Abstract
Intestinal homeostasis and regeneration are driven by intestinal stem cells (ISCs) lying in the crypt. In addition to the actively cycling ISCs that maintain daily homeostasis, accumulating evidence supports the existence of other pools of stem/progenitor cells with the capacity to repair damaged tissue and facilitate rapid restoration of intestinal integrity after injuries. Appropriate control of ISCs and other populations of intestinal epithelial cells with stem cell activity is essential for intestinal homeostasis and regeneration while their deregulation is implicated in colorectal tumorigenesis. In this review, we will summarize the recent findings about ISC identity and cellular plasticity in intestine, discuss regulatory mechanisms that control ISCs for intestinal homeostasis and regeneration, and put a particular emphasis on extrinsic niche-derived signaling and intrinsic epigenetic regulation. Moreover, we highlight several fundamental questions about the precise mechanisms conferring robust capacity for intestine to maintain physiological homeostasis and repair injuries.
Keywords: Intestinal stem cell, Homeostasis, Regeneration, Plasticity, Epigenetics, Nich
Introduction
Intestinal epithelium is one of the fastest renewing tissues in mammals. Owing to the constant exposure of its luminal surface to injurious factors, such as pathogens and toxins, the average turnover rate is every three to five days in mice to replenish damaged cells 1. These single-layered intestinal epithelial cells are organized into a crypt-villus structure and are predominantly composed of absorptive enterocytes and four secretory lineages known as enteroendocrine, Paneth, goblet, and tuft cells ( Figure 1). The thin intestinal epithelial sheet is responsible for nutrient absorption and stool compaction and also serves as a front-line barrier against microorganisms and infections 2. Maintenance and regeneration of this high-turnover tissue upon injury are fueled primarily by intestinal stem cells (ISCs) that reside at the bottom of the crypt, while differentiated absorptive and secretory cells are assembled as clusters or scattered along the crypt-villus axis 3, 4. More recently, considerable cellular plasticity was noted within intestine, and lineage-restricted progenitors or fully differentiated cells were able to replenish the tissue under certain conditions, such as DNA damage–induced injuries 5– 11.
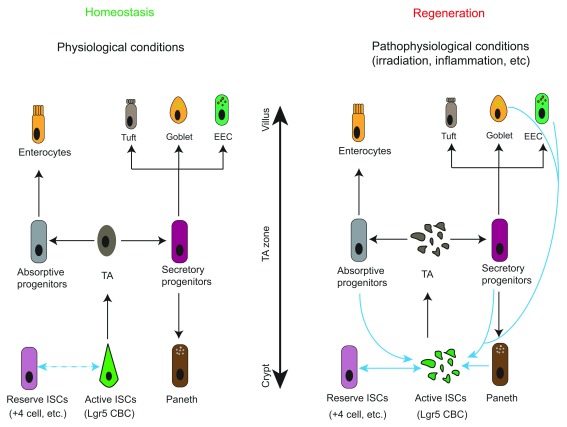
Figure 1. Hierarchy and plasticity of intestinal stem cells (ISCs).
Under homeostatic conditions, active ISCs such as Lgr5 crypt base columnar cells (CBCs) migrate upwards to become transit-amplifying (TA) cells in the TA zone. TA cells divide rapidly and specify into either absorptive or secretory progenitors. Absorptive progenitors further differentiate into large quantities of enterocytes, while secretory progenitors commit to the Paneth, goblet, enteroendocrine (EEC), or goblet cells. Interconversion between reserve ISCs and active ISCs occurs occasionally in this setting (demonstrated by double-headed dash line). In response to radio- or chemo-therapies, the highly proliferative Lgr5 CBCs and TA cells are ablated. Reserve ISCs enter the cell cycle to replenish CBCs for subsequent regenerative process. Plasticity of differentiated progenies, including secretory and absorptive progenitors as well as terminally differentiated EEC and Paneth cells, has been observed when CBCs are damaged. These cells can revert to active ISCs and give rise to all intestinal cell types. However, whether they can bypass CBCs to transdifferentiate directly into other intestinal lineages and their functional importance to intestinal regeneration upon injuries remain to be determined.
ISCs possess both self-renewing capacity and multipotency to give rise to all types of intestinal epithelial cells. They divide and migrate upward to the middle region of the crypt to convert into transit-amplifying (TA) cells that divide rapidly for massive expansion before specializing into absorptive or secretory lineages 4. Owing to the tremendous regenerative capacity and the simple anatomic structure, intestine and particularly its ISCs have become an elegant model system for studying homeostasis, regeneration, and oncogenic transformation of mammalian adult tissues 12, 13. Self-renewal and multipotency of ISCs and plasticity of intestinal epithelial cells are largely controlled by external signals emanated from neighboring niche cells and intrinsic molecular processes, including epigenetic regulation 14, 15. In support of a central role of extracellular niche factors, ISCs embedded in the crypt bottom are sustained by signals emanated from both epithelial and mesenchymal niches. In the in vitro 3D culture system, ISCs are able to self-organize into crypt-villus–like structures referred to as “organoids” (or precisely enteroids or colonoids if derived from small intestine or colon, respectively) in the presence of a defined set of growth factors 16. These organoids comprise self-renewing ISCs intermingled with Paneth cells at the base of budding crypt and various differentiated lineages at blunt villus-like compartments and can be grown and maintained for many passages without losing normal karyotype over time 17.
In this review, we summarize the latest advances in our understanding of ISC identity, cellular plasticity, the basis for intestinal homeostasis and regeneration as well as how ISC self-renewal and multipotency are regulated, with a particular focus on extrinsic niche-derived signaling and intrinsically epigenetic regulation . Considering such progress in the mechanistic understanding of intestinal homeostasis and regeneration as well as the development of new models and techniques to faithfully mimic intestinal pathophysiology, we envision a variety of potent and effective therapeutic approaches for the treatment of intestinal diseases.
Intestinal stem cells and cellular plasticity in intestine
For decades, crypts have been known as compartments comprising cellular sources for continuous intestinal homeostasis and robust post-injury regeneration 18. However, the cellular basis and nature of ISCs that fuel the rapid renewal of intestine have been among the mysteries in the field of adult stem cell biology. It has long been assumed that mammalian tissue-resident adult stem cells, including ISCs, predominantly reside out of the cell cycle in a relatively quiescent G 0 state so that genomic integrity can be sustained in response to genotoxic insults 2, 19. However, this prevailing notion has been amended by the identification of long-lived yet rapidly dividing intestinal crypt base columnar cells (CBCs) with relatively specific expression of Lgr5 20. They self-renew and are capable of differentiating into all types of intestinal epithelial cells in in vivo, as evidenced by lineage-tracing studies of in vivo and in vitro cultured organoids 16, 20, 21. Owing to their mitotically active feature, Lgr5 CBCs were termed active ISCs and thought to sustain physiological homeostasis of the rapid renewing intestine 3. Intriguingly, a subset of epithelial cells residing specifically at +4 position relative to the base of crypts was observed to share some properties of tissue-resident adult stem cells, such as the ability of long-term DNA label retention and a strong resistance to stress, including chemotherapy and irradiation 19, 22, 23, and thus had been postulated to represent ISCs long before Lgr5 CBCs were identified.
