomics has the potential to guide nursing research exploring contributions to individual variability. This article is meant to serve as an introduction to the novel methods available through genomics for addressing this critical issue and includes a review of methodological considerations for selected genomic approaches.
Approach:
This review presents essential concepts in genetics and genomics that will allow readers to identify upcoming trends in genomics nursing research and improve research practice. It introduces general principles of genomic research and provides an overview of the research process. It also highlights selected nursing studies that serve as clinical examples of the use of genomic technologies. Finally, the authors provide suggestions about how to apply genomic technology in nursing research along with directions for future research.
Conclusions:
Using genomic approaches in nursing research can advance the understanding of the complex pathophysiology of disease susceptibility and different patient responses to interventions. Nurses should be incorporating genomics into education, clinical practice, and research as the influence of genomics in health-care research and practice continues to grow. Nurses are also well placed to translate genomic discoveries into improved methods for patient assessment and intervention.
Keywords: epigenomics, genomics, nursing research
The leading causes of death worldwide, including heart disease, cerebrovascular diseases, lower respiratory infections, and diabetes (World Health Organization, n.d.), involve multifaceted interactions among genetic and environmental factors. To understand the complexities that underlie the risks and outcomes for these common disorders, it is essential to examine the comprehensive biological mechanisms at a molecular level. Genomics can increase our understanding of these diseases by helping us to decipher the interactions of all components in the genome with one another and the environment. Analysis of the genome and its interaction with the environment is also imperative for developing precision health approaches for routine clinical care that will increase our ability to prevent and treat disease and disability (Feero, Guttmacher, & Collins, 2010). Genomics is the study, not just of single genes, but of the functions and interactions of all the genes in the genome (Guttmacher & Collins, 2004) at the deoxyribonucleic acid (DNA), messenger ribonucleic acid (mRNA), or protein levels as well as their interactions with environmental factors.
In early 2013, the Genomic Nursing State of the Science Initiative (Calzone et al., 2013) established a blueprint for genomic nursing science that encompassed a broad range of genomic nursing care and research topics that are mapped to the four major areas of the National Institute of Nursing Research Strategic Plan: health promotion and disease prevention, advancing the quality of life, innovation, and training. In order to meet the goals of this ambitious initiative, nurse scientists must integrate recent genomic technologies into their research. Such a practice should lead to an improved ability to identify patients at greater risk for diseases and to promote health and well-being through a more comprehensive and systematic approach to characterizing patients’ phenotypes, the observable manifestations of their genotypes.
With their broad and comprehensive view of health and disease, nurses have traditionally bridged gaps among health-care providers, individuals, families, and communities. They have the ability to translate complex biological concepts and uses of front-end technologies into clinically relevant practice. In order to remain integral members of the health-care team, however, nurses must begin to incorporate genomics into their research and to translate genomic findings into clinical practice. The purpose of this article is to review general principles of genomics and recent genomic methods in an effort to support nurse educators, researchers, and clinicians as they strive to integrate these concepts into their own work. We also use the current state of the science to suggest future directions for research. Authors of previous reviews have suggested ways to integrate genomics into nursing research and practice (Beery & Hern, 2004; Cashion, Driscoll, & Sabek, 2004; Lea, 2009; Loescher & Merkle, 2005), yet none have focused on methodological considerations or recent genomic technologies. Therefore, we present here a comprehensive methodological review of recent genomic research along with the description of exemplary nursing studies involving neurological disorders. Online Supplementary Table 1 provides definitions of and brief discussions about terms and concepts we use in this review.
General Principles of Genomics
Complex Biomolecular Interactions: More Than Making Proteins
Genetic information encoded in DNA is transcribed to various types of RNA molecules. The most commonly known type of RNA, mRNA, is translated into polypeptides, which subsequently results in producing the proteins that the gene codes. However, recent research has revealed important functions for RNA molecules beyond mRNA’s role in the production of proteins (Strachan & Read, 2010).
RNA that does not serve as the template for encoding proteins, or noncoding RNA (ncRNA), influences phenotypes and/or disease state by influencing the regulatory process of gene expression. MicroRNAs (miRNAs) are one type of ncRNAs that can control the function of DNA, RNA, and protein, influencing the chain of events (Chen & Rajewsky, 2007). Initially, the very short miRNA (21–25 nucleotides long on average) was omitted from the analyses of the human genome. When miRNA binds to the base-pairing site of the target mRNA, it degrades the mRNA, reduces expression of the target gene, and eventually inhibits translation. MiRNA along with the several other known types of ncRNAs, such as transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nucleolar RNAs (snoRNAs), and small interfering RNAs (siRNAs), dramatically increase the complexity of genomics, as we now know that it involves a multitude of regulatory factors functioning differently when faced with different circumstances (for descriptions of characteristics and functions of diverse small ncRNAs, please see previous reviews such as the one by, e.g., Li and Liu, 2011).
Several other small biomolecular mechanisms increase genomic complexity and regulate gene expression. Epigenetic markers, for example, have garnered significant attention in clinical studies because their actions result in modified phenotypes or disease risk without changing the genotype. The epigenetic pattern is potentially dynamic throughout life and thus has the potential to serve as a therapeutic target (Bjornsson, Daniele Fallin, & Feinberg, 2004). Along with miRNA, DNA methylation and histone modification are the major epigenetic mechanisms.