Lgr5 CBCs are mitotically active and can regenerate whole intestinal epithelium under homeostatic conditions 20. However, owing to their exquisite sensitivity to genotoxic stresses, Lgr5 CBCs are rapidly lost upon radio-/chemo-induced damage and thus could not account for the robust regenerative potential of post-injury intestine 24. Moreover, studies with genetic ablation of Lgr5 CBCs by diphtheria toxin (DT) treatment of mice harboring Lgr5-driven DT receptor (DTR) allele revealed that these cells are dispensable for normal intestinal homeostasis, implying the existence of other epithelial cells with both stem cell activity and DNA damage–resistant capacity to replace Lgr5 CBC loss for intestinal regeneration 25. Multiple populations of rare crypt cells marked by Bmi1 26, Hopx 26, mTert 27, Krt19 28, Lrig1 29, Sox9 30, Mex3a 31, or Prox1 6 have been found to reside at approximately +4 position by short-term CreER-activated cell fate mapping assay. In sharp contrast to Lgr5 CBCs, most cells labeled by these reporter alleles are slowly cycling and injury-resistant and can give rise to clonal lineage-tracing events albeit at much lower frequency than Lgr5 CBCs 5. In light of the above features, these reporter-marked, predominantly +4 resident cells were defined as reserve ISCs in the literature 3.
In contrast to their unique spatial localization noted in genetic-marked reporter assays, transcriptomic analyses revealed that endogenous Bmi1, mTert, and Hopx are broadly expressed throughout crypt cells, even in the active Lgr5 CBCs, reflecting a certain inconsistency between reporter activity and actual mRNA expression of the endogenous alleles 32– 34. Multiple reasons could underlie this discrepancy, such as (1) difference in the 3′ untranslated region (UTR) sequence between CreER reporter and endogenous alleles. A direct comparison between the mRNA level of CreER reporter and endogenous alleles among distinct populations of crypt cells could determine whether CreER reporter can faithfully recapitulate expression of its endogenous counterpart at transcriptional and post-transcriptional levels. (2) As activation of genetic reporters in lineage-tracing studies requires reaching a certain threshold of CreER activity, cells marked by genetic reporters following short-term tamoxifen administration may point to a stronger CreER activity in these cells than in other populations of crypt cells, which could result from differential levels of CreER protein or tamoxifen permeability in distinct types of crypt cells. Ideally, development of immunohistological grade antibodies that can specially recognize the endogenous protein of reported markers for ISCs will resolve these discrepancies and help determine their actual distribution pattern throughout crypts.
As reserve ISCs marked by Bmi1 +, Hopx +, mTert, Lrig1, and so on can give rise to all types of epithelial lineages, including the active Lgr5 CBCs in lineage-tracing studies, they have initially been posited to sit at the apex of the cellular hierarchy in intestine 25, 26. However, single- or bulk-cell transcriptomic profiling analyses have invariably detected the expression of a few enteroendocrine markers within reserve populations of ISCs, indicating some common features between reserve ISCs and committed enteroendocrine cells or hinting at a potential developmental plasticity of lineage-restricted secretory progenitors in intestine 35. It should be mentioned that definition of reserve ISCs hinges solely on the functional criteria, including injury resistance, multipotency, cell cycle entry from G 0 to G 1 upon damage, and long-term maintenance, all of which can be illustrated by Cre recombinase–induced lineage-tracing assays 3, 5. Cells marked with Bmi1 +, Hopx +, mTert, and so on fulfill these functionally defining criteria and could act as bona fide reserve ISCs for tissue regeneration 24, 26, 27, 36. The fact that stem cells can express multiple-lineage genes in a fluctuating way, assumed as priming, has been reported in adult stem cells 37 as well as in pluripotent embryonic stem cells, which express a significant level of representative genes for primordial germ cells in an undifferentiated pluripotent state 38– 40. Therefore, caution should be exercised when defining ISC identity simply on the basis of transcriptomic analysis. Nevertheless, more intense studies to further investigate the heterogeneity of +4 epithelial cells with single-cell high-throughput strategies are required to deepen our understanding of the identity of reserve stem cells in intestine.
Cellular plasticity has been observed in several mammalian tissues and could act as an additional mechanism for tissue regeneration 41. Direct evidence showing developmental plasticity of enteroendocrine progenitors comes from lineage tracing of cells marked with Prox1, which is a transcription factor expressed predominantly in mature enteroendocrine cells and is essential for its commitment from ISCs 6, 42– 45. Prox1 enteroendocrine cells can function as reserve ISCs as they assume both homeostatic and injury-inducible stem activity in lineage-tracing reporter assays 6. Besides mature enteroendocrine cells, considerable plasticity and clonal lineage-tracing events have been noted in other progenies of Lgr5 CBCs, such as enteroendocrine progenitors marked by Bmi1 7, Alpi + absorptive progenitors 8, secretory precursors expressing Atoh1 10 or Dll1 11, CD69 + CD274 + goblet precursor 7, fully differentiated Paneth cells 46, 47, and a small population of Dclk1 + tuft cells 48 in certain situations, such as inflammation, radio-/chemo-induced intestinal injuries, or upon loss of Lgr5 CBCs by DT administration ( Figure 1). However, the functional robustness and contribution of these lineage-committed cells to intestinal homeostasis and restoration upon tissue damage remain unclear and will be worth future investigation. Furthermore, whether these differentiated epithelia can bypass Lgr5 CBCs and transdifferentiate directly into other epithelial lineages is also instrumental for understanding the mechanisms of regenerative process and awaits clarification in forthcoming studies. Another concern to be considered is that haploinsufficiency in the knocked-in allele used in current genetic tracing assays was recently reported to result in a misleading phenotype as seen in pancreatic systems 49.
Extrinsic niche regulation of intestinal stem cells
The activity of ISCs is stringently controlled to ensure proper proliferation and differentiation. Tight regulation of ISCs is achieved primarily through extrinsic signaling molecules emanated from their surrounding cells that altogether constitute a unique niche microenvironment 15, 50. Recent intensive studies have led to gradual identification of vital niche components that include both epithelial progenies of ISCs and mesenchymal cells, such as Paneth cells, fibroblasts, immune cells, enteric neurons, and endothelial cells 51, 52. When perturbed by injurious factors, these intestinal niche cells can be rewired for coordinated production of cytokines and growth factors to activate ISCs for rapid regeneration. To date, various niche factor–derived signaling pathways have been identified to be essential for ISC activity, intestinal homeostasis, and regeneration. In this section, we will briefly summarize the central roles of Wnt, bone morphogenetic protein (BMP), Notch, epidermal growth factor (EGF), and Hippo signaling in ISC regulation.
In the Wnt pathway, binding of Wnt ligands to their Frizzled receptor on targeted cells induces collapse of cytoplasmic APC destruction complex and subsequent nuclear translocation of β-catenin to activate Wnt target genes through association with T-cell factor (TCF) transcription factors 53. Wnt signaling is progressively reduced from crypt to villus axis and is indispensable for ISC maintenance and intestinal regeneration 32, 54, 55. Its abrogation via inactivating Tcf1/2 mutation 56, Tcf4 deletion 57, or exogenous expression of Wnt inhibitor Dkk1 58, 59 invariably leads to Lgr5 CBC loss and decreased crypt cell proliferation. Augmentation of activity of this pathway by Apc inactivating mutation, constitutive activating mutation in β-catenin, or simultaneous deletion of two E3 ligases (Rnf43 and Znrf3) targeting Wnt ligand receptors for degradation unanimously result in crypt expansion and rapid appearance of intestinal adenomas in mice 60– 64. Although the functional importance of Wnt signaling and its contribution to intestinal regulation have been well appreciated 55, the cellular source for Wnt ligands remains incompletely understood. Various types of niche cells have been found to express a significant level of Wnt ligands, such as small intestinal Paneth cells 51, colonic Reg4 + 65 or cKit + 66 secretory cells, and numerous subsets of stromal cells 67, 68. Although epithelial Wnts can promote expansion of Lgr5 CBCs during in vitro organoid culture, their ablation does not demonstrate a notable impact on crypt cell proliferation and intestinal homeostasis in vivo, suggesting a significant redundancy of cellular origin of Wnt ligands in sustaining intestinal integrity 67– 70. In line with this assumption, recent studies found that Foxl1 + 71, 72, Pdgfrα + 73, Gli1 + 55, 74, and CD34 + Gp38 + αSMA – 75 mesenchymal cells serve as major sources of Wnt activity and play pivotal roles in sustaining intestinal homeostasis in vivo. Abolition of Wnt secretion in Foxl1 +, Gli1 +, or Pdgfrα + cells by genetic excision of porcupine ( Porcn) impairs proliferation of Lgr5 CBCs and leads to corruption of intestinal integrity 71, 73, 74. Whether these mesenchymal cells with distinct markers overlap with each other to some extent or even represent the same subsets of non-epithelial cells is currently unclear. Moreover, how Wnt activity in these mesenchymal cells is regulated in response to intestinal injuries, such as inflammation and chemo-/radio-toxicity, remains unanswered. Future investigations combining genetic approaches with single-cell RNA-sequencing technology will address these critical questions.
BMP signaling assumes an increasing gradient along the intestinal crypt-villus axis and serves as a critical inducer for ISC differentiation, thereby playing a vital role in balancing the effect of Wnt signaling on intestinal homeostasis 76, 77. BMPs belong to the transforming growth factor-beta superfamily of ligands and can induce phosphorylation of cytoplasmic R-Smads (Smad1/5/8) through binding to the membrane-embedded serine/threonine kinase type I and II receptors. Phosphorylated R-Smads form a complex with Co-Smad (Smad4) and subsequently enter the nucleus, where they regulate expression of targeted genes through association with a variety of co-factors 78, 79. The initial experimental evidence implicating an inhibitory role of BMP signaling in ISC self-renewal comes from conditional deletion of BMP receptor Bmpr1a and transgenic expression of BMP antagonist Noggin in mice, both of which unanimously lead to development of multiple polyposis in small intestine 80– 82. Similarly, aberrant expression of BMP antagonist Gremlin1 was noted in patients with hereditary mixed polyposis syndrome and its transgenic expression in mice leads to the appearance of ectopic crypts and subsequent oncogenic transformation 83, 84. Gene expression and in situ hybridization analyses demonstrated that BMP ligands and BMP antagonists are produced primarily by mesenchymes residing at different regions of the crypt-villus unit. BMP2 and BMP4 are secreted by intravillus and intercrypt mesenchymal cells, whereas Noggin, Gremlin1, Gremlin2, and chordin-like 1 antagonists are expressed by cryptal myofibroblasts and smooth muscle cells beneath the crypt bottom 80, 85. To date, little is known about the role and cellular source of other BMP ligands in intestine and whether ablation of these niche cells has an impact on intestinal integrity and tumorigenesis. Future studies are needed to address questions in this regard.
Notch receptors are single-pass type I transmembrane heterodimer proteins that comprise functional extracellular truncation, transmembrane, and intracellular (NICD) domain. Activation of Notch receptors begins with their binding to Notch ligands presented on an adjacent cell, which leads to cytoplasmic proteolytic cleavage of Notch receptors by γ-secretase complex and subsequent nuclear translocation of intracellular NICD to regulate transcription of target genes 86. Notch signaling has been shown to contribute to Lgr5 CBC maintenance and proliferation 87– 89 and its inhibition skews intestinal differentiation toward goblet lineage 90– 92. Paneth cells express and present Notch ligands Dll1 and Dll4 to adjacent Lgr5 CBCs to facilitate their self-renewal 51. Moreover, activation of Notch signaling in Paneth cells by forced expression of NICD can result in acquisition of a stem-like property in these cells 46. Notch signaling is also involved in controlling secretory and absorptive lineage determination when CBCs start to differentiate. Its activation stimulates Hes1 expression for transcriptional silencing of Atoh1, a master transcription factor for secretory lineage determination, thereby blocking secretory differentiation but promoting enterocyte specification of Notch receptor activated cells 93. Expression of Notch ligand Dll1 on secretory cells prevents a secretory fate of neighboring cells but drives these cells toward absorptive lineage through Notch signaling activation, which is a biological event referred to as lateral inhibition 88. Although Notch signaling contributes to CBC proliferation and intestinal homeostasis, the role of niche cells expressing Notch ligands remains unclear.
The Hippo pathway functions as a vital regulator of tissue homeostasis and organ size and its deregulation is implicated in the development of multiple types of cancer in humans. In the presence of extracellular stimuli, transcriptional activator YAP/TAZ complex is phosphorylated by upstream serine–threonine kinase MST1/MST2/Sav1-LATS1/2/MOB1A/B cascade, leading to either cytoplasmic sequestration or proteasomal degradation of YAP/TAZ. Without Hippo activation, non-phosphorylated YAP/TAZ enters the nucleus and acts as a co-activator for the TEAD transcription factor to regulate expression of genes associated with cell growth and proliferation 94, 95. The Hippo pathway is constitutively active in intestine under homeostatic conditions and its genetic inactivation via transgenic expression of YAP/TAZ or conditional deletion of Sav1, an upstream negative regulator of YAP, increases ISC proliferation and leads to crypt hyperplasia 96, 97. Hippo signaling is also indispensable for intestinal regeneration in mouse models of dextran sodium sulfate (DSS)-induced colitis and radiation-induced injury 97, 98. Transgenic expression of YAP induces an EGF signaling–dependent regenerative program to facilitate intestinal regeneration 98. Appropriate activation of Wnt signaling is essential for intestinal recovery following injury. However, its aberrant activation could lead to oncogenic transformation or reduce ISC survival through increasing radio-sensitivity of ISCs to DNA damage during intestinal regeneration 99. In addition to the transcriptional regulatory activity of nuclear YAP/TAP complex, unphosphorylated cytoplasmic complexes are integral components of β-catenin destruction complex and thus play a critical role in restricting Wnt signaling activity 100. Paradoxically, it has been noted that conditional deletion of YAP in intestinal epithelia augments Wnt activity and causes crypt hyperplasia and overgrowth throughout small intestine and colon after radiation-induced injury 101. These findings suggest that the Hippo pathway has dual roles in regulating both Wnt and Egf signaling and its impact on intestinal regeneration is context-dependent and could be determined by net activity of these two pathways. More work is needed to deepen our understanding of the molecular mechanisms that control the balanced activation of Wnt and Egf pathways by Hippo signaling during intestinal regeneration.