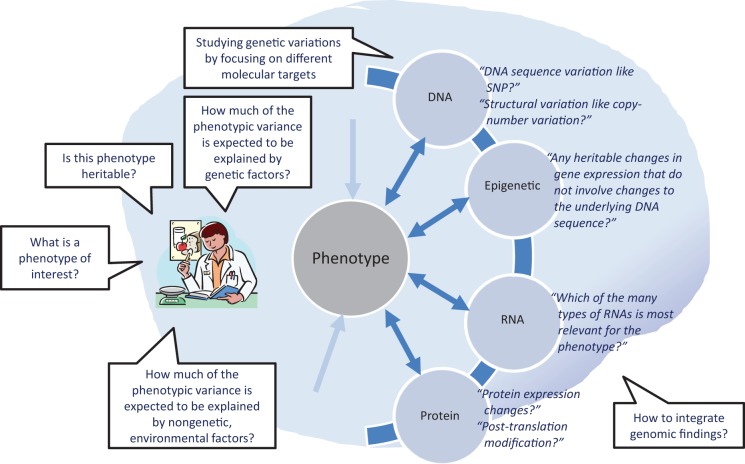
Along with these epigenetic markers, the three main genetic components (DNA, RNA, and proteins) interact in such a way that they are also able to mediate an individual’s response to environmental factors (e.g., pathogens, pollutants). Thus, study of these interactions can provide insights into the intra- and interindividual variability in common responses to environmental threats. The genome, transcriptome, and proteome interconnect in binary and ternary interactions with each other, particularly through RNA-, protein-, and small-molecule-mediated regulatory interactions (Bhartiya et al., 2012). In Figure 1, we incorporated these complex biomolecular interactions to illustrate the process of determining the most appropriate genomic nursing research approach. We describe the major components of this process in more detail below.
Figure 1.
Process for determining the genomic approaches for research. In order to determine the most appropriate genomic approach when incorporating the complex biomolecular interactions among genome, transcriptome, and proteome into their research, nurse scientists should consider questions such as those shown in this figure. SNP = single-nucleotide polymorphism.
Phenotype in Genomic Research
A person’s genome interacts with internal and external factors to create phenotypes such as height, physical appearance, and personality. These phenotypes may be explained, in part, by variations in the genomic sequence, but interactions among genetic and environmental factors also play a role. As explained above, DNA, RNA, protein, and epigenetic factors can all affect phenotypes; for instance, they can affect disease progression, onset or nonrecovery and symptomatology, and even drug metabolism and tolerance.
In some instances, the genome sequence plays a major role in the variation of the phenotype, while for other phenotypes, it contributes minimally or not at all to the variation. The likelihood of finding genetic variations that are responsible for a specific phenotype increases when a genome sequence contributes to the phenotype more than environmental factors do. Therefore, careful and accurate quantification and measurement of the genomic underpinning of the phenotype or symptom is a crucial step in this research. For instance, the National Institutes of Health–Symptom Science Model (NIH-SSM) describes a process where a complex symptom or symptom cluster is characterized into a phenotype consisting of biological and clinical data, followed by the biomarker discovery using genomic or other applications, and then subsequent application of the findings as therapeutic targets in clinical practice (Cashion & Grady, 2015).
Heritability and Relative Risk of Phenotype
One of the important concepts related to phenotypic variation is heritability, which estimates the proportion of phenotypic variance within a population that genetic inheritance explains (Strachan & Read, 2010). A researcher can identify the extent to which genomic variation contributes to the height differences within a population by calculating a heritability estimate of human height. Heritability estimates, albeit with some limitations, can also be used to predict disease risk and provide a guide to altering clinical care according to the risk.
Another parameter used to estimate the contribution of genomic factors to phenotypes is relative risk, which refers to the difference in the risk of developing a condition for people with a family history compared to those without a history (Strachan & Read, 2010). For example, the relative risk of breast cancer is higher in women with first-degree relatives (e.g., mother–daughter relationship), who have had breast cancer compared to the general population. When the heritability estimate or relative risk of a phenotype is low, the influence of the genome sequence is considered to be relatively smaller than the influence of other factors such as environment, and the genomic influence can be easily masked or have a negligible impact.
Intermediate Phenotype (Endophenotype)
Most human diseases are non-Mendelian and rather complex, involving many genes and their interactions as well as nongenetic factors. Intermediate phenotypes, or endophenotypes, offer an approach to overcoming the limitation of finding genomic contributions to complex diseases. Endophenotypes characterize disease in a molecular or genetic manner rather than using a clinical diagnosis to define the phenotype. A fundamental assumption of this construct is that variation in an endophenotype depends on variation in a fewer number of genes than does the more complex clinical traits, therefore making it easier to identify genetic contributions (Gottesman & Shields, 1973). As endophenotypes lie on the spectrum between genes and disease, they are better able than clinically based phenotypes to differentiate diagnoses that present with similar symptoms.
In Table 1, we list studies that serve as examples of the use of the particular genomic technologies employed. For instance, some nurse scientists have used endophenotypes such as transcriptome profiles (a set of differently expressed mRNAs) from peripheral whole blood to reveal underlying genetic contributions to traumatic brain injury (Barr et al., 2010) and ischemic stroke (Kim et al., 2013).
Table 1.