EGF communicates with target cells through the EGF receptor, which is a member of the ErbB family of tyrosine receptor kinases 102. Paneth cells express EGF and sustain proliferation of Lgr5 CBCs through its receptor ErbB 103. EGF supplementation of culture medium dramatically increases the efficiency of organoid formation in vitro 17. Lrig1 is a negative feedback regulator of ErbB receptor and its ablation causes crypt expansion in mice 29, 104. Although these findings support a crucial role of EGF signaling in CBC proliferation, the additional cellular source of EGF growth factor except for Paneth cells in intestine remains ambiguous.
Intrinsically epigenetic regulation of intestinal stem cells
Intestinal homeostasis and regeneration are accompanied by drastic transcriptional alterations that are achieved through the cooperation between extrinsic niche signaling–controlled transcription factors/co-activators and intrinsic epigenetic regulators. Epigenetics refers to the inherited alteration in gene expression and phenotype that occur without changes in DNA sequence. It consists primarily of DNA methylation, histone modification, and chromatin remodeling 105, 106. In contrast to the extensively studied extrinsic niche factors, the role of intrinsically epigenetic mechanisms in intestinal homeostasis and regeneration is poorly understood. In this section, we will briefly discuss the currently limited knowledge of epigenetic mechanisms in intestinal regulation.
With the tremendous advancements in fluorescence-activated cell sorting, identification of lineage-specific markers, and next-generation sequencing technology, epigenetic characterization of distinct types of intestinal epithelial cells has recently become feasible. Initial studies with DNase I mapping for accessible chromatin regions and H3K4me2 and H3K27ac ChIP-seq (chromatin immunoprecipitation sequencing) for active enhancers showed that ISCs and enterocyte and secretory progenitors assume remarkable similarity in distribution pattern of open chromatin elements and active enhancers throughout the genome 107. However, these observations are not in line with the subsequent studies that employ assay for transposase-accessible chromatin using sequencing (ATAC-seq) to profile open chromatin regions among ISCs and enterocyte and secretory progenitors. The latter studies identified thousands of unique open chromatin regions among these populations and found that accessible chromatin states in secretory progenitors shift to resemble ISCs during dedifferentiation upon injuries ( Figure 2) 7. Although the discrepancy between the two studies can be explained by cellular heterogeneity and the different sensitivity of technologies employed, the functional importance of those unique open chromatin elements in secretory lineage specification, dedifferentiation, and regeneration has not been determined experimentally. In addition, comprehensive profiling of distinct histone modifications in purer populations of intestinal lineages will determine whether the epigenetic landscape varies during intestinal homeostasis and regeneration.
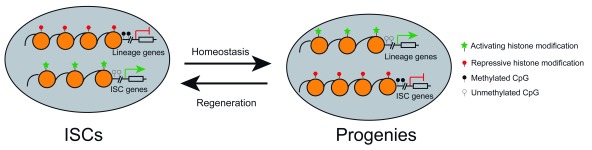
Figure 2. Epigenetic reprogramming for intestinal homeostasis and regeneration.
During homeostasis, intestinal stem cells (ISCs) adopt a unique epigenetic signature and chromatin accessibility that collaboratively lead to expression of ISC-related genes and shutdown of lineage-specific factors. Lineage specification of ISCs is accompanied by epigenetic remodeling and chromatin accessibility changes that turn off ISCs genes while activating lineage-restricted genes. During the regenerative process, epigenetic landscape and chromatin accessibility are reconfigured to resemble ISCs during dedifferentiation of ISC progenies.
Initial bisulfite-sequencing of ISCs and their differentiated progenies in villus revealed minimal dynamics of DNA methylome during intestinal differentiation 108. This study used a cutoff of a minimal differential methylation of 40% to identify differentially methylated regions (DMRs) between ISCs and villous cells. As the distal regulatory regions are low-methylated at an overall methylation level of 30% in intestinal cells and display average change of about 15% in DNA methylation during intestinal differentiation, the cutoff criteria of analysis used in the study eliminates the identification of DMRs during differentiation of ISCs 108, 109. Subsequent bisulfite-sequencing studies with distinct analytic standards identified many DMRs at enhancers and disclosed a tight correlation between the DMRs and transcriptional alterations between two populations of cells, implying that the DNA methylation may have a vital role in intestinal renewal 110. This viewpoint has been proven by genetic ablation of Dnmt1 and Tet1 in intestine, which are maintenance methyltransferase and hydroxylase for DNA methylation, respectively 106. Dnmt1 is essential for intestinal development in newborn mice as inducible deletion of Dnmt1 at perinatal stage causes genomic instability, premature differentiation, apoptosis, loss of villi, and decreased proliferation of crypt cells 111, 112. The essential role of DNA methylation for intestinal development is also reflected in Tet1-deleted mice. Tet1-null mice exhibit retarded growth, shorter intestines, weaker capability to form in vitro organoids, and reduced postnatal viability 113. Acute ablation of Dnmt1 in adult intestinal epithelia causes a slight expansion of the proliferative crypt zone. However the crypt morphology recovers and DNA methylation restores to normal level several days after Dnmt1 deficiency. A subsequent study revealed that de novo methyltransferase Dnmt3b can compensate for Dnmt1 loss to maintain intestinal integrity as ablation of both enzymes leads to genome demethylation, genomic instability, increased apoptosis, and decreased survival 114. Taken together, these pieces of genetic evidence strongly support that dynamic regulation of DNA methylation acts as an essential epigenetic mechanism underlying intestinal development and homeostasis.
Post-translational modification of histones, such as methylation and ubiquitination, constitutes an additional component of epigenetic mechanism in transcriptional regulation, embryonic development, and adult tissue homeostasis 115. Monoubiquitination of lysine 119 on H2A (H2A119 mUb) and methylation of lysine 27 on H3 (H3K27me) correlate with transcriptional repression and are implemented by polycomb repressive complex 1 (PRC1) and polycomb repressive complex 2 (PRC2), respectively 116. The activity of PRC1 and PRC2 stems from Ring1a/1b E3 ligase and Ezh1/2 methyltransferases and has been shown to be essential for normal intestinal homeostasis or regeneration following injuries. Loss of function of the total PRC1 activity via Ring1a/b double deletion in intestinal epithelia compromises ISC self-renewal and intestinal integrity and results in morbidity through de-repression of a number of transcription factors that negatively regulate Wnt signaling 117.
Loss of PRC2 activity via Ah Cre or VillinCreER-induced epithelial deletion of the Eed, a scaffold protein of the complex, leads to a clear defect in cell proliferation in crypts, a marked increase in the number of goblet cells, mislocalized Paneth cells, and compromised regenerative capacity 118– 120. The homeostatic and regenerative defect of intestine in the absence of PRC2 results at least in part from aberrant upregulation of Cdkn2a and some master regulators for secretory lineages that are bivalently marked, normally targeted, and repressed by this complex 118. Conditional ablation of catalytic subunit of PRC2 complex Ezh2 does not show any abnormalities in the intestinal homeostasis, indicating that the other H3K27 methyltransferase Ezh1 could compensate for Ezh2 loss to maintain intestinal integrity 120. Further studies are needed to know the role and target genes of individual non-canonic PRC1 and PRC2 complexes in intestinal renewal and regeneration.
Conclusions and future perspectives
Over the past decade, remarkable progress has been made in our understanding of intestinal biology. It has become evident that active Lgr5 CBCs and reserve ISCs work in a coordinated manner to maintain intestinal homeostasis and replenish the tissue upon injuries. In addition, differentiated epithelial cells possess considerable plasticity and can dedifferentiate to ISCs for intestinal regeneration in mouse models when active CBCs are damaged or artificially removed. Many niche-derived factors and a few epigenetic regulators have been identified and characterized for their roles in intestinal morphogenesis, homeostasis, and regeneration. Despite these considerable advances in recent years, several fundamental questions remain to be addressed about the precise molecular mechanisms controlling self-renewal, lineage commitment, and plasticity of ISCs. First, the cellular source of distinct environmental factors crucial for ISC function and the role of immune cells, peripheral nerve cells, and other ISC-proximal cells remain poorly understood; thus, extensive studies of these niche components will offer more insight into the extrinsic regulatory mechanisms for ISC function. Second, comprehensive characterization of the epigenetic landscape of various intestinal cells coupled with conditional ablation of key chromatin regulators in murine models will help determine the molecular basis underlying the remarkable plasticity of intestine in response to injuries. Third, the physiological function and contribution of differentiated crypt cells to radio-/chemo-induced intestinal regeneration remain to be experimentally evaluated. Fourth, the molecular mechanisms that reserve ISCs and differentiated crypt cells use to sense and replenish lost CBCs are largely unexplored; future investigation in this regard will help design effective therapeutic approaches to facilitate intestinal recovery after damage. Lastly, does aging-associated loss of regenerative potential of ISCs result from alterations in niche factors or epigenetic regulation or both 121– 123? The more we understand the above fundamental questions, the better we can employ ISCs for regenerative medicine.
Acknowledgments
We are grateful to Karen Tannen (Stowers Institute for Medical Research, Kansas City, MO, 64110, USA) for proofreading and editing. We apologize to colleagues whose studies were not cited in this review due to space limitations.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Ramesh A Shivdasani, Department of Medicine, Dana-Farber Cancer Institute and Harvard Medical School, Boston, USA
Eduard Batlle, Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Barcelona, Spain
Rongwen Xi, National Institute of Biological Sciences (NIBS), Beijing, China
Funding Statement
DH was supported by grants from the National Natural Science Foundation of China (31872825) and the Natural Science Foundation of Tianjin (18JCYBJC42400). Study in LL’s lab was partially supported by National Institutes of Health grant U01DK085507, which is a collaborative research project of the Intestinal Stem Cell Consortium funded by the National Institute of Diabetes and Digestive and Kidney Diseases and the National Institute of Allergy and Infectious Diseases.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 3 approved]
References
- 1. Beumer J, Clevers H: Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development. 2016;143(20):3639–49. 10.1242/dev.133132 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 2. Clevers H: The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154(2):274–84. 10.1016/j.cell.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 3. Li L, Clevers H: Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327(5965):542–5. 10.1126/science.1180794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barker N, van de Wetering M, Clevers H: The intestinal stem cell. Genes Dev. 2008;22(14):1856–64. 10.1101/gad.1674008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yousefi M, Li L, Lengner CJ: Hierarchy and Plasticity in the Intestinal Stem Cell Compartment. Trends Cell Biol. 2017;27(10):753–64. 10.1016/j.tcb.2017.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yan KS, Gevaert O, Zheng GXY, et al. : Intestinal Enteroendocrine Lineage Cells Possess Homeostatic and Injury-Inducible Stem Cell Activity. Cell Stem Cell. 2017;21(1):78–90.e6. 10.1016/j.stem.2017.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jadhav U, Saxena M, O'Neill NK, et al. : Dynamic Reorganization of Chromatin Accessibility Signatures during Dedifferentiation of Secretory Precursors into Lgr5+ Intestinal Stem Cells. Cell Stem Cell. 2017;21(1):65–77.e5. 10.1016/j.stem.2017.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 8. Tetteh PW, Basak O, Farin HF, et al. : Replacement of Lost Lgr5-Positive Stem Cells through Plasticity of Their Enterocyte-Lineage Daughters. Cell Stem Cell. 2016;18(2):203–13. 10.1016/j.stem.2016.01.001 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 9. Ishibashi F, Shimizu H, Nakata T, et al. : Contribution of ATOH1 + Cells to the Homeostasis, Repair, and Tumorigenesis of the Colonic Epithelium. Stem Cell Reports. 2018;10(1):27–42. 10.1016/j.stemcr.2017.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 10. Tomic G, Morrissey E, Kozar S, et al. : Phospho-regulation of ATOH1 Is Required for Plasticity of Secretory Progenitors and Tissue Regeneration. Cell Stem Cell. 2018;23(3):436–443.e7. 10.1016/j.stem.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 11. van Es JH, Sato T, van de Wetering M, et al. : Dll1 + secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14(10):1099–104. 10.1038/ncb2581 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 12. Potten CS, Booth C, Hargreaves D: The small intestine as a model for evaluating adult tissue stem cell drug targets. Cell Prolif. 2003;36(3):115–29. 10.1046/j.1365-2184.2003.00264.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang F, Scoville D, He XC, et al. : Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay. Gastroenterology. 2013;145(2):383–395.e21. 10.1053/j.gastro.2013.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roostaee A, Benoit YD, Boudjadi S, et al. : Epigenetics in Intestinal Epithelial Cell Renewal. J Cell Physiol. 2016;231(11):2361–7. 10.1002/jcp.25401 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 15. Sailaja BS, He XC, Li L: The regulatory niche of intestinal stem cells. J Physiol. 2016;594(17):4827–36. 10.1113/JP271931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sato T, Vries RG, Snippert HJ, et al. : Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–5. 10.1038/nature07935 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 17. Sato T, Clevers H: Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340(6137):1190–4. 10.1126/science.1234852 [DOI] [PubMed] [Google Scholar]