Selected Genomic Approaches and Technologies in Increasing Use.
| Molecular Target and Methods | Description | Technology | Examples From Existing Research |
|---|---|---|---|
| DNA: Measuring DNA sequence (e.g., SNP), copy number or epigenetic variation by analyzing genotypes with specific phenotypes, either symptom or disease (e.g., blood pressure or disease prevalence), in a group of individuals | |||
| a. Candidate gene association study | Testing selected genes, based on their known biological function, for a correlation between disease phenotype and genetic variation | Fluorescent 5′ exonuclease assay (e.g., TaqMan system): Using primer and probe sets for genotyping for multiple individual SNPs from selected small genomic regions like specific genes | Exploration of genotypes of neuroglobin encoding gene (NGB) in brain-injury patients (Chuang et al., 2010) |
| b. Genome-wide association study | Testing the entire genome for a correlation between disease phenotype and common genetic variation | Microarray assay: Using probes designed for millions of preselected sequence variations and arrayed on a small surface, hybridizing samples and scanning fluorescent tags on the hybridized probes to generate genotype calls | Study of an association between SNPs and transcranial Doppler signals, a measure of vasospasm in subarachnoid hemorrhage patients (Kim et al., 2013) |
| c. Epigenetic study | Identifying functionally relevant modification of the genome that causes enduring and heritable changes in gene expression without a change in DNA sequence (e.g., DNA methylation, histone modification) | Genome-wide DNA methylation analysis: Bisulfite or chromatin immunoprecipitation treatment followed by microarray scanning or sequencing | Study of differential methylation of inflammatory regulatory genes in patients with PTSD (Smith et al., 2011) |
| Study of hyperglycemia-mediated induction of genes and pathways by genome-wide histone (H3K9, H3K14) hyperacetylation and DNA methylation (methyl CpG) analysis methods (Pirola et al., 2011) | |||
| RNA: Measuring gene expression as a synthesis of encoded RNA (transcription) for the eventual synthesis of proteins (translation) related to specific phenotypes | |||
| a. Targeted analysis | — | Northern blotting: Detecting and quantifying mRNA isolated and separated by gel electrophoresis, which is mostly appropriate to address the presence/absence of a specific RNA molecule | Study of clinical markers of acute rejection after pancreas transplantation by evaluation of differential gene expression using RT-qPCR (Cashion et al., 2006) |
| RT-qPCR: Semiquantifying specific mRNAs to detect the level of expression of a specific gene by amplifying and measuring fluorescence or fluorescent report-probed targets simultaneously. The fluorescence is compared to that of reference or housekeeping genes | |||
| b. Genome-wide analysis | — | Microarray assay: Simultaneous determination of levels of all RNA transcripts in a sample compared to those of reference genes as an analysis of global RNA levels and transcriptional profiling. Arrays contain multiple and densely mapped probes of protein-coding genes to reduce any potential selection bias and to be more sensitive in detecting the expression level of splicing variants | Study of transcriptome profiles using peripheral whole blood to reveal underlying genetic contributions to traumatic brain injury (Barr et al., 2010) |
| Protein: Determining protein concentration or expression pattern and localization in order to detect the presence of disease or abnormal physiological conditions | |||
| a. Antibody-based methods | Using antibody probes that react with specific proteins, producing an image of the protein’s location | Enzyme-linked immunosorbent assay: Proteins from sample are attached to the surface of a plate, where a specific antibody is applied and bound to the proteins. The antibody is then linked to an enzyme, and the enzyme’s substrate is added to produce a detectable color image | Study of use of inflammatory proteins for predicting future major adverse coronary events in patients with heart disease who underwent elective coronary stent insertion (Frazier, Vaughn, Willerson, Ballantyne, & Boerwinkle, 2009) |
| Study to investigate hypothalamus–pituitary–adrenal axis dysregulation and immune function alterations in PTSD (Gill, Vythilingam, & Page, 2008) | |||
| b. Mass spectrometry | Detecting and characterizing proteins based on their mass after fractionating the proteins or peptides using two-dimensional gel electrophoresis or high-performance liquid chromatography | — | Study to investigate cord/maternal transfer ratios for buprenorphine and norbuprenorphine in women at delivery by measuring drug concentrations in maternal and cord serum samples using liquid chromatography-tandem mass spectrometry (Bartu, Ilett, Hackett, Doherty, & Hamilton, 2012) |
| c. Nanoproteomic study | Rapidly detecting low-abundance biomarkers with the application of nanoparticles for early detection of diseases | — | Study to develop porous, buoyant, core-shell hydrogel nanoparticles that contain high-affinity reactive chemical baits for better protein and peptide harvesting, concentration, and preservation in bodily fluids (Tamburro et al., 2011) |
Note. SNP = single-nucleotide polymorphism; PTSD = post-traumatic stress disorder; RT-qPCR = real-time reverse-transcription polymerase chain reaction.
Genomic Technologies
Candidate Gene Approach
The ultimate goal of genomic studies is to better understand the biological basis of disease, including early indicators of disease, which should result in a greater ability to prevent disease, individualize pharmacological treatment, and improve the quality of life through better management of symptoms. Researchers have widely employed candidate gene association (CGA) to achieve this goal by analyzing associations between genotypes and specific phenotypes. As described in Table 1, CGA is mainly based on, and limited by, existing knowledge in biology, yet it can be easily applied to genetic studies with less time, effort, and financial costs than need to be expended for a genome-wide approach, which we discuss in the next section. To conduct a CGA study, researchers select a small number of candidate genes or regions in the human genome to test a hypothesis. Their selection is based on the literature and prior hypotheses, experimental findings in animal models, or empirical information. Investigation of genetic variations or the function of genomic components in specific genes or regions can be conducted using various biomolecular techniques, including, for one, real-time reverse-transcription polymerase chain reaction (RT-qPCR), a method used to amplify, detect, and quantify targeted DNA or gene copies. Individual single-nucleotide polymorphism (SNP) genotyping is another popular genomic technology because of the high frequency and relatively uniform distribution of SNPs across the human genome (more than 30 million across the genome). SNP genotyping focuses on targeted SNPs at specific genetic loci to explore genetic variation.