- 18. Bjerknes M, Cheng H: Intestinal epithelial stem cells and progenitors. Methods Enzymol. 2006;419:337–383. 10.1016/S0076-6879(06)19014-X [DOI] [PubMed] [Google Scholar]
- 19. Potten CS, Kovacs L, Hamilton E: Continuous labelling studies on mouse skin and intestine. Cell Prolif. 1974;7(3):271–83. 10.1111/j.1365-2184.1974.tb00907.x [DOI] [PubMed] [Google Scholar]
- 20. Barker N, van Es JH, Kuipers J, et al. : Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–7. 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 21. Barker N, van Es JH, Jaks V, et al. : Very long-term self-renewal of small intestine, colon, and hair follicles from cycling Lgr5 +ve stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:351–6. 10.1101/sqb.2008.72.003 [DOI] [PubMed] [Google Scholar]
- 22. Potten CS, Owen G, Booth D: Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115(Pt 11):2381–8. [DOI] [PubMed] [Google Scholar]
- 23. Potten CS: Extreme sensitivity of some intestinal crypt cells to X and gamma irradiation. Nature. 1977;269(5628):518–21. 10.1038/269518a0 [DOI] [PubMed] [Google Scholar]
- 24. Yan KS, Chia LA, Li X, et al. : The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109(2):466–71. 10.1073/pnas.1118857109 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. Tian H, Biehs B, Warming S, et al. : A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478(7368):255–9. 10.1038/nature10408 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 26. Takeda N, Jain R, LeBoeuf MR, et al. : Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334(6061):1420–4. 10.1126/science.1213214 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 27. Montgomery RK, Carlone DL, Richmond CA, et al. : Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci U S A. 2011;108(1):179–84. 10.1073/pnas.1013004108 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 28. Asfaha S, Hayakawa Y, Muley A, et al. : Krt19+/ Lgr5− Cells Are Radioresistant Cancer-Initiating Stem Cells in the Colon and Intestine. Cell Stem Cell. 2015;16(6):627–38. 10.1016/j.stem.2015.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Powell AE, Wang Y, Li Y, et al. : The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149(1):146–58. 10.1016/j.cell.2012.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Roche KC, Gracz AD, Liu XF, et al. : SOX9 maintains reserve stem cells and preserves radioresistance in mouse small intestine. Gastroenterology. 2015;149(6):1553–1563.e10. 10.1053/j.gastro.2015.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barriga FM, Montagni E, Mana M, et al. : Mex3a Marks a Slowly Dividing Subpopulation of Lgr5+ Intestinal Stem Cells. Cell Stem Cell. 2017;20(6):801–816.e7. 10.1016/j.stem.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Muñoz J, Stange DE, Schepers AG, et al. : The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent '+4' cell markers. EMBO J. 2012;31(14):3079–91. 10.1038/emboj.2012.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Itzkovitz S, Lyubimova A, Blat IC, et al. : Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat Cell Biol. 2011;14(1):106–14. 10.1038/ncb2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li N, Yousefi M, Nakauka-Ddamba A, et al. : Single-cell analysis of proxy reporter allele-marked epithelial cells establishes intestinal stem cell hierarchy. Stem Cell Reports. 2014;3(5):876–91. 10.1016/j.stemcr.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Buczacki SJ, Zecchini HI, Nicholson AM, et al. : Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495(7439):65–9. 10.1038/nature11965 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 36. Sangiorgi E, Capecchi MR: Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40(7):915–20. 10.1038/ng.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Akashi K, He X, Chen J, et al. : Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood. 2003;101(2):383–9. 10.1182/blood-2002-06-1780 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 38. Yamaji M, Ueda J, Hayashi K, et al. : PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell. 2013;12(3):368–82. 10.1016/j.stem.2012.12.012 [DOI] [PubMed] [Google Scholar]
- 39. Bao S, Leitch HG, Gillich A, et al. : The germ cell determinant Blimp1 is not required for derivation of pluripotent stem cells. Cell Stem Cell. 2012;11(1):110–7. 10.1016/j.stem.2012.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chu LF, Surani MA, Jaenisch R, et al. : Blimp1 expression predicts embryonic stem cell development in vitro. Curr Biol. 2011;21(20):1759–65. 10.1016/j.cub.2011.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 41. Tetteh PW, Farin HF, Clevers H: Plasticity within stem cell hierarchies in mammalian epithelia. Trends Cell Biol. 2015;25(2):100–8. 10.1016/j.tcb.2014.09.003 [DOI] [PubMed] [Google Scholar]
- 42. Petrova TV, Nykänen A, Norrmén C, et al. : Transcription factor PROX1 induces colon cancer progression by promoting the transition from benign to highly dysplastic phenotype. Cancer Cell. 2008;13(5):407–19. 10.1016/j.ccr.2008.02.020 [DOI] [PubMed] [Google Scholar]
- 43. Egerod KL, Engelstoft MS, Grunddal KV, et al. : A major lineage of enteroendocrine cells coexpress CCK, secretin, GIP, GLP-1, PYY, and neurotensin but not somatostatin. Endocrinology. 2012;153(12):5782–95. 10.1210/en.2012-1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Engelstoft MS, Egerod KL, Lund ML, et al. : Enteroendocrine cell types revisited. Curr Opin Pharmacol. 2013;13(6):912–21. 10.1016/j.coph.2013.09.018 [DOI] [PubMed] [Google Scholar]
- 45. Habib AM, Richards P, Cairns LS, et al. : Overlap of endocrine hormone expression in the mouse intestine revealed by transcriptional profiling and flow cytometry. Endocrinology. 2012;153(7):3054–65. 10.1210/en.2011-2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu S, Tong K, Zhao Y, et al. : Paneth Cell Multipotency Induced by Notch Activation following Injury. Cell Stem Cell. 2018;23(1):46–59.e5. 10.1016/j.stem.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 47. Schmitt M, Schewe M, Sacchetti A, et al. : Paneth Cells Respond to Inflammation and Contribute to Tissue Regeneration by Acquiring Stem-like Features through SCF/c-Kit Signaling. Cell Rep. 2018;24(9):2312–2328.e7. 10.1016/j.celrep.2018.07.085 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 48. Westphalen CB, Asfaha S, Hayakawa Y, et al. : Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J Clin Invest. 2014;124(3):1283–95. 10.1172/JCI73434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shen J, Ha DP, Zhu G, et al. : GRP78 haploinsufficiency suppresses acinar-to-ductal metaplasia, signaling, and mutant Kras -driven pancreatic tumorigenesis in mice. Proc Natl Acad Sci. 2017;114:E4020–E4029. 10.1073/pnas.1616060114 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 50. Chacón-Martínez CA, Koester J, Wickström SA: Signaling in the stem cell niche: regulating cell fate, function and plasticity. Development. 2018;145(15): pii: dev165399. 10.1242/dev.165399 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 51. Sato T, van Es JH, Snippert HJ, et al. : Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469(7330):415–8. 10.1038/nature09637 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 52. Roulis M, Flavell RA: Fibroblasts and myofibroblasts of the intestinal lamina propria in physiology and disease. Differentiation. 2016;92(3):116–31. 10.1016/j.diff.2016.05.002 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 53. Clevers H, Nusse R: Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–205. 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- 54. Batlle E, Henderson JT, Beghtel H, et al. : Beta-catenin and TCF mediate cell positioning in the intestinal epithelium by controlling the expression of EphB/ephrinB. Cell. 2002;111(2):251–63. 10.1016/S0092-8674%2802%2901015-2 [DOI] [PubMed] [Google Scholar]
- 55. Valenta T, Degirmenci B, Moor AE, et al. : Wnt Ligands Secreted by Subepithelial Mesenchymal Cells Are Essential for the Survival of Intestinal Stem Cells and Gut Homeostasis. Cell Rep. 2016;15(5):911–8. 10.1016/j.celrep.2016.03.088 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 56. Korinek V, Barker N, Willert K, et al. : Two members of the Tcf family implicated in Wnt/beta-catenin signaling during embryogenesis in the mouse. Mol Cell Biol. 1998;18(3):1248–56. 10.1128/MCB.18.3.1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. van Es JH, Haegebarth A, Kujala P, et al. : A critical role for the Wnt effector Tcf4 in adult intestinal homeostatic self-renewal. Mol Cell Biol. 2012;32(10):1918–27. 10.1128/MCB.06288-11 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 58. Kuhnert F, Davis CR, Wang HT, et al. : Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A. 2004;101(1):266–71. 10.1073/pnas.2536800100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pinto D, Gregorieff A, Begthel H, et al. : Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17(14):1709–13. 10.1101/gad.267103 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 60. Koo BK, Spit M, Jordens I, et al. : Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488(7413):665–9. 10.1038/nature11308 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 61. Hao HX, Xie Y, Zhang Y, et al. : ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485(7397):195–200. 10.1038/nature11019 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 62. Jiang X, Charlat O, Zamponi R, et al. : Dishevelled promotes Wnt receptor degradation through recruitment of ZNRF3/RNF43 E3 ubiquitin ligases. Mol Cell. 2015;58(3):522–33. 10.1016/j.molcel.2015.03.015 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 63. Korinek V, Barker N, Morin PJ, et al. : Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC -/- colon carcinoma. Science. 1997;275(5307):1784–7. 10.1126/science.275.5307.1784 [DOI] [PubMed] [Google Scholar]
- 64. Morin PJ, Sparks AB, Korinek V, et al. : Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275(5307):1787–90. 10.1126/science.275.5307.1787 [DOI] [PubMed] [Google Scholar]
- 65. Sasaki N, Sachs N, Wiebrands K, et al. : Reg4 + deep crypt secretory cells function as epithelial niche for Lgr5 + stem cells in colon. Proc Natl Acad Sci U S A. 2016;113(37):E5399–407. 10.1073/pnas.1607327113 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 66. Rothenberg ME, Nusse Y, Kalisky T, et al. : Identification of a cKit + colonic crypt base secretory cell that supports Lgr5 + stem cells in mice. Gastroenterology. 2012;142(5):1195–1205.e6. 10.1053/j.gastro.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 67. Farin HF, van Es JH, Clevers H: Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology. 2012;143(6):1518–1529.e7. 10.1053/j.gastro.2012.08.031 [DOI] [PubMed] [Google Scholar]
- 68. Kabiri Z, Greicius G, Madan B, et al. : Stroma provides an intestinal stem cell niche in the absence of epithelial Wnts. Development. 2014;141(11):2206–15. 10.1242/dev.104976 [DOI] [PubMed] [Google Scholar]
- 69. Durand A, Donahue B, Peignon G, et al. : Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1). Proc Natl Acad Sci U S A. 2012;109(23):8965–70. 10.1073/pnas.1201652109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. San Roman AK, Jayewickreme CD, Murtaugh LC, et al. : Wnt secretion from epithelial cells and subepithelial myofibroblasts is not required in the mouse intestinal stem cell niche in vivo. Stem Cell Reports. 2014;2(2):127–34. 10.1016/j.stemcr.2013.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shoshkes-Carmel M, Wang YJ, Wangensteen KJ, et al. : Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature. 2018;557(7704):242–6. 10.1038/s41586-018-0084-4 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 72. Aoki R, Shoshkes-Carmel M, Gao N, et al. : Foxl1-expressing mesenchymal cells constitute the intestinal stem cell niche. Cell Mol Gastroenterol Hepatol. 2016;2(2):175–88. 10.1016/j.jcmgh.2015.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 73. Greicius G, Kabiri Z, Sigmundsson K, et al. : PDGFRα + pericryptal stromal cells are the critical source of Wnts and RSPO3 for murine intestinal stem cells in vivo. Proc Natl Acad Sci U S A. 2018;115(14):E3173–E3181. 10.1073/pnas.1713510115 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 74. Degirmenci B, Valenta T, Dimitrieva S, et al. : GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature. 2018;558(7710):449–53. 10.1038/s41586-018-0190-3 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 75. Stzepourginski I, Nigro G, Jacob JM, et al. : CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc Natl Acad Sci U S A. 2017;114(4):E506–E513. 10.1073/pnas.1620059114 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 76. Hardwick JCH, van den Brink GR, Bleuming SA, et al. : Bone morphogenetic protein 2 is expressed by, and acts upon, mature epithelial cells in the colon. Gastroenterology. 2004;126(1):111–21. 10.1053/j.gastro.2003.10.067 [DOI] [PubMed] [Google Scholar]
- 77. Qi Z, Li Y, Zhao B, et al. : BMP restricts stemness of intestinal Lgr5 + stem cells by directly suppressing their signature genes. Nat Commun. 2017;8:13824. 10.1038/ncomms13824 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 78. David CJ, Massagué J: Contextual determinants of TGFβ action in development, immunity and cancer. Nat Rev Mol Cell Biol. 2018;19(7):419–35. 10.1038/s41580-018-0007-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Feng XH, Derynck R: Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–93. 10.1146/annurev.cellbio.21.022404.142018 [DOI] [PubMed] [Google Scholar]
- 80. He XC, Zhang J, Tong WG, et al. : BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat Genet. 2004;36(10):1117–21. 10.1038/ng1430 [DOI] [PubMed] [Google Scholar]
- 81. Haramis AP, Begthel H, van den Born M, et al. : De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 2004;303(5664):1684–6. 10.1126/science.1093587 [DOI] [PubMed] [Google Scholar]
- 82. Batts LE, Polk DB, Dubois RN, et al. : Bmp signaling is required for intestinal growth and morphogenesis. Dev Dyn. 2006;235(6):1563–70. 10.1002/dvdy.20741 [DOI] [PubMed] [Google Scholar]
- 83. Davis H, Irshad S, Bansal M, et al. : Aberrant epithelial GREM1 expression initiates colonic tumorigenesis from cells outside the stem cell niche. Nat Med. 2015;21(1):62–70. 10.1038/nm.3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Jaeger E, Leedham S, Lewis A, et al. : Hereditary mixed polyposis syndrome is caused by a 40-kb upstream duplication that leads to increased and ectopic expression of the BMP antagonist GREM1. Nat Genet. 2012;44(6):699–703. 10.1038/ng.2263 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 85. Kosinski C, Li VS, Chan AS, et al. : Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci U S A. 2007;104(39):15418–23. 10.1073/pnas.0707210104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kopan R, Ilagan MX: The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–33. 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Pellegrinet L, Rodilla V, Liu Z, et al. : Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology. 2011;140(4):1230–1240.e1-7. 10.1053/j.gastro.2011.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. VanDussen KL, Carulli AJ, Keeley TM, et al. : Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139(3):488–97. 10.1242/dev.070763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Tian H, Biehs B, Chiu C, et al. : Opposing activities of Notch and Wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Rep. 2015;11(1):33–42. 10.1016/j.celrep.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Milano J, McKay J, Dagenais C, et al. : Modulation of notch processing by gamma-secretase inhibitors causes intestinal goblet cell metaplasia and induction of genes known to specify gut secretory lineage differentiation. Toxicol Sci. 2004;82(1):341–58. 10.1093/toxsci/kfh254 [DOI] [PubMed] [Google Scholar]