Genome-Wide Approach
Even with the recent achievement of the Encyclopedia of DNA Elements (ENCODE) project, which uncovered the roles of the functional elements within the human genome (Maher, 2012), CGA is still limited by our current and incomplete understanding of the biological significance of the human genome. Consequently, the genome-wide association (GWA) approach continues to gain in popularity because it tests 200,000–400,000 known RNA transcripts simultaneously or millions of SNPs across the whole genome without the bias inherent in the initial selection process of the CGA (Wang, Kim, Wang, & Dionne, 2012). In GWA studies, researchers typically use microarray technology to index human genetic variation such as SNP variations (affecting single-nucleotide bases of DNA sequence) and copy number variations (structural variations with sizes up to several megabases of DNA sequence) as well as expression levels of genes or magnitude of epigenetic modifications using matched probes on a small surface or membrane (Grant & Hakonarson, 2008). The probes of an array are designed to examine preselected sequence variations, transcripts, or epigenetic changes. After samples are hybridized to an array, fluorescence on the hybridized probes is scanned to generate genotype results, gene-expression level, or magnitude of epigenetic modifications.
To establish biological parameters associated with disease states, researchers have investigated biomarkers or pathways that can be used to monitor responses to therapeutics by focusing on the specific tissues in which the response is expected to occur. As shown in Table 1, the technique of transcriptome profiling is ideal for discovering molecular targets and/or biological pathways that contribute to advancing clinical diagnosis, predicting outcomes, or delineating therapeutic treatment options (Pongrac, Middleton, Lewis, Levitt, & Mirnics, 2002). The transcriptome is partially controlled by the epigenome, which causes enduring and heritable modification of the genome without changing the DNA sequence. Major epigenetic markers include DNA methylation and histone modifications, which are key identifiers of transcriptional output. These markers provide unique signatures of cellular identity and control cell fates. In particular, DNA methylation, which is one of the most investigated of the epigenetic changes, induces gene silencing (Brenet et al., 2011). Reductions in methylation may indicate overactivated gene functions. In Table 1, we provide an example of an analysis method used to detect DNA methylation associated with disease phenotype. Another epigenetic marker, histone proteins modification, relies on the assumption that a particular combination of histone modifications controls the activity of the DNA coiled around the histone proteins. Histone acetylation, for example, is known to increase gene expression in most cases. However, this description is overly simplistic because no single histone modification is predictive for DNA activity.
Next-Generation Sequencing: Producing More High-Throughput Data
The use of next-generation sequencing (NGS) technologies provides high-throughput sequence data in a relatively short amount of time for moderate cost, which has led to an increase in investigators’ ability to examine the genetic components of disease. The cost of sequencing a genome has dropped exponentially since the introduction of NGS technologies that are replacing the traditional microarray. In GWA studies, whole-genome sequencing is becoming more and more popular. Whole-genome sequencing is now a standard tool of the GWA study. Next-generation sequencers can replace almost all assays based on the microarray platforms. The transcriptome and epigenome can be analyzed with NGS through RNA sequencing (RNA-Seq) and chromatin immunoprecipitation sequencing (ChIP-Seq) (characterizing protein–DNA interactions involved in gene regulation or chromatin organization) analyses, respectively.
Methodological Considerations
Population Stratification: Gender, Ethnicity, and Cell Type
Many complex diseases have distinct demographic features that contribute to an increase in risk for development of the disease, while genetic variations also have distinct patterns/characteristics across gender and ethnic groups. If more than one gender or ethnic group are included in a sample, it can result in selection bias due to an imbalance between the control and affected subgroups. In such cases, false positives or negative associations between genotypes and phenotypes may not be avoidable. In animal models, researchers can easily avoid selection bias by using genetically homogeneous inbred strains. Among humans, one of the best approaches to avoid selection bias is to sample from as homogeneous group as possible (e.g., in gender and ethnicity). Alternatively, researchers may use well-matched patient and control sampling in terms of gender and ethnicity. In gene-expression profiling or epigenome profiling, homogeneity is required not only at the population level but also at the biological level (i.e., the type of cells collected). Even though the genome comprises the same sequence for more than 200 different human cell types, the list of expressed genes and the expression pattern differs widely among cell type, which induces different cell behaviors and eventually complex traits. For example, the gene-expression profile of peripheral blood cells is distinct from that of epithelial cells in the large intestine and may not be as valuable in diagnosis and prognosis of colon cancer. Thus, researchers must carefully consider which tissues will provide the best information to address the research question.
Validation Is Necessary
There is always a risk of error in the multiple steps of laboratory and analysis procedures. Therefore, validation of findings on a small scale with different platforms or different types of assays is always required. For example, findings of a microarray gene-expression analysis can be validated with RT-qPCR. Findings of sequence variation in a next-generation sequencer can be validated in a targeted sequencing of a genomic region with a sequencer based on different chemistry including a traditional capillary sequencer.
Statistical Considerations for Genomic Data Analysis
With advances in technologies, including microarray and next-generation sequencers, come advances in statistical approaches to interpreting the data. When the technology generates higher throughput data, researchers require more complex statistical approaches to analyze those data. Particularly with NGS technology, hundreds of millions to billions of data points are generated from one subject. Therefore, traditional statistical tools and computing power are not sufficient for handling these high-throughput data efficiently. Because multiple hypotheses are tested in genomic studies, the traditional threshold for statistical significance, established with a p value of .05, is not applicable. For example, in a study testing 1 million SNPs for genetic association, 50,000 false positives will be found when the p value is set at .05. Therefore, the researcher must apply multiple test correction to adjust for these false positives. One of the most conservative ways to correct inflation from multiple tests is the Bonferroni method, which divides a p value threshold by the number of hypotheses tested. For example, for 1 million SNP arrays, the statistically significant p value threshold would be .05/1,000,000 = 5 × 10−8. However, this method may overcorrect because SNPs that are tested are not all independent, thus leading to higher false-negative rates (Chanock et al., 2007). Less stringent alternatives such as the false discovery rate (FDR) test or permutation-based methods are thus preferable for multiple-test correction in genomic studies. For differential gene-expression analyses, researchers can use probe-set filtering methods to reduce the number of tested hypotheses and increase statistical power.
Increasing Need for Better Computing Power and Complicated Tools
Because the actual read length of most next-generation sequencers is usually short (ranging between 100 and 500 bases) and cannot cover the entire chromosome or genome, these reads must be aligned and compared to a reference genome to make a sequence. It is recommended to sequence each base at least 30 times for nonsomatic variant finding (30× coverage for sequence alignment), which requires 90 billion base reads for one human genome (i.e., 3 billion × 30 times). Therefore, researchers require a large amount of digital storage, up to several terabytes, are needed for a typical NGS project. For genome-wide somatic mutation findings, at least 80× coverage is recommended, which increases the required storage space by about 3 times compared to the nonsomatic-variant finding.
With the tremendous amount of raw data generated in a sequencer, sequence alignment requires extensive computing, power, and time. Network speed is also important as researchers will be required to transfer data frequently from the storage space to the operating computer or vice versa. After the alignment, sequence data are statistically analyzed by multiple layers of filtering. Existing software can be modified with additional features called plug-ins to conduct this task. Researchers have also used cloud computing to address the requirements of computing power and storage space as well as data sharing. Currently, researchers are using both Amazon and Google Cloud for genomics data analysis. For example, researchers involved in the 1000 Genome Project are sharing data using Amazon S3 cloud space, and the Cancer Genome Atlas (TCGA) data are available via both Amazon and Google Cloud.
Linking Genome to Proteome
Challenges in Proteomics
To understand the complex biological processes that underlie disease onset and influence recovery, it is vital to understand how proteins function in and around cells. Proteins determine differential cell functioning and contribute to individual variability. The most common studies in proteomics focus on determining protein concentrations in blood, saliva, sweat, hair, cerebral spinal fluid, or other types of tissues using enzyme-linked immunosorbent assay (ELISA) or mass spectrometry. As discussed earlier, although the fundamental pathways from DNA to RNA to protein production are well described, there are many complexities that can alter these processes and may relate to disease and recovery. One of these complexities is the overwhelming number of proteins that are yet to be identified. The human genome contains ∼20,000 protein-encoding genes, but the total number of proteins is at least 10 times higher. Another challenge is that amino acids, the base units of proteins, are so small that their identification is difficult. Lastly, many proteins that regulate central processes of health may be found only in peripheral circulation and at very low concentrations, which restricts researchers’ ability to measure them accurately. However, recent proteomic technologies (e.g., a novel ultrasensitive single-molecule ELISA) make the detection of miniscule circulating peripheral biomarkers possible, including, for example, plasma total tau, which is linked to compromised neurological function and cognitive decline after a brain injury (Olivera et al., 2015).
A New Wave in Proteomics: Proteogenomics and Nanoproteomics
Technological advances that combine proteomics and genomics can overcome the limits of proteomics and provide new opportunities to determine proteomic mechanisms of disease. The integrative approach of proteogenomics will be instrumental in developing novel intervention methods (Sarwal, Sigdel, & Salomon, 2011). Another advancing area of science that will revolutionize proteomic research is the application of nanoparticles to biomarker identification and measurement. Many low-abundance biomarkers that can be used for early detection of disease are invisible to mass spectrometry because they exist in bodily fluids at very low concentrations, are masked by high-abundance proteins such as albumin and immunoglobulins, and are very labile. Nanoproteomics offers techniques to overcome these barriers. Using nanoparticles provides a robust analytical platform for real-time and sensitive detection of low-abundance proteins. Nanoproteomics offers several advantages, such as ultralow detection levels, short assay time, high-throughput capability, and low sample consumption. Nanotechnology more generally offers the promise of new disease therapies that act through molecular mimicking methods to result in selective neuronal regeneration (Kubinová & Syková, 2009). The application and further development of nanoproteomics will thus offer new avenues for identifying biological processes that underlie diseases and developing novel therapeutics to minimize the risks of the diseases.
A Final Step: Integrative Genomics
The Human Genome Project, completed in 2003 (National Human Genome Research Institute, 2003), has greatly enriched our knowledge of the human genome and also led to the development and refinement of many genomic technologies. However, the project, itself, revealed very little about the functional relevance of the genes in the genome, some of which has been gradually uncovered by later international projects (e.g., the HapMap or ENCODE projects).
Genetic variations, epigenetic changes, expression of genes and proteins, as well as nongenomic factors interact in complex processes that the research community is only beginning to understand. Very few details about these complex interactions are currently known, providing a challenge to researchers in the field to characterize these biological processes that mediate morbidity and mortality risks. A systemic approach to research that integrates information from DNA, RNA, and protein along with environmental factors will thus most effectively contribute to our understanding of the complexity of human health and disease.
The human microbiome constantly interacts with environmental microbiomes in every ecosystem including soil, the ocean, and the atmosphere (Biteen et al., 2016). These microbiomes affect health and risks for particular diseases (Johnson & Versalovic, 2012). For instance, disruption of microbe–host interactions by exposure to certain diets or chemicals (e.g., antibiotics) can lead to dysbiosis in the host. Research that increases our understanding of metabolites and small molecules found in the bodily tissues, organs, and cells has advanced microbiome studies. Therefore, using different genomic approaches with the same biological samples can be useful. In addition, comparing findings from different species, which allows consideration of research questions through gene homology, model organisms, and comparative biology, will help construct more valid mappings of complex traits (Bubier & Chesler, 2012).
Regardless of its promising future, the science of integrative genomics is still very limited, especially because of the lack of appropriate analysis tools. Considering high-throughput data, for instance, those generated by GWAs that easily reach millions of variables, integrative genomic analysis needs to handle millions × millions of interactions among variables. Also, a lack of consensus regarding aspects of study design such as population stratification, sample size, and multiple test corrections adds confusion to the interpretation of published results. To overcome these limitations, the NIH launched a data-sharing network and initiated data collection from all NIH-funded genomic studies, such as the Gene Expression Omnibus (GEO) and Sequence Read Archive (SRA) databases. NIH’s Genomic Data Sharing policy mandates that investigators share all genomic data (anonymized) generated via NIH-funded studies through publicly available databases effective January 25, 2015.
Conclusion
Because the fields of public health and clinical care are increasingly integrating genomic information into practice (Feero et al., 2010), health-care practitioners and researchers have an increasing need to understand the major genomic technologies and related general principles. Genomic and proteomic technologies have progressed very rapidly, and the accumulation of genomic knowledge is outpacing its absorption and uptake by health-care professionals, including nurses. In this article, we described key concepts of the current genomic approaches as well as methodological considerations in order to provide a quick road map for nurses initiating or considering genomic nursing.
Research has identified thousands of DNA, RNA, and protein biomarkers along with their function and association with human phenotypes. However, we still do not have enough knowledge to map the genome and show all the links among the associated biological processes. Interactions among these molecules can be much more powerful than the simple mathematical summation of components. These interactions influence phenotypes and the complex processes underlying morbidity and mortality.
One basis of nursing is the individualization of care for patients exhibiting symptoms. These patients’ clinical symptoms have underlying bimolecular functions, which usually accompany complex bimolecular interactions. Therefore, nurses are in a key position for transforming personalized health care by translating genomic information as they counsel patients regarding their disease risks. Nurse scientists should, thus, participate in interdisciplinary research not only to promote biologic discovery but also to increase understanding of symptom biology and clinical outcomes as well as nurses’ ability help patients and their families in their clinical decision-making (Williams, Tripp-Reimer, Daack-Hirsch, & DeBerg, 2016). In order to facilitate this type of research, investigators may also outsource clinical biomarker assays and analyses, which may save time and reduce costs (Tsou, 2016).
Acquiring and continually updating their basic knowledge of genomics and related technologies are crucial first steps for nurses attempting to build a foundation to provide competent personalized health care with more effective treatments for precision medicine (White House, 2015) and, ultimately, to become leaders in this promising and developing area of health care (Lea, 2009). The American Nurses Association lists genomics as an essential nursing competency and expects proficiency in the incorporation of genetic and genomic information into practice (Consensus Panel on Genetics/Genomics Nursing Competencies, 2006). Therefore, nursing education at every level, for both current and future nurses, should incorporate genomic technologies and approaches, an imperative that nursing faculty generally acknowledge (Jenkins & Calzone, 2012). Nurses in all areas of practice should also expect to participate in genetic risk assessments, assume a pivotal role in explaining genetic risk and genetic testing to patients, and support informed health decisions and opportunities for early intervention. Given nursing’s important role in the integration of genomic research into health care and the dissemination of genomic information to patients, nurse scientists should continue to extend the scope and methods of nursing research to genomic approaches for integrating biological and behavioral factors and critical health outcomes into a cohesive, more holistic model for nursing care.
Supplementary Material
Footnotes
Authors’ Note: The opinions and assertions in this article are those of the authors and do not necessarily represent those of the U.S. government.
Author Contributions: Hyunhwa Lee contributed to conception, design, data acquisition, data analysis, and interpretation; drafted manuscript; critically revised manuscript; gave final approval; and agrees to be held accountable for all aspects of work ensuring integrity and accuracy. Jessica Gill contributed to conception, data acquisition, data analysis, and interpretation; drafted manuscript; critically revised manuscript; gave final approval; and agrees to be held accountable for all aspects of work ensuring integrity and accuracy. Taura Barr contributed to conception, data acquisition, data analysis, and interpretation; drafted manuscript; gave final approval; and agrees to be held accountable for all aspects of work ensuring integrity and accuracy. Sijung Yun contributed to conception, data acquisition, data analysis, and interpretation; drafted manuscript; critically revised manuscript; gave final approval; and agrees to be held accountable for all aspects of work ensuring integrity and accuracy. Hyungsuk Kim contributed to conception, design, data acquisition, data analysis, and interpretation; drafted manuscript; critically revised manuscript; gave final approval; and agrees to be held accountable for all aspects of work ensuring integrity and accuracy.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Supplemental Material: The online [appendices/data supplements/etc.] are available at http://journals.sagepub.com/doi/suppl/10.1177/1099800416689822.
References
- Barr T. L., Conley Y., Ding J., Dillman A., Warach S., Singlenton A., Matarin M. (2010). Genomic biomarkers and cellular pathways of ischemic stroke by RNA gene expression profiling. Neurology, 75, 1009–1014. doi:10.1212/WNL.0b013e3181f2b37f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartu A. E., Ilett K. F., Hackett L. P., Doherty D. A., Hamilton D. (2012). Buprenorphine exposure in infants of opioid-dependent mothers at birth. Australian & New Zealand Journal of Obstetrics & Gynaecology, 52, 342–347. doi:10.1111/j.1479-828X.2012.01424.x [DOI] [PubMed] [Google Scholar]
- Beery T. A., Hern M. J. (2004). Genetic practice, education, and research: An overview for advanced practice nurses. Clinical Nurse Specialist, 18, 126–132. [DOI] [PubMed] [Google Scholar]
- Bhartiya D., Kapoor S., Jalali S., Sati S., Kaushik K., Sachidanandan C.…Scaria V. (2012). Conceptual approaches for lncRNA drug discovery and future strategies. Expert Opinion on Drug Discovery, 7, 503–513. doi:10.1517/17460441.2012.682055 [DOI] [PubMed] [Google Scholar]
- Biteen J. S., Blainey P. C., Cardon Z. G., Chun M., Church G. M., Dorrestein P. C.…Young T. D. (2016). Tools for the microbiome: Nano and beyond. ACS Nano, 10, 6–37. doi:10.1021/acsnano.5b07826 [DOI] [PubMed] [Google Scholar]
- Bjornsson H. T., Daniele Fallin M., Feinberg A. P. (2004). An integrated epigenetic and genetic approach to common human disease. Trends in Genetics, 20, 350–358. doi:10.1016/j.tig.2004.06.009 [DOI] [PubMed] [Google Scholar]
- Brenet F., Moh M., Funk P., Feierstein E., Viale A. J., Socci N. D., Scandura J. M. (2011). DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One, 6, e14524 doi:10.1371/journal.pone.0014524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubier J. A., Chesler E. J. (2012). Accelerating discovery for complex neurological and behavioral disorders through systems genetics and integrative genomics in the laboratory mouse. Neurotherapeutics, 9, 338–348. doi:10.1007/s13311-012-0111-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzone K. A., Jenkins J., Bakos A. D., Cashion A., Donaldson N., Feero W. G.…Webb J. A. (2013). A blueprint for genomic nursing science. Journal of Nursing Scholarship, 45, 96–104. doi:10.1111/jnu.12007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashion A., Sabek O., Driscoll C., Gaber L., Kotb M., Gaber O. (2006). Correlation of genetic markers of rejection with biopsy findings following human pancreas transplant. Clinical Transplantation, 20, 106–112. doi:10.1111/j.1399-0012.2005.00450.x [DOI] [PubMed] [Google Scholar]
- Cashion A., Driscoll C., Sabek O. (2004). Emerging genetic technologies in clinical and research settings. Biological Research for Nursing, 5, 159–167. doi:10.1177/1099800403257458 [DOI] [PubMed] [Google Scholar]
- Cashion A., Grady P. (2015). The National Institutes of Health/National Institutes of Nursing Research intramural research program and the development of the National Institutes of Health Symptom Science Model. Nursing Outlook, 63, 484–487. doi:10.1016/j.outlook.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanock S. J., Manolio T., Boehnke M., Boerwinkle E., Hunter D. J., Thomas G.…Collins F. S. (2007). Replicating genotype-phenotype associations. Nature, 447, 655–660. doi:10.1038/447655a [DOI] [PubMed] [Google Scholar]
- Chen K., Rajewsky N. (2007). The evolution of gene regulation by transcription factors and microRNAs. Nature Reviews Genetics, 8, 93–103. doi:10.1038/nrg1990 [DOI] [PubMed] [Google Scholar]
- Chuang P.-Y., Conley Y. P., Poloyac S. M., Okonkwo D. O., Ren D., Sherwood P. R.…Alexander S. A. (2010). Neuroglobin genetic polymorphisms and their relationship to functional outcomes after traumatic brain injury. Journal of Neurotrauma, 27, 999–1006. doi:10.1089/neu.2009.1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consensus Panel on Genetics/Genomics Nursing Competencies. (2006). Essential nursing competencies and curricula guidelines in genetics and genomics. Silver Spring, MD: American Nurses Association. [Google Scholar]
- Feero W. G., Guttmacher A. E., Collins F. S. (2010). Genomic medicine—An updated primer. New England Journal of Medicine, 362, 2001–2011. doi:10.1056/NEJMra0907175 [DOI] [PubMed] [Google Scholar]
- Frazier L., Vaughn W. K., Willerson J. T., Ballantyne C. M., Boerwinkle E. (2009). Inflammatory protein levels and depression screening after coronary stenting predict major adverse coronary events. Biological Research for Nursing, 11, 163–173. doi:10.1177/1099800409332801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill J., Vythilingam M., Page G. G. (2008). Low cortisol, high DHEA, and high levels of stimulated TNF-α, and IL-6 in women with PTSD. Journal of Traumatic Stress, 21, 530–539. doi:10.1002/jts.20372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman I. I., Shields J. (1973). Genetic theorizing and schizophrenia. British Journal of Psychiatry, 122, 15–30. doi:10.1192/bjp.122.1.15 [DOI] [PubMed] [Google Scholar]
- Grant S. F. A., Hakonarson H. (2008). Microarray technology and applications in the arena of genome-wide association. Clinical Chemistry, 54, 1116–1124. doi:10.1373/clinchem.2008.105395 [DOI] [PubMed] [Google Scholar]
- Guttmacher A. E., Collins F. S. (2004). Genomic medicine—A primer In Guttmacher A. E., Collins F. S., Drazen J. M. (Eds.), Genomic medicine: Articles from the New England Journal of Medicine (pp. 3–13). Baltimore, MD: Johns Hopkins University Press. [Google Scholar]
- Jenkins J. F., Calzone K. A. (2012). Are nursing faculty ready to integrate genomic content into curricula? Nurse Educator, 37, 25–29. doi:10.1097/NNE.0b013e31823836ec [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. L., Versalovic J. (2012). The human microbiome and its potential importance to pediatrics. Pediatrics, 129, 950–960. doi:10.1542/peds.2011-2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Crago E., Kim M., Sherwood P., Conley Y., Poloyac S., Kerr M. (2013). Cerebral vasospasm after sub-arachnoid hemorrhage as a clinical predictor and phenotype for genetic association study. International Journal of Stroke, 8, 620–625. doi:10.1111/j.1747-4949.2012.00823.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubinová Š., Syková E. (2009). Nanotechnology for treatment of stroke and spinal cord injury. Nanomedicine, 5, 99–108. doi:10.2217/nnm.09.93 [DOI] [PubMed] [Google Scholar]
- Lea D. H. (2009). Basic genetics and genomics: A primer for nurses. OJIN: The Online Journal of Issues in Nursing, 14 doi:10.3912/OJIN.Vol14No02PPT01 [Google Scholar]
- Li L., Liu Y. (2011). Diverse small non-coding RNAs in RNA interference pathways. Methods in Molecular Biology, 764, 169–182. doi:10.1007/978-1-61779-188-8_11 [DOI] [PubMed] [Google Scholar]
- Loescher L. J., Merkle C. J. (2005). The interface of genomic technologies and nursing. Journal of Nursing Scholarship, 37, 111–119. doi:10.1111/j.1547-5069.2005.00022.x [DOI] [PubMed] [Google Scholar]
- Maher B. (2012). ENCODE: The human encyclopaedia. Nature, 489, 46–48. doi:10.1038/489046a [DOI] [PubMed] [Google Scholar]
- National Human Genome Research Institute. (2003). International consortium completes human genome project: All goals achieved; new vision for genome research unveiled. Retrieved from the National Human Genome Research Institute, National Institutes of Health Newsroom, at http://www.genome.gov/11006929
- Olivera A., Lejbman N., Jeromin A., French L. M., Kim H. S., Cashion A.…Gill J. (2015). Peripheral total tau in military personnel who sustain traumatic brain injuries during deployment. JAMA Neurology, 72, 1109–1116. doi:10.1001/jamaneurol.2015.1383 [DOI] [PubMed] [Google Scholar]
- Pirola L., Balcerczyk A., Tothill R. W., Haviv I., Kaspi A., Lunke S.…El-Osta A. (2011). Genome-wide analysis distinguishes hyperglycemia regulated epigenetic signatures of primary vascular cells. Genome Research, 21, 1601–1615. doi:10.1101/gr.116095.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongrac J., Middleton F. A., Lewis D. A., Levitt P., Mirnics K. (2002). Gene expression profiling with DNA microarrays: Advancing our understanding of psychiatric disorders. Neurochemical Research, 27, 1049–1063. doi:10.1023/A:1020904821237 [DOI] [PubMed] [Google Scholar]
- Sarwal M. M., Sigdel T. K., Salomon D. R. (2011). Functional proteogenomics—Embracing complexity. Seminars in Immunology, 23, 235–251. doi:10.1016/j.smim.2011.08.002 [DOI] [PubMed] [Google Scholar]
- Smith A. K., Conneely K. N., Kilaru V., Mercer K. B., Weiss T. E., Bradley B.…Ressler K. J. (2011). Differential immune system DNA methylation and cytokine regulation in post-traumatic stress disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics, 156, 700–708. doi:10.1002/ajmg.b.31212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan T., Read A. (2010). Human molecular genetics (4th ed). New York, NY: Garland Science. [Google Scholar]
- Tamburro D., Fredolini C., Espina V., Douglas T. A., Ranganathan A., Ilag L.…Luchini A. (2011). Multifunctional core–shell nanoparticles: Discovery of previously invisible biomarkers. Journal of the American Chemical Society, 133, 19178–19188. doi:10.1021/ja207515j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou J. A. (2016). Managing biomarker outsourcing: CRO evaluation, streamline outsource process, and quality management In Weiner R., Kelley M. (Eds.), Translating molecular biomarkers into clinical assays: Techniques and applications (pp. 189–199). Cham, Switzerland: Springer International. [Google Scholar]
- Wang D., Kim H., Wang X.-M., Dionne R. (2012). Genomic methods for clinical and translational pain research In Luo Z. D. (Ed.), Pain research: Methods and protocols (2nd ed, pp. 9–46). Totowa, NJ: Humana Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White House (Producer). (2015, January 30). President Obama speaks on the precision medicine initiative. Video retrieved from http://www.whitehouse.gov/photos-and-video/video/2015/01/30/president-obama-speaks-precision-medicine-initiative
- Williams J. K., Tripp-Reimer T., Daack-Hirsch S., DeBerg J. (2016). Five-year bibliometric review of genomic nursing science research. Journal of Nursing Scholarship, 48, 179–186. doi:10.1111/jnu.12196 [DOI] [PubMed] [Google Scholar]
- World Health Organization. (n.d.). The top 10 causes of death. Retrieved from http://www.who.int/mediacentre/factsheets/fs310/en/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.