- 91. van Es JH, van Gijn ME, Riccio O, et al. : Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435(7044):959–63. 10.1038/nature03659 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 92. van Es JH, de Geest N, van de Born M, et al. : Intestinal stem cells lacking the Math1 tumour suppressor are refractory to Notch inhibitors. Nat Commun. 2010;1(1):18. 10.1038/ncomms1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ueo T, Imayoshi I, Kobayashi T, et al. : The role of Hes genes in intestinal development, homeostasis and tumor formation. Development. 2012;139(6):1071–82. 10.1242/dev.069070 [DOI] [PubMed] [Google Scholar]
- 94. Fu V, Plouffe SW, Guan KL: The Hippo pathway in organ development, homeostasis, and regeneration. Curr Opin Cell Biol. 2017;49:99–107. 10.1016/j.ceb.2017.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 95. Pan D: The hippo signaling pathway in development and cancer. Dev Cell. 2010;19(4):491–505. 10.1016/j.devcel.2010.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Imajo M, Ebisuya M, Nishida E: Dual role of YAP and TAZ in renewal of the intestinal epithelium. Nat Cell Biol. 2015;17(1):7–19. 10.1038/ncb3084 [DOI] [PubMed] [Google Scholar]
- 97. Cai J, Zhang N, Zheng Y, et al. : The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010;24(21):2383–8. 10.1101/gad.1978810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Gregorieff A, Liu Y, Inanlou MR, et al. : Yap-dependent reprogramming of Lgr5 + stem cells drives intestinal regeneration and cancer. Nature. 2015;526(7575):715–8. 10.1038/nature15382 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 99. Tao S, Tang D, Morita Y, et al. : Wnt activity and basal niche position sensitize intestinal stem and progenitor cells to DNA damage. EMBO J. 2015;34(5):624–40. 10.15252/embj.201490700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Azzolin L, Panciera T, Soligo S, et al. : YAP/TAZ incorporation in the β-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158(1):157–70. 10.1016/j.cell.2014.06.013 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 101. Barry ER, Morikawa T, Butler BL, et al. : Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2013;493(7430):106–10. 10.1038/nature11693 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 102. Roskoski R, Jr: The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol Res. 2014;79:34–74. 10.1016/j.phrs.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 103. Schepers A, Clevers H: Wnt signaling, stem cells, and cancer of the gastrointestinal tract. Cold Spring Harb Perspect Biol. 2012;4(4):a007989. 10.1101/cshperspect.a007989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wong VW, Stange DE, Page ME, et al. : Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol. 2012;14(4):401–8. 10.1038/ncb2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Allis CD, Jenuwein T: The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17(8):487–500. 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 106. Hu D, Shilatifard A: Epigenetics of hematopoiesis and hematological malignancies. Genes Dev. 2016;30(18):2021–41. 10.1101/gad.284109.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Kim TH, Li F, Ferreiro-Neira I, et al. : Broadly permissive intestinal chromatin underlies lateral inhibition and cell plasticity. Nature. 2014;506(7489):511–5. 10.1038/nature12903 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 108. Kaaij LT, van de Wetering M, Fang F, et al. : DNA methylation dynamics during intestinal stem cell differentiation reveals enhancers driving gene expression in the villus. Genome Biol. 2013;14(5):R50. 10.1186/gb-2013-14-5-r50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Stadler MB, Murr R, Burger L, et al. : DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480(7378):490–5. 10.1038/nature10716 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 110. Sheaffer KL, Kim R, Aoki R, et al. : DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev. 2014;28(6):652–64. 10.1101/gad.230318.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Elliott EN, Sheaffer KL, Schug J, et al. : Dnmt1 is essential to maintain progenitors in the perinatal intestinal epithelium. Development. 2015;142(12):2163–72. 10.1242/dev.117341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Yu DH, Gadkari M, Zhou Q, et al. : Postnatal epigenetic regulation of intestinal stem cells requires DNA methylation and is guided by the microbiome. Genome Biol. 2015;16:211. 10.1186/s13059-015-0763-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kim R, Sheaffer KL, Choi I, et al. : Epigenetic regulation of intestinal stem cells by Tet1-mediated DNA hydroxymethylation. Genes Dev. 2016;30(21):2433–42. 10.1101/gad.288035.116 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 114. Elliott EN, Sheaffer KL, Kaestner KH: The ' de novo' DNA methyltransferase Dnmt3b compensates the Dnmt1-deficient intestinal epithelium. eLife. 2016;5: pii: e12975. 10.7554/eLife.12975 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 115. Shilatifard A: The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. 10.1146/annurev-biochem-051710-134100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Margueron R, Reinberg D: The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–9. 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Chiacchiera F, Rossi A, Jammula S, et al. : Polycomb Complex PRC1 Preserves Intestinal Stem Cell Identity by Sustaining Wnt/β-Catenin Transcriptional Activity. Cell Stem Cell. 2016;18(1):91–103. 10.1016/j.stem.2015.09.019 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 118. Chiacchiera F, Rossi A, Jammula S, et al. : PRC2 preserves intestinal progenitors and restricts secretory lineage commitment. EMBO J. 2016;35(21):2301–14. 10.15252/embj.201694550 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 119. Jadhav U, Nalapareddy K, Saxena M, et al. : Acquired Tissue-Specific Promoter Bivalency Is a Basis for PRC2 Necessity in Adult Cells. Cell. 2016;165(6):1389–400. 10.1016/j.cell.2016.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 120. Koppens MA, Bounova G, Gargiulo G, et al. : Deletion of Polycomb Repressive Complex 2 From Mouse Intestine Causes Loss of Stem Cells. Gastroenterology. 2016;151(4):684–697.e12. 10.1053/j.gastro.2016.06.020 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 121. Mihaylova MM, Cheng CW, Cao AQ, et al. : Fasting Activates Fatty Acid Oxidation to Enhance Intestinal Stem Cell Function during Homeostasis and Aging. Cell Stem Cell. 2018;22(5):769–778.e4. 10.1016/j.stem.2018.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 122. Potten CS, Martin K, Kirkwood TB: Ageing of murine small intestinal stem cells. Novartis Found Symp. 2001;235:66–79; discussion 79–84, 101–4. 10.1002/0470868694.ch7 [DOI] [PubMed] [Google Scholar]
- 123. Nalapareddy K, Nattamai KJ, Kumar RS, et al. : Canonical Wnt Signaling Ameliorates Aging of Intestinal Stem Cells. Cell Rep. 2017;18(11):2608–21. 10.1016/j.celrep.2017.02.056 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation